Abstract
Mre11 is a critical participant in upkeep of nuclear DNA, its repair, replication, meiosis, and maintenance of telomeres. The upkeep of mitochondrial DNA (mtDNA) is less well characterized, and whether Mre11 participates has been unknown. We previously found that high NaCl causes some of the Mre11 to leave the nucleus, but we did not then attempt to localize it within the cytoplasm. In the present studies, we find Mre11 in mitochondria isolated from primary renal cells and show that the amount of Mre11 in mitochondria increases with elevation of extracellular NaCl. We confirm the presence of Mre11 in the mitochondria of cells by confocal microscopy and show that some of the Mre11 colocalizes with mtDNA. Furthermore, crosslinking of Mre11 to DNA followed by Mre11 immunoprecipitation directly demonstrates that some Mre11 binds to mtDNA. Abundant Mre11 is also present in tissue sections from normal mouse kidneys, colocalized with mitochondria of proximal tubule and thick ascending limb cells. To explore whether distribution of Mre11 changes with cell differentiation, we used an experimental model of tubule formation by culturing primary kidney cells in Matrigel matrix. In nondifferentiated cells, Mre11 is mostly in the nucleus, but it becomes mostly cytoplasmic upon cell differentiation. We conclude that Mre11 is present in mitochondria where it binds to mtDNA and that the amount in mitochondria varies depending on cellular stress and differentiation. Our results suggest a role for Mre11 in the maintenance of genome integrity in mitochondria in addition to its previously known role in maintenance of nuclear DNA.
Keywords: mitochondrial DNA damage, osmotic stress, NaCl, bleomycin
the mre11 complex has three components: Mre11, Rad50, and Nbs1. It is involved in pathways critical for DNA damage repair, DNA replication, meiosis, and telomere maintenance. The complex senses breaks in DNA, signals for activation of cell cycle checkpoints, and plays a central role in the repair of double-strand breaks (DSBs) by both nonhomologous end joining and homologous recombination. In addition to DNA repair, the Mre11 complex prevents formation of DSBs during DNA replication in both normal and stressed cells (reviewed in Ref. 4), and a recent study demonstrated that the complex is also involved in base excision repair (38). Mre11 is the core member of the complex. It interacts both with itself and with Rad50 and Nbs1. Mre11 has endonuclease and 3′-5′ exonuclease activities that process DNA ends for recognition by other DNA repair and cell cycle checkpoint proteins (23, 27, 31). In addition to its role in DNA processing, the Mre11 complex tethers DNA (8, 45). It has been proposed that Mre11 complex has an essential role in the repair of replication fork-associated lesions, which prevents accumulation of DNA damage (reviewed in Refs. 4 and 42). Considering its broad activities, the Mre11 complex is recognized to play a central role in all aspects of DNA metabolism and in maintenance of genomic stability (44, 45). The central role of the Mre11 complex in these processes is further supported by observations that null mutations in any of its three proteins leads to embryonic lethality in mice (49). Also, mutation in humans of any of the genes encoding the components of Mre11 complex lead to Nijmegen breakage syndrome and ataxia telangiectasia-like disorder, which are characterized by checkpoint deficiencies, chromosome instability, radio-resistant DNA synthesis, hypersensitivity to ionizing radiation and increased incidence of cancer (35, 39).
NaCl is a major solute in extracellular fluids. In peripheral blood it accounts for most of the osmolality, which normally is ∼300 mosmol/kg. In renal medullary interstitial fluid, however, NaCl normally is much higher, serving there to drive the urinary concentrating mechanism. Such high NaCl increases DNA breaks, not only in the renal medulla (13), but also in a variety of cells in culture (11, 24), in C. elegans (14), and in marine invertebrates (15). In addition to increasing DNA breaks, high NaCl also inhibits the repair of DNA breaks caused by high NaCl and other stresses, like radiation, and alters the function of DNA damage response proteins (10, 11).
Mre11 has been most studied as a component of the nuclear DNA repair network and is generally thought of as a nuclear protein. However, Mre11 has also been observed in the cytoplasm. Thermal stress (50) and high NaCl (9, 13) cause translocation of some of the nuclear Mre11 into the cytoplasm. However, the location and function of Mre11 in the cytoplasm has remained unknown. Since Mre11 is present in the proteome of yeast mitochondria (36), we suggested that it might also be present in mammalian mitochondria. Also, a possible role for Mre11 in mitochondria was suggested by observations that Mre11 binds to broken nuclear DNA and processes it for repair (40) and that mitochondria contain DNA (mtDNA) that is constantly being damaged by the reactive oxygen species (ROS) produced there (1, 3, 46). Mammalian mitochondria possess DNA repair pathways similar to those in the nucleus, namely base excision repair, mismatch repair, nonhomologous end joining, and homologous recombination (reviewed in Ref. 28). Although the pathways have been identified in mitochondria, the molecular mechanisms involved are less well characterized than in the nucleus. Human mitochondria contain multiple copies of a 16.5-kb circular DNA that encodes 13 polypeptides required for oxidative phosphorylation, 22 tRNAs, and two rRNAs. All other mitochondria proteins are encoded in nuclear DNA and are posttranslationally imported into mitochondria (30). The multiple mtDNA molecules are organized into discrete protein-DNA complexes called nucleoids (21, 37).
In the present studies, we find that Mre11 is present in the mitochondria of mammalian cells, both in culture and in the mouse kidney in vivo. We demonstrate that within mitochondria a fraction of Mre11 colocalizes with mitochondria nucleoids and binds to mtDNA. We suggest that Mre11 plays a role in the maintenance of mitochondrial genome integrity similar to its function in maintenance of nuclear DNA.
MATERIALS AND METHODS
Cell Cultures
mIMCD3 cells.
Subconfluent cultures of mIMCD3 cells (33) were used in passages 13–17 and were grown in medium containing 45% DMEM low glucose (Invitrogen, Carlsbad, CA), 45% F12 Coon's modification (cat. no. F6636; Sigma, St. Louis, MO), 10% fetal bovine serum (HyClone, Logan, UT), 2 mM l-glutamine, 10,000 U/ml penicillin G and 10,000 U/ml streptomycin sulfate. Osmolality of control medium, was 300–320 mosmol/kg. Hypertonic medium, prepared by adding NaCl, was substituted for the control medium, as indicated. Cells were incubated at 37°C and gassed with 5% CO2-95% air.
Preparation of mouse kidney primary cells.
Whole kidneys from one 2- to 3-mo-old mouse were cut in 2 × 2-mm pieces and digested in DMEM without phenol red (Invitrogen), containing 10% FBS (HyClone), 2 mM l-glutamine, 10,000 U/ml penicillin G, and 10,000 U/ml streptomycin sulfate, and supplemented with 2 mg/ml collagenase B (Roche Applied Science). The pieces of renal tissue were incubated for 90 min at 37°C in a humidified incubator (5% CO2-95% O2). The tissue suspension was mixed every 15 min by pipetting up and down 10 times. The resulting suspension was centrifuged at 160 g for 1 min and washed 3 times with DMEM containing 10% fetal bovine serum, 2 mM l-glutamine, 10,000 U/ml penicillin G, and 10,000 U/ml streptomycin sulfate. The cell suspension obtained from two kidneys was plated in four 10-cm plastic dishes. After they became confluent (3–4 days), cells were harvested by trypsinization and used for experiments.
Growing mouse kidney primary cells on Matrigel matrix.
Primary kidney cells, prepared as described above, were grown in a 1-mm layer of Matrigel basement membrane matrix (cat. no. 356234; BD Biosciences, Bedford, MA). After 7 days, some cells formed tubules and other cells remained undifferentiated. Cells and tubules were recovered from the Matrigel matrix using cell recovery solution (cat. no. 354253; BD Biosciences) and stained for Mre11 as described below. Also, some cells were exposed to high-NaCl media for 1 h before recovery from the Matrigel.
Mice
Mice were purchased at age of 2–3 mo (cat. nos. 129S6; Taconic Farms, Hudson, NY) and housed in the National Heart, Lung, and Blood Institute (NHLBI) animal facility. All mouse study protocols were reviewed and approved by the NHLBI, and mice were housed in an Association for Assessment and Accreditation of Laboratory Animal Care-accredited facility.
Immunohistochemical Detection of Mre11 in Paraffin Kidney Sections
Mouse kidneys were fixed overnight in 4% paraformaldehyde at 4°C, and then embedded in paraffin. Sections were cut and mounted on silanized slides by American Histolabs (Gaithersburg, MD). Sections were stained with anti-Mre11 (cat. no. 4895; Cell Signaling Technology, Danvers, MA) as previously described (12). A Nikon E800 Widefield Microscope was used for photography.
Immunofluorescent Detection of Mre11 and Mytochondia in Frozen Kidney Sections
Mouse kidneys were embedded in cryoembedding medium by the face down cryoembedding technique (32) (Pathology Innovations, Wyckoff, NJ). Sections were cut using a Leica cryostat and put on microscope slides. Sections were fixed with 2.5% formaldehyde (cat. no. 18814; Polyscence, Warrington, PA) for 10 min, washed 3 times, 5 min each, with 0.1% Triton X-100 in PBS (PBST) and treated with 3% H2O2 in PBS for 10 min to block endogenous peroxidase. Sections were blocked with 3% BSA in PBST for 1 h, then incubated overnight at 4°C with anti-Mre11 (rabbit) (cat. no. 4895; Cell Signaling Technology) and human anti-mitochondrial antigen (cat. no. HMS-0100; Immunovision) antibodies, followed by 1 h incubation with secondary antibodies, anti-rabbit-horseradish peroxidase (HRP), and anti-human Alexa Fluor 633 (Invitrogen). HRP was detected using tyramide signal amplification kit (cat. no. T20924; Invitrogen) in which thyramide Alexa Fluor 568 deposit is produced by reaction with HRP. After two washes with PBS, cells were stained with DAPI (DNA stain) (Invitrogen), mounted with ProLong Gold antifade reagent (Invitrogen), and subjected to microscopy (Leica SP1 laser scanning confocal microscope).
Immunofluorescent Detection of Mre11 in Cells and Tubules Recovered from Matrigel Matrix
Pelleted cells and tubules were fixed with 2.5% formaldehyde (cat. no. 18814; Polyscence) for 10 min and then resuspended in 100% methanol at −20°C. A drop of this methanol suspension was placed on microscope slide and air dried. Slides were washed, blocked, and stained with anti-Mre11 antibodies (cat. no. 4895; Cell Signaling Technology) using Tyramide signal amplification kit (cat. no. T20924; Invitrogen) and DAPI, as described for staining of frozen kidney sections. Confocal microscopy images were taken with Leica SP1 laser scanning confocal microscope (Leica Microsystems, Manheim, Germany). Series of images along the z-axis were collected through the depth of the sample and reconstructed as three-dimensional images using Imaris 7.2 (Bitplane, Zurich, Switzerland) software. To assess changes in distribution over cytoplasm and nucleus, the intensity line profile feature of Bitplane software was used: confocal images were analyzed by drawing a line through cells in transverse section, and fluorescence intensities along this line of each channel were plotted as histograms. The profile of the DAPI channel was used for nucleus positioning as previously described (43).
Immunofluorescent Detection of Mre11 in Primary Kidney Cells and mIMCD3 Cells
Cells grown on eight chamber slides were fixed for 10 min in 2% formaldehyde (cat. no. 18814; Polysciences) at room temperature, washed with PBS, permeabilized with 0.1% Triton X-100 in PBS and blocked with 3% bovine serum albumin for 1 h at room temperature. Slides were incubated with primary antibodies for rabbit Mre11 (cat. no. 4895; Cell Signaling Technology) and human mitochondrial antigen (cat. no. HMS-0100; Immunovision, Sptingdale, AR) at 4°C overnight, followed by secondary antibodies, anti-rabbit labeled with Alexa Fluor 488 nm (green emission), and anti-human Alexa Fluor 630 (red emission; Invitrogen) at room temperature for 1 h. After two washes with PBS, cells were stained with 2.5 μg/ml DAPI (DNA stain; Invitrogen) and mounted with ProLong Gold antifade reagent (Invitrogen). Samples were examined by confocal microscopy using a Zeiss LSM 510 microscope (Carl Zeiss MicroImaging, Jena, Germany) with a ×63 NA1.4 oil-immersion objective. Stacks of fluorescence images were captured sequentially to avoid bleed through, by using a 405-nm excitation and 385- to 470-nm emission for DAPI (2 channels were set: normal laser and detector gain to image nuclear DNA and higher laser and detector gain to image the weaker signal from mitochondrial DNA); 488-nm excitation and 505- to 550-nm emission for Alexa Fluor 488; and a 633-nm excitation and emission over 650 nm for Alexa Fluor 633. Images were deconvolved using Huygens software (Scientific Volume Imaging, Hilversum, Netherlands), prior to three-dimensional reconstruction and colocalization analyzes performed with Imaris 7.2 software (Bitplane). The degree of colocalization of Mre11 (green) and with mitochondria (red) was quantified in the three-dimensional data sets. In addition, voxels from the DNA channel were used to create a mask (white) excluding nuclear DNA. This mask was used as a region of interest to analyze colocalization of mtDNA (white) with Mre11 (green). Colocalized pixels (voxels in three-dimension) are displayed as a yellow (Mre11/mitochondria) or white (Mre11/mtDNA), respectively, overlapping the fluorescence channels over the images. The volume of Mre11 colocalized with mtDNA was compared between experimental conditions. Three series of images for each condition were analyzed in three separate experiments.
In Situ Fractionation Before Immunofluorescent Detection of Mre11
The cells, grown on eight well slides, were fixed with 2% formaldehyde and then treated for 10 min with either 0.1% Triton X-100 to permeabilize plasma membranes or with 1% Triton X-100 to also extract some soluble cytoplasmic and nuclear proteins. The cells were immunostained with rabbit anti-Mre11 (cat. no. 4895; Cell Signaling Technology) as described above.
Extraction and Quantification of Nuclear and Cytoplasmic Proteins
Nuclear and cytoplasmic proteins were extracted separately, using an NE-Per kit (cat. no. 78833; Pierce, Rockford, IL). Western blot analysis was performed by immunoblot of proteins separated by SDS-PAGE. Immunoblots used primary antibodies against Mre11 (cat. no. 4895; Cell Signaling Technology) and secondary antibodies labeled with Alexa Fluor 680 nm dye (Invitrogen). Immunoblots were scanned and integral fluorescence from each band was measured using the Odyssey infrared imaging system (Li-COR Biosciences, Lincoln, NE). To calculate the percentage of Mre11 in the cytoplasmic fraction, the amount of Mre11 in cytoplasmic fraction {IFcyto [Vcyto(total)/ Vcyto(loaded)]} was divided by the total amount of Mre11 in cytoplasmic and nuclear fractions {IFcyto [Vcyto (total)/ Vcyto (loaded)]+ IFnucl [Vnucl (total)/ Vnucl (loaded)]}, where V(loaded) is volume of the sample loaded on the gel, V(total) is volume of entire sample, and IF is integral fluorescence measured from the corresponding band on immunoblot.
Western Blot Analysis of Mitochondria Proteins
Mitochondria were isolated with a mitochondria isolation kit according to the manufacturer's instructions by using a reagent-based method (cat. no. 89874; Pierce, Rockford, IL). Samples were separated by SDS-PAGE. Immunoblots used primary antibodies against Mre11 (cat. no. 4895; Cell Signaling Technology), human mitochondrial antigen (cat. no. HMS-0100; Immunovision), golgin-97 (cat. no. A-21270; Invitrogen), histone H2A (cat. no. 2572; Cell Signaling Technology), mtTFA (cat. no. sc-23588; Santa Cruz Biotechnology, Santa Cruz, CA), and secondary antibodies labeled with Alexa Fluor 680 nm dye (Invitrogen). Immunoblots were scanned, and integral fluorescence from each band was measured using Odyssey infrared imaging system (Li-COR Biosciences).
Analysis of Mre11 Binding to mtDNA by mtDNA Immunoprecipitation
Analysis of Mre11 binding to mtDNA was by mtDNA immunoprecipitation (see also Fig. 7 for an overview the method). mIMCD3 cells were grown on 15-cm dishes. Mitochondria were isolated from two dishes per condition with a mitochondria isolation kit according to the manufacturer's instructions by using the reagent-based method (cat. no. 89874; Pierce). Isolated mitochondria were fixed with 1% formaldehyde for 5 min on ice, spun down at 12,000 g for 10 min at 4°C, washed 2 times with ice-cold PBS, and lysed for 10 min on ice with 50 μl of chromatin immunoprecipitation lysis buffer (1% SDS), 50 mM Tris·HCl (pH 8.0), 10 mM EDTA. The Mre11 mtDNA-immunoprecipitation was performed using reagents from the enzymatic chromatin immunoprecipitation kit (cat. no. 9003; Cell Signaling Technology). The volume of each lysate was adjusted to 500 μl by the addition of 450 μl of chromatin immunoprecipitation buffer. For each sample, 50 μl of the lysate was saved for analysis of input mtDNA. Then 200 μl of the mitochondrial lysate was immunoprecipitated for 2 h with 4 μg of anti-Mre11 antibody (cat. no. 4895; Cell Signaling Technology) or normal rabbit IgG (cat. no. 2729; Cell Signaling Technology), followed by 1-h incubation with 30 μl of protein G magnetic beads (cat. no. 9006; Cell Signaling Technology). Captured Mre11 complexes were eluted from the beads, the proteins and RNA in the samples were enzymatically digested, and the DNA was purified by phenol-chloroform extraction in 1.5-ml phase lock gel light tubes (cat. no. 2900306; 5 Prime, Gaithersburg, MD), followed by ethanol precipitation. mtDNA in input samples and in Mre11 and IgG IP samples was quantified by real-time PCR with SYBR-Green PCR Kit (cat. no. 204054; QIAGEN, Valencia, CA). Eight primer pairs specific for mtDNA were designed using Primer-BLAST tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). Sequences of the primers (5′-/-3′): forward: CAAACCGGGCCCCCTTCGAC; reverse: CGAATGGGCCGGCTGCGTAT; forward: CCACACCCCCACGGGACTCA; reverse: GTATGACCGCGGTGGCTGGC; forward: CCAGTGCTAGCCGCAGGCAT; reverse: TTGGGTCCCCTCCTCCAGCG; forward: TAGCCACACCCCCACGGGAC; reverse: TCGTATGACCGCGGTGGCTG; forward: TCGGAAGCCTCGCCCTCACA; reverse: GGCTCAGGCGTTGGTGTTGC; forward: CCCCCACGGGACTCAGCAGT; reverse: TATGACCGCGGTGGCTGGCA; forward: ACCCCCACGGGACTCAGCAG; reverse: CGCGGTGGCTGGCACGAAAT; forward: CGGCAAACAAGAACCCCGCC; reverse: CACGGTCAGGATACCGCGGC. Specificity of the primers was verified by gel electrophoresis of the PCR products, each producing a single band.
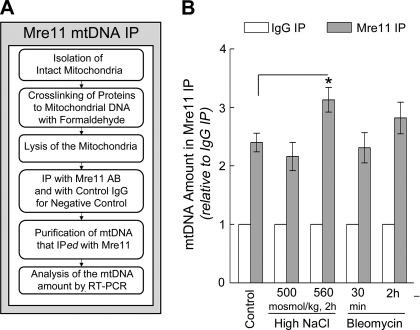
Fig. 7.
Mre11 binds to mtDNA. A: overview of the method used to detect binding of Mre11 to mtDNA (see also Analysis of Mre11 Binding to mtDNA by mtDNA Immunoprecipitation). B: osmolality bathing mIMCD cells was increased by adding NaCl or bleomycin (20 μg/ml), as shown. Mitochondria were isolated and mtDNA bound to Mre11 was immunoprecipitated, purified, and quantified by RT-PCR as described in A and materials and methods. Data are presented as mtDNA bound to Mre11 relative to mtDNA in IgG control (3–4 experiments, 8 mtDNA primers in each: means ± SE, n = 24–32, *P < 0.05, t-test). Mre11 is bound to mtDNA in controls (300 mosmol/kg) and binding increases when NaCl increases.
RESULTS
Occurrence of Mre11 in Mitochondria
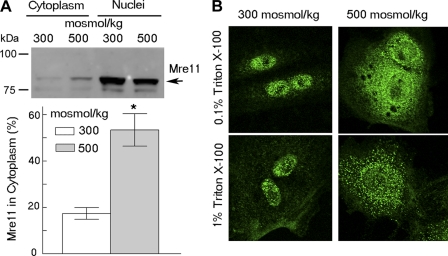
We previously found that high-NaCl treatment of cell lines, including mIMCD3 cells, mouse embryonic and dermal fibroblasts (9), Hela cells (11), Cos7 and HT116 cells (Dmitrieva NI, unpublished observations), causes some nuclear Mre11 to translocate to cytoplasm. In the present studies, to test whether Mre11 translocation into cytoplasm after treatment with high NaCl extends beyond immortalized cells in culture, we tested primary kidney cells. We find that Mre11 is translocated to the cytoplasm after treatment of primary mouse kidney cells with high NaCl (Fig. 1, A and B top), similar to the result in transformed and/or immortalized cells. In addition, even when NaCl is not high (i.e., at 300 mosmol/kg), some Mre11 already exists in the cytoplasm, arranged in a punctuate pattern (Fig. 1). After treatment with high NaCl, the amount of Mre11 in cytoplasm increases, leading to diffuse bright staining. To determine whether after treatment with high NaCl, Mre11 in cytoplasm is soluble or confined within cytoplasmic bodies, we performed in situ fractionation. We extracted soluble proteins by treatment of cells grown on slides with 1% Triton X-100. This leaves punctuate staining of Mre11 in the cytoplasm (Fig. 1B, bottom). These results suggested that Mre11 exists in the cytoplasm within organelles, and that, after treatment with high NaCl, additional Mre11 translocates from nucleus to cytoplasm, where it exists both freely dissolved and confined within organelles. In the studies that follow we identify mitochondria as a cytoplasmic organelle containing Mre11.
Fig. 1.
High NaCl causes movement of Mre11 into the cytoplasm of primary renal cells where it exists in 2 pools: a soluble fraction distributed throughout cytoplasm and a second fraction within organelles. A: Western blot analysis of Mre11 translocation from nucleus to cytoplasm after treatment for 2 h, either increasing NaCl (final osmolality 500 mosmol/kg) or exchanging to medium at 300 mosmol/kg. Top: representative Western blot; bottom: % of total Mre11 in cytoplasmic fraction, calculated based on densitometry, volumes of nuclear and cytoplasmic protein fractions, and volumes loaded on the gel (means ± SE, n = 4, *P < 0.05) (See materials and methods for the calculation). B: analysis by immunocytochemistry and in situ cell fractionation of the subcellular localization of Mre11. Cells were exposed for 2 h to high NaCl (final osmolality 500 mosmol/kg) or kept at 300 mosmol/kg, fixed with 2% formaldehyde, and then treated with 0.1% or 1% of Triton X-100 in PBS for 10 min. Top: 0.1% Triton X-100 permeabilizes plasma membranes; bottom: 1% Triton X-100 extracts soluble cytoplasmic proteins.
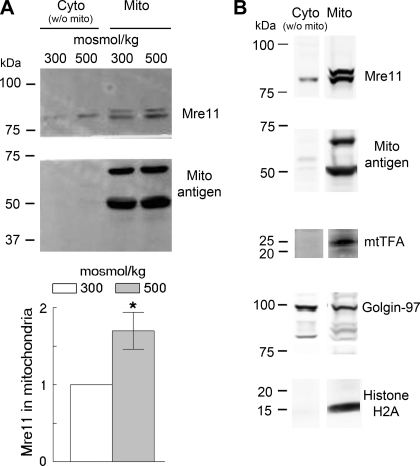
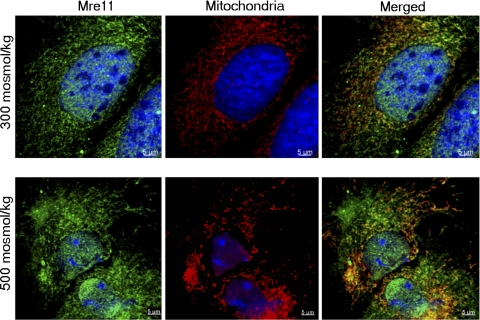
We screened by Western blot analysis for Mre11 among proteins extracted from mitochondria isolated with a mitochondria isolation kit. Mitochondria from primary renal cells apparently contain Mre11, and its abundance apparently increases within 2 h after NaCl is increased (Fig. 2A). However, this result is of itself inconclusive because the mitochondrial preparation is contaminated with proteins from other organelles, including golgin-97 (from golgi) and histone H2A (from chromatin) (Fig. 2B). Therefore, we used confocal immunofluorescence microscopy to test further for Mre11 in mitochondria. Mre11 colocalizes with anti-mitochondrial antigen both at 300 and 500 mosmol/kg (Fig. 3), confirming that it is present in mitochondria. When NaCl is elevated, some Mre11 is also seen in cytoplasm outside of mitochondria (Fig. 3 bottom), consistent with the Western blot analysis result (Fig. 2A).
Fig. 2.
Western blot analysis shows high NaCl induced increase of Mre11 in mitochondria prepared from primary renal cells. A: primary renal cells were exposed for 2 h to high NaCl (final osmolality 500 mosmol/kg) or kept at 300 mosmol/kg, then mitochondria were isolated. Blots were probed with anti-Mre11 and anti-mitochondrial antigen antibodies (Mito). Top: representative Western blot; bottom: quantitation of Mre11 relative to 300 mosmol/kg, normalized by mitochondria antigen (means ± SE, n = 4, *P < 0.05, t-test). B: contamination of the isolated mitochondria by proteins from other organelles. Mitochondria were isolated from mIMCD3 cells at 300 mosmol/kg. Blots were probed for Mre11, mitochondrial antigen, mtTFA, golgin, and histone H2A. The mitochondria-enriched fraction is contaminated with golgi and chromatin.
Fig. 3.
Detection of Mre11 in mitochondria of primary renal cells by immunocytochemistry. Primary kidney cells were exposed for 2 h to high NaCl (final osmolality 500 mosmol/kg) or kept at 300 mosmol/kg. The cells were immunostained for Mre11 (green) and mitochondrial antigen (red). DNA was stained with DAPI (blue). Mre11 is present in mitochondria (colocalization) both at 300 and 500 mosmol/kg. When NaCl is high, some of the Mre11 in cytoplasm is separate from mitochondria, consistent with the Western blot analysis result shown in Fig 2.
Location of Mre11 Within Mouse Kidney Cells In Vivo
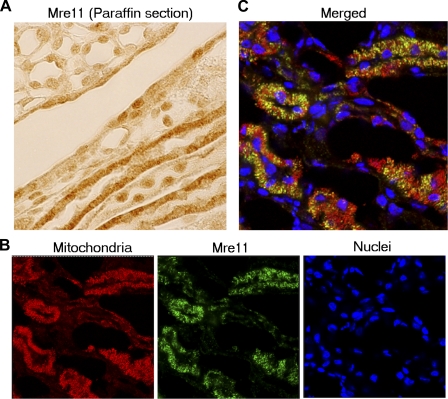
Immunohistochemical analysis of Mre11 in paraffin-embedded sections from normal mouse kidney shows subcellular localization of Mre11 that differs between cell types (Fig. 4). Some cells have only nuclear staining, while others exhibit strong cytoplasmic staining (Fig. 4A). Double staining for Mre11 and mitochondria in frozen kidney sections shows that cytoplasmic localization of Mre11 occurs mostly in mitochondrial-rich cells and that the Mre11 colocalizes with mitochondria (Fig. 4, B and C).
Fig. 4.
In mouse kidney, Mre11 is mostly cytoplasmic in metabolically active mitochondria-rich cells where it colocalizes with mitochondria. Analysis of subcellular localization of Mre11 in normal mouse kidney. A: immunohistochemistry of Mre11 in paraffin sections of normal mouse kidney. The subcellular distribution of Mre11 differs between cell types. Mre11 is mostly in the cytoplasm of some cells, but mostly in the nucleus of other cells. B and C: immunocytochemistry of Mre11 in frozen sections from normal mouse kidney. Sections are immunostained with anti-Mre11 (green) and anti-mitochondrial antigen (red) antibodies. DNA is stained with DAPI (blue). B: images scanned sequentially at different wave lengths. C: merged image of B. Much of Mre11 colocalizes with mitochondria.
Our observation that Mre11 is preferentially located in mitochondria of mitochondrial-rich cells of the kidney in vivo, but is mostly located in nuclei of proliferating cells in culture, suggested to us that the divergent distributions of Mre11 might depend on differentiation and metabolism. Cells tend to dedifferentiate in continuous culture. Also, mtDNA damage could be greater in mitochondrial-rich cells of proximal tubules and thick ascending limbs. Elevated oxidative metabolism in those cells is associated with rapid production of ROS in their mitochondria, and the ROS could damage their DNA (1, 3, 46). In our next experiments we tested those possibilities.
Effect of Cellular Differentiation on Subcellular Location of Mre11
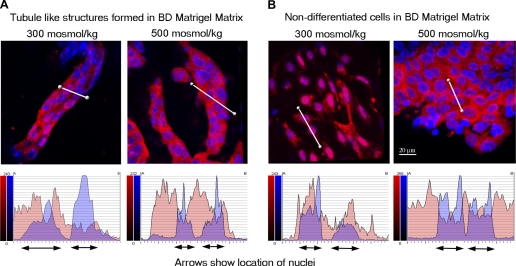
The Mre11 complex is important for DNA replication because it rapidly repairs replication fork-associated lesions, preventing accumulation of DNA damage (reviewed in Refs. 4 and 42). This role might favor retention of Mre11 in the nuclei of actively replicating cells, but not in less-rapidly proliferating differentiated cells. To test for a possible role of cellular differentiation on subcellular localization of Mre11, we grew primary kidney cells in BD Matrigel matrix, which at room temperature polymerizes to produce biologically active matrix material resembling the mammalian cellular basement membrane. When primary kidney cells are grown in Matrigel matrix, some differentiate and form tubules, while others continue to proliferate. We analyzed the subcellular location of Mre11 at different stages of differentiation and at different levels of NaCl (Fig. 5). At 300 mosmol/kg Mre11 is mostly in the nucleus of nondifferentiated cells (Fig. 5B, left), but it is mostly in the cytoplasm of cells that have formed tubules (Fig. 5A, left). These results are consistent with idea that active replication favors retention of Mre11 in the nucleus, but slowing of replication as a result of differentiation favors localization of Mre11 in mitochondria where active metabolic processes can damage mtDNA. When NaCl is increased, Mre11 occurs in the cytoplasm of both nondifferentiated and differentiated cells (Fig. 5, A and B, right). High NaCl increases ROS of mitochondrial origin (47, 48). We suggest that high NaCl causes Mre11 to move into mitochondria in response to the resulting mtDNA damage.
Fig. 5.
Differentiation of primary kidney cells into tubules causes Mre11 to translocate from nucleus to cytoplasm. Primary kidney cells were grown in Matrigel matrix. After 7 days, some cells formed tubules, other cells stayed undifferentiated. Tubules and cells were recovered from the matrix using matrix solubilization solution and immunostained with anti-Mre11 (red). Some tubules and cells were treated with high-NaCl medium for 1 h before recovery from the matrix. Nuclei were stained with DAPI (blue), and confocal images were collected through the depth of the structures. Three-dimensional images reconstructed from a series of images along the z-axis (see materials and methods for details) of tubule-like structures (A) and of nondifferentiated cells (B) are displayed (top). A semiquantitative analysis of Mre11 nuclear cytoplasmic redistribution was performed using line profile of Bitplane software (bottom). Fluorescence intensities along the white lines sectioning the cells through the cytoplasm and nucleus are plotted for both Mre11 (red) and DAPI (blue); DAPI locates the position of the nuclei, and red indicates corresponding nuclear Mre11. The plots show marked differences in the ratio of cytoplasmic/nuclear Mre11 with differentiation. A: in tubule-like structures, Mre11 is mostly in the cytoplasm both at 300 and 500 mosmol/kg. B: in nondifferentiated cells, Mre11 is mostly in nuclei at 300 mosmol/kg and translocates to cytoplasm after treatment with high NaCl.
Effect of mtDNA Damage on Colocalization of Mre11 with Mitochondrial DNA
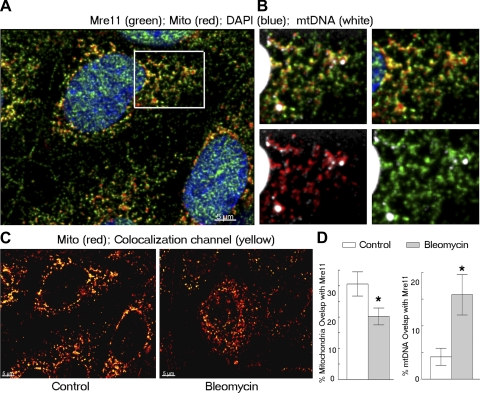
To test the possibility that Mre11 participates in the response to mitochondrial DNA damage, we visualized mitochondrial Mre11 and DNA by confocal microscopy in mIMCD3 cells (Fig. 6). Mre11 staining forms foci within mitochondria (Fig. 6A). mtDNA is organized into discrete protein-DNA complexes called nucleoids (21, 37), which appear in cytoplasm as relatively large particles (DAPI staining, Fig. 6B). Some Mre11 foci colocalize with mtDNA (Fig. 6B), consistent with participation of Mre11 in mtDNA processing. Bleomycin induces DSBs in mitochondrial DNA, which the mitochondria repair within several hours (29, 34). We determined the effect of bleomycin on the location of Mre11 within mitochondria and the degree of its colocalization with mtDNA (Fig. 6, C and D). Bleomycin decreases the fraction of mitochondria volume occupied by Mre11 compared with control conditions (Fig. 6D, left). Visually Mre11 staining appears in more distinct foci within mitocondria in bleomycin-treated cells compared with control (Fig. 6C). In addition, bleomycin treatment increased a fraction of mtDNA that is colocalyzed with Mre11 (Fig. 6D, right). Taken together, our analyses indicates that within mitochondria Mre11 partially colocalizes with nucleoids and bleomycin treatment causes Mre11 to translocate to mtDNA, where it presumably participates in repair of the damage induced by bleomycin. Nevertheless, colocalization of Mre11 with nucleoids does not prove physical binding of Mre11 to mtDNA, and redistribution of Mre11 within mitochondria could also be caused by changes in mitochondrial structure after bleomycin treatment. Therefore, we tested more directly for binding of Mre11 to mtDNA.
Fig. 6.
Bleomycin changes distribution of Mre11 within mitochondria, increasing its overlap with mtDNA. mIMCD3 cells were treated for 30 min with bleomycin (10 μg/ml), known to induce double-strand breaks in mtDNA. The cells were immunostained for Mre11 (green) and mitochondrial antigen (red). mtDNA was stained, by using a high concentration of DAPI (white). Images through the depth of the cells (z-axis), obtained by confocal microscopy, were deconvolved with Hyugens software and quantitative volumetric colocalization was analyzed with Imaris software. A: representative merged image showing colocalization (yellow) of Mre11 (green) with mitochondria (red) in cells under control conditions. B: enlargements of the boxed-area in A. Merged images are top left: Mre11 (green), mitochondria (red) and mtDNA (white, high gain). Top right: Mre11 and mitochondria; bottom left: mitochondria and mtDNA; bottom right: Mre11 and mtDNA. C: representative images showing colocalization (yellow) of Mre11 with mitochondria (red) in the control condition and after treatment with bleomycin. D: quantitative analysis of % of mitochondrial volume (left) or mtDNA volume (right) colocalized with Mre11 (means ± SE, n = 6–9, *P < 0.05). Bleomycin decreases the volume of mitochondria occupied by Mre11 and increases the volume of mtDNA colocalized with Mre11.
mtDNA-Immunoprecipitation Confirms Binding of Mre11 to mtDNA
We analyzed binding of Mre11 to mtDNA by a method similar to chromatin immunoprecipitation but performed on mtDNA. Proteins in isolated mitochondria were crosslinked to mtDNA by formaldehyde, mitochondria were lysed, and Mre11-mtDNA complexes were immunoprecipitated with anti-Mre11. mtDNA in Mre11 immunoprecipitates was quantified by real-time PCR (see materials and methods and Fig. 7A). We call the method “mtDNA-IP.” Some Mre11 is already bound to mtDNA at 300 mosmol/kg, and raising NaCl significantly increases binding of Mre11 to mtDNA (Fig. 7B). However, the increased binding following bleomycin is not statistically significant (Fig. 7B).
DISCUSSION
Mitochondria are known to repair many types of damage to their DNA (26), but it has not been clear what DNA damage-response proteins are involved. Recombinational repair mechanisms apparently exist since in vitro studies demonstrated homologous recombination and end-joining activities in mammalian mitochondrial extracts (6, 25, 41). When DSBs are induced in mitochondria by restriction endonucleases, both intramolecular and intermolecular recombination products with large deletions appear (2, 19). The deletions are most likely mediated by DNA repair involving homologous recombination and nonhomologous end joining. Thus, mitochondria apparently repair DNA DSBs. However, few of the molecular components involved in this repair are known. Possible candidates are retinoblastoma protein (18), BRCA1 (5), and p53 (17), all of which have been identified in mitochondria. Our identification of Mre11 adds another. These proteins presumably are involved in maintaining the stability not only of the nuclear genome, but of the mitochondrial genome, as well.
We find Mre11 in mitochondria of unstressed, as well stressed, mammalian cells. The presence even in unstressed cells is not surprising since mitochondria constantly produce ROS, an unavoidable byproduct of oxidative phosphorylation. Proximity to mtDNA could lead to the higher steady-state level of oxidative damage in mitochondrial, compared with nuclear, DNA (1, 22, 46). Oxidized DNA bases can be a source of DSBs for which function of Mre11 is well studied, because DSBs are generated by the collapse of replication forks when the replication machinery encounters the single-strand breaks that are intermediates in repair of oxidative damage (7). In addition to the well-described role of Mre11 in repair of DSBs, it also participates in base excision repair (38). Base excision repair is a major repair pathway for 8-oxoguanine lesions, the form of base damage most frequently induced by hydrogen peroxide, which is produced in mitochondria. Therefore, repair of mtDNA damage is a likely candidate for function of Mre11 in mitochondria. This notion is supported by several observations from our study. First, Mre11 is highly expressed in mitochondria of mitochondrial-rich cells of proximal tubules and thick ascending limbs in normal mouse kidney (Fig. 4), where mtDNA damage could be greater due to the elevated oxidative metabolism in those cells that is associated with rapid production of ROS in their mitochondria. Second, Mre11 is mostly in the nucleus of nondifferentiated cells, but it is mostly in the cytoplasm of cells that have formed tubules (Fig. 5). These results are consistent with the idea that active replication, which increases DNA breaks in nuclear DNA, favors retention of Mre11 in the nucleus, but slowing of replication as a result of differentiation favors localization of Mre11 in mitochondria, where active metabolic processes still can damage mtDNA. Third, high NaCl increases Mre11 in mitochondria (Fig. 2). That also might be caused by increased mtDNA damage, since mitochondrial ROS production is increased by high NaCl (47, 48). Fourth, colocalization of Mre11 with nucleoids within mitochondria (Fig. 6), which bleomycin increases, supports the idea that Mre11 participates in mtDNA metabolism and repair (Fig. 6). Fifth, physical binding of Mre11 to mtDNA, demonstrated by mtDNA-IP (Fig. 7), strengthens the inference that Mre11 participates in repair of mtDNA.
We conclude that Mre11 is present in mitochondria where it binds to mtDNA and that the amount in mitochondria varies, depending on cellular stress and differentiation. Our results suggest a role for Mre11 in the maintenance of genome integrity in mitochondria, in addition to its previously known role in maintenance of nuclear DNA.
Perspectives and Significance
Mutations in mtDNA have been linked to a number of human diseases. Among them are such common pathological conditions as neurogenerative disorders, diabetes mellitus, and cancer (reviewed in Ref. 16). Also, accumulation of damage to the mitochondrial genome is central in the mitochondrial theory of aging (1, 20, 22). DNA repair serves to reduce accumulation of mutations in mtDNA, but the mechanisms of mtDNA repair are not nearly as well understood as those of nuclear DNA repair. Many questions remain to be answered. Identification of the components of mtDNA repair pathways and understanding the mechanisms of the repair of different types of mtDNA damage will help understand how such pathological states develop and how they can be prevented or delayed. Finding that Mre11, a major contributor to nuclear DNA repair, is associated with mitochondrial DNA is a step in this direction.
GRANTS
This work was supported by the National Institutes of Health Intramural Research Program.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
ACKNOWLEDGMENTS
We thank Matthew Gastinger (Bitplane, Zurich, Switzerland) for assistance with colocalization analysis.
REFERENCES
- 1. Alexeyev MF. Is there more to aging than mitochondrial DNA and reactive oxygen species? FEBS J 276: 5768–5787, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bacman SR, Williams SL, Moraes CT. Intra- and inter-molecular recombination of mitochondrial DNA after in vivo induction of multiple double-strand breaks. Nucleic Acids Res 37: 4218–4226, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berneburg M, Kamenisch Y, Krutmann J, Rocken M. ‘To repair or not to repair - no longer a question’: repair of mitochondrial DNA shielding against age and cancer. Exp Dermatol 15: 1005–1015, 2006 [DOI] [PubMed] [Google Scholar]
- 4. Borde V, Cobb J. Double functions for the Mre11 complex during DNA double-strand break repair and replication. Int J Biochem Cell Biol 41: 1249–1253, 2009 [DOI] [PubMed] [Google Scholar]
- 5. Coene ED, Hollinshead MS, Waeytens AA, Schelfhout VR, Eechaute WP, Shaw MK, Van Oostveldt PM, Vaux DJ. Phosphorylated BRCA1 is predominantly located in the nucleus and mitochondria. Mol Biol Cell 16: 997–1010, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Coffey G, Lakshmipathy U, Campbell C. Mammalian mitochondrial extracts possess DNA end-binding activity. Nucleic Acids Res 27: 3348–3354, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Collis SJ, DeWeese TL, Jeggo PA, Parker AR. The life and death of DNA-PK. Oncogene 24: 949–961, 2005 [DOI] [PubMed] [Google Scholar]
- 8. deJager M, van NJ, van G, Dekker C, Kanaar R, Wyman C. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell 8: 1129–1135, 2001 [DOI] [PubMed] [Google Scholar]
- 9. Dmitrieva NI, Bulavin DV, Burg MB. High NaCl causes Mre11 to leave the nucleus, disrupting DNA damage signaling and repair. Am J Physiol Renal Physiol 285: F266–F274, 2003 [DOI] [PubMed] [Google Scholar]
- 10. Dmitrieva NI, Burg MB. Osmotic stress and DNA damage. Methods Enzymol 428: 241–252, 2007 [DOI] [PubMed] [Google Scholar]
- 11. Dmitrieva NI, Burg MB. Analysis of DNA breaks, DNA damage response, and apoptosis produced by high NaCl. Am J Physiol Renal Physiol 295: F1678–F1688, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dmitrieva NI, Burg MB. High NaCl promotes cellular senescence. Cell Cycle 6: 3108–3113, 2007 [DOI] [PubMed] [Google Scholar]
- 13. Dmitrieva NI, Cai Q, Burg MB. Cells adapted to high NaCl have many DNA breaks and impaired DNA repair both in cell culture and in vivo. Proc Natl Acad Sci USA 101: 2317–2322, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dmitrieva NI, Celeste A, Nussenzweig A, Burg MB. Ku86 preserves chromatin integrity in cells adapted to high NaCl. Proc Natl Acad Sci USA 102: 10730–10735, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dmitrieva NI, Ferraris JD, Norenburg JL, Burg MB. The saltiness of the sea breaks DNA in marine invertebrates: possible implications for animal evolution. Cell Cycle 5: 1320–1323, 2006 [DOI] [PubMed] [Google Scholar]
- 16. Druzhyna NM, Wilson GL, LeDoux SP. Mitochondrial DNA repair in aging and disease. Mech Ageing Dev 129: 383–390, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ferecatu I, Bergeaud M, Rodriguez-Enfedaque A, Le FN, Oliver L, Rincheval V, Renaud F, Vallette FM, Mignotte B, Vayssiere JL. Mitochondrial localization of the low level p53 protein in proliferative cells. Biochem Biophys Res Commun 387: 772–777, 2009 [DOI] [PubMed] [Google Scholar]
- 18. Ferecatu I, Le FN, Bergeaud M, Rodriguez-Enfedaque A, Rincheval V, Oliver L, Vallette FM, Mignotte B, Vayssiere JL. Evidence for a mitochondrial localization of the retinoblastoma protein. BMC Cell Biol 10: 50, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fukui H, Moraes CT. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum Mol Genet 18: 1028–1036, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gredilla R, Bohr VA, Stevnsner T. Mitochondrial DNA repair and association with aging–an update. Exp Gerontol 45: 478–488, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Holt IJ, He J, Mao CC, Boyd-Kirkup JD, Martinsson P, Sembongi H, Reyes A, Spelbrink JN. Mammalian mitochondrial nucleoids: organizing an independently minded genome. Mitochondrion 7: 311–321, 2007 [DOI] [PubMed] [Google Scholar]
- 22. Hudson EK, Hogue BA, Souza-Pinto NC, Croteau DL, Anson RM, Bohr VA, Hansford RG. Age-associated change in mitochondrial DNA damage. Free Radic Res 29: 573–579, 1998 [DOI] [PubMed] [Google Scholar]
- 23. Jazayeri A, Balestrini A, Garner E, Haber JE, Costanzo V. Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. EMBO J 27: 1953–1962, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kultz D, Chakravarty D. Hyperosmolality in the form of elevated NaCl but not urea causes DNA damage in murine kidney cells. Proc Natl Acad Sci USA 98: 1999–2004, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lakshmipathy U, Campbell C. Double strand break rejoining by mammalian mitochondrial extracts. Nucleic Acids Res 27: 1198–1204, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Larsen NB, Rasmussen M, Rasmussen LJ. Nuclear and mitochondrial DNA repair: similar pathways? Mitochondrion 5: 89–108, 2005 [DOI] [PubMed] [Google Scholar]
- 27. Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308: 551–554, 2005 [DOI] [PubMed] [Google Scholar]
- 28. Liu P, Demple B. DNA repair in mammalian mitochondria: much more than we thought? Environ Mol Mutagen 51: 417–426, 2010 [DOI] [PubMed] [Google Scholar]
- 29. Morel F, Renoux M, Lachaume P, Alziari S. Bleomycin-induced double-strand breaks in mitochondrial DNA of Drosophila cells are repaired. Mutat Res 637: 111–117, 2008 [DOI] [PubMed] [Google Scholar]
- 30. Neupert W. Protein import into mitochondria. Annu Rev Biochem 66: 863–917, 1997 [DOI] [PubMed] [Google Scholar]
- 31. Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol Cell 1: 969–979, 1998 [DOI] [PubMed] [Google Scholar]
- 32. Peters SR. The art of embedding tissue for frozen section. Part I: a system for face down cryoembedding of tissues using freezing temperature embedding. J Histotechnol 26: 11–19, 2003 [Google Scholar]
- 33. Rauchman MI, Nigam SK, Delpire E, Gullans SR. An osmotically tolerant inner medullary collecting duct cell line from an SV40 transgenic mouse. Am J Physiol Renal Fluid Electrolyte Physiol 265: F416–F424, 1993 [DOI] [PubMed] [Google Scholar]
- 34. Shen CC, Wertelecki W, Driggers WJ, LeDoux SP, Wilson GL. Repair of mitochondrial DNA damage induced by bleomycin in human cells. Mutat Res 337: 19–23, 1995 [DOI] [PubMed] [Google Scholar]
- 35. Shiloh Y. Ataxia-telangiectasia and the Nijmegen breakage syndrome: related disorders but genes apart. Annu Rev Genet 31: 635–662, 1997 [DOI] [PubMed] [Google Scholar]
- 36. Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schönfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci USA 100: 13207–13212, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Spelbrink JN. Functional organization of mammalian mitochondrial DNA in nucleoids: history, recent developments, and future challenges. IUBMB Life 62: 19–32, 2010 [DOI] [PubMed] [Google Scholar]
- 38. Steininger S, Ahne F, Winkler K, Kleinschmidt A, Eckardt-Schupp F, Moertl S. A novel function for the Mre11-Rad50-Xrs2 complex in base excision repair. Nucleic Acids Res 38: 1853–1865, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99: 577–587, 1999 [DOI] [PubMed] [Google Scholar]
- 40. Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol 12: 90–103, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thyagarajan B, Padua RA, Campbell C. Mammalian mitochondria possess homologous DNA recombination activity. J Biol Chem 271: 27536–27543, 1996 [DOI] [PubMed] [Google Scholar]
- 42. Trenz K, Smith E, Smith S, Costanzo V. ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J 25: 1764–1774, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wan Z, Zhi N, Wong S, Keyvanfar K, Liu D, Raghavachari N, Munson PJ, Su S, Malide D, Kajigaya S, Young NS. Human parvovirus B19 causes cell cycle arrest of human erythroid progenitors via deregulation of the E2F family of transcription factors. J Clin Invest 120: 3530–3544, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Williams GJ, Lees-Miller SP, Tainer JA. Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair (Amst) 9: 1299–1306, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol 85: 509–520, 2007 [DOI] [PubMed] [Google Scholar]
- 46. Yakes FM, Van HB. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci USA 94: 514–519, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Z, Dmitrieva NI, Park JH, Levine RL, Burg MB. High urea and NaCl carbonylate proteins in renal cells in culture and in vivo, and high urea causes 8-oxoguanine lesions in their DNA. Proc Natl Acad Sci USA 101: 9491–9496, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhou X, Ferraris JD, Burg MB. Mitochondrial reactive oxygen species contribute to high NaCl-induced activation of the transcription factor TonEBP/OREBP. Am J Physiol Renal Physiol 290: F1169–F1176, 2006 [DOI] [PubMed] [Google Scholar]
- 49. Zhu J, Petersen S, Tessarollo L, Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol 11: 105–109, 2001 [DOI] [PubMed] [Google Scholar]
- 50. Zhu WG, Seno JD, Beck BD, Dynlacht JR. Translocation of MRE11 from the nucleus to the cytoplasm as a mechanism of radiosensitization by heat. Radiat Res 156: 95–102, 2001 [DOI] [PubMed] [Google Scholar]