Abstract
Genome editing appears poised to enter an exciting new era. Targeted double-stranded breaks due to custom restriction enzymes are powerful nucleating events for the induction of local changes in the genome. The zinc finger nuclease (ZFN) platform established the potential of this approach for the zebrafish, but access to high quality reagents has been a major bottleneck for the field. However, two groups recently report successful somatic and germline gene modification using a new nuclease architecture, transcription activator-like effector nucleases (TALENs). TALEN construction is simpler, potentially more reliable, and in the few cases examined, shows fewer off-target effects than corresponding ZFNs. TALENs promise to bring gene targeting to the majority of zebrafish laboratories.
Introduction
The zebrafish (Danio rerio) has a completed genome (www.sanger.ac.uk/Projects/D_rerio/Zv9_assembly_information.shtml), but mutations in only about 10% of the genome are currently available to the zebrafish researcher. Custom zinc finger nuclease (ZFN) technology has proved to be a viable approach for the generation of targeted mutations in this animal model,1–3 with dozens of mutant alleles published in the past 2 years.4 The main challenge to large-scale use of ZFNs has been the difficulty in manufacture and the diverse range of targeting efficacy of these reagents.
Transcription Activator-Like Effector Nucleases
Recent advances in DNA targeting have focused on transcription activator-like effector (TALE) sequence-specific DNA binding domain proteins from plant pathogenic bacteria.5,6 When fused to the FokI nuclease domain (the same nuclease used in ZFNs), TALE nucleases (TALENs) recognize specific DNA sequences using a straightforward DNA base recognition cipher (Fig. 1). Binding of two TALENs to DNA allows FokI to dimerize and create a targeted chromosome break.
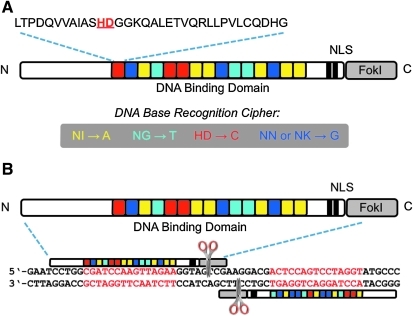
FIG. 1.
Engineered transcription activator-like effector Nuclease (TALEN) cartoons. (A) Engineered TALE DNA binding domain recognizes specific bases by using a known cipher involving two key amino acid residues. These amino acids are embedded in a 33–35 amino acid repeat. Nuclear access is provided by the native nuclear localization signal (NLS). (B) Total sequence specificity and formation of a fully functional nuclease is achieved from each of the two TALEN monomers after dimerization on the target sequence in the genome.
Zebrafish Gene Targeting Using TALENs
Two recent reports show that TALENs can effectively recognize targeted loci in both somatic7 and germline8 cells in the zebrafish. Germline frequencies from this initial report appear comparable to that seen by ZFNs.8 Importantly, for those sites successfully targeted by ZFNs, TALENs appear to be readily able to induce cleavage and subsequent mutations.7 Thus, switching a sequence-specific DNA binding platform from zinc fingers to TALEs appears to be a straightforward technology transition.
Gene Targeting for the Masses?
In addition to this very promising functional utility for constructed TALENs, four groups have reported molecular toolboxes for the ready construction of TALENs.9–12 The DNA base recognition cipher for TALE proteins is much simpler than zinc fingers (Fig. 1A), with an individual base recognized by two amino acids in an individual TALE repeat unit. Rapid assembly of custom TALENs is very accessible to even small laboratories, as the entire platform can be initiated with clones from a single 96-well plate, and construction takes less than 1 week once the system is operational.
Pioneering Data from ZFNs—A Cautionary TALE?
The future looks bright for TALENs and their use in zebrafish and other model organisms, and the current trajectory is heavily dependent on the rich history of ZFN science. ZFNs have been the leading technology for custom nuclease-catalyzed genome editing applications. However, ZFNs are not without limitations. Several papers also report that ZFNs can induce unintended changes within the host genomes in addition to the target location.13–15 Notably, with unbiased discovery approaches, some of the double-stranded breaks were not predicted based on in silico predictions.15 Whether such issues will be encountered with TALENs is only beginning to be investigated, but it does appear that they, too, will create off-target mutations at some frequency.16 However, one recent TALEN paper reports reduced off-target effects for a TALEN compared with ZFNs that recognize the same chromosome site.17 For genetics research, the take-home message is that having multiple alleles at the same locus is a worthy goal even with the potential for complications due to background off-target effects.
Acknowledgments
The authors thank Victoria Bedell and Gary Moulder for constructive comments. This article was supported in part by grants from NIH (GM63904 and DA14546 to S.C.E.) and the State of Minnesota (H001274506 to D.F.V. and S.C.E.).
Disclosure Statement
D.F. Voytas consults for Cellectis, a company that markets TALENS, and he is a named inventor on a TALEN patent.
References
- 1.Doyon Y. McCammon JM. Miller JC. Faraji F. Ngo C. Katibah GE, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meng X. Noyes MB. Zhu LJ. Lawson ND. Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foley JE. Yeh JR. Maeder ML. Reyon D. Sander JD. Peterson RT, et al. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN) PLoS One. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawson ND. Wolfe SA. Forward and reverse genetic approaches for the analysis of vertebrate development in the zebrafish. Dev Cell. 2011;21:48–64. doi: 10.1016/j.devcel.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Moscou MJ. Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science (New York, NY) 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 6.Boch J. Scholze H. Schornack S. Landgraf A. Hahn S. Kay S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science (New York, NY) 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 7.Sander JD. Cade L. Khayter C. Reyon D. Peterson RT. Joung JK, et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang P. Xiao A. Zhou M. Zhu Z. Lin S. Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 9.Cermak T. Doyle EL. Christian M. Wang L. Zhang Y. Schmidt C, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang F. Cong L. Lodato S. Kosuri S. Church GM. Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morbitzer R. Elsaesser J. Hausner J. Lahaye T. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res. 2011;39:5790–5799. doi: 10.1093/nar/gkr151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li T. Huang S. Zhao X. Wright DA. Carpenter S. Spalding MH, et al. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011;39:6315–6325. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabriel R. Lombardo A. Arens A. Miller JC. Genovese P. Kaeppel C, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol. 2011 doi: 10.1038/nbt.1948. [Epb ahead of print]; [DOI] [PubMed] [Google Scholar]
- 14.Gupta A. Meng X. Zhu LJ. Lawson ND. Wolfe SA. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res. 2011;39:381–392. doi: 10.1093/nar/gkq787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pattanayak V. Ramirez CL. Joung JK. Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011 doi: 10.1038/nmeth. [Epub ahead of print]; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hockemeyer D. Wang H. Kiani S. Lai CS. Gao Q. Cassady JP, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mussolino C. Morbitzer R. Lutge F. Dannemann N. Lahaye T. Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr597. [Epub ahead of print]; [DOI] [PMC free article] [PubMed] [Google Scholar]