Abstract
Inhibition of bacterial transcription represents an effective and clinically validated anti-infective chemotherapeutic strategy. We describe the evolution of our approach to the streptolydigin class of antibiotics that target bacterial RNA polymerases (RNAPs). This effort resulted in the synthesis and biological evaluation of streptolydigin, streptolydiginone, streptolic acid and a series of new streptolydigin-based agents. Subsequent biochemical evaluation of RNAP inhibition demonstrated that the presence of both streptolic acid and tetramic acid subunits was required for activity of this class of antibiotics. In addition, we identified 10,11-dihydrostreptolydigin as a new RNAP-targeting agent, which was assembled with high synthetic efficiency of 15 steps in the longest linear sequence. Dihydrostreptolydigin inhibited three representative bacterial RNAPs and displayed in vitro antibacterial activity against S. salivarius. The overall increase in synthetic efficiency combined with substantial antibacterial activity of this fully synthetic antibiotic demonstrates the power of organic synthesis in enabling design and comprehensive in vitro pharmacological evaluation of new chemical agents that target bacterial transcription.
Introduction
Cellular RNA polymerase (RNAP) is a multisubunit enzyme that catalyzes the synthesis of RNA from the corresponding DNA template. Despite overall structural and functional similarities, bacterial RNAP sequences are substantially different from those of the corresponding eukaryotic enzymes, which explains why rifamycin antibiotics can be used to selectively block bacterial RNAPs, while having no effect on eukaryotic RNAPs.1 As a result of potent RNAP inhibition, rifamycins display broad-spectrum antibacterial activity.2 Three semisynthetic derivatives of rifamycin A, including rifampin, rifapentine and rifabutin, are currently in clinical use for treatment of Mycobacterial infections, including tuberculosis and leprosy.3 Despite the high potency, low toxicity and broad antibacterial spectrum of rifamycins, pathogens develop resistance to this class of antibiotics at a relatively high rate by substitution of the amino-acid residues in the rifamycin-binding site of bacterial RNAP.4 Rapid onset of bacterial resistance is the primary reason why current use of rifamycins is restricted to combinations with other drugs, such as isoniazid, or to clinical emergencies. Thus, there is a significant need for the development of new antibiotics that target bacterial RNAPs by different biochemical mechanisms and display broad-spectrum antibacterial activity. Several other classes of natural products have been shown to inhibit bacterial RNAPs by binding to alternative regions of this multisubunit protein, which typically yields notable antibiotic activity. Such compounds were found to be effective against rifamycin-resistant RNAPs and strains.5
Streptolydigin (1, Figure 1A) is a dienoyl tetramic acid antibiotic,6 which elicits its antibacterial activity by inhibiting initiation, elongation and pyrophosphorylation steps of bacterial RNAP.7 High-resolution X-ray crystallographic characterization of the streptolydigin-RNAP complex revealed a unique biochemical mechanism of RNAP inhibition.8 Streptolydigin (1) traps the bridge-helix of the RNAP in a straight conformation and induces opening of the trigger-loop of the enzyme. As a result, streptolydigin (1) stabilizes the catalytically inactive substrate-bound transcription intermediate and blocks structural isomerization of RNAP into a fully active state, which requires conformational changes of both the bridge-helix and the trigger-loop moieties.8 The streptolydigin-binding region is located 20 Å away from the rifamycins binding site.9 As a result of this unique biochemical mechanism of RNAP inhibition and a distinct binding site, streptolydigin (1) and rifamycins exhibit only minimal cross-resistance.8b,10
Figure 1.
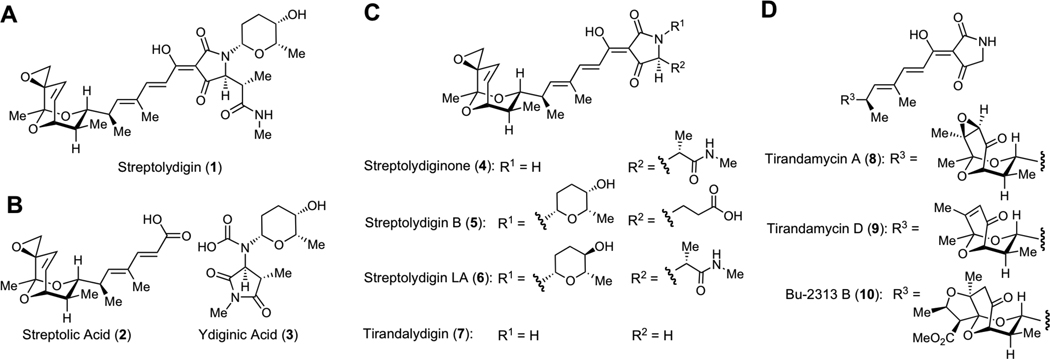
Structures of Dienoyl Tetramic Acids Antibiotics and Degradation Fragments. A. Structure of streptolydigin (1), the parent member of this antibiotic family, which was isolated from S. lydicus. B. Structures of streptolic acid (2) and ydiginic acid (3), which were obtained by chemical degradation of streptolydigin (1). C. Structures of several derivatives of streptolydigin, which were biosynthetically produced in re-engineered S. lydicus strains and structure of tirandalydigin (7), which was isolated from S. tirandis. D. Structures of two representative tirandamycins (8 and 9), which were isolated from S. tirandis and marine-derived Streptolyces species, as well as Bu-2313B (10), which was isolated from oligosporic actinomycete strain, No. E864-61.
The structure of streptolydigin (1) features an epoxide-containing bicyclic ketal connected by a polyene spacer to a higly substituted, glycosylated acyl tetramic acid. Structure elucidation of streptolydigin (1) entailed initial oxidative degradation of the natural product into two simplified subunits, streptolic acid (2) and ydiginic acid (3), which derived from the bicyclic ketal fragment and the tetramic acid subunit of the natural product, respectively (Figure 1B).11 Complete stereochemical assignment of streptolic acid (2) was ultimately secured by X-ray crystallographic analysis.12 Re-engineering of streptolydigin biosynthesis in S. lydicus enabled recent production of several new antibiotics shown in Figure 1C, including streptolydiginone (4),13 which represents a streptolydigin aglycone, as well as streptolyidigin B (5)14 and streptolydigin LA (6).13 Following the initial report on isolation of streptolydigin in 1956,6 several other members of the dienoyl tetramic acid antibiotic family have been identified, including tirandalydigin (7),15 tirandamycins (i.e., 8 and 9),16 Bu-2312B (10)17 and nocamycins, which were found to be structurally analogous to 10.18 The bicyclic ketal subunit of tirandalydigin (7) is identical to that of streptolydigin (Figure 1C). However, the tetramic acid moiety of this metabolite lacks L-rhodinose and the amide containing side-chain. Tirandamycins (8 and 9) and Bu-2312B (10) possess the same unsubstituted acyl tetramic acid subunit of tirandalydigin (7), but differ in the substitution of the ketal moiety (Figure 1D). Despite isolation and biosynthetic production of a number of structurally homologous dienoyl tetramic acid antibiotics over the years, streptolydigin (1) features the most elaborate structure and the highest antimicrobial activity recorded within this class.
Evaluation of the antibiotic activity of streptolydigin (1) against a broad panel of microbial strains using standard broth dilution testing revealed that this natural product elicited notable activity against a number of Gram-positive organisms.6a Inhibition of several representative Clostridium and Streptococcus species by 1 was particularly potent with minimum inhibitory concentrations (MICs) as low as 0.04 µg/mL. In addition, inhibition of Mycobacterium strains, including M. tuberculosis, was also observed.6a Streptolydigin (1) was shown to be generally more potent than tirandalydigin (7) and tirandamycin A (8) in a panel of several anaerobic and aerobic gram-positive bacteria.19 While streptolydiginone (4) and streptolydigin B (5) inhibited growth of S. albus, they were found to be less active compared to streptolydigin (1) and streptolydigin LA (6).13,14
Streptolydigin (1) was also evaluated in a series of in vivo studies conducted in mice.6c The maximum tolerated doses of 1 were 1800 mg/kg/day when administered orally and 500 mg/kg/day when administered subcutaneously.6c Importantly, administration of streptolydigin (1) at subcutaneous doses of 100–330 mg/kg/day protected animals against infection with S. haemolyticus, S. pneumoniae and P. multocida.6c Similarly, Bu-2313B (10) was effective in protecting mice against P. fragilis and C. perfingens when administered by both oral and subcutaneous routes.17a
While several total syntheses of tirandamycin A (8)20 have been developed over the years and the preparation of two advanced fragments of streptolydigin (1) has been described,21 development of viable synthetic approaches to streptolydigin (1) and closely related tirandalydigin (7) remained challenging in part due to the presence of a highly labile alkenyl epoxide fragment, which is absent in tirandamycin A (8). Indeed, while a recent synthetic effort enabled assembly of a protected tirandalydigin (7), the final acid-mediated deprotection step was not successful presumably due to the instability of the epoxide moiety.22
Following our initial communication of the synthesis of streptolydigin (1),23 which afforded this natural product with a longest linear sequence of 24 steps, we describe here the evolution of our synthetic approach to this important class of RNAP-targeting antibiotics. This effort resulted in the assembly and biological evaluation of a series of streptolydigin-based agents, which demonstrated that the presence of both streptolic acid and tetramic acid subunits was required for activity of streptolydigin antibiotics. In addition, we identified a new RNAP-targeting synthetic antibiotic, 10,11-dihydrostreptolydigin, which was assembled with the high synthetic efficiency of 15 steps in the longest linear sequence. Dihydrostreptolydigin inhibited three representative bacterial RNAPs and displayed in vitro antibacterial activity against S. salivarius. The overall increase in synthetic efficiency combined with substantial antibacterial efficacy of this synthetic congener of streptolydigin demonstrates the power of organic synthesis in enabling design and comprehensive in vitro pharmacological evaluation of new antibiotics that target bacterial transcription.
Results and Discussion
Syntheses of Streptolic Acid, Methyl Streptolate and Streptolydiginone
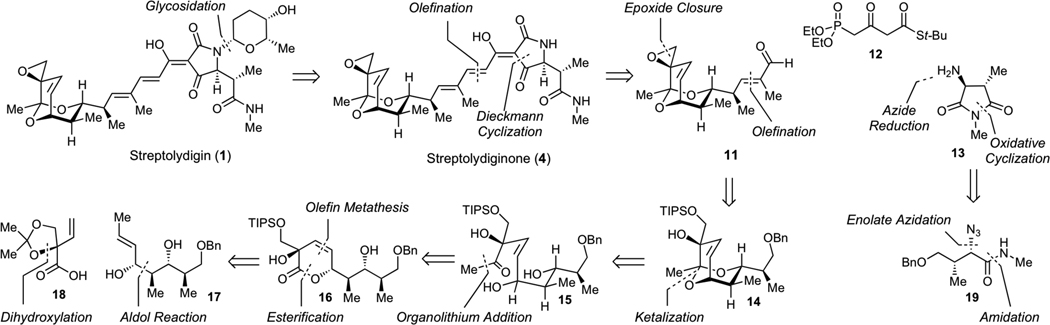
Guided by the logic of the streptolydigin biogenesis, which entails final-stage glycosylation of streptolydiginone (4) with L-rhodinose,13 we devised our initial synthetic strategy shown in Scheme 1. Streptolydiginone (4) would derive from three simplified fragments, including aldehyde 11, diethylphosphono-3-oxobutanthioate 12 and imide 13. The assembly process would entail the Horner-Wadsworth-Emmons olefination of aldehyde 11 with phosphonate 12, followed by N-acylation of imide 13 and Dieckmann cyclization under the protocol developed by Rinehart.24 The imide 13 would derive from azide 19, which would be in turn assembled by the Evans diastereoselective enolate azidation.25 The assembly of aldehyde 11 would entail late-stage installation of the epoxide moiety starting with appropriately protected vicial diol 14 containing the requisite bicyclic core of the streptolic acid, which would be derived from the corresponding diol 15. To control the cis-geometry of alkene 15, we envisioned that this acyclic product would be prepared starting from lactone 16 by a controlled carbonyl addition. The synthesis of lactone 16 would entail chemoselective acylation of diol 17 with carboxylic acid 18, followed by a ring-closing metathesis.
Scheme 1.
Synthetic strategy to streptolydiginone (4) and projected conversion to streptolydigin (1).
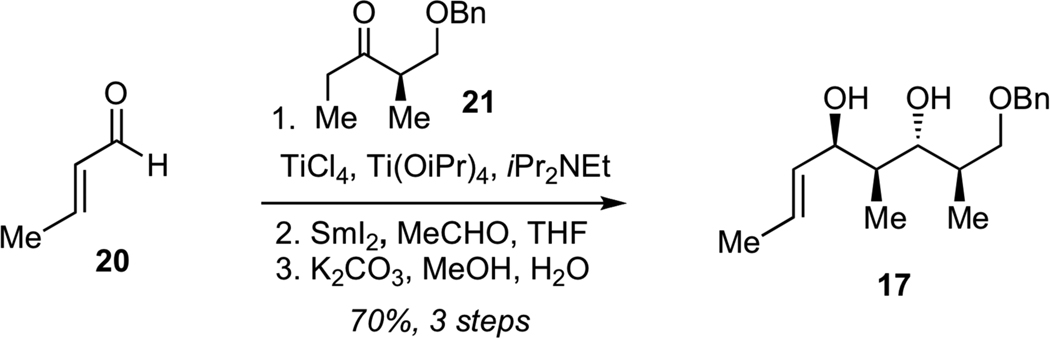
Preparation of diol 17 (Scheme 2) commenced with the aldol reaction of crotonaldehyde (20) with titanium enolate derived from benzyloxyketone 2126 to give the corresponding syn aldol product with excellent diastereoselectivity (dr >95:5). Subsequent anti-selective Evans-Tishchenko reduction27 and saponification of the resulting acetate delivered the target diol 17 in 70% yield for 3 steps as a single diastereomer.
Scheme 2.
Synthesis of Diol 17
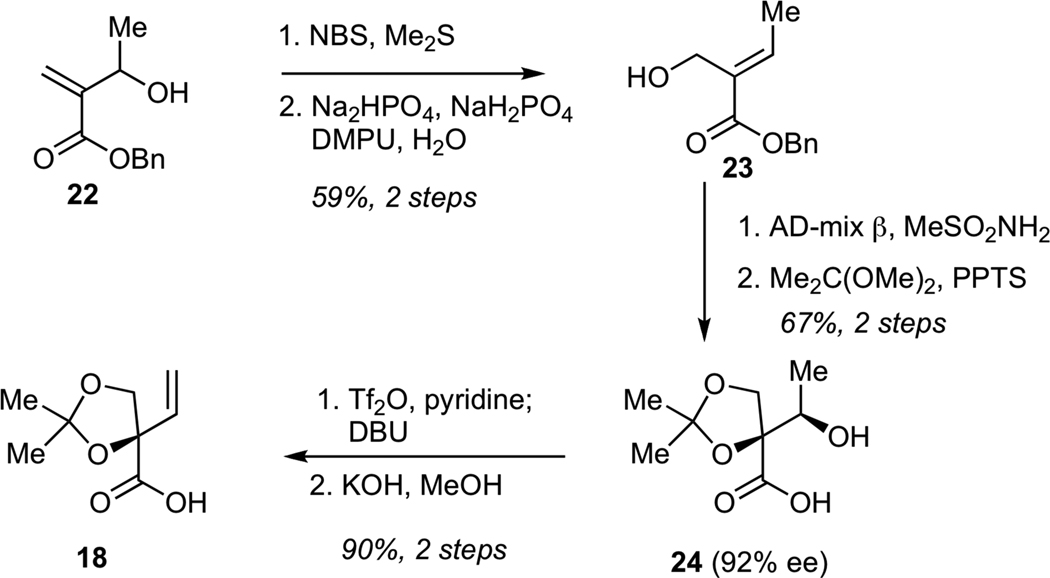
The synthesis of carboxylic acid 18 is shown in Scheme 3. The sequence began from hydroxy enoate 22, which was readily prepared by Baylis-Hillman reaction of benzyl acrylate with acetaldehyde. Alcohol transposition in 22 was achieved via its initial conversion into the corresponding allylic bromide, which underwent hydrolysis in aqueous buffer to give primary alcohol 23. Subjection of 23 to Sharpless dihydroxylation,28 followed by chemoselective protection of the corresponding triol as the acetonide delivered secondary alcohol 24 in 92% ee. Conversion of alcohol 24 into the corresponding triflate, followed by elimination with DBU and hydrolysis of the benzyl ester gave the requisite carboxylic acid 18 containing a termial alkene.
Scheme 3.
Synthesis of Carboxylic Acid 18
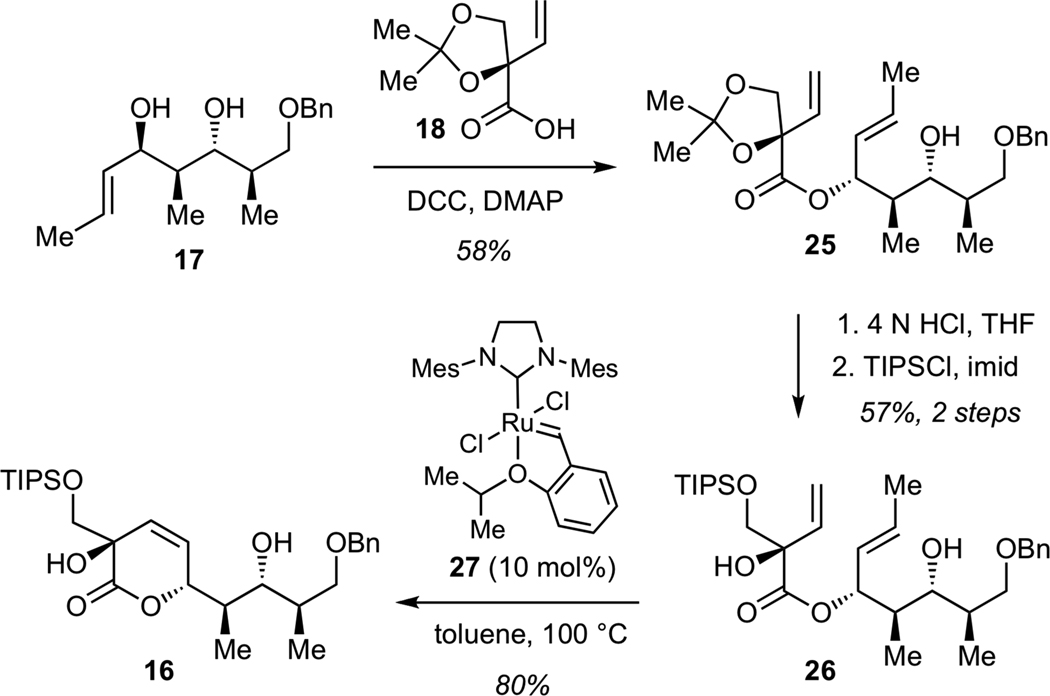
Chemoselective acylation of alcohol 17 with acid 18 was achieved in the presence of dicyclohexyl carbodiimide (DCC) and dimethylaminopyridine (DMAP) to give ester 25 (Scheme 4). Subjection of 25 directly to a series of Ru-based alkene metathesis catalysts afforded only low yields of the desired unsaturated lactone, presumably due to considerable steric bulk presented by the acetonide moiety. Acid-promoted removal of acetonide (4N aq. HCl in THF), followed by protection of the primary alcohol as a triisoprolylsilyl (TIPS) ether afforded diene 26. Ring-closure of 26 to lactone 16 proved to be superior to that using the acetonide 25. The optimum efficiency was achived using Ru catalyst 2729 to give lactone 16 in 80% yield.
Scheme 4.
Synthesis of Lactone 16
We next examined the conversion of lactone 16 to the requisite bicyclic ketal. While direct conversion of 16 to the corresponding methyl ketone using methylithium proved unsuccessful, this transformation could be efficiently achived in a two-step fashion involving initial formation of the Weinreb amide 28, followed by methyllithium addition (Scheme 5). Subsequent acid-catalyzed intramolecular acetalization afforded bicyclic ketal 14 in 72% yield for 3 steps. Reductive deprotection of benzyl ether, followed by Dess-Martin oxidation of the resulting alcohol and Wittig olefination furnished ester 29, a key structural subunit for the assembly of streptolydigin antibiotics.
Scheme 5.
Synthesis of Bicyclic Ester 29
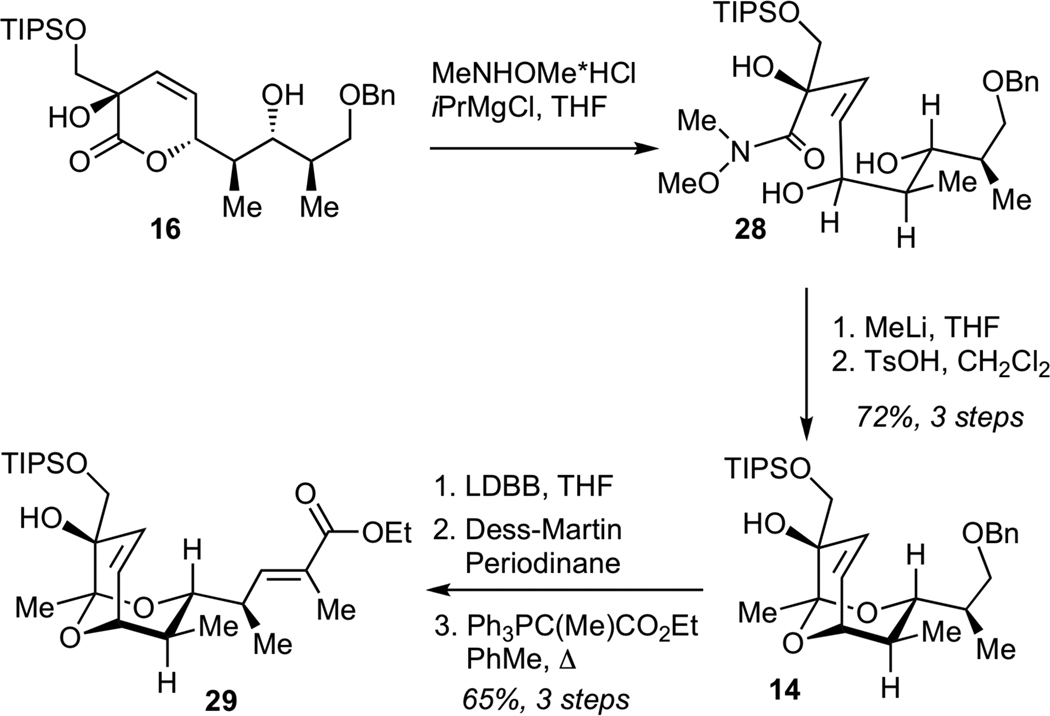
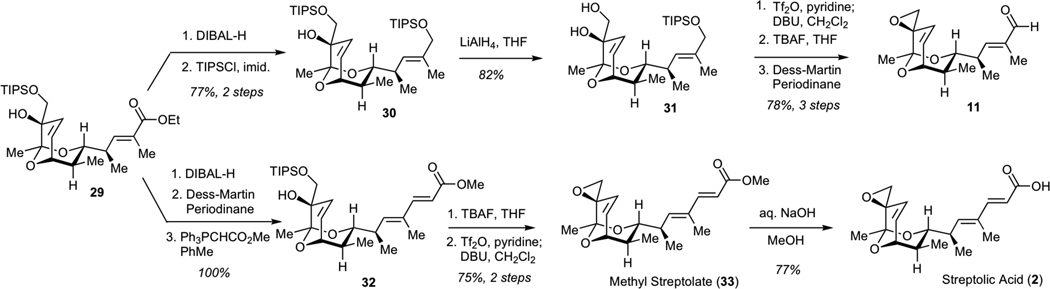
Subsequent elaboration of 29 required installation of the exocyclic epoxide and conversion of the ester to the aldehyde moiety. The optimized sequence for this functional group transformation began with 1,2-reduction of unsaturated ester 29 using DIBAL-H, followed by protection of the resulting allylic alcohol as a bis-TIPS ether 30 (Scheme 6). Chemoselective removal of the TIPS ether adjacent to the unprotected tertiary alcohol was efficiently achived by subjecting 30 to LiAlH4, presumably due to the participation of the neighboring alkoxide in the desilylation step. The resulting diol 31 was converted to the epoxide via intermediacy of the corresponding primary triflate. Finally, deprotection of the silyl ether and Dess-Martin oxidation efficiently afforded aldehyde 11.
Scheme 6.
Synthesis of Methyl Streptolate (33), Streptolic Acid (2) and Aldehyde 11
We also examined the conversion of ester 29 to streptolic acid (2). While streptolic acid represents a product of chemical degradation of streptolydigin, the importance of this synthetic effort was two-fold. First, this conversion would enable us to verify the correct structure of ester 29 by correlating analytical data of the synthetically produced streptolic acid (2) with those present in the literature. Second, the synthesis of compounds containing the fully functionalized bicyclic ketal core of streptolydigin, which was connected to the dienoyl moiety, would enable us to test the importance of the streptolic acid subunit for biological activity of the parent natural product (vide infra). This transformation began with the reduction of ester 29 with DIBAL-H, followed by the Dess-Martin oxidation of the resulting alcohol and Wittig olefination to give chain-extended ester 32 with excellent efficiency (Scheme 6). Fluoride-mediated desilylation of 32, followed by conversion of the resulting diol to an epoxide via the intermediacy of the corresponding primary triflate gave methyl streptolate (33). Subjection of 33 to basic hydrolysis afforded streptolic acid (2), which proved to be identical to the corresponding degradation product of streptolydigin, thus securing the chemical structure of all prior synthetic intermediates.11,21c
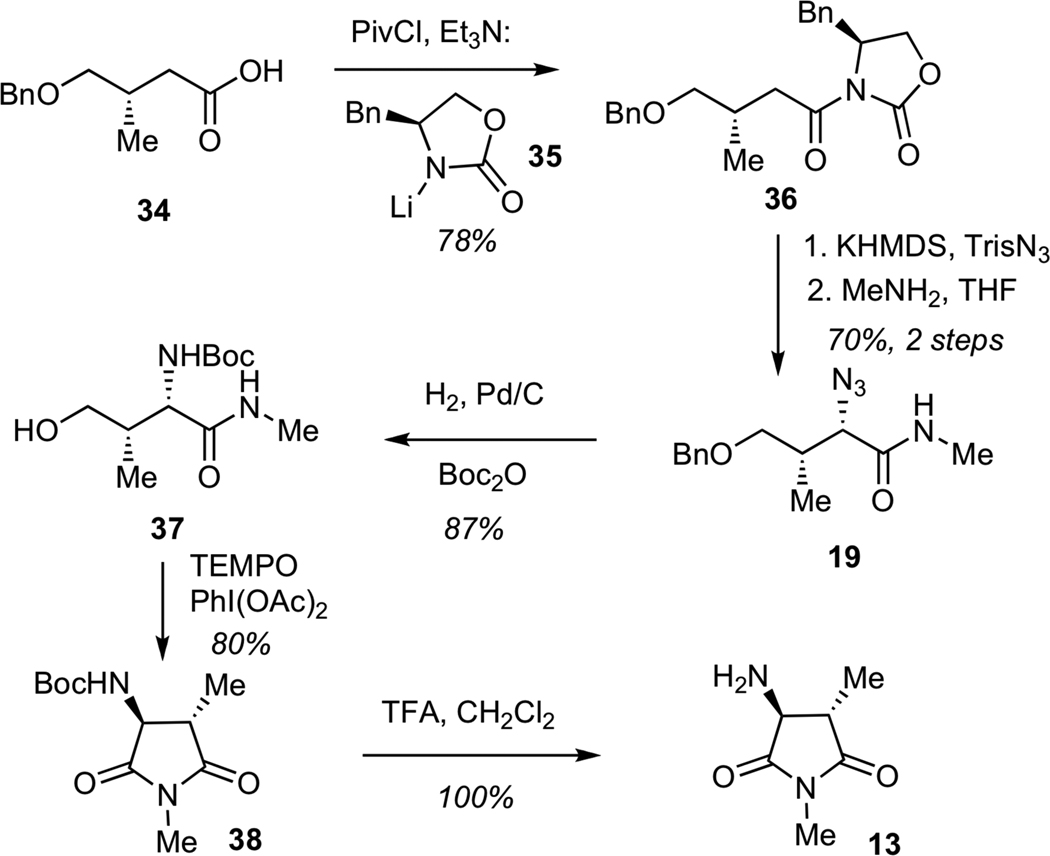
Having secured synthetic access to the streptolic acid moiety, we turned our attention to the construction of the tetramic acid fragment, which would arise from aspartimide 13 (Scheme 7). The assembly process began with the conversion of known acid 34 to the corresponding imide 36 by addition of lithium oxazolidinone 35 to the mixed anhydride derived from 34. The diasteoselective azide transfer was performed according to the Evans protocol,25 which entailed generation of the potassium enolate, followed by treatment with 2,4,6-triisopropylbenzenesulfonyl azide (TrisN3). The resulting imide was treated with methylamine to give amide 19. Reduction of azide 19 under a hydrogen atmosphere with simulatenous hydrogenolysis of the benzyl ether, followed by in situ Boc-protection of the resulting amine furnished carbamate 37. Subjection of amide 37 to a TEMPO-mediated oxidative cyclization afforded the corresponding aspartimide 38, which was treated with trifluoroacetic acid (TFA) to enable Boc-deprotection to give primary amine 13.
Scheme 7.
Synthesis of Imide 13
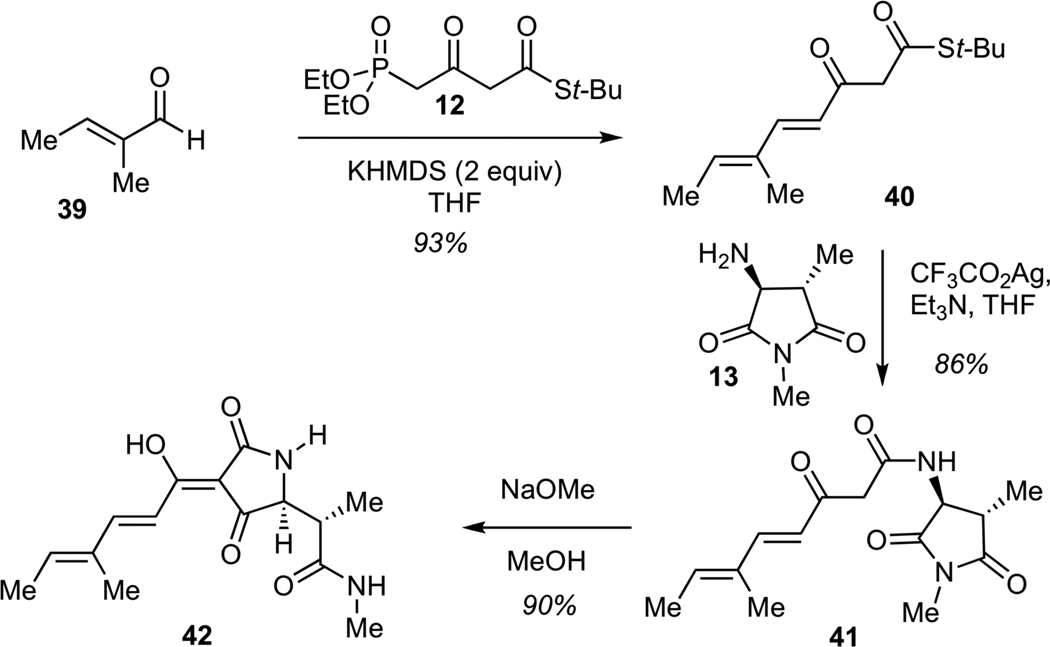
We next examined the assembly of the simplified methyl hexadienoyl tetramic acid 42, which corresponded to the structure of streptolydiginone (4) with a fully truncated bicyclic ketal (Scheme 8). This sequence was designed to model the final stages of the streptolydiginone synthesis and would also enable us to test the importance of the tetramic acid subunit of this agent on its biological activity (vide infra). The sequence began with the Horner-Wadsworth-Emmons olefination of unsaturated aldehyde 39 with known phosphonate 1230 to give keto thioester 40. Treatment of 40 with silver (I) trifluoroacetate in the presence of amine 13 delivered ketoamide 41, which underwent Dieckmann cyclization12 upon exposure to methanolic sodium methoxide to give tetramic acid 42 in 90% yield.
Scheme 8.
Synthesis of 4-Methyl-2,4-Hexadienoyl Tetramic Acid (42)
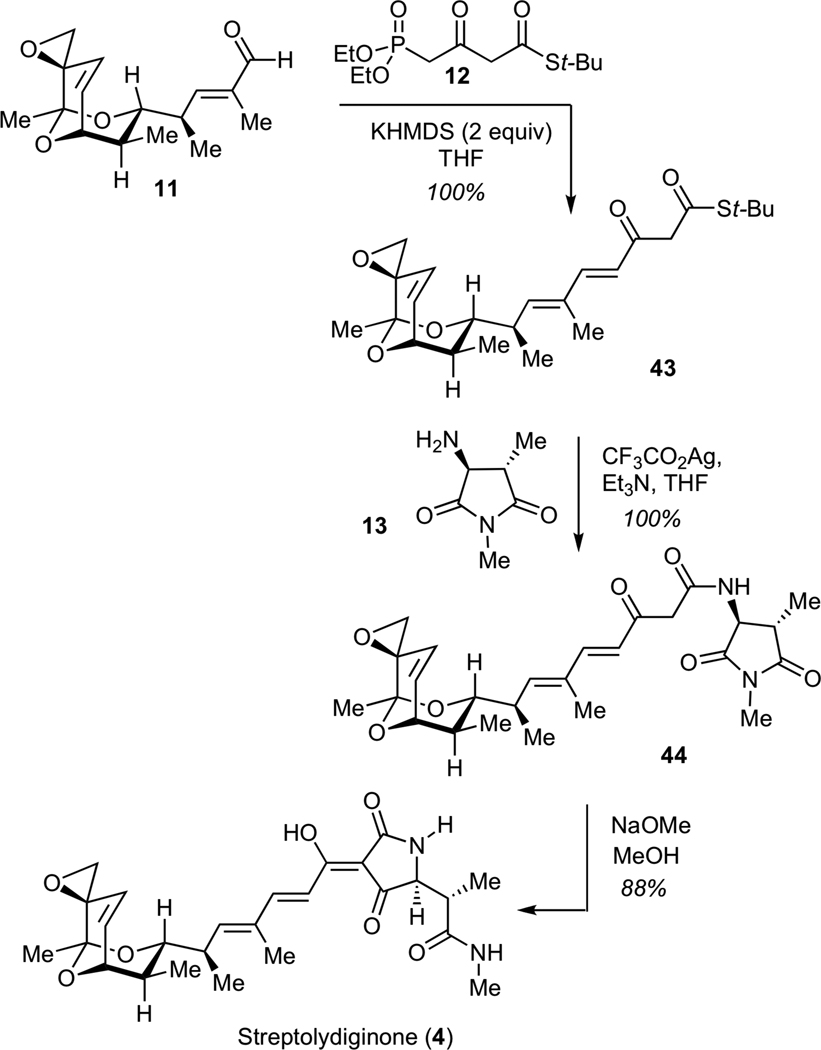
The stage was now set for completing the assembly of streptolyidiginone (4). To this end, we subjected aldehyde 11 to the Horner-Wadsworth-Emmons olefination with phosphonate 1230 (Scheme 9). This transformation efficiently delivered the desired keto thioester 43. Subsequent treatment of 43 with silver (I) trifluoroacetate in the presence of amine 13 resulted in formation of ketoamide 44. Under such conditions, both steps proceeded with quantitative efficiency without any detectable degradation of the labile epoxide moiety. The next step entailed the Dieckmann cyclization of 44 using a methanolic solution of sodium methoxide, according to the Rinehart protocol,12 to deliver streptolydiginone (4) in 88% yield and a longest linear sequence of 26 steps. The structure of 4 was confirmed by comparison of the 500 MHz 1H NMR and 125 MHz 13C NMR spectral data for the sodium salt of synthetic streptolydiginone with the corresponding NMR spectra for this aglycon, which was isolated by the Salas group from the re-engineered S. lydicus strain with suppressed glycosylation of streptolydigin.13
Scheme 9.
Synthesis of Streptolydiginone (4)
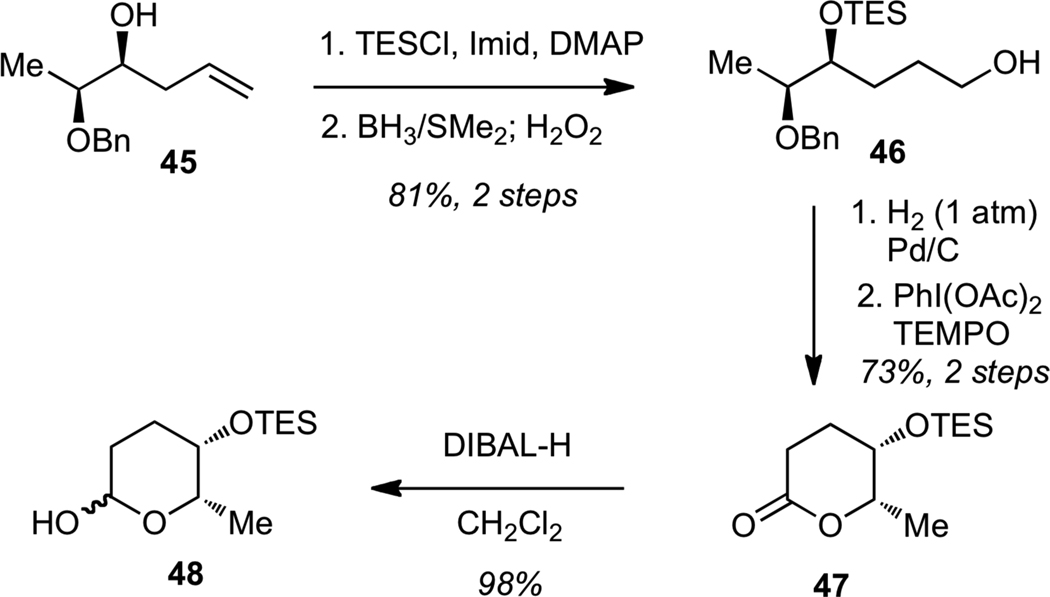
We next examined the glycosylation of streptolydiginone (4). The synthesis of the L-rhodinose moiety of streptolydigin is depicted in Scheme 10. Protection of the known homoallylic alcohol 4530 as a triethylsilyl (TES) ether, followed by hydroboration-oxidation of the alkene gave alcohol 46. Hydrogenolysis of the benzyl ether, followed by PhI(OAc)2-mediated oxidative cyclization of the resulting diol afforded lactone 47. Reduction of 47 with DIBAL-H delivered the desired lactol 48.
Scheme 10.
Synthesis of Protected L-Rhodinose (48)
Our exploratory studies revealed that simple acyl tetramic acids could be efficiently glycosylated with protected L-rhodinose derivatives by treatment of the corresponding lactol or glycal with tetramic acid in choloroform at 20 °C in the presence of a catalytic amount of p-TsOH. However, glycosylation of the streptolydiginone’s tetramic acid moiety could not be achieved under the same reaction conditions presumably due to the increased steric bulk of the branched alkyl substituent in the direct proximity to the reactive N-H bond of the tetramic acid.
Synthesis of Streptolydigin
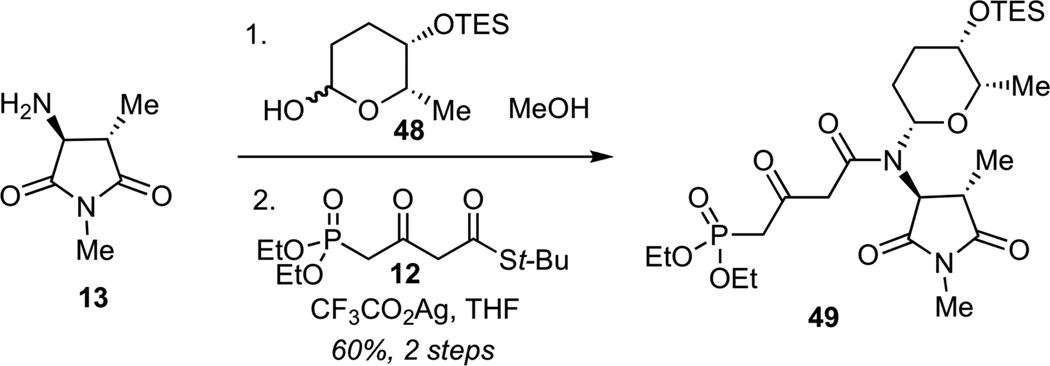
Due to the inability to glycosylate streptolydiginone (4), we examined an alternaive synthetic approach to streptolydigin (1), which entailed installation of the glycoside moiety prior to the formation of the final tetramic acid (Scheme 11). The key disconnection of streptolydigin (1) was designed to unite two advanced fragments, including bicyclic aldehyde 11 and phosphonate 49. The latter would be assembled by N-glycosylation and N-acylation starting with three fragments 12, 13 and 48, which were previously prepared in our route to streptolydiginone (4). It is noteworthy that phosphonates similar to 49 were prepared by both the Boeckman and Schlessinger laboratories.21a,b However, no attempts to use such fragments for elaboration of streptolydigin have been reported. The main challenge was to ensure that the projected assembly process would be compatible with the highly labile terminal epoxide moiety of aldehyde 11.
Scheme 11.
Revised Synthetic Approach to Streptolydigin (1)
Following a series of exploratory studies, we devised a simple, two-step sequence for the assembly of N-glycosyl phosphonate 49 (Scheme 12). Direct treatment of amine 13 with TES-protected rhodinose 48 afforded the desired N-glycosylated product as a single requisite diastereomer. Subsequent Ag-promoted N-acylation of this glycosyl amine with thioester 1231 afforded phosphonate 49 in 60% yield for 2 steps.
Scheme 12.
Synthesis of Phosphonate 49
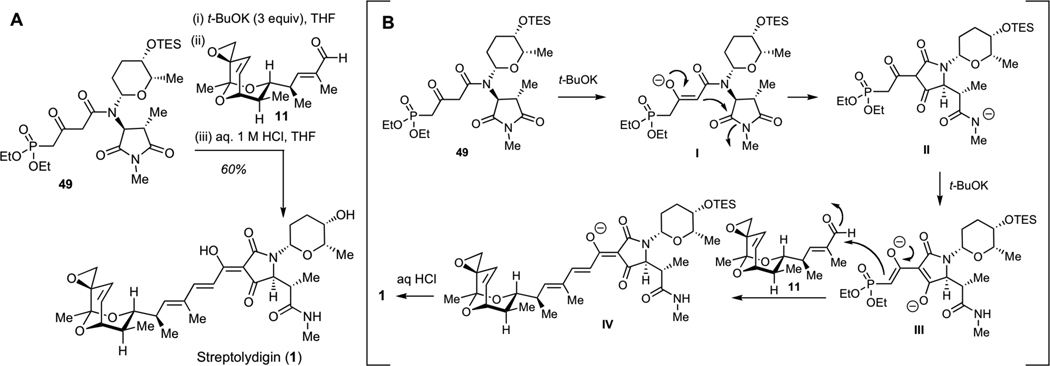
After significant optimization, we ultimately developed a one-flask protocol for synthesis of streptolydigin (1), which entailed treatment of the phosphonate 49 with 3 equivalents of t-BuOK, followed by the addition of aldehyde 2 and a rapid acidic work-up with 1 M aqueous solution of HCl at 0 °C (Figure 2A). This one-pot process entailed several consecutive steps, including initial deprotonation of 49 to produce enolate I (Figure 2B), Dieckmann cylization to give tetramic acid II, subsequent abstraction of two acidic protons to give III, followed by the Horner-Wadsworth-Emmons olefination and final neutralization with concomitant acid-mediated TES deprotection to furnish 1. Synthetic streptolydigin (1) proved to be identical in all respect to the authentic sample of the natural product.32 This sequence enabled the assembly of this natural product with a longest linear sequence of 24 steps and a higher level of convergency compared to our initial route, which afforded streptolydiginone (4) in 26 chemical steps.
Figure 2.
Completion of the Streptolydigin Synthesis. A. Optimized one-flask protocol for conversion of phosphonate 49 and aldehyde 11 to streptolydigin (1). B. Mechanism of streptolydigin formation, which entails initial Dieckmann cyclization to give II, Horner-Wadsworth-Emmons olefination of aldehyde 11 with III, followed by subsequent acidification of IV with concomitant removal of the TES group to give 1.
RNAP Inhibitory Activity of Streptolydigin, Streptolydiginone, Streptolic Acid and Tetramic Acid
Our next objective was to examine the inhibition of bacterial RNAP by streptolydigin (1), streptolydiginone (4), as well as other compounds prepared during our initial synthetic studies. While antibacterial activity of several members of this antibiotic family has been reported, the RNAP inhibitory profile has only been studied for streptolydigin (1)7,8 and tyrandamycin A (8).32 These studies demonstrated that tyrandamycin A was a substantially less potent bacterial RNAP inhibitor.33 The antimicrobial data of dienoyl tetramic acid antibiotics have been obtained using only a small number of compounds, isolated from various microbial producers, which provided only limited structure-activity relationship data. While notable success in re-engineering streptolydigin biosynthesis has been achieved, this approach has thus far provided only a few compounds with modified glycoside and tetramic acid subunits of the parent natural product.13,14
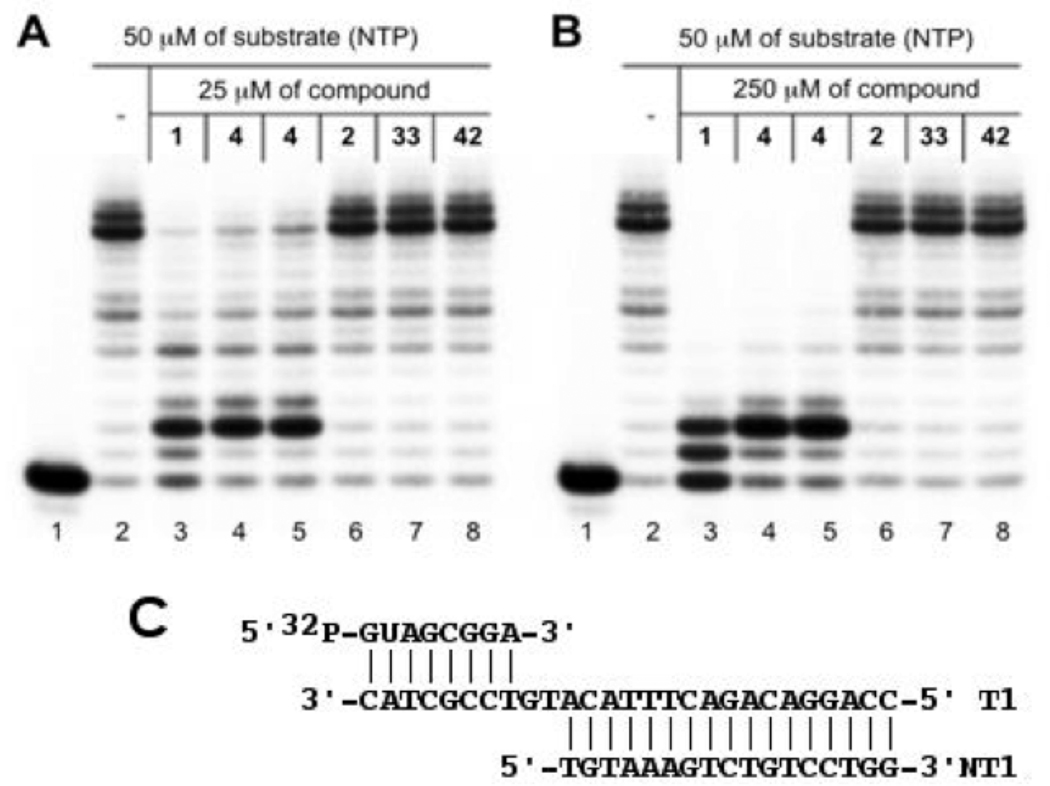
Our study was aimed at evaluating the importance of two major structural subunits of streptolydigin (1), including streptolic acid and the tetramic acid fragments. To this end, we examined inhibition of a well-characterized Thermus aquaticus RNAP34 by following in vitro transcription in the presence of 50 µM of nucleotide triphosphates (NTPs) and two inhibitor concentrations (25 µM and 250 µM, Figures 3A and 3B, respectively). The assay consisted of assembling T. aquaticus RNAP complexes with nucleic acid scaffolds that mimic the structure of nucleic acids in the transcription elongation complex (Figure 3C). It was previously shown that artifical transcription elongation complexes assembled on nucleic acid scaffolds are functional35 and can be used to monitor the effects of various inhibitors of transcription elongation.36 To monitor RNAP-catalyzed elongation of the 8-nucleotide-long RNA component of the scaffold, it was radioactively labeled at the 5’ end, allowing its visualization after denaturing gel-electrophoresis (lane 1). The addition of NTPs led to the appearance of a run-off product, which resulted from RNAP-catalyzed extension of the nascent transcript until the end of the template (lane 2). The addition of synthetic streptolydigin (1, 25 µM) inhibited the appearance of run-off transcript (Figure 3A, lane 3), an expected result for a transcription elongation inhibitor. A shorter transcript that has undergone only two cycles of nucleotide addition was formed. At 250 µM of inhibitor, the amount of the extended transcript decreased and signficant amount of the initial nascent transcript remained intact (a product of a single nucleotide addition was also observed, Figure 3B, lane 3). The same pattern of transcription elongation inhibition was observed when an authentic sample of 1, which was isolated from S. lydicus, was used (data not shown). We also found that streptolydiginone (4), both as a free tetramic acid (lane 4) and as a corresponding sodium salt (lane 5), was effective at inhibiting T. aquaticus RNAP. At the highest concentration tested, the observed RNAP inhibitory activity of 4 was visibly lower than streptolydigin’s (compare lanes 3 with lanes 4 and 5, Figure 3B). This observation is correlated with weaker antibacterial activity of streptolydiginone (4) against S. albus, which was reported previously.13
Figure 3.
In vitro inhibition of bacterial transcription catalyzed by T. aquaticus RNAP. Artificial transcription elongation complexes containing radioactively labeled 8-nt RNA primer were assembled on nucleic-acid scaffolds (C) using T. aquaticus RNAP core enzyme. Where indicated, reaction mixtures were supplemented with 25 (A) or 250 µM (B) of small-molecule inhibitors and transcription was initiated by the addition of NTPs. Reaction products were resolved by denaturing polyacrylamide gel-elctrophoresis and revealed by autoradioagraphy.
We were unable to observe any RNAP inhibition by either the streptolic acid (2, lane 6) or the corresponding methyl ester (33, lane 7). Furthermore, the truncated tetramic acid subunit of streptolydiginone, which is represented by compound 42, was also inactive at both concentrations tested (lane 8). These results demonstrate that the presence of both structural subunits of streptolydigin is essential for activity of this class of antibiotics. This conclusion is also in agreement with previous crystallographic studies of streptolydigin-RNAP complexes, which established that both streptolic and tetramic fragments of the natural product interacted with the bridge-helix and the trigger-loop regions of the protein, respectively.8 Thus, removal of each of the individual structural subunits of streptolydigin would result in substantial decrease of the binding efficiency and loss of inhibitory activity.
Rationally Simplified Streptolydigin Analogs: Design, Synthesis and RNAP Inhibition
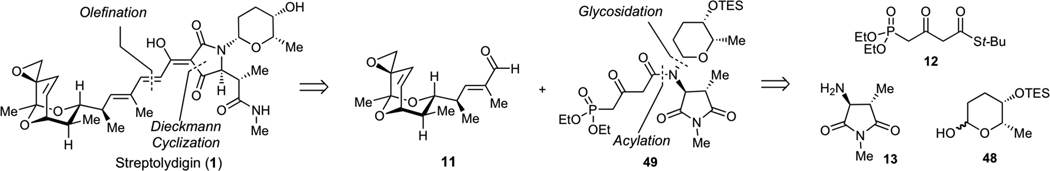
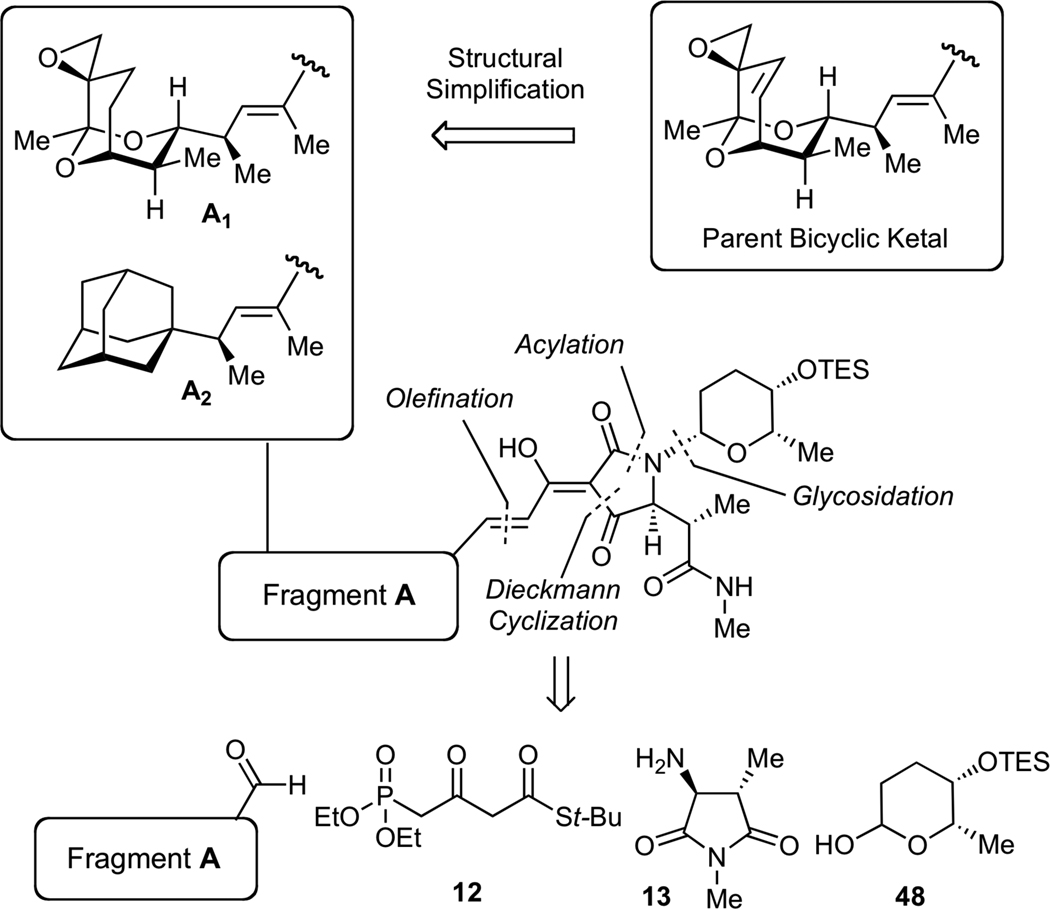
Our initial biochemical analysis revealed that truncation of either streptolic or tetramic acid subunits of streptolydigin resulted in complete loss of RNAP inhibitory activity. We envisioned that additional modifications could be introduced into the streptolic subunit of the natural product provided that the fully functionalized tetramic acid fragment remains intact (Figure 4). Specifically, we planned to remove the C(10)-C(11) alkene moiety in the bicyclic ketal core of the natural product (Fragment A1, Figure 4). This structural alteration was expected to considerably increase the efficiency of our assembly process (vide infra). In addition, we intended to substitute the entire bicyclic ketal moiety with an adamantyl fragment, which was expected to preserve the overall shape and most of the physicochemical properties of this relatively hydrophobic subunit (Fragment A2, Figure 4). Each new compound would derive from a highly convergent sequence that would entail a condensation of four building blocks following our previously established synthetic sequences (Figure 4). Due to structural simplification of fragment A, we envisioned that such derivatives would be assembled with significantly higher synthetic efficiency compared to our original routes to streptolydigin (1) and streptolydiginone (4), which required 24 and 26 steps, respectively.
Figure 4.
General Strategy to Simplified Analogs of Streptolydigin. The synthetic approach would entail a sequential condensation of four reaction partners, including aldehyde-containing fragment A, phosphonate 12, imide 13 and protected L-rhodinose 48. The projected assembly process entails N-glycosidation, N-acylation, Dieckman cyclization, olefination and final deprotection.
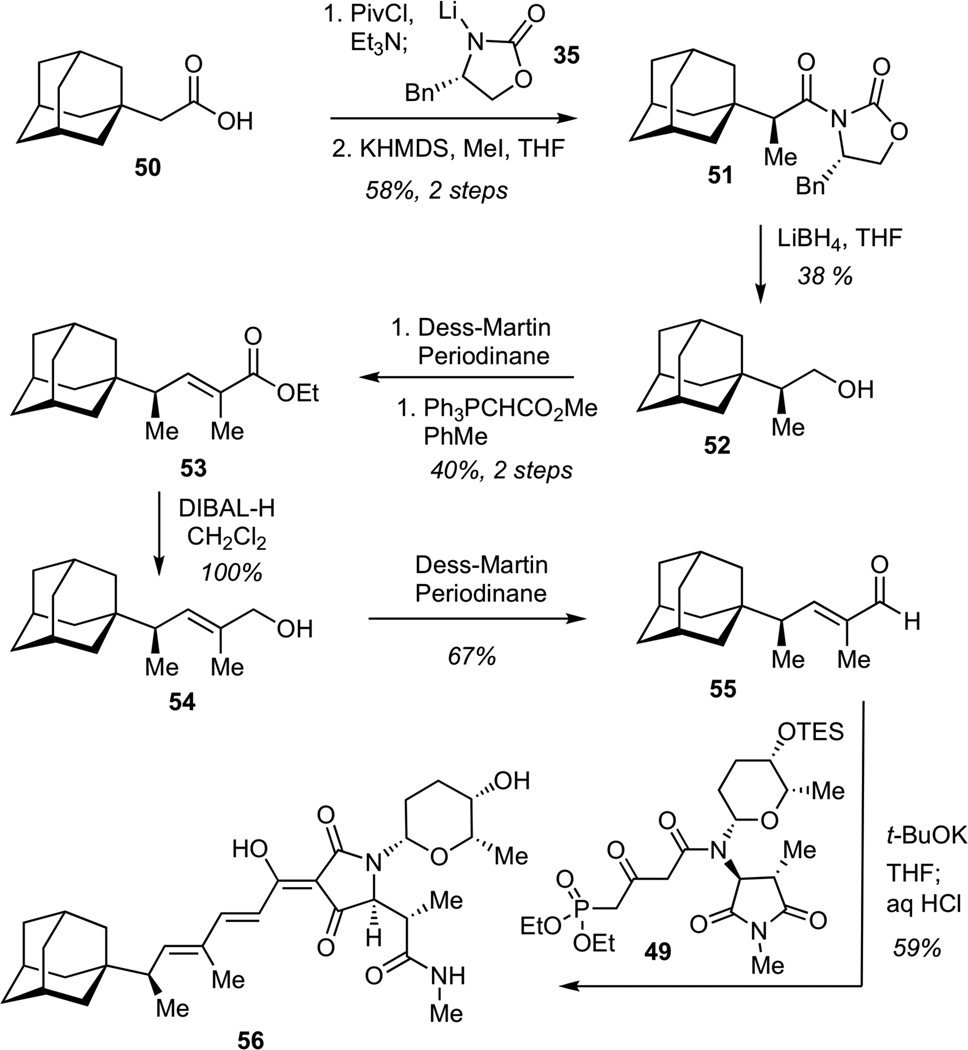
The synthesis of the adamantane-bearing derivative began with asymmetric Evans alkylation37 of chiral imide derived from the commercially available carboxylic acid 50 (Scheme 13). This process delivered the requisite alkylation product in good efficiency and excellent diastereomeric purity. Subsequent elaboration entailed reduction of imide 51 with LiBH4, followed by Dess-Martin oxidation of alcohol 52, and Wittig olefination of the resulting aldehyde to give unsaturated ester 53. Reduction of 53 with DIBAL-H, followed by Dess-Martin oxidation of the allylic alcohol 54 afforded aldehyde 55. Treatment of aldehyde 55 with phosphonate 49 in the presence of excess of t-BuOK using our previously developed one-flask protocol for sequential olefination-cyclization-deprotection afforded requisite adamantane-containing tetramic acid 56 in 59% yield (13-step longest linear sequence from commercially available precursors).
Scheme 13.
Synthesis of Adamantane-Substituted Dienoyl Tetramic Acid (56)
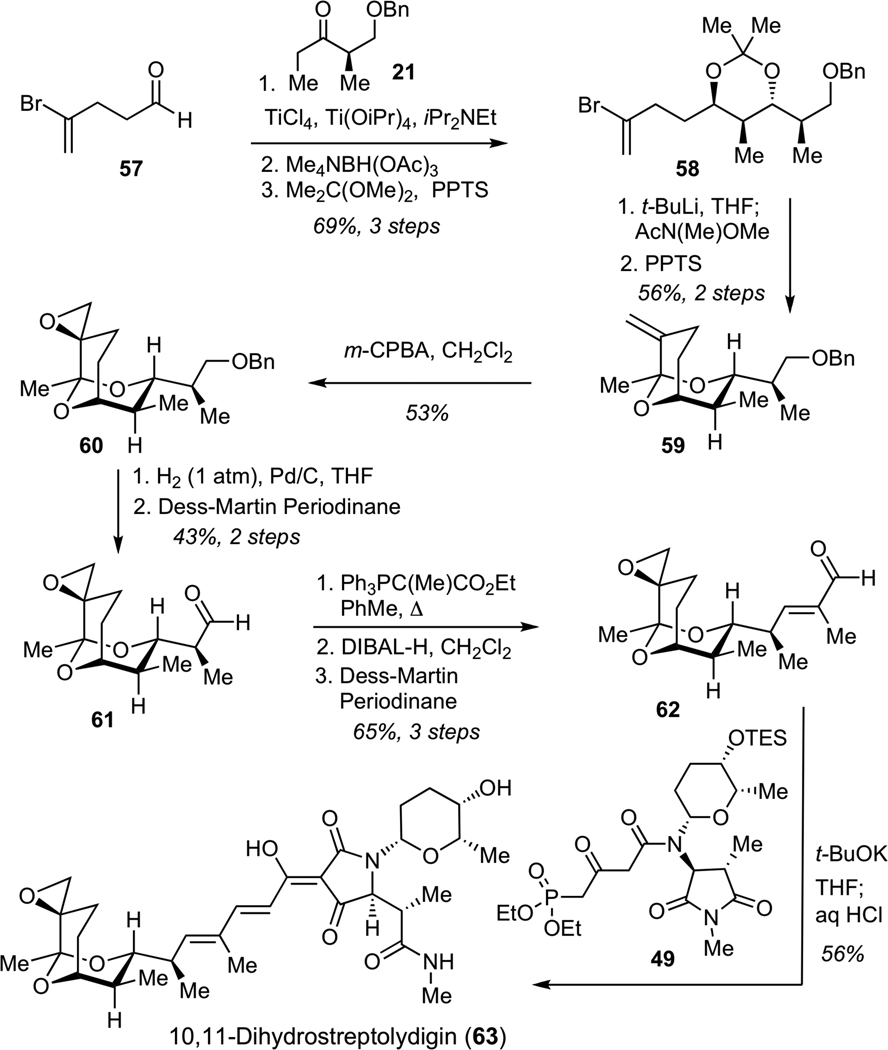
Construction of 10,11-dihydrostreptolydigin (63) commenced with a Ti-mediated aldol reaction between ketone 21 and aldehyde 57, followed by an anti-reduction of the corresponding hydroxyketone to afford diol, which was protected as acetonide 58 (Scheme 14). Bromine-lithium exchange and subsequent acylation gave the expected enone, which was treated with PPTS in methanol to generate bicyclic ketal 59. Concave-face epoxidation of alkene 59 delivered desired epoxide 60. Hydrogenolysis of 60, followed by Dess-Martin oxidation of the resulting alcohol afforded aldehyde 61. Subjection of 61 to a Wittig olefination furnished an unsaturated ester, which was subjected to reduction with DIBAL-H, followed by Dess-Martin oxidation to deliver aldehyde 62. Final assembly of 10,11-dihydrostreptolydigin (63) was performed under the conditions developed for the preparation of streptolydigin. Treatment of ketoamide 49 with excess potassium tert-butoxide, followed by addition of aldehyde 62 and acidic work-up afforded 10,11-dihydrostreptolydigin (63). Removal of the C(10)-C(11) alkene moiety enabled significant increase in the overall synthetic efficiency of this route (15-step longest linear sequence) compared to our original approach to streptolydigin, which required 24 synthetic steps.
Scheme 14.
Synthess of 10,11-Dihydrostreptolydigin (63)
Having completed the syntheses of compounds 56 and 63, we examined their ability to inhibit transcription catalyzed by T. aquaticus RNAP (Figure 5) using a nucleic acid scaffold similar to the one presented in Figure 3C but having a longer downstream (transcribed) segment. Streptolydigin (1) was included as a positive control (lane 2, Figure 5A). We found that 10,11-dihydrostreptolydigin (63) inhibited T. aquaticus RNAP (lane 3, Figure 5A) very similarly to streptolydigin (1), with transcript elongation blocked after incorporation of two nucleotides into the growing RNA strand. This observation suggested that removal of the C(10)-C(11) alkene preserved a substantial amount of the parent activity from the parent natural product 1 (lane 2, Figure 5A). Interestingly, we did not observe any inhibition of RNAP by adamantane-containing analog 56 at 100 µM (lane 4, Figure 5A) or 1 mM (data not shown) concentrations. This finding indicated that the adamantane core of 56 did not provide a viable replacement for a bicyclic ketal moiety of streptolydigin (1).
Figure 5.
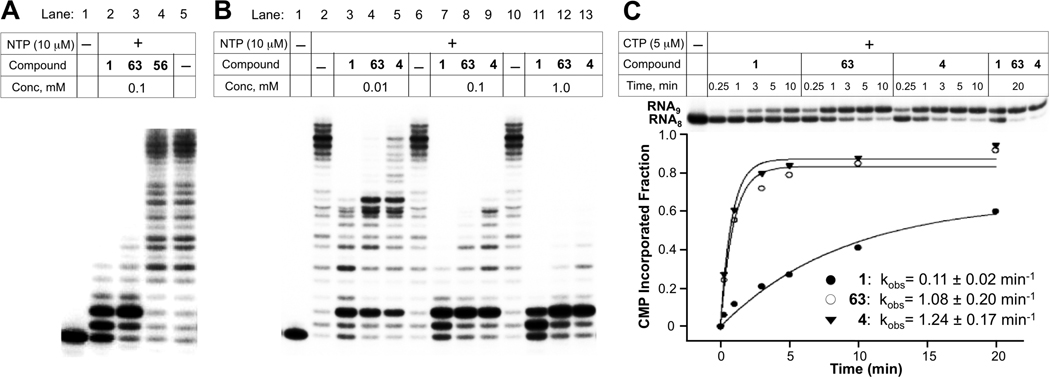
In vitro inhibition of transcription catalyzed by T. aquaticus RNAP by streptolydigin derivatives. A. A comparison of RNAP inhibitory activity of streptolydigin (1), 10,11-dihydrostreptolydigin (63), and adamantane derivative 56 using 10 µM NTP and 0.1 mM of each inhibitor. B. Dose-dependent RNAP inhibition by streptolydigin (1), streptolydiginone (4) and 10,11-dihydrostreptolydigin (63). C. Quantitative assessment of inhibitory effects of streptolydigin (1), streptolydiginone (4) and 10,11-dihydrostreptolydigin (63) on the rate of CMP incorporation into nascent 8-nt starting RNA.
We next compared the relative efficacy of T. aquaticus RNAP inhibition by three compounds, which were shown to elicit activity, including streptolydigin (1), streptolydiginone (4), and 10,11-dihydrostreptolydigin (63). The results are presented in Figure 5B. At the two highest concentrations of streptolydigin (1) (0.1 and 1.0 mM), the transcription was blocked either completely or following the addition of one or two nucleotides (Figure 5B, lanes 7 and 11). At the same concentrations of streptolydiginone (4) and 10,11-dihydrostreptolydigin (63), inhibition occurred almost exclusively after incorporation of two nucleotides (Figure 5B, lanes 8, 9, 12, and 13). These results once again confirmed that 10,11-dihydrostreptolydigin (63) and streptolydiginone (4) were effective at inhibiting transcription in vitro. This conclusion was further supported by an experiment conducted in the presence of subsaturating (10 µM) concentrations of inhibitors (Figure 5B, lanes 3, 4 and 5). Similar results were obtained in experiments using a closely related T. thermophilus RNAP, as well as RNAPs from M. smegmatis (data not shown). RNAPs from Gram-negative organisms such as Francisella tularensis and E. coli required much higher concentrations of inhibitors to acheive the same extent of inhibition (data not shown), confirming earlier observations that RNAPs from Gram-negative bacteria are significantly more resistant to streptolydigin than their counterparts from Gram-positive organisms.
We also followed the rate of single nucleotide incorporation catalyzed by T. aquaticus RNAP in the presence of streptolydigin (1), streptolydiginone (4), and 10,11-dihydrostreptolydigin (63). This study provided a quantitative basis for assessing the difference in inhibitory activities of each of the three compounds tested (Figure 5C). In this assay, transcription complexes containing radioactively-labeled nascent transcript were supplemented with CTP, which was necessary to extend the transcript by only one nucleotide. The concentration of CTP (5 µM) and the reaction temperature (25 °C) were kept low to slow down the RNAP catalysis rate, which enabled following the CMP addition in real time in the presence of inibitors. In the absence of inhibitors, the reaction was complete (full conversion of 8-nt starting RNA to 9-nt product) at the first time point sampled (15 sec, data not shown). In the presence of inhibitors, the reaction rate was slowed down considerably, with full extension achieved at 5 minutes for streptolydiginone (4) and 10,11-dihydrostreptolydigin (63), which appeared to inhibit nucleotide addition with similar efficiency. Streptolydigin (1) inhibition was more potent, with nascent transcript extension reaction being ~50% complete after 20 minutes of incubation. The data obtained were used to derive the apparent rates of RNAP-catalyzed single nucleotide extension, which were found to be 0.11 min−1, 1.08 min−1, and 1.24 min−1 in the presence of streptolydigin (1), 10,11-dihydrostreptolydigin (63), and streptolydiginone (4), respectively. Overall, the results of in vitro transcription assays establish that removal of either the glycoside moiety of streptolydigin or the C(10)-C(11) alkene preserved RNAP inhibitory activity of the resulting compounds, while weakening their in vitro potency by about 10 fold.
Antimicrobial Activity against S. salivarius
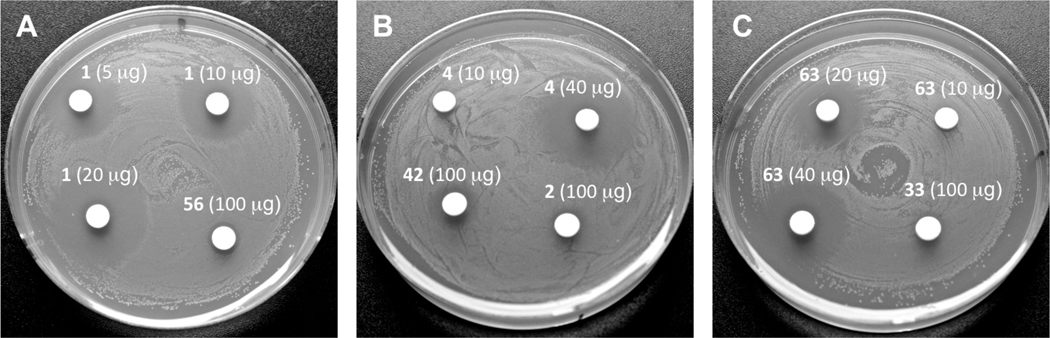
We also examined antimicrobial activity of all streptolydigin-based compounds prepared in our study. Originally, streptolydigin (1) was found to be effective in inhibiting several Streptococcus species, including S. faecalis, S. hemolyticus, S. lactis, S. mitis, S. viridans and S. pneumoniae with MICs in the range of 0.19–3.12 µg/mL.6a Therefore, we selected S. salivarius, a representative Streptococcus strain, which typically colonizes human oral and nasopharyngeal epithelia and only becomes harmful to the host if the immune status is altered or there is a loss of control of epithelial cell sensing and discriminatory systems.38 Our main obective was to use this model, normally nonpathogenic organism in order to correlate the initially observed RNAP inhibitory activity of all compounds with their antibacterial action. The results of this study, which employed a standard disk-diffusion protocol, are shown in Figure 6. As expected, streptolydigin (1) inhibited growth of S. salivarius in dose-dependent manner at concentrations of 5 µg, 10 µg and 20 µg per disk (Figure 6A). Streptolydiginone (4) was also found to elicit antibacterial activity, albeit not as potent as that of streptolydigin (1). While substantial growth inhibition was observed at 40 µg of 4 per disk, no effect on bacterial proliferation occurred at a lower dose of 10 µg per disk (Figure 6B). Evaluation of 10,11-dihydrostreptolydigine (63) revealed dose-dependent antibacterial action at concentrations of 20 µg and 40 µg per disk (Figure 6C). Similar to streptolydiginone (4), no antibacterial activity was observed with 63 at 10 µg per disk (Figure 6C). Overall, the observed antibacterial activities of 1, 4 and 63 were in excellent correlation with their RNAP inhibitory profiles. In addition, streptolic acid (2) and methyl streptolate (33) were found to be completely inactive even at high concentration of 100 µg/disk (Figure 6B and 6C). The adamantane-containing derivative 56 and tetramic acid 42 elicited only weak antibacterial action at 100 µg/disk (Figure 6A and 6B) and no activity at lower concentrations tested (data not shown). The lack of activity of 2, 33, 42, and 56 was also in excellent agreement with inability of such compounds to inhibit T. aquaticus RNAP. This study demonstrated once again that the presence of both streptolic and tetramic subunits of streptolydigin is required for its antibiotic activity and established that RNAP inhibition of streptolydigin-based compounds was well correlated with their antibacterial profile.
Figure 6.
Antimicrobial Activity of Synthetic Streptolydigin Antibiotics against S. salivarius. Each compound was tested according to the general protocol described in the Experimental Procedures. Most potent compounds, including 1, 4 and 63 were tested at several concentrations shown. All other compounds were tested at a concentration of 100 µg/disk.
Conclusions
We described the development of a general synthetic strategy to streptolydigin antibiotics, as well as their comprehensive biochemical and antimicrobial evaluation. Our initial studies resulted in the total synthesis of streptolydiginone (4) and streptolydigin (1) and enabled the assembly of a series of simplified derivatives of the parent natural products, which were designed to assess the importance of the individual streptolic and tetramic acid subunits. Indeed, the presence of both major fragments of streptolydigin was found to be essential for its activity. This finding was in agreement with previous X-ray crystallographic characterization of streptolydigin-RNAP complexes. In addition, our effort enabled a 15-step synthesis of 10,11-dihydrostreptolydigin, which inhibited three bacterial RNAPs and elicited antimicrobial activity against S. salivarius. The ability to access efficiently simplified analogs of streptolydigin that elicit substantial antibacterial activity demonstrates the power of our general synthetic approach, which could be employed for subsequent rational design and development of new streptolydigin-based antibiotics with enhanced potency and improved pharmacological profile.
Experimental Procedures
Streptolic acid (2)
A solution of methyl streptolate 33 (13.0 mg, 0.039 mmol) in a mixture of methanol (1 mL) and water (0.1 mL) was treated with sodium hydroxide (16.0 mg, 0.39 mmol). Reaction mixture was left overnight at room temperature, concentrated in vacuo and treated with aqueous HCl (0.4 mL of 1 N solution). The resulting solution was extracted twice with ethyl acetate. Combined organic extracts were washed with brine, dried over MgSO4 and concentrated in vacuo. Preparative TLC (development with ethyl acetate:methanol 20:1) afforded 10.0 mg (77% yield) of streptolic acid (2). [α]24D = +135.0 (c = 0.7, EtOH); 1H NMR (500 MHz, CDCl3) δ 0.69 (d, 3H, J = 7.0 Hz), 1.04 (d, 3H, J = 7.0 Hz), 1.23 (s, 3H), 1.80 (s, 3H), 1.94 (m, 1H), 2.73 (m, 1H), 2.81 (d, 1H, J = 5.0 Hz), 2.98 (d, 1H, J = 5.0 Hz), 3.63 (d, 1H, J = 10.5 Hz), 4.35 (t, 1H, J = 4.5 Hz), 5.62 (d, 1H, J = 10.0 Hz), 5.81 (d, 1H, J = 15.5 Hz), 6.13 (d, 1H, J = 10.0 Hz), 6.34 (dd, 1H, J = 10.0, 5.0 Hz), 7.44 (d, 1H, J = 15.5 Hz); 13C NMR (125 MHz, CDCl3) δ 12.2, 12.5, 17.1, 22.2, 33.7, 35.0, 50.5, 55.0, 71.4, 76.1, 98.8, 115.1, 130.5, 132.6, 133.8, 143.3, 152.0, 172.5; HRMS (ESI) calculated for C18H25O5 [M+H]+ 321.1702, found 321.1702.
Streptolydiginone (4)
A stirred solution of ketoamide 44 (16.0 mg, 0.033 mmol) in methanol (0.7 mL) was treated with sodium methoxide (0.33 mL of 0.5 M solution in methanol, 0.165 mmol) at 0 °C. Reaction mixture was stirred overnight at room temperature and concentrated in vacuo. The residue was dissolved in water (1 mL), treated with aqueous HCl (0.17 mL of 1 N solution) at 0 °C, and extracted twice with ethyl acetate. The combined organic extracts were washed with water, brine, then dried over MgSO4 and concentrated in vacuo to afford 14.0 mg (88% yield) of streptolydiginone (4). [α]25D = +24.9 (c = 0.3, CHCl3); 1H NMR (500 MHz, CDCl3) δ 0.70 (m, 3H), 1.05 (m, 6H), 1.23 (s, 3H), 1.90 (s, 3H), 1.93 (m, 1H), 2.73–2.91 (m, 6H), 2.98 (d, 1H, J = 5.0 Hz), 3.65 (d, 1H, J = 10.5 Hz), 4.10 (s, 1H), 4.35 (t, 1H, J = 4.5 Hz), 5.63 (d, 1H, J = 10.1 Hz), 5.67 (s, 1H), 6.26 (d, 1H, J = 9.9 Hz), 6.34 (dd, 1H, J = 10.1, 4.7 Hz), 7.10 (d, 1H, J = 15.6 Hz), 7.58 (d, 1H, J = 15.6 Hz); 13C NMR (125 MHz, CDCl3) δ 11.4, 12.2, 12.5, 17.1, 22.2, 26.4, 34.1, 35.2, 40.9, 50.5, 55.0, 62.9, 71.4, 76.1, 98.9, 100.3, 116.2, 130.6, 133.8, 133.9, 146.0, 150.3, 175.0, 175.5, 175.7, 194.0; HRMS (ESI) calculated for C26H35N2O7 [M+H]+ 487.2444, found 487.2442. A portion of the product was treated with dilute aqueous NaHCO3, concentrated in vacuo and azeotroped three times with chloroform. The residue was extracted with chloroform, solution phase was concentrated in vacuo and azeotroped with chloroform to afford a sodium salt of streptolydiginone. 1H NMR (500 MHz, (CD3)2SO) δ 0.65 (d, 3H, J = 6.9 Hz), 0.78 (d, 3H, J = 6.9 Hz), 0.98 (d, 3H, J = 6.8 Hz), 1.08 (s, 3H), 1.78 (s, 3H), 1.78 (m, 1H), 2.57 (d, 3H, J = 4.2 Hz), 2.66 (q, 1H, J = 6.9 Hz), 2.75 (m, 1H), 2.89 (d, 1H, J = 5.0 Hz), 2.94 (d, 1H, J = 5.0 Hz), 3.58 (m, 2H), 4.10 (s, 1H), 4.33 (t, 1H, J = 4.6 Hz), 5.62 (d, 1H, J = 10.1 Hz), 5.76 (d, 1H, J = 9.7 Hz), 6.08 (s, 1H), 6.39 (dd, 1H, J = 10.0, 4.6 Hz), 7.01 (d, 1H, J = 15.6 Hz), 7.67 (d, 1H, J = 15.6 Hz), 7.84 (q, 1H, J = 4.3 Hz); 13C NMR (125 MHz, (CD3)2SO) δ 10.5, 12.2, 12.6, 17.3, 22.3, 25.6, 32.9, 34.7, 40.3, 49.8, 54.8, 60.4, 70.4, 75.9, 98.2, 102.0, 126.8, 130.2, 133.6, 134.0, 137.5, 141.1, 175.0, 176.4, 181.4, 193.9.
Streptolydigin (1)
A stirred solution of phosphonate 49 (30.0 mg, 0.05 mmol) in THF (0.4 mL) was treated with potassium tert-butoxide (0.15 mL of 1 M solution in THF, 0.15 mmol) at 0 °C. After 40 min, aldehyde 11 (7.0 mg, 0.0252 mmol) was added in one portion. The reaction was warmed up to room temperature and left overnight. THF (0.5 mL) was added, and the reaction was quenched with aqueous HCl (0.18 mL of 1 M solution, 0.18 mmol, slight excess with respect to potassium tert-butoxide) at 0 °C. After 1 h, ethyl acetate and water were added. Phases were separated, and the organic phase was washed three times with water and once with brine, dried over MgSO4, and concentrated in vacuo. Purification by flash chromatography on silica gel (chloroform, followed by the elution with chloroform:methanol 100:3) afforded 9.0 mg (60% yield) of streptolydigin (1). 1H NMR (500 MHz, CDCl3) d 0.70 (d, 3H, J = 6.9 Hz), 1.05 (d, 3H, J = 6.9 Hz), 1.11 (m, 6H), 1.23 (s, 3H), 1.77 (m, 1H), 1.90 (s, 3H), 1.94 (m, 1H), 2.07 (d, 1H, J = 13.3 Hz), 2.50 (q, 1H, J = 11.7 Hz), 2.78 (m, 1H), 2.81 (d, 1H, J = 5 Hz), 2.89 (d, 3H, J = 4.4 Hz), 2.98 (d, 1H, J = 5 Hz), 3.05 (q, 1H, J = 6.9 Hz), 3.41 (s, 1H), 3.65 (m, 2H), 4.35 (t, 1H, J = 4.6 Hz), 4.86 (s, 1H), 5.59 (d, 1H, J = 11.2 Hz), 5.62 (d, 1H, J = 10.1 Hz), 5.85 (m, 1H), 6.25 (d, 1H, J = 9.9 Hz), 6.34 (dd, 1H, J = 10.1, 4.8 Hz), 7.16 (d, 1H, J = 15.6 Hz), 7.57 (d, 1H, J = 15.6 Hz); 13C NMR (125 MHz, CDCl3) d 10.1, 12.2, 12.5, 17.1, 17.2, 21.2, 22.2, 26.8, 30.2, 34.1, 35.2, 42.1, 50.5, 55.0, 62.8, 66.3, 71.4, 76.1, 76.5, 78.9, 98.9, 99.7, 116.2, 130.5, 133.8, 134.0, 146.1, 150.5, 173.7, 175.0, 175.2, 193.9; HRMS (ESI) calculated for C32H45N2O9 [M+H]+ 601.3125, found 601.3121. The optical rotation of synthetic sample of 41 was determined to be [α]22D = −16.2 (c = 0.7, CHCl3), which was in good agreement with that of an authentic sample of streptolydigin obtained from ChemCon GmbH.
Adamantane-Containing Tetramic Acid (56)
A stirred solution of phosphonate 49 (23.0 mg, 0.039 mmol) in THF (0.3 mL) was treated with potassium tert-butoxide (0.12 mL of 1 M solution in THF, 0.12 mmol) at 0 °C. The resulting solution was stirred 30 min and transferred into a solution of aldehyde 55 (6.0 mg, 0.0259 mmol) in THF (0.1 mL) at 0 °C. The reaction was warmed up to room temperature and left overnight. THF (0.4 mL) was added, and the reaction was quenched with aqueous HCl (0.15 mL of 1 M solution, 0.15 mmol, slight excess with respect to potassium tert-butoxide) at 0 °C. After 1 h, ethyl acetate and water were added. Phases were separated, and the organic phase was washed three times with water and once with brine, dried over MgSO4, and concentrated in vacuo. Purification by flash chromatography on silica gel (chloroform, followed by the elution with chloroform:methanol 100:3) afforded 8.5 mg (59% yield) of 56. [α]25D = − 96.4 (c = 0.8, CHCl3); 1H NMR (500 MHz, CDCl3) d 0.90 (d, 3H, J = 7.0 Hz), 1.10 (m, 6H), 1.46 (d, 3H, J = 11.5 Hz), 1.54 (d, 3H, J = 11.5 Hz), 1.60 (d, 3H, J = 12.0 Hz), 1.67 (d, 3H, J = 12.0 Hz), 1.77 (m, 1H), 1.88 (s, 3H), 1.95 (s, 3H), 2.04 (m, 1H), 2.23 (m, 1H), 2.50 (q, 1H, J = 12.0 Hz), 2.89 (d, 3H, J = 4.5 Hz), 3.05 (q, 1H, J = 7.0 Hz), 3.40 (s, 1H), 3.64 (m, 1H), 4.86 (s, 1H), 5.57 (d, 1H, J = 11.0 Hz), 5.85 (s, 1H), 6.05 (d, 1H, J = 10.5 Hz), 7.12 (d, 1H, J = 15.5 Hz), 7.55 (d, 1H, J = 15.5 Hz); 13C NMR (125 MHz, CDCl3) d 10.1, 12.5, 13.3, 17.2, 21.2, 26.8, 28.7, 30.2, 36.0, 37.2, 39.8, 42.1, 44.0, 62.8, 66.3, 76.5, 78.9, 99.5, 115.5, 133.6, 150.6, 151.2, 173.8, 175.1, 175.4, 193.8.
10,11-Dihydrostreptolydigin (63)
A stirred solution of phosphonate 49 (23.6 mg, 0.04 mmol) in THF (0.3 mL) was treated with potassium tert-butoxide (0.12 mL of 1 M solution in THF, 0.12 mmol) at 0 °C. The resulting solution was stirred 30 min and transferred into a solution of aldehyde 62 (7.5 mg, 0.0268 mmol) in THF (0.1 mL) at 0 °C. The reaction was warmed up to room temperature and left overnight. THF (0.4 mL) was added, and the reaction was quenched with aqueous HCl (0.15 mL of 1 M solution, 0.15 mmol, slight excess with respect to potassium tert-butoxide) at 0 °C. After 1 h, ethyl acetate and water were added. Phases were separated, and the organic phase was washed three times with water and once with brine, dried over MgSO4, and concentrated in vacuo. Purification by flash chromatography on silica gel (chloroform, followed by the elution with chloroform:methanol 100:3) afforded 9.0 mg (56% yield) of 10,11-dihydrostreptolydigin (63). [α]25D = − 53.3 (c = 0.9, CHCl3); 1H NMR (500 MHz, CDCl3) d 0.76 (d, 3H, J = 7.0 Hz), 1.07–1.12 (m, 9H), 1.22 (s, 3H), 1.73–1.82 (m, 4H), 1.90 (s, 3H), 1.93 (m, 1H), 2.04 (m, 1H), 2.14 (m, 1H), 2.51 (q, 1H, J = 12.0 Hz), 2.64 (d, 1H, J = 5.0 Hz), 2.81 (d, 1H, J = 5.0 Hz), 2.84 (m, 1H), 2.89 (d, 3H, J = 4.5 Hz), 3.05 (q, 1H, J = 7.0 Hz), 3.40 (s, 1H), 3.65 (m, 1H), 3.82 (d, 1H, J = 11.0 Hz), 3.98 (m, 1H), 4.86 (s, 1H), 5.59 (d, 1H, J = 11.0 Hz), 5.85 (d, 1H, J = 4.0 Hz), 6.24 (d, 1H, J = 10.0 Hz), 7.15 (d, 1H, J = 15.5 Hz), 7.56 (d, 1H, J = 16.0 Hz); 13C NMR (125 MHz, CDCl3) d 10.1, 12.3, 12.7, 17.2, 21.0, 21.2, 22.5, 26.8, 27.2, 27.4, 30.2, 30.4, 35.1, 35.3, 42.1, 51.3, 58.5, 62.8, 66.3, 70.3, 76.5, 78.9, 98.2, 99.7, 116.1, 134.3, 146.3, 150.6, 173.7, 175.0,
RNAP and Scaffolds
For in vitro transcription experiments, recombinant T. aquaticus core RNAP was purified as described earlier.34 To assemble a previously described35 nucleic acid scaffold used in Figure 3 experiment, the following oligonucleotide strands were used: RNA (5'-GUAGCGGA-3'), template DNA (3'-CATCGCCTGTACATTTCAGACAGGACC-5'), and non-template DNA (5'-TGTAAAGTCTGTCCTGG-3'). To assemble a scaffold with longer transcribed part used in Figure 5, the same RNA was combined with 3'-CATCGCCTGTACATTTCAGACAGGACCAGGTGTTGGG-5' template DNA and 5'-TGTAAAGTCTGTCCTGGTCCACAACCC-3' non-template DNA. To visualize transcription products, RNA was 32P-labelled at the 5'-end with T4 Polynucleotide Kinase before scaffold assembly.
Transcription assays
Transcription elongation complexes were reconstituted by combining 200 nM RNAP and 50 nM nucleic acids scaffold in 10 µL of transcription buffer (25 mM Tris-HCl, 50 mM NaCl, 5 mM MgCl2, 0.5 mM DTT, pH 7.9) and incubation at 37 °C for 10 min. Following the addition of streptolydigin derivatives and a 10-min incubation at 37 °C, transcription was initiated by the addition of NTPs at concentration indicated in the figures. After a 3-min incubation at 37 °C, reactions were terminated by the addition of 1 volume of formamide loading buffer. After heating at 100 °C for 1 min, samples were separated by 20% PAGE with 7M Urea, followed by PhosphorImager analysis. For single-nuceotide addition experiments, the concentration of CTP was 5 µM and reaction incubation temperature was 25 °C. Reaction incubation times varied and are indicated in Figure 5C. For data analysis, the results of single-nucleotide addition experiments were fitted to a single exponent described by equation: P=A*(1-exp(-kt)), where P is the fraction of the 9-nt product, A is the fraction of the product at t->∞, and k and t are the rate and time of the transcription reaction. The fitting was performed by nonlinear regression using the program SigmaPlot (Version 8.0).
Bacterial strains and media
Streptococcus salivarius Andrewes and Horder was obtained from ATCC (9758) and was grown in Bacto Tryptic Soy Broth Medium at 37 °C.
Antibacterial Testing
The antibiotic activity of all compounds was analyzed via disk diffusion assay. Several drops of S. salivarius culture were spread in a Petri dish containing Difco Tryptic Soy Agar. Paper disks of 5 mm diameter were used. Each disk was treated with a solution of each individual compound dissolved in ethanol (2–15 µL depending on the sample). Following evaporation of the ethanol, the disks were placed on top of agar. Plates were incubated at 37 °C for 24 h.
Supplementary Material
Acknowledgements
This work was supported in part by National Institutes of Health grants GM64530 (to K.S.), AI090558 (to K.K.) and Molecular and Cell Biology Program grant from the Russian Academy of Sciences (to K.S.). We also thank Dmitry Vassyliev for helpful discussions and Rodrigo J. Carbajo for providing copies of NMR spectra of 4.
Footnotes
Supporting Information Available. The contents of Supporting Information include detailed experimental procedures as well as analytical and spectral characterization data for all new compounds. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Floss HG, Yu T-W. Chem. Rev. 2005;105:621–632. doi: 10.1021/cr030112j. [DOI] [PubMed] [Google Scholar]
- 2.Wehrli W, Staehelin M. Bacteriol. Rev. 1971;35:290–309. doi: 10.1128/br.35.3.290-309.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shinnick T, editor. Current Topics in Microbiology and Immunology. New York: Academic Press; 1996. [Google Scholar]
- 4.(a) Wehrli W. Rev. Infect. Dis. 1983;5 Suppl. 3:S407–S411. doi: 10.1093/clinids/5.supplement_3.s407. [DOI] [PubMed] [Google Scholar]; (b) David HL. Appl. Microbiol. 1970;20:810–814. doi: 10.1128/am.20.5.810-814.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Gillespie SH. Antimicrob. Agents Chemother. 2002;46:267–274. doi: 10.1128/AAC.46.2.267-274.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Chopra I. Curr. Opin, Investig. Drugs. 2007;8:600–607. [PubMed] [Google Scholar]; (b) Darst SA. Trends Biochem. Sci. 2004;29:159–160. doi: 10.1016/j.tibs.2004.02.005. [DOI] [PubMed] [Google Scholar]; (c) Villain-Guillot P, Bastide L, Gulatieri M, Leonetti J. Drug Discovery Today. 2007;12:159–160. doi: 10.1016/j.drudis.2007.01.005. [DOI] [PubMed] [Google Scholar]; (d) Ho MX, Hudson BP, Das K, Arnold E, Ebright RH. Curr. Opin. Struct. Biol. 2009;19:715–723. doi: 10.1016/j.sbi.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) DeBoer C, Dietz A, Silver WS, Savage GM. Antibiotics Ann. 1956:886–892. [PubMed] [Google Scholar]; (b) Eble TE, Large CM, DeVries WH, Crum GF, Shell JW. Antibiotics Ann. 1956:893–896. [PubMed] [Google Scholar]; (c) Lewis C, Wilkins JR, Schwartz DF, Nikitas CT. Antibiotics Ann. 1956:897–902. [PubMed] [Google Scholar]
- 7.(a) Siddhikol C, Erbstoeszer JW, Weiblum B. J. Bacteriol. 1969;99:151–155. doi: 10.1128/jb.99.1.151-155.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]; (a) Schleif R. Nature. 1969;223:1068–1069. doi: 10.1038/2231068a0. [DOI] [PubMed] [Google Scholar]; (c) McClure WR. J. Biol. Chem. 1980;255:1610–1616. [PubMed] [Google Scholar]
- 8.(a) Temiakov D, Zenkin N, Vassylyeva MN, Perederina A, Tahirov TH, Kashkina E, Savkina M, Zorov S, Nikiforov V, Igarashi N, Matsugaki N, Wakatsuki S, Severinov K, Vassylyev DG. Mol. Cell. 2005;19:655–666. doi: 10.1016/j.molcel.2005.07.020. [DOI] [PubMed] [Google Scholar]; (b) Tuske S, Sarafianos SG, Wang X, Hudson B, Sineva E, Mukhopadhyay J, Birktoft JJ, Leroy O, Ismail S, Clark AD, Dharia C, Napoli A, Laptenko O, Lee J, Borukhov S, Ebright RH, Arnold E. Cell. 2005;122:541–552. doi: 10.1016/j.cell.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Nature. 2007;448:163–169. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]; (d) Miropolskaya N, Artsimovitch I, Klimasauskas S, Nikiforov V, Kulbachinskiy A. Proc. Natl. Acad. Sci USA. 2009;106:18942–18947. doi: 10.1073/pnas.0905402106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]; (b) Artsimovitch I, Vassylyeva MN, Svetlov D, Svetlov V, Perederina A, Igarashi N, Matsugaki N, Wakatsuki S, Tahirov TH, Vassylyev DG. Cell. 2005;122:351–363. doi: 10.1016/j.cell.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 10.(a) Xu M, Zhou YN, Goldstein BP, Jin DJ. J. Bacteriol. 2005;187:2783–2792. doi: 10.1128/JB.187.8.2783-2792.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) O’Neill A, Oliva B, Storey C, Hoyle A, Fishwick C, Chopra I. Antimicrob. Agents Chemother. 2000;44:3163–3166. doi: 10.1128/aac.44.11.3163-3166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Heisler LM, Suzuki H, Landick R, Gross CA. J. Biol. Chem. 1993;268:25369–25375. [PubMed] [Google Scholar]; (d) Severinov K, Markov D, Severinova E, Nikiforov V, Landrick R, Darst SA, Goldfarb A. J. Biol. Chem. 1995;270:23926–23929. doi: 10.1074/jbc.270.41.23926. [DOI] [PubMed] [Google Scholar]; (e) Yang X, Price CW. J. Biol. Chem. 1995;270:23930–23933. doi: 10.1074/jbc.270.41.23930. [DOI] [PubMed] [Google Scholar]
- 11.(a) Rinehart KL, Jr., Beck JR, Epstein WW, Spicer LD. J. Am. Chem. Soc. 1963;85:4035–4037. [Google Scholar]; (b) Rinehart KL, Jr., Borders DB. J. Am. Chem. Soc. 1963;85:4037–4038. [Google Scholar]; (c) Rinehart KL, Jr., Beck JR, Borders DB, Kinstle TH, Krauss D. J. Am. Chem. Soc. 1963;85:4038–4039. [Google Scholar]
- 12.Duchamp DJ, Branfman AR, Button AC, Rinehart KL., Jr. J. Am. Chem. Soc. 1973;95:4077–4078. doi: 10.1021/ja00793a058. [DOI] [PubMed] [Google Scholar]
- 13.Olano, Gómez CC, Pérez M, Palomino M, Pineda-Lucena A, Carbajo RJ, Braña AB, Méndez C, Salas JA. Chem. Biol. 2009;16:1031–1044. doi: 10.1016/j.chembiol.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 14.Horna DH, Gómez C, Olano C, Palomino-Schätzlein M, Pineda-Lucena A, Carbajo RJ, Braña AF, Méndez C, Salas JA. J. Bacteriol. 2011;193:2647–2651. doi: 10.1128/JB.00108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brill GM, McAlpine JB, Whittern D. J. Antibiot. 1988;41:36–44. doi: 10.7164/antibiotics.41.36. [DOI] [PubMed] [Google Scholar]
- 16.(a) Meyer CE. J. Antibiot. 1971;24:558–560. doi: 10.7164/antibiotics.24.558. [DOI] [PubMed] [Google Scholar]; (b) Hagenmaier H, Jaschke KH, Santo L, Scheer M, Zahner H. Arch. Microbiol. 1976;109:65–74. doi: 10.1007/BF00425114. [DOI] [PubMed] [Google Scholar]; (c) Carlson JC, Li S, Burr DA, Sherman DH. J. Nat. Prod. 2009;72:2076–2079. doi: 10.1021/np9005597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.(a) Tsukiura H, Tomita K, Hanada M, Kobaru S, Tsunakawa M, Fujisawa K, Kawaguchi H. J. Antibiot. 1980;33:157–165. doi: 10.7164/antibiotics.33.157. [DOI] [PubMed] [Google Scholar]; (b) Tsunakawa M, Toda S, Okita T, Hanada M, Nakagawa S, Tsukiura H, Naito T, Kawaguchi H. J. Antibiot. 1980;33:166–172. doi: 10.7164/antibiotics.33.166. [DOI] [PubMed] [Google Scholar]; (c) Tomita K, Hoshino Y, Sasahira T, Hasegawa K, Akiyama M, Tsukiura H, Kawaguchi H. J. Antibiot. 1980;33:1491–1501. doi: 10.7164/antibiotics.33.1491. [DOI] [PubMed] [Google Scholar]; (d) Bansal MB, Dhawan VK, Thadepalli H. Chemotherapy. 1982;28:200–203. doi: 10.1159/000238076. [DOI] [PubMed] [Google Scholar]
- 18.(a) Gause GF, Sveshnikova MA, Ukholina RS, Komarova GN, Bazhanov VS. Antibiotiki. 1977;22:483–486. [PubMed] [Google Scholar]; (b) Brazhnikova MG, Konstantinova NV, Potapova NP, Tolstykh IV, Rubasheva LM, Rozynov BV, Horvath G. Bioorg. Khim. 1981;7:298. [Google Scholar]
- 19.Karwowski JP, Jackson M, Theriault RJ, Barlow GJ, Coen L, Hensey DM, Humphrey PE. J. Antibiot. 1992;45:1125–1132. doi: 10.7164/antibiotics.45.1125. [DOI] [PubMed] [Google Scholar]
- 20.(a) Schlessinger RH, Bebernitz GR, Lin P, Poss AJ. J. Am. Chem. Soc. 1985;107:1777–1778. [Google Scholar]; (b) DeShong P, Ramesh S, Elango V, Perez JJ. J. Am. Chem. Soc. 1985;107:5219–5224. [Google Scholar]; (c) Boeckman RK, Starrett JE, Nickell DG, Sum PE. J. Am. Chem. Soc. 1986;108:5549–5559. [Google Scholar]; (d) Neukom C, Richardson DP, Myerson JH, Bartlett PA. J. Am. Chem. Soc. 1986;108:5559–5568. [Google Scholar]; (e) Shimshock SJ, Waltermire RE, DeShong P. J. Am. Chem. Soc. 1991;113:8791–8796. [Google Scholar]
- 21.(a) Boeckman RK, Jr., Potenza JC, Enholm EJ. J. Org. Chem. 1987;52:469–472. [Google Scholar]; (b) Schlessinger RH, Graves DD. Tetrahedron Lett. 1987;28:4385–4388. [Google Scholar]; (c) Ireland RE, Smith MG. J. Am. Chem. Soc. 1988;110:854–860. [Google Scholar]
- 22.Iwata Y, Maekawara N, Tanino K, Miyashita M. Angew. Chem. Int. Ed. 2005;44:1532–1536. doi: 10.1002/anie.200462300. [DOI] [PubMed] [Google Scholar]
- 23.Pronin SV, Kozmin SA. J. Am. Chem. Soc. 2010;132:14394–14396. doi: 10.1021/ja107190w. [DOI] [PubMed] [Google Scholar]
- 24.Cartwright D, Lee VJ, Rinehart KL. J. Am. Chem. Soc. 1978;100:4237–4239. [Google Scholar]
- 25.Evans DA, Britton TC. J. Am. Chem. Soc. 1987;109:6881–6883. [Google Scholar]
- 26.Solsona JG, Nebot J, Romea P, Urpi F. J. Org. Chem. 2005;70:6533–6536. doi: 10.1021/jo050792l. [DOI] [PubMed] [Google Scholar]
- 27.Evans DA, Hoveyda A. J. Am. Chem. Soc. 1990;112:6447–6449. [Google Scholar]
- 28.Kolb HC, VanNieuwenhze MS, Sharpless KB. Chem. Rev. 1994;94:2483–2547. [Google Scholar]
- 29.Garber SV, Kingsbury JS, Gray BL, Hoveyda AH. J. Am. Chem. Soc. 2000;122:8168–8179. [Google Scholar]
- 30.Schlessinger RH, Graves D. Tetrahedron Lett. 1987;28:4381–4384. [Google Scholar]
- 31.(a) Ley SV, Woodward PR. Tetrahedron Lett. 1987;28:345–346. [Google Scholar]; (b) Ley SV, Smith SC, Woodward PR. Tetrahedron. 1992;48:1145–1174. [Google Scholar]
- 32.This includes comparison of 500 MHz 1H NMR, 125 MHz 13C NMR, HRMS and optical rotation. For original detailed 13C NMR analysis, see: Lee VJ, Rinehart KL. J. Antibiotics. 1980;33:408–415. doi: 10.7164/antibiotics.33.408.
- 33.(a) Reusser F. Infect. Immun. 1970;2:77–81. doi: 10.1128/iai.2.1.77-81.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Reusser F. Antimicrob. Agents Chemother. 1976;10:618–622. doi: 10.1128/aac.10.4.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuznedelov K, Minakhin L, Severinov K. Methods Enzymol. 2003;370:94–108. doi: 10.1016/S0076-6879(03)70009-3. [DOI] [PubMed] [Google Scholar]
- 35.Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, Darst SA. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- 36.Kuznedelov K, Semenova E, Knappe TA, Mukhamedyarov D, Srivastava A, Chatterjee S, Ebright RH, Marahiel MA, Severinov K. J. Mol. Biol. 2011 Mar 15; doi: 10.1016/j.jmb.2011.02.060. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans DA, Ennis MD, Mathre DJ. J. Am. Chem. Soc. 1982;104:1737–1739. [Google Scholar]
- 38.Sherman JM, Niven CF, Smiley KL. J. Bacteriol. 1943;45:249–263. doi: 10.1128/jb.45.3.249-263.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.