Abstract
Stem cells are a potential key strategy for treating neurodegenerative diseases in which the generation of new neurons is critical. A better understanding of the characteristics and molecular properties of neural stem cells (NSCs) and differentiated neurons can help with assessing neuronal maturity and, possibly, in devising better therapeutic strategies. We have performed an in-depth gene expression profiling study of murine NSCs and primary neurons derived from embryonic mouse brains. Microarray analysis revealed a neuron-specific gene expression signature that distinguishes primary neurons from NSCs, with elevated levels of transcripts involved in neuronal functions, such as neurite development and axon guidance in primary neurons and decreased levels of multiple cytokine transcripts. Among the differentially expressed genes, we found a statistically significant enrichment of genes in the ephrin, neurotrophin, CDK5, and actin pathways, which control multiple neuronal-specific functions. We then artificially blocked the cell cycle of NSCs with mitomycin C (MMC) and examined cellular morphology and gene expression signatures. Although these MMC-treated NSCs displayed a neuronal morphology and expressed some neuronal differentiation marker genes, their gene expression patterns were very different from primary neurons. We conclude that 1) fully differentiated mouse primary neurons display a specific neuronal gene expression signature; 2) cell cycle block at the S phase in NSCs with MMC does not induce the formation of fully differentiated neurons; 3) cytokines change their expression pattern during differentiation of NSCs into neurons; and 4) signaling pathways of ephrin, neurotrophin, CDK5, and actin, related to major neuronal features, are dynamically enriched in genes showing changes in expression level.
Keywords: neural stem cell, primary neuron, cell cycle block
neural stem cells (NSCs) exist in various regions of the central nervous system throughout the life span in mammals (7, 21, 26, 31, 46). For example, the subventricular zone and subgranular zone of the hippocampus contain a relatively high density of NSCs. These areas are considered neurogenic since they continue to produce neurons throughout adult life, in contrast to other brain regions where neurogenesis is not observed in the adult (20, 25, 62). The division and differentiation of these endogenous NSCs can be regulated by both physiological stimuli and pathological conditions (4, 28).
To better understand NSC biology, various cell lines were derived (2, 13, 19, 30, 34, 35, 40, 45). NSC lines were typically screened for the presence of NSC markers and subsequently used for a variety of applications such as 1) transplantations for the purpose of alleviating brain damage after certain diseases, 2) their ability to promote neuronal survival and neurite outgrowth, and 3) the secretion of a variety of growth factors (9, 30, 34–36, 38, 40, 43, 48, 50, 51, 53, 56, 58).
Furthermore, to better understand NSC biology and the response of these cells to different conditions, which can help better design regenerative strategies, various NSCs have been used to study the effect of many compounds in vitro (6, 8, 24, 32, 67) and their effect on differentiation (17, 60, 67). Therefore, it becomes crucial to appreciate how representative such neurons are compared with primary neurons (PNs), as data obtained with these studies will impact clinical applications.
We were particularly interested in the possibility of generating neurons through a nonspecific arrest in cell cycle. Since NSCs would arrest in the G0/G1 phase prior to differentiation (5, 37), we wanted to determine whether an NSC line arrested in other phases of cell cycle, in particular in the S phase, can lead to differentiated neurons. To this end, we hypothesized that defining the characteristics of embryonic PNs will provide a basic neuronal signature that can be used to assess the differentiation status of in vitro-differentiated NSCs. We also wanted to shed light on factors that allow/facilitate the transition from the NSC line into neurons, as such lines are often used for various pharmacological in vitro testing. Therefore, we first performed an in-depth gene expression profiling analysis of NSCs and PNs. Comparison of PN with NSC gene expression patterns revealed an upregulation of genes involved in many neuronal-specific functions, which constitute a neuronal signature. Genes involved in cell division were among the most significantly downregulated in PNs. We then treated the C17.2 NSC line with mitomycin C (MMC) to artificially induce a arrest in cell cycle in the S phase. Our results revealed that NSCs treated with MMC results in cells that have a neuronal morphology and express certain neuronal markers but does not result in fully differentiated neurons following arrest in the S phase, i.e., cells that have only a partial PN gene expression profile.
MATERIALS AND METHODS
C17.2 NSC culture.
The C17.2 NSCs were generously donated by Dr. Evan Snyder from the Sanford/Burnham Institute for Biomedical Research, La Jolla, CA (52, 63). Cells were passaged at 1:10 dilution once a week and maintained in DMEM (high glucose + l-glutamine + sodium pyruvate), 10% fetal bovine serum, 5% horse serum, 2 mM glutamine, 1% penicillin/streptomycin/fungizome (all from Gibco). Standard cultures were maintained at 37°C- 5% CO2 in a humidified incubator.
Differentiation of C17.2 NSCs into neurons.
C17.2 NSCs at ∼70% confluency were treated with MMC (final concentration 0.4 μg/ml; Sigma) under normoxic conditions for 48 h. Culture medium was then progressively replaced by changing half of the medium every 2 days. Treated cells were cultured for 6 days before they reached a stable neuronal morphology. They were then cultured for an additional 2 days before experimental use.
Primary embryonic neuronal culture.
Time-pregnant C57BL/6J female mice were obtained from Jackson Laboratories (Bar Harbor, ME). At embryonic day 17, embryos were extracted, and brains were isolated and placed in sterile neurobasal medium (Gibco) on ice. Cortexes were carefully dissected out and incubated with 0.25% trypsin (Gibco) for 15 min at 37°C. An equal volume of trypsin inhibitor (Gibco) was added, and cortexes were incubated for 5 min at 37°C, triturated, and centrifuged at 1,400 rpm for 5 min at 4°C. Pellets were then resuspended in plating medium consisting of neurobasal medium (Gibco), B27 1× final (Gibco), glutamine 500 μM (Gibco), penicillin/streptomycin 100 U/ml, 100 μg/ml (Gibco), and glutamic acid 25 μM (Sigma). Cells were counted by trypan blue extrusion and diluted to 106 cells/ml. One milliliter of cells were added onto a 35-mm plate previously coated with poly-d-lysine > 300 kDa (Sigma) at 0.15 mg/ml and cultured in 5% CO2 at 37°C (day 0). After 24 h (day 1), 1 ml of plating medium was added. Then every 4 days (day 4, day 8) 1 ml of the plating medium was replaced with an equal volume of maintaining medium (same composition as plating medium without glutamic acid). Cells were grown for 6 days before experimental use.
Microscopy and cell death.
The morphology of all cultured cells was assessed with phase contrast microscopy, and cell death was evaluated by propidium iodide staining (5 μg/μl; Sigma). Images were acquired with a Zeiss microscope with AxioCam MRm camera using the Axiovision Rel 4.5 software.
Immunocytochemistry staining.
Immunocytochemistry was performed on NSCs, MMC-treated NSCs, and PNs to detect the NSC marker nestin (intermediate filament), early neuronal marker MAP2 (microtubule-associated protein 2), and late neuronal marker NeuN (neuronal nuclei) by using the following primary antibodies: rabbit anti-nestin 1:250, mouse anti-nestin 1:200, mouse anti-NeuN 1:200, rabbit anti-MAP2 1:250 (all from Chemicon), and secondary antibodies: goat anti-rabbit Texas Red 1:100, goat anti-mouse TRITC 1:100, goat anti-rabbit fluorescein 1:100, and goat anti-mouse fluorescein 1:100 (all from Chemicon). All antibodies were diluted in 3% normal goat serum (in 1× PBS). Staining was performed on cells grown in petri dishes. Briefly, cells were washed twice with PBS and fixed for 20 min at room temperature in 4% paraformaldehyde pH 7.4, permeabilized using 0.2% Triton X-100 in 1× PBS for 15 min at room temperature, and then blocked with 3% normal goat serum for 1 h at room temperature. Cells were then incubated with primary antibodies at 4°C overnight, washed with 0.1% Triton X-100 in 1× PBS, incubated with secondary antibodies at room temperature for 2.5 h, and washed again. DAPI was then added, and cells were observed under fluorescent microscope (Zeiss). Images were acquired with AxioCan MRm camera (Zeiss) using the Axiovision Rel 4.5 software. Magnifications used were ×100 and ×400.
Protein quantification.
To verify the protein levels of certain markers (nestin, MAP2, NeuN), protein extraction was performed using HEPES buffer (Ca, Mg, Na-free; 200 mM mannitol, 80 mM HEPES, 41 mM KOH), and samples were homogenized for 30 s on ice. Thirty micrograms of protein per sample were loaded on gel. Membranes were blocked with 5% milk and then probed with the following primary antibodies: rabbit anti-MAP2, mouse anti-NeuN, goat anti-nestin, and goat anti-actin (all from Chemicon) and then treated with secondary antibodies: rabbit anti-goat-horseradish peroxidase (HRP), goat anti-rabbit-HRP, and rabbit anti-mouse-HRP (all from Zymed).
Microarray analysis.
Total RNA was extracted from NSC, PN, and MMC-treated NSCs (n = 3 each) by using the RNeasy kit (Qiagen) according to the manufacturer's instructions. Spectrophotometer readings are taken with the NanoDrop ND-1000. The RNA integrity was checked with the Agilent 2100 BioAnalyzer system (Agilent Technologies, Santa Clara, CA). Total RNA (300 ng) was used to synthesize a biotin-labeled complementary RNA (cRNA) probes using Illumina RNA amplification kit (Ambion, Austin, TX) as previously described (43). Illumina sentrix mouse-6 expression GeneChips (Illumina, San Diego, CA) were used to determine differences in gene expression. Biotin-labeled cRNA (1.5 μg) was added to the chip and incubated for 16–20 h at 55°C. The bound biotin-labeled cRNA was then stained with streptavidin-Cy3. After hybridization, the GeneChips were washed, dried, and scanned by the BeadArray Reader (Illumina, San Diego, CA). The absolute intensity of each probe on the image was generated with BeadStudio software (Illumina). For analysis, intensities are normalized using modified LOESS as previously described (54). A permutation-based F-test statistic was then computed on the normalized Log2 expression signals using the multitest package (18). Pairwise fold changes were computed for the MMC-treated NSCs and the PNs using the NSC expression values as a baseline. We then determined differentially expressed transcripts by a permutation-based F-test (P < 0.05) and at least a twofold change in at least one of the above two comparisons. These probes were then clustered by array (using Euclidean distance metric) and by probe (using cosangle distance metric) with the hierarchical ordered partitioning and collapsing hybrid (HOPACH) clustering algorithm (63). Clusters and comparison groups were annotated with statistically significant GO term overrepresentation using the MappFinder algorithm and GO-Elite software packages (14). Functional categorization of gene alterations was created with Ingenuity Pathway's (Redwood City, CA) analysis program (68). The P value for each network or function was calculated with a right-tailed Fisher's exact test. The score for each network of function was shown as −log10 [P value], which indicates the likelihood of finding a set of focus genes in the network or function by random chance. The significance threshold was set to a score of 1.3 (i.e., P ≤ 0.05). All microarray data is MIAME compliant and the raw data has been deposited in the Gene Expression Ominibus database at http://www.ncbi.nlm.nih.gov/geo and can be retrieved using access number GSE24116.
Research ethics.
The experimentations requiring animal use were submitted to the Institutional Animal Care and Use Committee at the University of California San Diego, San Diego, CA, and approval was obtained (no. S05534). According to the committee regulations, the use of the NSC line C17.2 for in vitro studies does not pose an ethical issue and therefore did not require approval.
Statistical analysis.
Statistical analysis was performed using Student's t-test. Mean values were considered statistically significant if P ≤ 0.05.
RESULTS
Morphological features of C17.2 NSCs and PNs.
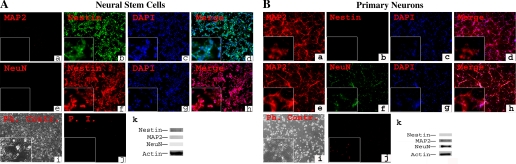
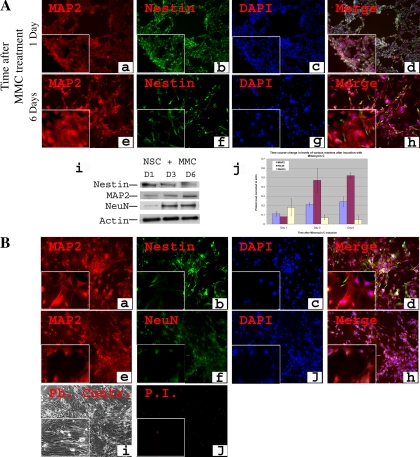
All cultured NSCs expressed the neural progenitor marker nestin (Fig. 1, A, b, f) and showed no expression of neuronal markers such as MAP2 (Fig. 1, A, a) and NeuN (Fig. 1, A, e), as confirmed by Western blot analysis (Fig. 1, A, k), indicating no spontaneous differentiation had occurred by our culture conditions. NSCs showed a predominantly pyramidal cell shape (Fig. 1, i) with a low baseline cell death (Fig. 1, A, j). Primary neurons (Fig. 1, B, i) also showed baseline cell death throughout the study (Fig. 1, B, j) and immunocytochemistry revealed the presence of MAP2 and NeuN and the absence of nestin 8 days in culture (Fig. 1, B, a, b, e, f, j), as verified by Western blot analysis (Fig. 1, B, k).
Fig. 1.
Characteristics of neural stem cells (NSCs) and neurons. A: NSCs were cultured in 21% O2-5% CO2. Cells were costained by using antibodies directed against mouse stem cell marker nestin and neuronal markers MAP2 (a–d) or against nestin and the neuronal marker NeuN (e–h). All cells expressed nestin (b, f), but there was no detectable MAP2 or NeuN (a, e). Phase contrast (Ph. Contr.) microscopy revealed a pyramidal shape of the cells (i), and only very few dead cells were observed or were seen with propidium iodide (PI) labeling (j). Magnification: ×100; insets, ×400. Western blot analysis confirmed the absence of MAP2 and NeuN from NSCs (k). B: primary neurons were cultured from embryonic day 17 mouse cortex in normoxia for 8 days. Immunocytochemistry was performed for nestin, MAP2, and NeuN. There was a complete absence of nestin (b). MAP2 (a, e), and NeuN (f) were detected in all cells. Phase contrast microscopy revealed typical neuronal shape with processes and synapses (i). Magnification: ×100; insets, ×400. Western blot analysis confirmed the presence of MAP2 and NeuN and the absence of nestin from primary neurons.
Microarray analysis reveals a neuronal-specific gene expression profile.
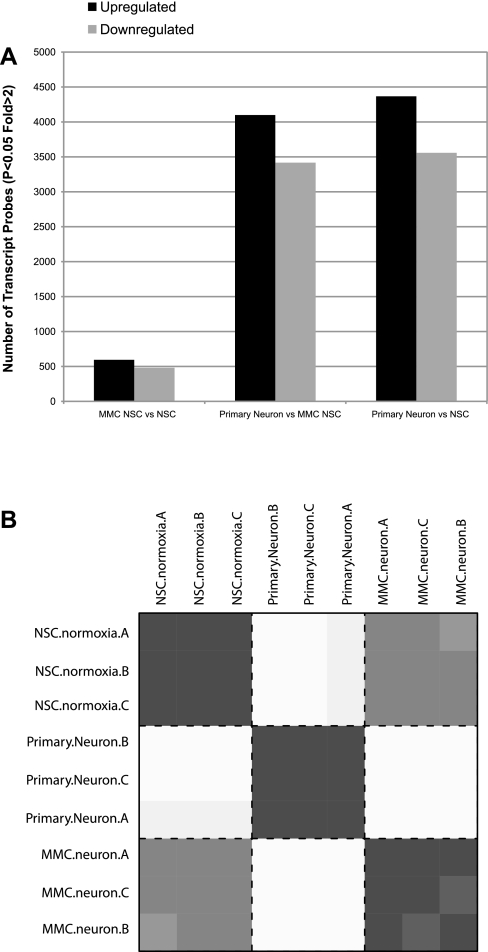
The microarray expression profile revealed that 7,925 genes changed more than twofold (P < 0.05) in PNs relative to NSCs. There were 4,367 upregulated genes and 3,558 downregulated genes. (Fig. 2A). Heat map HOPACH correlation confirmed the reproducibility between samples of the same cell type (Fig. 2B).
Fig. 2.
Microarray analysis of NSCs and neurons. A: number of microarray probes up- and downregulated comparing NSC ± MMC and primary isolated neurons. MMC, mitomycin C. B: heat plot representing all possible pairwise hierarchical ordered partitioning and collapsing hybrid (HOPACH) correlation (euclidean) values. Darkest values indicate a high level of correlation (= 1 across the diagonal), and lighter gray indicates lower correlation values.
Next, we examined the general expression profile of the NSCs to verify whether there is any profile indicative of a glial or neuronal differentiation phenotype and found none, confirming that this cell line does not show any spontaneous differentiation into glia or neurons (data available online at the Omnibus database; see materials and methods).
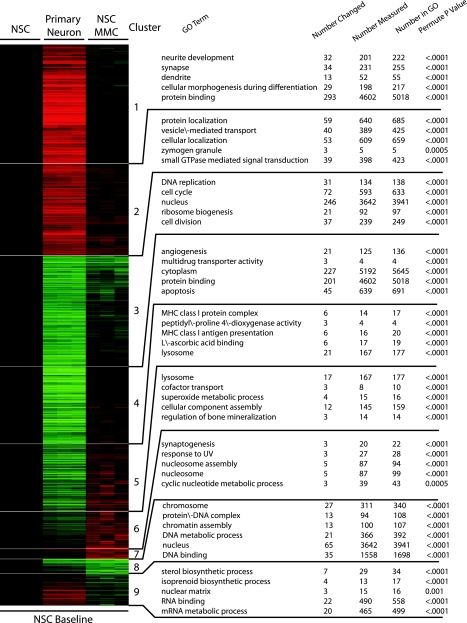
We then used Gene Ontology (GO) to functionally annotate differentially expressed transcripts. Among the downregulated genes, the most significantly changed belong to intracellular parts, organelles, cell cycle, DNA replication, and catalytic activity, but also other functions were affected, such as organelle organization and biogenesis, apoptosis, angiogenesis, cytokine production, matrix components, and some metabolic constituents (Table 1). Among the upregulated genes, the most significantly enriched biological processes belong to neuron-specific functions, such as neurite development, synapse, and neurite projection (Table 1, Figs. 3 and 6). Genes related to the Notch pathway were upregulated in PNs relative to NSCs. (Table 1).
Table 1.
Cellular functions with genes changed in primary neurons relative to neural stem cells
| GO Type | GO Name | Number Changed | Number Measured | Number in GO | Adjusted P Value |
|---|---|---|---|---|---|
| Downregulated | |||||
| C | Intracellular part | 1188 | 7905 | 8617 | <0.0001 |
| C | Organelle | 1011 | 6576 | 7174 | <0.0001 |
| P | Cell cycle | 156 | 593 | 633 | <0.0001 |
| P | DNA replication | 55 | 134 | 138 | <0.0001 |
| P | Cell division | 72 | 239 | 249 | <0.0001 |
| F | Catalytic activity | 657 | 4431 | 4851 | <0.0001 |
| P | Organelle organization and biogenesis | 179 | 955 | 1028 | <0.0001 |
| P | Ribonucleoprotein complex biogenesis and assembly | 39 | 134 | 143 | <0.0001 |
| P | Apoptosis | 124 | 639 | 691 | <0.0001 |
| C | Protein/DNA complex | 30 | 94 | 108 | <0.0001 |
| P | Chromosome segregation | 18 | 45 | 50 | <0.0001 |
| C | Basement membrane | 19 | 49 | 54 | <0.0001 |
| P | Hexose metabolic process | 31 | 110 | 123 | <0.0001 |
| P | Angiogenesis | 33 | 125 | 136 | <0.0001 |
| P | DNA geometric change | 5 | 7 | 7 | <0.0001 |
| P | Ribonucleoside metabolic process | 5 | 7 | 9 | <0.0001 |
| P | Nucleoside monophosphate biosynthetic process | 8 | 16 | 17 | <0.0001 |
| P | IMP metabolic process | 3 | 3 | 3 | 0.0195 |
| P | 4-Hydroxyproline metabolic process | 3 | 3 | 3 | 0.0465 |
| P | SMAD protein nuclear translocation | 3 | 3 | 3 | 0.0195 |
| P | Translation | 59 | 285 | 351 | <0.0001 |
| F | Structural constituent of ribosome | 29 | 111 | 134 | <0.0001 |
| P | Embryonic development ending in birth or egg Hatching | 61 | 300 | 325 | <0.0001 |
| F | Adenyl nucleotide binding | 192 | 1212 | 1314 | 0.0195 |
| F | L-ascorbic acid binding | 8 | 17 | 19 | 0.0195 |
| P | Tube morphogenesis | 34 | 142 | 155 | <0.0001 |
| C | MHC class I protein complex | 7 | 14 | 17 | <0.0001 |
| P | Response to DNA damage stimulus | 48 | 226 | 241 | <0.0001 |
| P | Immune system process | 166 | 1031 | 1133 | <0.0001 |
| P | rRNA metabolic process | 18 | 61 | 64 | <0.0001 |
| P | Cell proliferation | 90 | 507 | 553 | <0.0001 |
| P | Peptidyl-histidine modification | 4 | 6 | 6 | 0.0465 |
| P | Folic acid and derivative biosynthetic process | 4 | 6 | 7 | 0.034 |
| P | Transcription factor import into nucleus | 6 | 12 | 14 | <0.0001 |
| F | Calcium-dependent phospholipid binding | 8 | 19 | 20 | <0.0001 |
| C | Integrin complex | 9 | 23 | 25 | 0.0195 |
| P | Liver development | 10 | 28 | 31 | 0.0195 |
| P | Positive regulation of NF-κB transcription factor activity | 6 | 13 | 16 | <0.0001 |
| P | Endoplasmic reticulum unfolded protein response | 5 | 10 | 12 | 0.034 |
| P | Homeostasis of number of cells | 56 | 301 | 327 | 0.0195 |
| P | Oxidation reduction | 149 | 961 | 1040 | 0.0465 |
| P | Nonapoptotic programmed cell death | 72 | 412 | 444 | 0.0195 |
| P | Protein folding | 24 | 105 | 117 | 0.034 |
| P | Regulation of angiogenesis | 9 | 27 | 30 | 0.034 |
| P | Cytokine production | 26 | 118 | 127 | <0.0001 |
| P | Regulation of cell size | 25 | 113 | 119 | <0.0001 |
| P | Extracellular matrix organization and biogenesis | 14 | 53 | 59 | 0.034 |
| C | Intracellular part | 1188 | 7905 | 8617 | <0.0001 |
| Upregulated | |||||
| P | Neurite development | 95 | 201 | 222 | <0.0001 |
| C | Synapse | 102 | 231 | 255 | <0.0001 |
| C | Neuron projection | 66 | 131 | 141 | <0.0001 |
| F | Protein binding | 916 | 4602 | 5018 | <0.0001 |
| P | Synaptic transmission | 82 | 188 | 205 | <0.0001 |
| P | Localization | 546 | 2607 | 2814 | <0.0001 |
| P | Small GTPase mediated signal transduction | 122 | 398 | 423 | <0.0001 |
| C | Cell junction | 121 | 401 | 432 | <0.0001 |
| C | Synaptic vesicle | 30 | 52 | 55 | <0.0001 |
| P | Telencephalon development | 25 | 51 | 55 | <0.0001 |
| P | Periplasmic space organization and biogenesis | 63 | 196 | 223 | <0.0001 |
| P | Neuron recognition | 10 | 12 | 12 | <0.0001 |
| P | Microtubule-based process | 61 | 197 | 211 | <0.0001 |
| P | Biological adhesion | 131 | 537 | 589 | <0.0001 |
| F | Kinase activity | 172 | 760 | 836 | <0.0001 |
| F | Ion transmembrane transporter activity | 138 | 585 | 643 | <0.0001 |
| P | Learning and/or memory | 28 | 69 | 76 | <0.0001 |
| C | Golgi apparatus | 134 | 570 | 607 | <0.0001 |
| C | Synaptosome | 24 | 56 | 59 | <0.0001 |
| F | Gated channel activity | 69 | 246 | 267 | <0.0001 |
| F | Calcium ion binding | 167 | 753 | 810 | <0.0001 |
| P | Neurite regeneration | 17 | 34 | 39 | <0.0001 |
| P | Negative regulation of neurogenesis | 16 | 31 | 33 | <0.0001 |
| C | Microtubule cytoskeleton | 73 | 268 | 294 | <0.0001 |
| F | GTPase regulator activity | 78 | 293 | 317 | <0.0001 |
| P | Neuromuscular process | 32 | 88 | 105 | <0.0001 |
| P | Regulation of adenylate cyclase activity | 9 | 13 | 14 | <0.0001 |
| F | α-N-acetylneuraminate α-2,8-Sialyltransferase activity | 5 | 5 | 5 | <0.0001 |
| P | Posttranslational protein modification | 188 | 899 | 972 | <0.0001 |
| C | Cell soma | 25 | 68 | 73 | <0.0001 |
| C | Ionotropic glutamate receptor complex | 8 | 12 | 13 | <0.0001 |
| F | GTP binding | 75 | 298 | 314 | <0.0001 |
| F | Glutamate receptor activity | 12 | 23 | 30 | <0.0001 |
| P | Membrane depolarization | 13 | 27 | 30 | <0.0001 |
| P | Central nervous system neuron differentiation | 15 | 34 | 36 | <0.0001 |
| P | Cerebellar granular layer development | 4 | 4 | 5 | <0.0001 |
| F | Calcium channel regulator activity | 6 | 8 | 9 | <0.0001 |
| C | Heterotrimeric G protein complex | 14 | 31 | 34 | <0.0001 |
| C | Coated pit | 13 | 28 | 29 | <0.0001 |
| C | Site of polarized growth | 10 | 19 | 21 | <0.0001 |
| P | Positive regulation of lyase activity | 7 | 11 | 12 | <0.0001 |
| P | Positive regulation of cyclase activity | 7 | 11 | 12 | <0.0001 |
| P | Synapse organization and biogenesis | 19 | 51 | 56 | <0.0001 |
| F | AMP binding | 8 | 14 | 16 | <0.0001 |
| F | l-Amino acid transmembrane transporter activity | 11 | 23 | 26 | 0.0135 |
| P | Sensory organ development | 60 | 244 | 276 | <0.0001 |
| C | cAMP-dependent protein kinase complex | 5 | 7 | 8 | 0.0135 |
| F | Kinase regulator activity | 19 | 54 | 58 | <0.0001 |
| P | Action potential propagation | 8 | 15 | 18 | <0.0001 |
| P | Enzyme-linked receptor protein signaling pathway | 66 | 280 | 299 | <0.0001 |
| C | Coated membrane | 16 | 43 | 51 | <0.0001 |
| F | UDP-galactose:β-N-Acetylglucosamine β-1,3-Galactosyltransferase activity | 4 | 5 | 5 | 0.025 |
| P | Regulation of neuron differentiation | 15 | 40 | 42 | <0.0001 |
| P | Regulation of cellular component size | 16 | 44 | 45 | <0.0001 |
| P | Membrane organization and biogenesis | 66 | 284 | 311 | <0.0001 |
| P | Cerebral cortex neuron differentiation | 7 | 13 | 13 | 0.025 |
| P | Response to organic substance | 17 | 49 | 55 | <0.0001 |
| P | Regulation of GTPase activity | 21 | 66 | 72 | <0.0001 |
| P | Wnt receptor signaling pathway | 43 | 170 | 182 | <0.0001 |
| P | Multicellular organismal response to stress | 11 | 27 | 31 | 0.0135 |
| F | Adenylate cyclase activity | 6 | 11 | 11 | 0.044 |
| P | Protein polymerization | 16 | 47 | 51 | <0.0001 |
| P | Notch signaling pathway | 27 | 96 | 104 | <0.0001 |
| P | Adult behavior | 23 | 78 | 88 | <0.0001 |
| P | Sensory perception of pain | 10 | 25 | 26 | 0.025 |
| P | Regulation of protein polymerization | 8 | 18 | 20 | <0.0001 |
| P | Sterol biosynthetic process | 11 | 29 | 34 | <0.0001 |
| F | Potassium ion binding | 28 | 104 | 111 | <0.0001 |
| P | Metabotropic glutamate receptor Signaling pathway | 13 | 38 | 44 | <0.0001 |
| P | Peptidyl-serine modification | 16 | 51 | 55 | <0.0001 |
| F | Microtubule motor activity | 16 | 51 | 54 | 0.0135 |
| C | Voltage-gated potassium channel complex | 17 | 56 | 60 | 0.0135 |
| P | Photoreceptor cell differentiation | 29 | 113 | 121 | 0.044 |
| F | Receptor signaling protein activity | 22 | 81 | 86 | 0.025 |
| C | Perinuclear region of cytoplasm | 12 | 37 | 39 | 0.044 |
| C | Leading edge | 19 | 69 | 77 | 0.0135 |
| P | Calcium-mediated signaling | 18 | 65 | 73 | 0.044 |
| P | Regulation of action potential | 12 | 38 | 45 | 0.044 |
| P | Neuropeptide signaling pathway | 19 | 70 | 77 | 0.0345 |
| P | Dopamine receptor signaling pathway | 10 | 30 | 33 | 0.044 |
GO, Gene Ontology; P, biological process; C, cellular component; F, molecular function.
Fig. 3.
Functional annotation of transcripts differentially expressed between NSC ± MMC and primary neurons. HOPACH cluster of the 5,488 microarray probes that where differentially expressed (F-test statistic P < 0.05 and a 2-fold change in up- or downregulated compared with NSC control in any of the 2 comparisons). Clusters where annotated for statistical overrepresentation of transcript-associated Gene Ontology (GO) terms using MAPPFinder and GO-Elite (14) software.
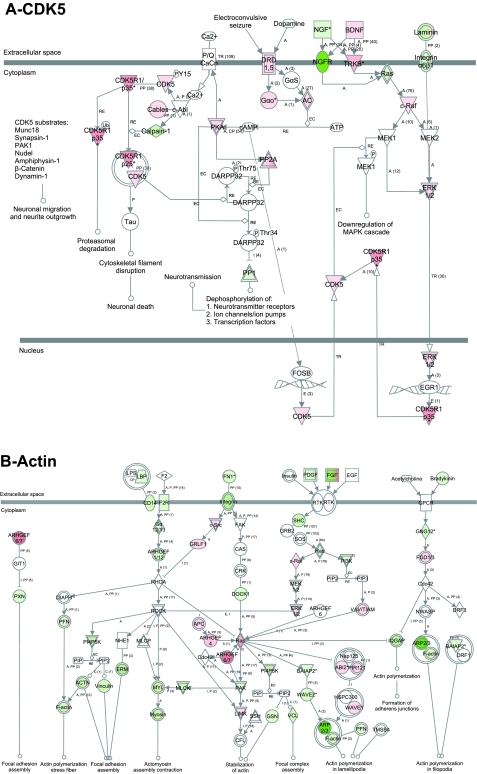
Fig. 6.
CDK5 (A) and actin (B) signaling pathway in primary neurons. Analysis using the Ingenuity Pathway Analysis software showed that CDK5 and actin signaling pathways are involved in multiple functions related to neurons, and a majority of the genes in those pathways have their expression level changed. Red, upregulated; green, downregulated; white, no change. CDK5: pathway shows upregulation of multiple genes related to neurite outgrowth, neurotransmitter receptors and ion channels. Actin: there is a dynamic change in the expression of genes related to actin polymerization and focal adhesion, suggesting dynamic instability of actin cytoskeleton, a feature known to be needed for neurotransmitter secretion, and axon guidance.
Interestingly, when the pathways with the most significantly downregulated genes were subjected to GO analysis, certain genes (e.g., cdc26, cdk2, cdk4, and Mcm) appeared involved in many functions, including intracellular organelles, cell cycle, and cell division. In pathways that had the most significantly upregulated genes, some genes (e.g., cdk5, BDNF, ANK3, Abi2) were involved in neurite development, neuron projection, protein binding, and localization, while Grik, nlgn, Gria, and slc were involved in synapse, protein binding, localization and cellular junction.
Functional categorization of genes differentially expressed in PNs relative to NSCs.
Data analysis using Ingenuity software revealed 53 functional gene families that significantly changed expression (Fig. 4A and Table 4). When these pathways were dissected to determine the relative contributions to neuronal function/phenotype, four pathways stood out: ephrin receptor pathway, CDK5 pathway, neurotrophin signaling pathway, and actin cytoskeleton signaling (Figs. 5 and 6). These pathways have very high numbers of genes with changed expression, and impact multiple neuronal functions.
Fig. 4.
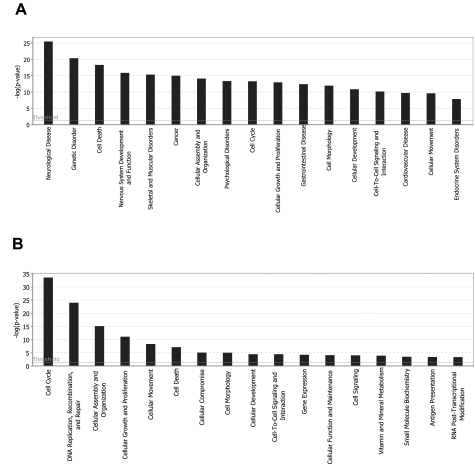
Biofunctional analysis of neuron gene expression profile relative to NSCs. Functional characterization was done using the Ingenuity Pathway Analysis software. Genes significantly altered were classified into associated functions (as depicted in the x-axis). Functions are listed from most to least significant (left to right). The y-axis depicts −log10 [P value]. Horizontal bar (threshold) indicates P < 0.05. The significance threshold was set to 1.3. Primary neurons: a total of 53 biofunctions were significantly changed (shown are the first 17 most significant pathways). MMC-treated NSCs; a total of 17 biofunctions were significantly changed.
Table 4.
Biofunction analysis of primary neurons gene expression profile relative to neural stem cells: functional categorization
| Biofunctions | |
|---|---|
| Neurological diseases | Connective tissue disorders |
| Genetic disorder | Cell signaling |
| Cell death | Molecular transport |
| Nervous system development and function | DNA replication, recombination and repair |
| Skeletal and muscle disorders | Amino-acid metabolism |
| Cancer | Posttranslational modification |
| Cellular assembly and organization | Small molecule biochemistry |
| Psychological disorders | Connective tissue development and function |
| Cell cycle | Protein synthesis |
| Cellular growth and proliferation | Renal and urological disease |
| Gastrointestinal disease | Tissue morphology |
| Cell morphology | Gene expression |
| Cellular development | Skeletal and muscular system development and function |
| Cell-to-cell signaling and interaction | Organismal injury and abnormalities |
| Cardiovascular disease | Developmental disorders |
| Cellular movement | Cellular compromise |
| Endocrine system disorders | Hepatic system disease |
| Metabolic disease | Embryonic development |
| Inflammatory disease | Endocrine system development and function |
| Respiratory disease | Lipid metabolism |
| Tissue development | Organismal development |
| Behavior | Reproductive system development and function |
| Organismal survival | Hair and skin development and function |
| Reproductive system disease | Organ development |
| Cellular function and maintenance | Tumor morphology |
| Immunological disease | Infection mechanism |
| Hematological disease |
Fig. 5.
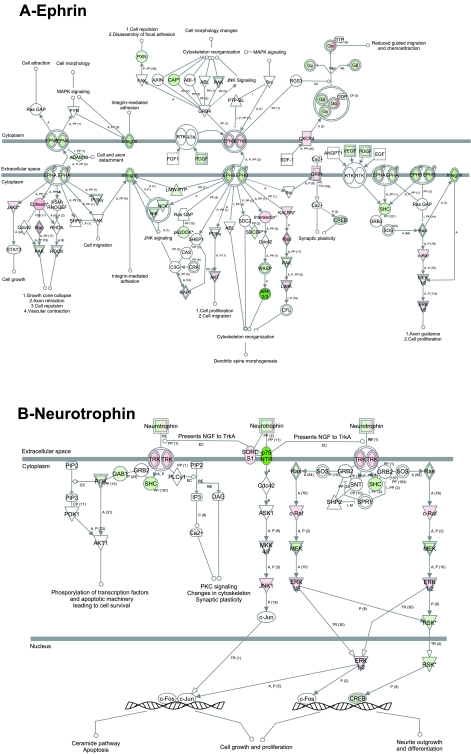
Ephrin and neurotrophin signaling pathways in primary neurons. Analysis using the Ingenuity Pathway Analysis software showed that ephrin (A) and neurotrophin (B) are among the pathways that control the highest number of neuronal-specific functions and showed a majority of genes have change in gene expression. Red, upregulated; green, downregulated; white, no change. Ephrin pathway: dynamic change in gene expression (up- and downregulation) is observed for molecules related to growth cone collapse, axon retraction, and cell repulsion, with upregulation of genes related to cell growth, dendritic spine morphogenesis, synaptic plasticity, and axon guidance. Neurotrophin pathway: there is a downregulation of genes related to apoptosis, and upregulation of genes related to synaptic plasticity; predominantly upregulation of genes related to neurite outgrowth.
The ephrin receptor pathway is involved in favoring growth cone collapse and axon retraction (PAK, Rac, ephrin A, Ephexin), cell proliferation and migration (ephrin B, AKT), and cell growth and axon guidance (ephrin B, integrin, c-raf) (Fig. 5A). Neurotrophic tyrosine kinase receptor signaling pathways control synaptic plasticity and cell survival, and neurite outgrowth (cAMP responsive element binding protein ribosomal protein S6 kinase) (Fig. 5B). The cyclin-dependent kinase 5 (CDK5) pathway showed upregulation of CDK5, and this is involved in neurite outgrowth (22). Genes involved in cytoskeletal filament disruption and proteasomal degradation were upregulated, providing dynamic changes needed for the formation/maintenance of neurites (Fig. 6A). Actin cytoskeleton signaling transcripts, including stress fibers and focal adhesion formation, were both up- and downregulated and involved in actin polymerization of lamellopodia and in stabilization of actin (Fig. 6B).
MMC-treatment of NSC induces a neuronal morphology.
After defining the neuronal signature characteristic of mature neurons, we investigated whether artificially blocking the cell cycle in the NSCs at the S phase can induce neuronal differentiation. In particular, since NSCs are known to differentiate when they are in G0/G1 (5, 37), we decided to investigate whether neuronal differentiation can take place when NSC are arrested in the S phase. We chose to use MMC as a nonspecific cell cycle blocker. MMC is an alkylating agent that blocks cell division mainly in the S phase (64). When C17.2 cells were treated with MMC (0.4 μg/ml), the majority of cells exhibited a neuronal morphology and stained positive for MAP2 (MAP2+ cells = 90.2 ± 3.1%, n = 8). MAP2 is a commonly utilized marker for mature neurons and is expressed mostly in dendrites (1, 47). Time course analysis from the time of induction up to 8 days revealed a change into neuronal-like cell shape (Fig. 7, A, d, h and B, d, h). In addition, there was a progressive increase in the expression of MAP2 protein that doubled from day1 to day 3 and then stabilized (Fig. 7, A, a, e, i, j and B, a, e). Nestin intensity decreased progressively, but was still detected (Fig. 7, A, b, f, i, j and B, b, f). Western blot analysis showed that the late neuronal marker NeuN had a significant (P < 0.005), approximately fivefold increase from day 1 to day 6 (Fig. 7, A, i, j). These cells showed only a low baseline level of cell death (Fig. 7, B, i, j). Since the neuronal phenotype obtained at day 6 remained stable afterwards (up to 3 wk, data not shown), as verified by various criteria described above, cells were incubated for a total of 8 days before microarray analysis was performed.
Fig. 7.
Generation and characterization of neuronal cells after MMC induction of NSCs. C17.2 NSCs were treated with 0.4 μg/ml of MMC under normoxic conditions, and immunostaining for the stem cell marker nestin and early neuronal marker NeuN is shown at 2 time points: 1 day and 6 days after treatment (A). Nestin was detected at day 1 (A, b) and decreased in intensity afterwards but remained detectable (A, f). MAP2 was weakly expressed at day 1 (A, a) and increased in intensity afterwards (A, e). Western blot analysis showed 2-fold increase in MAP2 from day 1 to day 3 and a significant (P < 0.005) ∼5-fold increase of NeuN from day 1 to day 6 (A, i, j). After 6 days of culture when the phenotype stabilized, cells were further cultured for an additional 2 days (total 8 days) and then characterized similarly. They were stained for nestin (B, b), MAP2 (B, a, e), and NeuN (B, f). Phase contrast microscopy confirmed the neuronal morphology (B, i). Mortality was only at a baseline level (B, j). Magnification: ×100; insets, ×400. Student's t-test, NeuN level at day 1 vs. day 6, P < 0.005.
Gene expression analysis of MMC-treated NSCs showed signs of partial neuronal differentiation and low differences from NSC.
Microarray data performed on MMC-treated NSCs revealed 1,076 genes with more than twofold change relative to NSCs, with 595 genes upregulated and 481 downregulated (Fig. 2A). Examination of all the possible pairwise Euclidian distance correlation values across each array indicated that MMC-treated NSC were much more similar to NSC than to PNs (Fig. 2B). This was also highlighted by examination of clustered transcripts (Fig. 3). The majority of the upregulated genes were related to functions such as growth factor activity, hormone activity, lysosome, antiapoptosis, and cell cycle and growth (Table 2), as well as Notch genes (Table 5). The majority of the downregulated genes were related to cell division and metabolic processes (Table 2, Fig. 3).
Table 2.
Cellular functions with genes changed in mitomycin-C-treated cells relative to neural stem cells
| GO Type | GO Name | Number Changed | Number Measured | Number in GO | Adjusted P Value |
|---|---|---|---|---|---|
| Downregulated | |||||
| C | Chromosome | 65 | 311 | 340 | <0.0001 |
| P | DNA replication | 39 | 134 | 138 | <0.0001 |
| P | Cell cycle | 82 | 593 | 633 | <0.0001 |
| P | Cell division | 45 | 239 | 249 | <0.0001 |
| C | Nucleus | 182 | 3642 | 3941 | <0.0001 |
| P | DNA packaging | 25 | 116 | 126 | <0.0001 |
| P | Chromosome organization and biogenesis | 40 | 325 | 346 | <0.0001 |
| C | Protein-DNA complex | 19 | 94 | 108 | <0.0001 |
| P | DNA repair | 25 | 175 | 189 | <0.0001 |
| P | DNA geometric change | 4 | 7 | 7 | <0.0001 |
| F | DNA clamp loader activity | 3 | 4 | 4 | <0.0001 |
| P | Deoxyribonucleotide biosynthetic process | 3 | 5 | 6 | 0.025 |
| F | DNA helicase activity | 5 | 16 | 17 | <0.0001 |
| P | DNA recombination | 11 | 76 | 84 | <0.0001 |
| F | Single-stranded DNA binding | 5 | 18 | 20 | <0.0001 |
| F | ATP binding | 49 | 1143 | 1237 | <0.0001 |
| P | Mitochondrial DNA metabolic process | 4 | 17 | 18 | 0.047 |
| C | Spindle | 6 | 36 | 40 | <0.0001 |
| P | Microtubule-based movement | 9 | 76 | 79 | <0.0001 |
| F | DNA-directed DNA polymerase activity | 5 | 27 | 27 | <0.0001 |
| F | Microtubule motor activity | 7 | 51 | 54 | <0.0001 |
| P | Nucleobase, nucleoside, nucleotide and nucleic acid transport | 7 | 56 | 57 | <0.0001 |
| P | Plastid DNA metabolic process | 5 | 32 | 34 | 0.025 |
| P | Isoprenoid biosynthetic process | 3 | 13 | 17 | 0.047 |
| F | Damaged DNA binding | 4 | 23 | 23 | 0.025 |
| C | Microtubule associated complex | 8 | 81 | 84 | <0.0001 |
| C | Microtubule | 13 | 187 | 206 | <0.0001 |
| P | RNA splicing | 12 | 170 | 183 | <0.0001 |
| P | Intracellular protein transport across a membrane | 6 | 57 | 58 | <0.0001 |
| P | mRNA processing | 13 | 218 | 234 | <0.0001 |
| F | ATP-dependent helicase activity | 6 | 62 | 66 | <0.0001 |
| P | Microtubule cytoskeleton organization and biogenesis | 6 | 68 | 75 | 0.025 |
| P | Purine salvage | 47 | 1717 | 1843 | <0.0001 |
| Upregulated | |||||
| C | Extracellular region | 90 | 1422 | 1562 | <0.0001 |
| F | Growth factor activity | 18 | 135 | 149 | <0.0001 |
| P | Collagen catabolic process | 5 | 16 | 16 | <0.0001 |
| P | Positive regulation of cell proliferation | 19 | 191 | 208 | <0.0001 |
| F | Hexosaminidase activity | 4 | 14 | 14 | 0.045 |
| F | Hormone activity | 11 | 104 | 111 | <0.0001 |
| P | Nucleosome assembly | 9 | 87 | 94 | 0.045 |
| C | Nucleosome | 9 | 87 | 99 | <0.0001 |
| P | Positive regulation of lymphocyte activation | 8 | 72 | 80 | <0.0001 |
| C | Lysosome | 13 | 167 | 177 | <0.0001 |
| P | Regulation of lymphocyte activation | 9 | 103 | 111 | <0.0001 |
| P | Antiapoptosis | 7 | 71 | 74 | <0.0001 |
| P | Cell cycle arrest | 5 | 43 | 46 | <0.0001 |
| P | Regulation of lymphocyte proliferation | 6 | 59 | 63 | 0.045 |
| P | Response to stress | 38 | 931 | 1034 | 0.045 |
| P | Negative regulation of programmed cell death | 11 | 175 | 188 | <0.0001 |
| P | Growth | 17 | 339 | 368 | 0.045 |
Abbreviations: GO, Gene Ontology; P, biological process; C, cellular component; F, molecular function.
Table 5.
Molecules functionally neuronal-related, commonly upregulated in primary neurons, and MMC-treated NSC
| Molecules Upregulated in PN and MMC-treated NSC |
|---|
| Cholinergic receptors |
| Neural cell adhesion molecule 2 |
| Neuron-glia CAM-related cell adhesion molecule |
| Neural growth factor |
| Insulin-like growth factor 1 |
| Matrix metalloproteases |
| Genes of the notch pathway |
PN, primary neurons; MMC, mitomycin C; NSCs, neural stem cells; CAM, cell adhesion molecule.
When the MMC-treated NSC genes were categorized based on GO, the changes in gene expression were involved in cell cycle and DNA packaging (Table 2). Functional grouping using the Ingenuity software showed 17 biofunctions with significant changes in gene expression (Fig. 4B). MMC-treated NSCs expressed a few neuronal molecules such as cholinergic receptors and molecules previously shown to be involved in neuronal differentiation (NGF) (11, 16, 61) and survival (IGF-1) (29).
Comparison of the differentially expressed transcripts between MMC-treated NSCs and NSCs versus PNs and NSCs showed that some cellular functions were shared between these two groups. Both groups contained upregulated transcripts involved in neurotransmitter secretion (e.g., protein and cellular localization), synaptogenesis, Notch signaling, nucleus and nucleotide metabolic processes, and downregulation of genes related to cell cycle/division and ribosomes, chromatin, and DNA binding (Fig. 3, Tables 3 and 5). Altogether, these data suggest that the MMC-treated NSCs show signs of partial neuronal differentiation, but they are not at the stage of a fully differentiated PNs (Fig. 3).
Table 3.
Cellular functions with genes changed in primary neurons relative to mitomycin C-treated cells
| GO Type | GO Name | Number Changed | Number Measured | Number in GO | Adjusted P Value |
|---|---|---|---|---|---|
| Downregulated | |||||
| C | Cytoplasm | 812 | 5192 | 5645 | <0.0001 |
| P | Immune system process | 196 | 1031 | 1133 | <0.0001 |
| P | Cell death | 141 | 674 | 730 | <0.0001 |
| F | Protein binding | 662 | 4602 | 5018 | <0.0001 |
| P | Blood vessel development | 54 | 197 | 215 | <0.0001 |
| F | Catalytic activity | 626 | 4431 | 4851 | <0.0001 |
| C | Extracellular matrix | 60 | 249 | 271 | <0.0001 |
| P | Metabolic process | 792 | 5907 | 6430 | 0.029 |
| F | l-Ascorbic acid binding | 10 | 17 | 19 | <0.0001 |
| P | Cell activation | 65 | 287 | 317 | <0.0001 |
| P | Homeostasis of number of cells | 67 | 301 | 327 | <0.0001 |
| P | Cell proliferation | 97 | 507 | 553 | <0.0001 |
| P | Actin filament-based process | 40 | 157 | 175 | <0.0001 |
| P | Response to molecule of bacterial origin | 12 | 26 | 31 | <0.0001 |
| P | Extracellular matrix organization and biogenesis | 18 | 53 | 59 | <0.0001 |
| P | Transcription factor import into nucleus | 7 | 12 | 14 | <0.0001 |
| C | Actin filament bundle | 8 | 15 | 20 | <0.0001 |
| P | Cytokine production | 31 | 118 | 127 | <0.0001 |
| P | Activation of NF-κB-inducing kinase activity | 5 | 7 | 7 | 0.016 |
| C | Cyclin-dependent protein kinase Holoenzyme complex | 5 | 7 | 7 | <0.0001 |
| P | Tube development | 45 | 201 | 219 | <0.0001 |
| P | Response to stress | 152 | 931 | 1034 | <0.0001 |
| P | Biological adhesion | 96 | 537 | 589 | <0.0001 |
| P | Mitochondrial depolarization | 3 | 3 | 3 | 0.029 |
| P | SMAD protein nuclear translocation | 3 | 3 | 3 | 0.0405 |
| P | Cardiac muscle growth | 7 | 13 | 13 | <0.0001 |
| P | Gliogenesis | 66 | 346 | 374 | <0.0001 |
| P | Regulation of muscle development | 7 | 14 | 14 | <0.0001 |
| C | MHC class I protein complex | 7 | 14 | 17 | <0.0001 |
| P | Muscle attachment | 7 | 14 | 14 | <0.0001 |
| P | Skeletal development | 42 | 195 | 211 | <0.0001 |
| P | Ossification | 33 | 143 | 152 | <0.0001 |
| P | Pharyngeal muscle development | 8 | 18 | 18 | <0.0001 |
| C | Cell-substrate adherens junction | 11 | 30 | 32 | <0.0001 |
| P | Epithelial cell differentiation | 22 | 83 | 89 | <0.0001 |
| P | G1/S transition checkpoint | 4 | 6 | 7 | 0.0405 |
| F | Calcium-dependent phospholipid binding | 8 | 19 | 20 | 0.016 |
| C | Integrin complex | 9 | 23 | 25 | <0.0001 |
| P | Cell growth | 26 | 108 | 114 | <0.0001 |
| P | Chordate embryonic development | 56 | 295 | 320 | <0.0001 |
| P | Regulation of intracellular protein transport | 7 | 16 | 18 | 0.016 |
| P | Vacuole organization and biogenesis | 7 | 16 | 18 | 0.016 |
| P | Liver development | 10 | 28 | 31 | 0.029 |
| P | Radial glial cell differentiation | 44 | 222 | 232 | <0.0001 |
| P | Cell migration | 54 | 291 | 321 | 0.016 |
| F | Copper ion binding | 13 | 45 | 52 | <0.0001 |
| P | Membrane invagination | 33 | 162 | 178 | <0.0001 |
| P | Regulation of angiogenesis | 9 | 27 | 30 | <0.0001 |
| P | Asexual reproduction | 13 | 47 | 49 | 0.029 |
| P | Brown fat cell differentiation | 14 | 53 | 56 | 0.016 |
| F | Transcription repressor activity | 30 | 148 | 161 | <0.0001 |
| P | Transition metal ion transport | 14 | 54 | 58 | 0.016 |
| F | Transcription activator activity | 34 | 178 | 190 | 0.016 |
| P | Developmental maturation | 23 | 115 | 125 | 0.016 |
| Upregulated | |||||
| C | Synapse | 97 | 231 | 255 | <0.0001 |
| P | Neurite development | 88 | 201 | 222 | <0.0001 |
| C | Neuron projection | 62 | 131 | 141 | <0.0001 |
| P | Synaptic transmission | 78 | 188 | 205 | <0.0001 |
| F | Protein binding | 846 | 4602 | 5018 | <0.0001 |
| C | Cell junction | 116 | 401 | 432 | <0.0001 |
| P | Localization | 496 | 2607 | 2814 | <0.0001 |
| C | Clathrin-coated vesicle | 31 | 68 | 72 | <0.0001 |
| P | Telencephalon development | 25 | 51 | 55 | <0.0001 |
| P | Small GTPase mediated signal transduction | 103 | 398 | 423 | <0.0001 |
| P | Learning and/or memory | 29 | 69 | 76 | <0.0001 |
| P | Biological adhesion | 126 | 537 | 589 | <0.0001 |
| P | Periplasmic space organization and biogenesis | 57 | 196 | 223 | <0.0001 |
| F | Calcium ion binding | 161 | 753 | 810 | <0.0001 |
| F | Kinase activity | 162 | 760 | 836 | <0.0001 |
| C | Synaptosome | 23 | 56 | 59 | <0.0001 |
| P | Microtubule-based process | 55 | 197 | 211 | <0.0001 |
| F | α-N-acetylneuraminate α-2,8-Sialyltransferase activity | 5 | 5 | 5 | <0.0001 |
| F | Substrate-specific transmembrane Transporter activity | 134 | 634 | 697 | <0.0001 |
| F | Gated channel activity | 63 | 246 | 267 | <0.0001 |
| F | Glutamate receptor activity | 12 | 23 | 30 | <0.0001 |
| P | Neuron recognition | 8 | 12 | 12 | <0.0001 |
| C | Cytoskeleton | 158 | 783 | 869 | <0.0001 |
| P | Membrane depolarization | 13 | 27 | 30 | 0.0145 |
| C | Cell soma | 24 | 68 | 73 | <0.0001 |
| P | Central nervous system neuron differentiation | 15 | 34 | 36 | <0.0001 |
| C | Heterotrimeric G protein complex | 14 | 31 | 34 | <0.0001 |
| F | GTP binding | 71 | 298 | 314 | <0.0001 |
| F | Calcium channel regulator activity | 6 | 8 | 9 | 0.0145 |
| P | Glial cell fate commitment | 4 | 4 | 5 | 0.0145 |
| P | Cerebellar granular layer development | 4 | 4 | 5 | <0.0001 |
| P | Regulation of adenylate cyclase activity | 8 | 13 | 14 | <0.0001 |
| P | Protein amino acid phosphorylation | 124 | 597 | 654 | <0.0001 |
| C | Site of polarized growth | 10 | 19 | 21 | <0.0001 |
| P | Synapse organization and biogenesis | 19 | 51 | 56 | <0.0001 |
| F | AMP binding | 8 | 14 | 16 | <0.0001 |
| P | Adult behavior | 25 | 78 | 88 | <0.0001 |
| P | Neurite regeneration | 14 | 34 | 39 | <0.0001 |
| P | Negative regulation of neurogenesis | 13 | 31 | 33 | <0.0001 |
| F | GTPase regulator activity | 67 | 293 | 317 | <0.0001 |
| C | Ionotropic glutamate receptor complex | 7 | 12 | 13 | <0.0001 |
| P | Action potential propagation | 8 | 15 | 18 | 0.0145 |
| F | Protein phosphatase type 2A regulator activity | 5 | 7 | 8 | <0.0001 |
| C | cAMP-dependent protein kinase complex | 5 | 7 | 8 | <0.0001 |
| P | Sterol biosynthetic process | 12 | 29 | 34 | <0.0001 |
| F | UDP-galactose:β-N-acetylglucosamine β-1,3-galactosyltransferase activity | 4 | 5 | 5 | 0.0145 |
| P | Cerebral cortex neuron differentiation | 7 | 13 | 13 | 0.0145 |
| F | Kinase regulator activity | 18 | 54 | 58 | <0.0001 |
| P | Wnt receptor signaling pathway | 42 | 170 | 182 | <0.0001 |
| P | Membrane organization and biogenesis | 63 | 284 | 311 | <0.0001 |
| C | Golgi apparatus | 112 | 570 | 607 | <0.0001 |
| P | Positive regulation of lyase activity | 6 | 11 | 12 | 0.0145 |
| P | Positive regulation of cyclase activity | 6 | 11 | 12 | 0.0145 |
| C | Clathrin coat | 10 | 25 | 31 | <0.0001 |
| C | Protein serine/threonine phosphatase complex | 10 | 25 | 28 | 0.0145 |
| P | Nerve growth factor receptor signaling pathway | 16 | 50 | 55 | 0.0145 |
| P | Peptidyl/serine modification | 16 | 51 | 55 | 0.0145 |
| P | Protein polymerization | 15 | 47 | 51 | 0.0145 |
| F | Potassium ion binding | 27 | 104 | 111 | <0.0001 |
| P | Morphogenesis of embryonic epithelium | 18 | 61 | 64 | 0.0145 |
| F | Diacylglycerol binding | 16 | 52 | 54 | <0.0001 |
| P | Multicellular organismal response to stress | 10 | 27 | 31 | 0.0375 |
| C | Anchored to membrane | 30 | 121 | 129 | <0.0001 |
| P | BMP signaling pathway | 21 | 76 | 84 | 0.0145 |
| P | Regulation of cellular component size | 14 | 44 | 45 | 0.0145 |
| P | Regulation of neuron differentiation | 13 | 40 | 42 | 0.026 |
| F | GTPase activity | 27 | 107 | 113 | <0.0001 |
| P | Response to organic substance | 15 | 49 | 55 | <0.0001 |
| P | Sensory organ development | 52 | 244 | 276 | <0.0001 |
| C | Coated pit | 10 | 28 | 29 | 0.0375 |
| P | G protein signaling, coupled to IP3 second messenger (phospholipase C activating) | 10 | 29 | 31 | 0.048 |
| P | Sensory perception of pain | 9 | 25 | 26 | 0.0375 |
| P | Notch signaling pathway | 24 | 96 | 104 | 0.026 |
| P | Nucleosome assembly | 22 | 87 | 94 | 0.0145 |
| P | Visual behavior | 8 | 22 | 26 | 0.0375 |
GO, Gene Ontology; P, biological process; C, cellular component; F, molecular function.
DISCUSSION
In the present study, we aimed to identify the gene expression profile that can be used as a universal marker for a fully differentiated neuron. Although the characteristics of neurons vary with their types, a common basic neuronal identity is expected to be shared among all neurons. We also wanted to uncover genes that are potentially involved in stem cell maintenance, and determine whether a cell cycle block in NSCs induces the formation of fully differentiated neurons.
Gene expression profiling reveals major characteristics of PNs and NSCs.
In the present study, we have 1) identified the gene expression neuronal signature and major signaling pathways that characterize the neuronal phenotype, 2) demonstrated that artificially blocking the C17.2 NSC line cell cycle in the S phase is not sufficient to result in full differentiation into neurons, and 3) identified genes as potential new candidates involved in the maintenance and/or function of NSC.
First, the microarray data revealed for the first time a detailed profile of the gene expression patterns of embryonic PNs, including genes involved in neuron-specific functions. The identification of genes highly expressed in PNs, but not NSCs, allowed us to group them based on the functions they contribute to through GO analysis (Table 1). This analysis revealed genes involved in the maturation and differentiation of neurons. (Table 7). For example, some of these genes are implicated in neurite development, involving neurite regeneration, neuron recognition, and regulation of dendrite development. Other genes are implicated in synapse, involving GABA receptor and glutamate receptor formation. Yet other genes are implicated in synaptic transmission, involving chemoreceptor signaling. All of these structures and components are needed to have a mature neuron characterized by the presence of axons and dendrites, special cell-cell communication (synapse), excitability, and release of neurotransmitter. All these features are basic characteristics of a fully differentiated neuron, and not present in a nondifferentiated NSC. From this, we conclude that these upregulated genes are neuronal specific, and therefore constitute a basic neuronal signature that characterizes fully differentiated neurons.
Table 7.
Neuronal-specific functions with enrichment of altered transcripts in primary neuron relative to neural stem cells, constituting the neuronal signature
| Neurite development |
|---|
| Synapse |
| Neuron projection |
| Synaptic transmission |
| Synaptic vesicle |
| Telencephalon development |
| Neuron recognition |
| Learning and memory |
A number of pathways with the most significant enrichment of altered transcripts were overrepresented. The ephrin receptor pathway is known to play a pivotal role in axon guidance (12, 27). Neurotrophins are involved in survival, axon outgrowth and synaptic plasticity (55). The CDK5 pathway has multiple changes affecting proteasomal degradation and cytoskeletal filament disruption, indicating a role of this pathway in the dynamic remodeling needed in mature neurons, complementing what has been previously described about the role of CDK5 in neurogenesis (22). The change observed in the actin pathway reflects local actin instability correlating with what is known about actin instability playing a role in axon polarization (64) and in secretion of neurotransmitter (10).
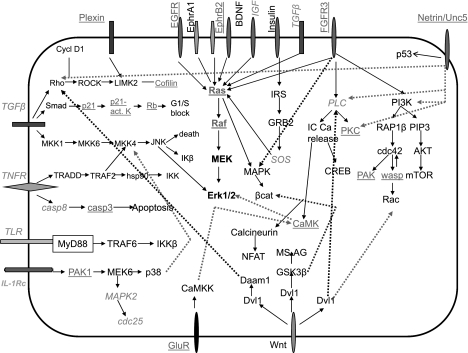
The general gene expression neuronal signature allowed us to draw a model reflecting how differentiated neurons stabilize their phenotype (Fig. 8). Growth factors, glutamate receptors, and netrin, which are mostly upregulated, activate the Ras and PLC pathways, resulting in signaling cascades leading to various cellular activities involved in axon formation, guidance, remodeling, attraction of growth cones, synapse, stress fiber activation, and cytoskeletal remodeling, as well as global effects such as long-term potentiation, learning, and memory. Furthermore, multiple cytokines are downregulated; although the specific function of most of these in neuronal and stem cell physiology remains unclear, we hypothesize that they play a role in preserving the stem cell phenotype and/or in regulating differentiation into neurons.
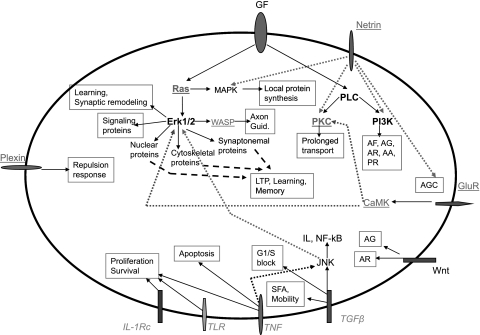
Fig. 8.
Maintenance of neuronal phenotype in a primary neuron: suggested model. Molecules at the cell surface represent receptors and/or ligands, as indicated. Molecules underlined are upregulated, and molecules in italics are downregulated, in microarray analysis. Some major neuronal-specific functions and the upstream signals are involved in its maintenance. Growth factors are indicated in boldface type, since some are up- and some are downregulated (see Table 3). All other molecules are not changed on the microarray analysis. GF, growth factors; AF, axon formation; AG, axon guidance; AR, axon remodeling; AA, anti-apotptosis; PR, proliferation response; axon guid, axon guidance; LTP, long-term potentiation; SFA, stress fiber activation; GluR. glutamate receptor; AGC, attraction of growth cone.
Second, to verify whether an artificial block in the S phase of the cell cycle in NSCs is sufficient to allow full differentiation into neurons, we examined the gene expression profile of C17.2 NSCs treated with MMC. Microarray analysis using these cells showed some aspects of neuronal differentiation. For example, these cells 1) express molecules such as cholinergic receptors (Table 5); 2) upregulate NGF, which has been previously shown to play a role in neuronal differentiation (11, 16, 61); 3) upregulate IGF-1, known to be important for neuronal survival (3, 11); 4) metalloproteases are upregulated and tissue inhibitors of metalloproteases are downregulated, correlating with a known role in migration/differentiation (23); and 5) Notch genes are upregulated (Table 5), and these are important in the function of mature neurons and, in particular, neurite remodeling, synapses, learning, and memory (33, 49). These cells, however, lack most of the genes observed in PNs (Table 3), suggesting incomplete differentiation. These data demonstrate that artificially arresting the NSC cycle in the S phase is not sufficient on its own to produce fully differentiated neurons; other factors seem to be required, such as the presence of astrocytes (59).
The results of our study add deep and broader knowledge to a previous study using the same NSC line. In that study by Mi et al (44), there was a comparison of certain characteristics of the NSC line with primary NSCs (primary cortical NSC and cerebellar granule cells). There, the authors, using neurosphere cultures and restricted stem cell microarray, showed that, while primary NSCs share common features, they do, however, show minor differences, suggesting that stem cells from different brain locations can have different properties. In addition, they compared the primary NSC with the NSC line, and showed similarities in the stem cell characteristics. At the same time, the cell line displayed a broader cytokine array than the primary NSCs. Given the similarities between the C17.2 NSC line and primary NSCs, we elected to use that NSC line since this provides insight for potential applications related to regenerative medicine where the use of cell lines is more feasible than primary NSCs.
In our present study, we studied the gene expression profile using the complete mouse expression microarray of the NSCs and compared it to that of cortical neurons to elucidate the major characteristics of mature neurons at gene expression level, which allowed us to determine the gene expression neuronal signature that defines mature neurons. We also blocked the cell cycle of the NSC at the S phase and analyzed the phenotype of the resulting cells. Since such cells had neuronal morphology and expressed known neuronal markers, we analyzed the gene expression profile compared with cortical neurons. To our surprise, we found that the differentiated cells lack most of the neuron-specific gene expression, suggesting that, while some cells can display a neuronal morphology and express some neuronal markers, they are not necessarily mature neurons. Thus, we suggest the use of the gene expression neuronal signature as the means to define whether some cells are mature neurons, instead of relying on their microscopic features.
Third, in an attempt to discover new genes that could play a role in stem cell biology, we examined the genes that downregulated in PNs compared with NSCs and which were not involved in housekeeping function and cell division. The cytokine group attracted our attention because there is evidence in the literature that cytokines play a role in self renewal (3). Our findings add new candidates to the families of TNF, tumor growth factor (TGF), IFN, and interleukins as playing a role in the stem cell maintenance and/or function (Table 6). Indeed, a previous study (42) has provided evidence for the role of cytokines in neural stem and progenitor cells.
Table 6.
Cytokine and cytokine-related molecules down-regulated in primary neurons relative to neural stem cells
| IL-1, -12, -15, -17 receptors |
|---|
| IL-28 receptor-α |
| IL-1, -12 Rc associated protein |
| TLR2, TLR6 |
| IFN-α, -β, IFN Rc2, IFN-γ Rc-1, IFN regulating factor 1 |
| TGF-β1, -β3, -β-induced, |
| TGF-induced factor homeobox 1 |
| TNF-Rc-10β, TNF-associated protein 1 |
| TNF-ligand 12 |
TGF, tumor growth factor; TLR, toll-like receptor.
Maintaining the phenotype in a differentiated neuron: intrinsic regulation.
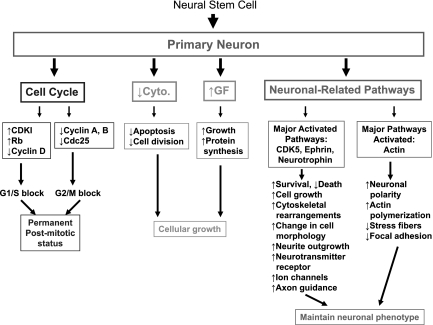
Since a wide array of genes had changes in expression levels in PNs compared with the NSCs, we wanted to use the microarray data to construct a model of how a fully differentiated neuron maintains its phenotype in an in vitro setting, i.e., independently of the presence of glial cells or other exogenous factors.
The first question is related to the gene profile that maintains neurons in a differentiated state. By examining genes involved in a cell cycle, we were able to generate a model revealing that changes in the expression of various genes can actually lock the cell in a nondividing state (Fig. 9). The model is rather straightforward: molecules that promote cell division, such as cyclins, MAPK, and CDKs, were downregulated, in particular, cyclins B and -A, and cdc25, which can result in/maintain a G2/M block. In addition, CDK4, and cyclin D were downregulated, while molecules opposing cell division, such as Rb and p21, were upregulated, maintaining a G1/S arrest. So the cell cycle in a PN is stopped at the level of two check points. Furthermore, when we examine the effect of growth factors on a cell cycle, although they normally activate molecules involved in cell division, these molecules, such as cyclins, are downregulated, therefore securing a cell cycle block and locking the neuron in a nondividing status.
Fig. 9.
Maintenance of cell cycle block in primary neurons. Microarray analysis revealed changes in genes expression of molecules involved in cell cycle that were either downregulated (italics) or upregulated (underlined). Downregulated molecules include factors involved in promoting cell cycle, such as cyclins and CDKs, and upregulated molecules were mostly antiproliferative, such as Rb and p21. The pattern of change in the level of these molecules can maintain a block in cell cycle at two levels: G1/S block and G2/M block, which will keep the cell nondividing, despite the presence of stimulatory growth factors.
We then wanted to understand what gene expression profiles can define the role of growth factors and cytokines in the differentiation of neurons. The microarray data analysis allowed us to generate a model related to the function of the cytokines and growth factors in neuronal differentiation (Fig. 10). For example, multiple downregulated cytokines are normally involved in cell division (IL-1, TGF-β) or inducing cell death (TNF). However, these cytokines are downregulated in PNs, thus reducing the probability for apoptosis, and preventing cell division, thus maintaining the nondividing status of PNs. Although TGF-β itself is downregulated, molecules downstream of it, such as p21 and RB, are upregulated, therefore participating in maintaining a postmitotic status.
Fig. 10.
Major pathways involved in survival and growth in primary neurons. Microarray analysis revealed changes in gene expression levels of many cytokines (that were mainly downregulated, in italics), and growth factors that were mainly upregulated (underlined). The data showed molecules that can induce apoptosis and cell division were downregulated, and those that can induce cell cycle block were upregulated. Many growth-promoting factors were upregulated, leading to downstream pathways that favor protein synthesis, cellular growth, and cytoskeleton rearrangements. IKβ, inhibitor of κB; IKKβ, inhibitor of κB kinase.
There were a number of molecules in growth factor pathways that had a gene expression level that was changed in a direction favoring the activation of Ras/Erk pathway. For example, EGFR, ephrin B2, FGFR3, Ras, and Raf, were all upregulated, while an inhibitor of Ras, ephrin A1, was downregulated. Erk pathway is known to induce/maintain differentiation, so the expression pattern of molecules involved in this pathway is in favor of activating this pathway, in addition to CamK, an Erk1/2 activator, which was also upregulated. At the same time, there is an upregulation of netrin and its receptor Unc5. This receptor activates multiple pathways including PKC, PI3K, and Rho. PKC is involved in various functions such as cell survival, differentiation, and protein synthesis. PI3K is involved in survival, and Rho is involved in cytoskeletal remodeling. Altogether, these molecular expression changes illustrate some of the mechanisms of how neurons maintain basic functions related to differentiation and survival, i.e., by upregulating molecules in favor of such status and downregulating the corresponding inhibitors.
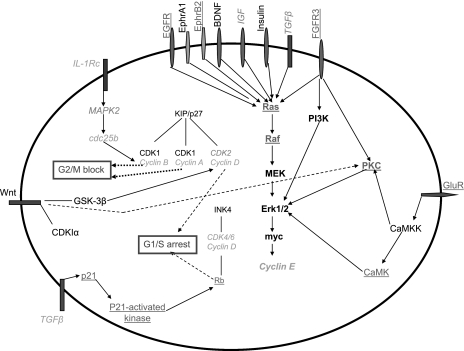
Finally, we asked how the neuron-specific characteristics are maintained, and examined the pathways of CDK5, ephrin, neurotrophin, and actin, as these pathways had a very significant enrichment of genes that changed their expression levels (Fig. 11). Pathways downstream of CDK5, in addition to minimizing neuronal death, favor neurite outgrowth, as well as the production of neurotransmitter receptors and ion channels. The ephrin pathway promotes cell growth and is also involved in axon guidance, cytoskeletal reorganization, and changes in cell morphology. These are important for maintaining the neuronal phenotype and for proper connectivity between neurons. Neurotrophins are also involved in neurite outgrowth and cytoskeletal changes, complementing the roles of CDK5 and ephrin. Finally, actin plays an important role in neuronal polarity and adhesion, both of which characterize the neuronal phenotype. Taken together, this analysis has revealed a mechanistic view as to how the phenotype of a mature neuron is maintained and demonstrated that gene expression profiling can give useful information to understand the major pathways involved in differentiation.
Fig. 11.
Gene expression changes maintain the phenotype of a differentiated neuron. A proposed model for how changes in gene expression that take place when a NSC differentiates into a neuron and maintains the phenotype of the mature neuron. Changes in cell cycle genes maintain a permanent block of cell division. Changes in cytokines (Cyto.) and growth factors (GF) assure the survival and growth of the cell; Finally, neuronal-related pathways are activated, and changes in gene expression of many genes in those pathways assure the presence of the basic constituents of a neuron, such as neurotransmitter, neurite, polarity, etc.
Neuronal differentiation and the interplay of multiple pathways.
From our results, we can extract a potential model that would integrate multiple pathways to allow for full neuronal differentiation. For a stem cell to become a differentiated nerve cell, the stem cell has to grow in size, undergo a series of morphological changes and cytoskeletal rearrangements, acquire polarity, and initiate neurite outgrowth, along with the expression of neurotransmitters and other molecules. Our data showed that the pathways of CDK5, ephrin, neurotrophin, and actin all participate in these processes. Blocking the cell cycle, at least in some contexts, does not provide enough signals needed to trigger all pathways required to form a mature neuron.
Concluding remarks.
The present study revealed details about the gene expression profile in PNs, which showed both expected and unexpected differences from NSCs. It also revealed that there are gene groups that are not known to be stem cell specific and yet are downregulated in PNs relative to stem cells (cytokines), pointing to a role in the NSCs. Our study has also allowed the elucidation of neuronal differentiation mechanisms.
Although this study has been conducted in an in vitro context, it has potential clinical implications. For example, in conditions where neurons are generated in vitro, the maturity of these neurons should be verified by the presence of the defined neuronal signature. This application can be further expanded to cases where NSCs are transplanted into recipient animals for regenerative purposes; neurons generated in vivo from these cells could eventually be extracted and verified for maturity. Comparison of the adult PNs with the embryonic PNs will also shed some light on the functional differences between mature neurons at various developmental stages.
Perspectives and Significance
Determining the various cues and triggers of the different stages in the formation of a mature neuron will allow better targeting of the neuronal differentiation. Time course analysis of gene expression of differentiating PNs will shed light on key changes needed for the formation of mature neurons.
GRANTS
This work was supported by National Institutes of Health (NIH) Grant P01-HD-032573 (to G. G. Haddad), American Heart Association Scientist Development Grant 10S-DG-2630130 and NIH Institutional Research and Academic Career Development Award GM-068524 (to A. C. Zambon), NIH Grant 1-DP2-OD006495–01 (to A. Muotri), a Parker B. Francis Fellowship Grant (to J. Xue), and American Heart Association Grant 0835188N (to D. Zhou).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
ACKNOWLEDGMENTS
We thank Shirley Reynolds for excellent laboratory management and assistance and Dennis Young for the flow cytometry acquisition and analysis. We also thank James Prague, Mila Angert, and Roman Sassik for performing the microarray experiments and pathway analysis.
REFERENCES
- 1. Abi Farah C, Leclerc N. HMWMAP2: new perspectives on a pathway to dendritic identity. Cell Motil Cytoskel 65: 515–527, 2008 [DOI] [PubMed] [Google Scholar]
- 2. Andres RH, Choi R, Steinberg GK, Guzman R. Potential for adult neural stem cells in stroke therapy. Regen Med 3: 893–905, 2008 [DOI] [PubMed] [Google Scholar]
- 3. Bauer S. Cytokine control of adult neural stem cells chronic versus acute exposure. Ann NY Acad Sci 1153: 48–56, 2009 [DOI] [PubMed] [Google Scholar]
- 4. Beatus P, Jhaveri DJ, Walker TL, Lacas PG, Rietze L, Cooper HM, Morikawa Y, Bartlett PF. Oncostatin M regulates neuronal precursor activity in the adult brain. Dev Neurobiol 71: 619–633, 2011 [DOI] [PubMed] [Google Scholar]
- 5. Becker EBE, Bonni A. Beyond proliferation-cell cycle control of neuronal survival and differentiation in the developing mammalian brain. Semin Cell Dev Biol 16: 439–448, 2005 [DOI] [PubMed] [Google Scholar]
- 6. Ben Menachem-Zidon O, Goshen I, Kreisel T, Ben Menahem Y, Reinhartz E, Ben Hur T, Yirmiya R. Intrahippocampal transplantation of transgenic neural precursor cells overexpressing interleukin 1 receptor antagonist blocks chronic isolation-induced impairment in memory and neurogenesis. Neuropsychopharmacology 33: 2251–2262, 2007 [DOI] [PubMed] [Google Scholar]
- 7. Bordey A. Adult neurogenesis–basic concept of signaling. Cell Cycle 5: 722–728, 2006 [DOI] [PubMed] [Google Scholar]
- 8. Buzanska L, Sypecka J, Nerini-Molteni S, Compagnoni A, Hogberg HT, del Torchio R, Domanska-Janik K, Zimmer J, Coecke S. A human stem cell-based model for identifying adverse effects of organic and inorganic chemicals on the developing nervous system. Stem Cells 27: 2591–2601, 2009 [DOI] [PubMed] [Google Scholar]
- 9. Chen HC, Ma HI, Sytwu HK, Wang HW, Chen CC, Liu SC, Chen CH, Chen HK, Wang CH. Neural stem cells secrete factors that promote auditory cell proliferation via a leukemia inhibitory factor signaling pathway. J Neurosci Res 88: 3308–3318, 2010 [DOI] [PubMed] [Google Scholar]
- 10. Cingolani LA, Goda Y. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat Rev 9: 344–356, 2008 [DOI] [PubMed] [Google Scholar]
- 11. Cirulli F, Alleva E. The NGF saga: from animal models of psychological stress to stress-related psychopathology. Front Neuroendocrinol 30: 379–395, 2009 [DOI] [PubMed] [Google Scholar]
- 12. Cooper HM. Axon guidance receptors direct growth cone pathfinding: rivalry at the leading edge. Int J Dev Biol 46: 621–631, 2002 [PubMed] [Google Scholar]
- 13. Daadi MM, Maag AL, Steinberg GK. Adherent self-renewable human embryonic stem cell-derived neural stem cell line: functional engraftment in experimental stroke model. PloS One 3: e1644, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene expression profile from microarray data. Genome Biol 4: R7, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dudoit S, Shater JP, Boldrick JC. Multiple hypothesis testing in microarray experiments. Statist Sci 18: 71–103, 2003 [Google Scholar]
- 16. Fiore M, Chaldakov GN, Aloe L. Nerve growth factor as a signaling molecule for nerve cells and also for the neuroendocrine-immune systems. Rev Neuosci 20: 133–145, 2009 [DOI] [PubMed] [Google Scholar]
- 17. Fritsche E, Cline JE, Nguyen NH, Scanlan TS, Abel J. Polychlorinated biphenyls disturb differentiation of normal human neural progenitor cells: clue for involvement of thyroid hormone receptors. Environ Health Perspect 113: 871–876, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gokhan S, Song Q, Mehler MF. Generation and regulation of developing immortalized neural cell lines. Methods 16: 345–358, 1998 [DOI] [PubMed] [Google Scholar]
- 20. Imayoshi I, Kageyama R. The role of notch signaling in adult neurogenesis. Mol Neurobiol. In press [DOI] [PubMed] [Google Scholar]
- 21. Imayoshi I, Sakamoto M, Kageyama R. Genetic methods to identify and manipulate neurons in the adult brain. Front Neurosci 5: 1–11, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jessberger S, Gage FH, Eisch AJ, Lagace DC. Making a neuron: Cdk5 in embryonic and adult neurogenesis. Trends Neurosci 32: 575–582, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kalluri HSG, Dempsey RJ. Growth factors, stem cells and stroke. Neurosurg Focus 24: E13, 2008 [DOI] [PubMed] [Google Scholar]
- 24. Kang KS, Park JE, Ryu DY, Lee YS. Effects and neuro-toxic mechanisms of 2,2′,4,4′,5,5′-hexachlorobiphenyl and endosulfan in neuronal stem cells. J Vet Med Sci 63: 1183–1190, 2001 [DOI] [PubMed] [Google Scholar]
- 25. Kempermann G. Adult Neurogenesis. New York: Oxford University Press, 2006 [Google Scholar]
- 26. Kennea NL, Mehmet H. Neural stem cells. J Pathol 197: 536–550, 2002 [DOI] [PubMed] [Google Scholar]
- 27. Koeberle PD, Bahr M. Growth and guidance cues for regenerating axons: where they have gone? J Neurobiol 59: 162–180, 2003 [DOI] [PubMed] [Google Scholar]
- 28. Kokaia Z, Lindvall O. Neurogenesis after ischaemic brain insults. Curr Opin Neurobiol 13: 127–132, 2003 [DOI] [PubMed] [Google Scholar]
- 29. Kooijman R, Sarre S, Michotte Y, De Keyser J. Insulin-like growth factor I: a potential neuroprotective compound for the treatment of acute ischemic stroke? Stroke 40: e83–e88, 2009 [DOI] [PubMed] [Google Scholar]
- 30. Kozlowska H, Jablonka J, Janowski M, Jurga M, Kossut M, Domańska-Janik K. Transplantation of a novel human cord blood-derived neural-like stem cell line in a rat model of cortical infarct. Stem Cells Dev 16: 481–488, 2007 [DOI] [PubMed] [Google Scholar]
- 31. Kriengstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci 32: 149–184, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lange C, Mix E, Frahm J, Glass A, Muller J, Schmitt O, Schmöle AC, Klemm K, Ortinau S, Hübner R, Frech MJ, Wree A, Rolfs A. Small molecule GSK-3 inhibitors increase neurogenesis of human neural progenitor cells. Neurosci Lett 488: 36–40, 2011 [DOI] [PubMed] [Google Scholar]
- 33. Lathia JD, Mattson MP, Cheng A. Notch: from neural development to neurological disorders. J Neurochem 107: 1471–1481, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee HJ, Kim KS, Kim ES, Choi HB, Lee KH, Park IH, Ko Y, Jeong SW, Kim SU. Brain transplantation of immortalized human neural stem cells promotes functional recovery in mouse intracerebral hemorrhage stroke model. Stem Cells 25: 1204–1212, 2007 [DOI] [PubMed] [Google Scholar]
- 35. Lee ST, Chu K, Jumg KH, Kim SJ, Kang M, Hong NH, Kim JH, Ban JJ, Park HK, Kim SU, Park CG, Lee SK, Kim M, Roh JK. Anti-inflammatory mechanism of intravascular neural stem cell transplantation in heamorrhagic stroke. Brain 131: 616–629, 2008 [DOI] [PubMed] [Google Scholar]
- 36. Li QJ, Tang YM, Liu J, Zhou DY, Li XP, Xiao SH, Jian DX, Xing YG. Treatment of Parkinson disease with C17.2 neural stem cells overexpressing NURR1 with a recombined republic-deficit adenovirus containing the NURR1 gene. Synapse 61: 971–977, 2007 [DOI] [PubMed] [Google Scholar]
- 37. Liu DZ, Ander BP, Sharp FR. Cell cycle inhibition without disruption of neurogenesis is a strategy for treatment of central nervous system diseases. Neurobiol Dis 37: 549–557, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu WG, Lu GQ, Li B, Chen SD. Dopaminergic neuroprotection by neurturin-expressing C17.2 neural stem cells in a rat model of Parkinson's disease. Parkinsonism Relat Disord 13: 77–88, 2007 [DOI] [PubMed] [Google Scholar]
- 40. Liu YP, Lang BT, Baskaya MK, Dempsey RJ, Vemuganti R. The potential of neural stem cells to repair stroke-induced brain damage. Acta Neuropathol 117: 469–480, 2009 [DOI] [PubMed] [Google Scholar]
- 41. Lu P, Jones LL, Snyder EY, Tuszynski MH. Neural stem cells constitutively secrete neurotrophic factors and promote extensive host axonal growth after spinal cord injury. Exp Neurol 181: 115–129, 2003 [DOI] [PubMed] [Google Scholar]
- 42. Luo Y, Cai J, Liu Y, Xue H, Chrest FJ, Wersto RP, Rao M. Microarray analysis of seleted genes in neural stem and progenitor cells. J Neurochem 83: 1481–1497, 2002 [DOI] [PubMed] [Google Scholar]
- 43. Lynch WP, Sharpe AH, Snyder EY. Neural stem cells as engraftable packaging lines can mediate gene delivery to microglia: evidence from studying retroviral env-related neurodegeneration. J Virol 73: 6841–6851, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mi R, Luo Y, Cai J, Limke TL, Rao MS, Höke A. Immortalized neural stem cells differ from nonimmortalized cortical neurospheres and cerebellar granule cell progenitors. Exp Neurol 194: 301–319, 2005 [DOI] [PubMed] [Google Scholar]
- 45. Neville CM, Huang AY, Shyu JY, Snyder EY, Hadlock TA, Sundback CA. Neural precursor cell lines promote neurite branching. Int J Neurosci 119: 15–39, 2009 [DOI] [PubMed] [Google Scholar]
- 46. Nixon K, Morris SA, Liput DJ, Kelso ML. Roles of neural stem cells and adult neurogenesis in adolescent alcohol use disorders. Alcohol 44: 39–56, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nunez J. Immature and mature variants of MAP2 and tau proteins and neuronal plasticity. TINS 11: 477–479, 1998 [DOI] [PubMed] [Google Scholar]
- 48. Ourednik J, Ourednik V, Lynch WP, Schachner M, Snyder EY. Neural stem cells display an inherent mechanism for rescuing dysfunctional neurons. Nat Biotechnol 20: 1103–1110, 2002 [DOI] [PubMed] [Google Scholar]
- 49. Pierfelice TJ, Schreck KC, Eberhart CG, Gaiano N. Notch, neural stem cells, and brain tumors. Cold Spring Harb Symp Quant Biol 73: 367–375, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Riess P, Zhang C, Saatman KE, Laurer HL, Longhi LG, Raghupathi R, Lenzlinger PM, Lifshitz J, Boockvar J, Neugebauer E, Snyder EY, McIntosh TK. Transplanted neural stem cells survive, differentiate, and improve neurological motor function after experimental traumatic brain injury. Neurosurgery 51: 1043–1052, 2002 [DOI] [PubMed] [Google Scholar]
- 51. Rosario CM, Yandava BD, Kosaras B, Zurakowski D, Sidman RL, Snyder EY. Differentiation of engrafted multipotent neural progenitors towards replacement of missing granule neurons in meander tail cerebellum may help determine the locus of mutant gene action. Development 124: 4213–4224, 1997 [DOI] [PubMed] [Google Scholar]
- 52. Ryder EF, Snyder EY, Cepko CL. Establishment and characterization of multipotent neural cell lines using retrovirus vector-mediated oncogene transfer. J Neurobiol 21: 356–375, 1990 [DOI] [PubMed] [Google Scholar]
- 53. Ryu MY, Lee MA, Ahn YH, Kim KS, Yoon SH, Snyder EY, Cho KG, Kim SU. Brain transplantation of neural stem cells cotransduced with tyrosine hydroxylase and GTP cyclohydrolase 1 in Parkinsonian rats. Cell Transpl 14: 193–202, 2005 [DOI] [PubMed] [Google Scholar]
- 54. Sasik R, Woelk CH, Corbeil J. Microarray truths and consequences. J Mol Endocrinol 33: 1–9, 2004 [DOI] [PubMed] [Google Scholar]
- 55. Schecterson LC, Bothwell M. Neurotrophin receptors: old friends with new partners. Dev Neurobiol 70: 332–338, 2010 [DOI] [PubMed] [Google Scholar]
- 56. Sharma R, Ottenhof T, Rzeczkowska PA, Niles LP. Epigenetic targets for melatonin: induction of histone H3 hyperacetylation and gene exression in C17.2 neural stem cells. J Pineal Res 45: 277–284, 2008 [DOI] [PubMed] [Google Scholar]
- 57. Snyder EY, Deichter DL, Walsh C, Arnold-Aldea S, Hartweig EA, Cepko PL. Multipotent neural cell lines can engraft and participate in development of mouse cerebellum. Cell 68: 33–51, 1992 [DOI] [PubMed] [Google Scholar]
- 58. Snyder EY, Yoon Flax JD, Macklis JD. Multipotent neural precursors can differentiate toward replacement of neurons undergoing targeted apoptotic degeneration in adult mouse neocortex. Proc Natl Acad Sci USA 94: 11663–11668, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Song H, Stevens CF, Gage FH. Astroglia induce neurogenesis from adult neural stem cells. Nature 417: 39–44, 2002 [DOI] [PubMed] [Google Scholar]
- 60. Sugiura M, Nagaoka M, Yabuuchi H, Akaike T. Overexpression of MCT8 enhances the differentiation of ES cells into neural progenitors. Biochem Biophys Res Commun 360: 741–745, 2007 [DOI] [PubMed] [Google Scholar]
- 61. Takei Y, Laskey R. Interpreting crosstalk between TNF-α and NGF: potential implications for disease. Trends Mol Med 14: 381–388, 2008 [DOI] [PubMed] [Google Scholar]
- 62. Temple S, Alvarez-Buylla A. Stem cells in the adult mammalian central nervous system. Curr Opin Neurobiol 9: 135–141, 1999 [DOI] [PubMed] [Google Scholar]
- 63. Van der Laan M, Pollard K. A new algorithm for hybrid hierarchical clustering with visualization and the bootstrap. J Statist Plan Infer 117: 275–303, 2003 [Google Scholar]
- 64. Witte H, Bradke F. The role of the cytoskeleton during neuronal polarization. Curr Opin Neurobiol 18: 479–487, 2008 [DOI] [PubMed] [Google Scholar]
- 65. Xi-Qiao W, Ying-Kai L, Chun Q, Shu-Liang L. A review of the effectiveness of antimitotic drug injections for hypertrophic scars and keloids. Ann Plas Surg 63: 688–692, 2009 [DOI] [PubMed] [Google Scholar]
- 66. Yang Z, Duan H, Mo L, Qiao H, Li X. The effect of the dosage of NT-3/chitosan carriers on the proliferation and differentiation of neural stem cells. Biomaterials 31: 4846–4854, 2010 [DOI] [PubMed] [Google Scholar]
- 67. Zang Y, Yu LF, Pang T, Fang LP, Feng X. AICAR induces astroglial differentiation of neural stem cells via activating the JAK/STAT3 pathway independently of AMP-activated protein kinase. J Biol Chem 283: 6201–6208, 2007 [DOI] [PubMed] [Google Scholar]
- 68. Zhou D, Wang J, Zapala MA, Xue J, Schork NJ, Haddad GG. Gene expression in mouse brain following chronic hypoxia: role of sarcospan in glial cell death. Physiol Genomics 32: 370–379, 2008 [DOI] [PubMed] [Google Scholar]