Abstract
Study Objectives:
Sleep loss triggers changes in inflammatory signaling pathways in the brain and periphery. The mechanisms that underlie these changes are ill-defined. The Toll-like receptor 4 (TLR4) activates inflammatory signaling cascades in response to endogenous and pathogen-associated ligands known to be elevated in association with sleep loss. TLR4 is therefore a possible mediator of some of the inflammation-related effects of sleep loss. Here we describe the baseline electroencephalographic sleep phenotype and the biochemical and electroencephalographic responses to sleep loss in TLR4-deficient mice.
Design, Measurements and Results:
TLR4-deficient mice and wild type controls were subjected to electroencephalographic and electromyographic recordings during spontaneous sleep/wake cycles and during and after sleep restriction sessions of 3, 6, and 24-h duration, during which sleep was disrupted by an automated sleep restriction system. Relative to wild type control mice, TLR4-deficient mice exhibited an increase in the duration of the primary daily waking bout occurring at dark onset in a light/dark cycle. The amount of time spent in non-rapid eye movement sleep by TLR4-deficient mice was reduced in proportion to increased wakefulness in the hours immediately after dark onset. Subsequent to sleep restriction, EEG measures of increased sleep drive were attenuated in TLR4-deficient mice relative to wild-type mice. TLR4 was enriched 10-fold in brain cells positive for the cell surface marker CD11b (cells of the monocyte lineage) relative to CD11b-negative cells in wild type mouse brains. To assess whether this population was affected selectively by TLR4 knockout, flow cytometry was used to count F4/80- and CD45-positive cells in the brains of sleep deprived and time of day control mice. While wild-type mice exhibited a significant reduction in the number of CD11b-positive cells in the brain after 24-h sleep restriction, TLR4-deficient mice did not.
Conclusion:
These data demonstrate that innate immune signaling pathways active in the monocyte lineage, including presumably microglia, detect and mediate in part the cerebral reaction to sleep loss.
Citation:
Wisor JP; Clegern WC; Schmidt MA. Toll-like receptor 4 is a regulator of monocyte and electroencephalographic responses to sleep loss. SLEEP 2011;34(10):1335–1345.
Keywords: Monocytes, microglia, CD11b, flow cytometry, innate immunity, cytokines
INTRODUCTION
Insufficient sleep has been statistically linked to disease states characterized by inflammation. Sleep deprivation (SDEP) modifies the synthesis of cytokine regulators of inflammation, among them tumor necrosis factor α (TNF) and interleukin-1β (IL-1β) in the brain1 and TNF, IL-1 β, and interleukin-6 (IL-6) in the periphery.2 It is thus likely that at least some health effects of SDEP are mediated by pro-inflammatory cytokines. In addition to influencing disease states, these cytokines act within the brain to promote sleep.3,4 Yet, the cellular signaling mechanisms that trigger changes in pro-inflammatory cytokine levels during SDEP remain to be determined.
Cerebral microglia, the innate immune cells of the brain, are a significant source of cytokines and other sleep regulatory molecules, such as nitric oxide and prostaglandins.5 Multiple lines of evidence indicate a role for these cells in detecting and reacting to neurochemical changes that occur in response to SDEP. Minocycline, a compound known to attenuate the reactivity of microglia to cerebral insults, attenuates the EEG response to SDEP1 and spontaneous wakefulness6 in mice and attenuates sleep related slow wave activity in humans.7 SDEP reduces the level of expression of the microglial cell-surface marker CD11b at the mRNA level in the brain.1 Microglia are enriched for purinergic receptors,8,9 which respond to sleep substances adenosine10 and ATP.11 Given this evidence that microglia sense and react to the changes in the neurochemical environment that occur with sleep loss, studies on the signaling pathways that mediate this reaction are warranted.
TLR4 is one of a family of receptors that recognize pathogen-associated ligands or “danger-associated” endogenous ligands (molecules indicative of cellular stress, death or injury12). Converging lines of evidence provide a compelling rationale for studies designed to assess the role of TLR4 as a mediator of cellular pro-inflammatory responses to SDEP. SDEP elevates cerebral levels of heat shock proteins13–15 which are thought to activate TLR4,16,17 although this notion remains controversial.18,19 TLR4 activation induces the synthesis and release of pro-inflammatory cytokines from innate immune cells.20 Pro-inflammatory cytokines amplify microglial activation through paracrine signaling.21 We reasoned that activation of TLR4 might underlie some of the biochemical and EEG changes that occur in association with spontaneous sleep or sleep loss. Accordingly, we tested the hypothesis that genetic inactivation of the tlr4 gene would alter sleep/wake states under baseline conditions or after sleep restriction. Additionally, as a follow-up to our previous observation that cd11b mRNA is downregulated by SDEP, we used flow cytometry to test the hypothesis that monocyte cell counts in the brain are reduced in number due to TLR4 activation during 24-h sleep restriction. Our results indicate that TLR4 mediates, in part, both EEG and biochemical changes in association with sleep loss.
MATERIALS AND METHODS
Animals and Surgery
Male TLR4-deficient (TLR4 KO; JAX strain name M,B6.B10ScN-Tlr4 strain # 7227) and wild-type (WT; JAX strain name C57BL/6; strain # 664) mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA) at age 6 weeks. They were kept on a 12:12 LD cycle with a temperature set point of 24.5°C and given ad libitum food and water throughout experimentation. All animal experimentation was approved by the Institutional Animal Care and Use Committee of Washington State University and adhered to the National Research Council Guide for the Care and Use of Laboratory Animals.22 Mice were surgically implanted under isoflurane anesthesia (5% induction, 1-3% maintenance) with a headmount (Pinnacle Technologies part # 8201, Lawrence KS, USA) composed of a plastic 6-pin connector glued to a printed circuit board (PCB). Three electroencephalographic (EEG) electrodes and 2 electromyographic (EMG) electrodes were affixed to the headmount. Stainless steel screws (length 0.1 inches; Pinnacle Technologies part # 8209) were fastened to the skull through 4 holes in the PCB. These screws served as electroencephalographic (EEG) leads. The opening in the PCB through which each screw passed was ringed with tin/lead solder, which then conducted to the 6-pin connector at the top of the headmount. Conductivity between EEG screws and the PCB was assured by application of silver-filled electrically conductive epoxy (Resinlab SEC1233, Ellsworth Adhesives) to the screw at the time of insertion. The 2 frontal screws were placed 1.5 mm lateral to the midline and 1 mm anterior to bregma. The left frontal screw served as an internal ground. The 2 parietal screws were placed 1.5 mm lateral to the midline and approximately 2 mm anterior to lambda. Parietal electrode locations relative to skull landmarks were approximate, as the placement of screw holes on the headmount was not adjustable. Two channels of EEG data were collected, one conveying the potential between the 2 parietal leads (channel 1), and the other the potential between the left parietal and right frontal leads (channel 2). Channel 2 was used for state classification. The 2 EMG electrodes consisted of stainless steel wires, 1.5 cm in length, attached to the circuit board on one end and terminating in a bolus of epoxy (roughly 1 mm diameter) at the other end. Insulating material was absent from approximately 2 mm of stainless steel immediately next to the bolus of epoxy; this exposed stainless steel served as the lead and was embedded in the neck musculature. The headmount was glued to the skull with high-viscosity cyanoacrylate (Fisher Scientific, part # NC9482241) and sealed with Ortho-Jet self-curing acrylic resin (Lang Dental manufacturing Co.). The incision on the head and neck was closed with Ethilon 5-0 nylon monofilament non-absorbable suture (Ethicon, NJ). The female contacts on the connector remained uncovered by the composite and skin. Animals were monitored closely following surgery until they were ambulatory. Buprenorphine was administered as an analgesic (0.05 mg/kg SC) at the end of surgery and on 3 subsequent days.
Recording Apparatus
After ≥ 10 days of recovery from surgery, mice were adapted to an EEG/EMG data collection system (Pinnacle Technologies part # 8200-SL). The headmount was attached via a 6-pin connector to a preamplifier (Pinnacle Technologies part # 8201). The pre-amplifier amplified the EEG and EMG signals 100-fold and high-pass filtered the signal (EEG at 0.5 Hz; EMG at 10 Hz). The preamplifier fed into six 15-cm wires, one for each EEG and EMG lead and one ground wire, which terminated in a male 6-pin connector. This connector was attached to a commutator (Pinnacle Technologies part # 8204), which conveyed potentials to a PC for data collection.
EEG Data Collection and Analysis
Mice were housed in a cylindrical acrylic plastic cage with a vertically oriented axis. The horizontal diameter of the cylinder was 25 cm, and the height of the cylinder was 20 cm. A metal bar 22 cm in length (used for automated sleep restriction) was attached to a post at the center of the base of the cage throughout experimentation. The bar was buried in bedding when not actively rotating (i.e., during baseline and post-sleep restriction recordings). EEG and EMG (digitization rate, 400 Hz) were collected with Pinnacle PAL-8200 software and processed for state classification and spectral analysis with Neuroscore 2.01 software (Data Sciences Inc, St Paul, MN). Digitized EEG from channel 2 (bandpass, 1-30 Hz) and integrated EMG (highpass, 10Hz) were processed in 10-s epochs, each of which was classified as wake, rapid eye movement sleep (REMS), or non-REMS (NREMS) by individuals expert in rodent sleep-state classification.
EEG data were subjected to power spectral analysis by an algorithm embedded in the Dataquest software. The algorithm performed Fourier transform-based power spectral measurements on 2-s intervals of EEG data. This analysis was repeated with a 50% overlap window across the entirety of each 10-s epoch. The resulting values were averaged to yield one final power value for the entire 10-s epoch. NREMS delta power (EEG power in the 0.5-4 Hz range) was calculated for each NREMS epoch and averaged over all NREMS epochs within each time interval subjected to analysis. To assess acute sleep restriction-induced changes in NREMS delta power, average NREMS delta power was calculated from the first 180 epochs of sleep subsequent to termination of each sleep restriction session and the analogous time of day on the baseline day.
Experiment 1: Effects of TLR4 KO on Baseline Sleep and the Compensatory Response to Sleep Restriction
Mice were first subjected to a 24-h adaptation period in the sleep recording environment beginning at the onset of the daily light phase and then to a 5-day recording session. They were subjected to a 24-h baseline measurement of EEG and EMG signals. They were then subjected to a 3-h sleep restriction session followed by 18 h of recovery and a 6-h sleep restriction session followed by 24 h of recovery. They were next subjected to a 24-h sleep restriction session followed by 24 h of recovery. All sleep restriction sessions terminated 8 h into the daily light phase. Sleep restriction was enforced by an automated sleep detection and intervention system. EEG and EMG were continuously monitored by software (Pinnacle Technologies part # 8229-M) that measured EEG delta power and integrated EMG on an instantaneous per-epoch (i.e., every 10 s) basis. When the EEG delta power exceeded a threshold value indicative of NREMS onset, and simultaneously the integrated EMG declined below a threshold value indicative of NREMS onset, the metal bar in the base of the cage began to rotate at 0.1 Hz, causing the animal to awaken at a frequency of 0.2 Hz. The thresholds triggering rotation were set by the experimenter based on the EEG delta power and integrated EMG values observed during baseline EEG/EMG recordings. Rotation continued until EEG delta power decreased, and integrated EMG increased, to values associated with wakefulness in baseline conditions. EEG and EMG data collected subsequent to sleep restriction sessions and during the analogous phase of the baseline day were subjected to analysis over a 6-h interval. For the purpose of calculating mean bout duration for sleep states in this 6-h interval, bout onset was defined by 3 consecutive epochs scored as a single state, and bout offset was defined by 2 consecutive epochs scored as any other state. These criteria were used to prevent brief state transitions, such as brief awakenings from sleep (≤ 10 s), from affecting average bout duration. When a bout was initiated in, but extended beyond, the end of the 6-h interval, the duration of that bout was measured and included in the calculation of average bout duration.
Experiment 2: Immunoaffinity Segregation of CD11b-positive and CD11b-negative Cell Pools for Assessment of Gene Expression
One week after completion of experiment 1, mice within each genotype group were divided into 2 groups for a terminal experiment. The sleep restriction group of mice (n = 6 TLR4 KO, n = 6 WT) were subjected to 24-h sleep restriction with the automated sleep restriction system and euthanized immediately at the end of sleep restriction, 8 h into the light phase of the LD12:12 cycle. Time of day (TOD) control mice (n = 5 TLR4 KO, n = 5 WT) were allowed to sleep spontaneously in their home cages and euthanized 8 h into the light phase of the LD12:12 cycle. Mice were killed by carbon dioxide asphyxiation followed by cervical dislocation. Brains were rinsed in Hanks Balanced Salt Solution and dissociated tryptically and mechanically at 37°C (gentleMACS Dissociator; Miltenyi Biotec, Inc., Auburn, CA). Dissociated brain preps were pulse centrifuged to concentrate cells and passed through a 70 μm filter. They were again pulse centrifuged, resuspended, and incubated with anti-myelin immunoglobulin-conjugated magnetic microbeads (myelin removal kit; Miltenyi Biotec, Inc., Auburn, CA) to remove myelin. Resulting cell suspensions were pelleted and resuspended in PBS.
A portion of the dissociated brain cell suspension was used on the day of euthanasia for the flow cytometry measurements performed in Experiment 3 (see below). The remainder was subjected to immunoaffinity-based sorting and gene expression assays. Anti-CD11b immunoglobulin-conjugated magnetic microbeads were used to isolate microglia and related cells of the monocyte lineage from other cerebral cell types, based on the expression of the cell-surface CD11b antigen in this lineage.23 Myelin-depleted dissociated brain suspensions were incubated with anti-CD11b immunoglobulin-conjugated microbeads and segregated into CD11b-positive and CD11b-negative cell pools by magnetic separation, using manufacturer protocols (Miltenyi Biotec, Inc., Auburn, CA). Cell numbers in the CD11b-positive and CD11b-negative cell pools were counted by visual hemacytometry. This procedure yields approximately 1.2-1.5 × 107 cells per dissociated brain in our experience, approximately 5% of which (6-8 × 105 cells) are CD11b-positive. This percentage is equivalent to the estimated percentage of brain cells that are microglia.24
Total RNA was isolated from CD11b-negative and CD11b-positive cell pools from sleep restriction and TOD mice using Trizol (Invitrogen, Carlsbad, CA, USA). cDNA synthesis was performed with the SuperScript First Strand Synthesis System (Invitrogen, Carlsbad, CA, USA). The resulting cDNA was stored in nuclease free water at −20°C. For real-time RT-PCR, each target cDNA of interest and a reference cDNA (18S) were simultaneously amplified in 8-well strip tubes (Bio-Rad, Hercules, CA, USA) on an MJ Research PTC200 Peltier Real Time Thermal Cycler coupled to a Bio-Rad Chromo4 continuous fluorescence detector. Taqman primer/probe sets were purchased from Applied Biosystems: cd11b (Mm00434455_m1),c-fos (Mm00487425_m1), interleukin-1β (il-1β; Mm00434228_m1), interleukin-6 (il-6; Mm00446190_m1), neuropeptide Y (npy; Mm00445771_m1) purinergic receptor 2y12 (p2y12; Mm00446026_m1), peripheral benzodiazepine receptor (pbr; Mm00437828_m1), tumor necrosis factor-α (tnf; Mm00443258_m1), and 18S rRNA (4319413E). Levels of the internal control transcript, 18S rRNA (VIC fluorophore), were measured simultaneously with each target transcript (FAM fluorophore). Real-time reaction products were subjected to normalization by the comparative CT method, also referred to as the 2-ΔΔCT method.25 A threshold fluorescence value (CT) was chosen by visual inspection of FAM and VIC fluorescence saturation curves for all reactions in a given assay run. This threshold was placed at a value that optimized the dynamic range of values across all samples. Fold-change of target transcript relative to 18S control was measured by the following formula: fold-change (i.e., 2-ΔΔCT) = (average per-sample CT for target gene in experimental group minus mean CT for 18S in experimental group)/(mean of all per-sample CT values for target gene in control group minus mean of all per-sample CT values for 18S in control group). CD11b-negative cell pools from TOD mice were used as the control group.
Experiment 3: Flow Cytometric Measurement of F4/80 and CD45 Immunopositivity
Myelin-depleted, dissociated brain tissues from sleep restriction and TOD control mice were incubated with fluorescein isothiocyanate (FITC)-conjugated anti-CD45.1 (eBiosciences, #11-0453-81) and PE (phycoerythrin)-conjugated anti-F4/80 (eBiosciences, #12-4801-80) antibodies. F4/80 is a marker enriched in the monocyte lineage, and CD45 discriminates between resident cells of the monocyte lineage and recently infiltrated cells.26 Antibodies were diluted 1:100 in flow cytometric buffer (phosphate-buffered saline with 5% fetal bovine serum). Cells were incubated with antibodies for 1 h at 4°C. Cells were washed and resuspended in antibody-free flow cytometric buffer. Flow cytometric measurement of fluorescence emission was performed on a total of 10,000 cells per dissociated brain by a Beckman/Coulter Epics XL-MCL flow cytometer with Expo 32 software (Brea, CA). Data were analyzed using FCS Express (DeNovo Software, Los Angeles, CA). In order to count the number of cells positive for a given antigen, it is necessary to establish threshold values, or gates, for each fluorescently tagged immunoglobulin. The gates used for counting cells as F4/80-positive or negative, and CD45-high and low were established based on previously published measurements of these parameters27 in dissociated brains. We established gates that yielded counts of F4/80-positive/CD45-high values in the wild-type TOD control mice equivalent to those published previously.27 The application of these gates to all other groups thus provided a measure of relative changes in expression of these cell surface markers in other groups relative to wild-type TOD control mice, if not an absolute measure of cell counts.
Statistics
Statistics were performed with Statistica 9.0 software (Statsoft, Inc., Tulsa, OK). Reported measures of variability are standard error of the mean.
RESULTS
TLR4 KO Alters Sleep Timing Under Baseline Conditions
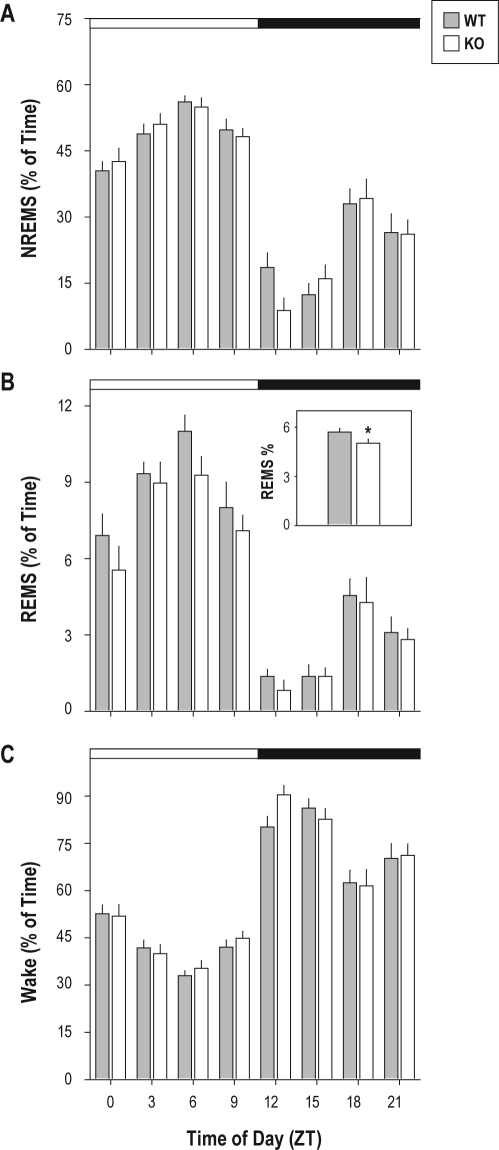
Under baseline conditions, daily rhythms of time spent asleep and awake were intact in animals of both genotypes (Figure 1). Repeated measures ANOVA, with genotype as grouping factor and time of day as repeating factor yielded significant effects of time of day on percentage of each 3-h bin spent in NREMS (F7,140 = 64.85,P < 0.001), REMS (F7,140 = 52.00,P < 0.001), and wake (F7,140 = 70.91,P < 0.001). None of these TOD effects was modulated by genotype (F ≤ 1.06, P ≥ 0.392). There was, however, a significant main effect of genotype on REMS as a percentage of time over the entire 24-h baseline (F1,20 = 4.82, P = 0.040). The percentage of time spent in REMS was significantly reduced by 12% over the entire 24-h baseline (5.0% in the KO vs. 5.7% in the wild-type; Figure 1B, inset) as a consequence of TLR4-deficiency.
Figure 1.
Sleep states during 24-h baseline recordings. Wild-type (gray) and TLR4 KO (white) mice were allowed to sleep spontaneously in an LD12:12 cycle. White and black bars at the top of each panel indicate the timing of the light/dark cycle. (A) NREMS, (B) REMS, and (C) wakefulness are reported as a percentage of each 3-h bin. The inset in panel B shows average percent of time spent in REMS across the entire 24-h recording session, the only sleep state variable for which a significant main effect of genotype was found. *P < 0.05 vs. wild-type, Student t.
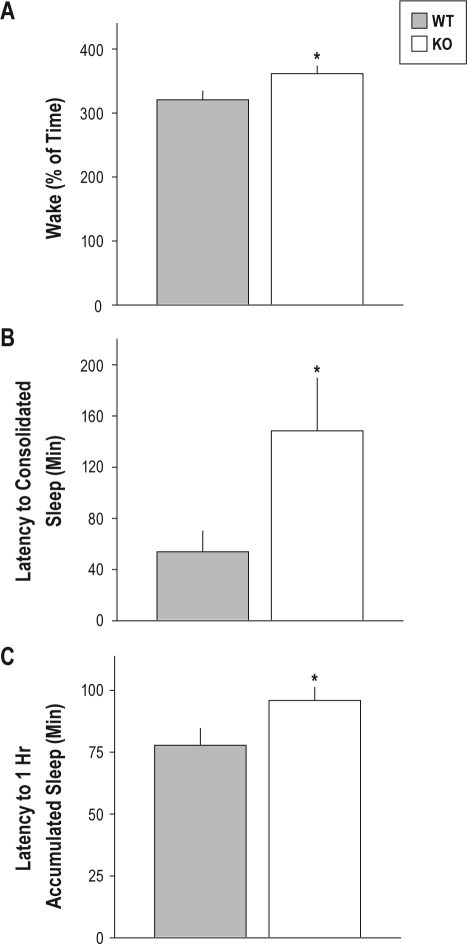
The surge of wakefulness initiated at dark onset under baseline conditions was of greater magnitude in TLR4 KO mice than wild-type controls by 3 measures. The time spent in wakefulness during the first 3 h after dark onset was significantly greater by 13% in TLR4 KO mice relative to wild-type mice (Figure 2A). The latency to consolidated sleep was 3-fold greater in TLR4 KO mice than in wild-type mice (Figure 2B). The latency to accumulation of 1 h of sleep was significantly greater by approximately 25% in TLR4 KO relative to wild-type mice (Figure 2C). The significant increase in wakefulness caused by TLR4-deficiency came at the expense of NREMS, not REMS: there was a significant effect of genotype on NREMS as a percentage of time during the 3-h interval beginning at dark onset. Wild-type mice exhibited a 2-fold increase in time spent in NREMS relative to TLR4 KO mice in this 3-h bin (P = 0.0172, Student t; data not shown). ANOVA also yielded a significant effect of genotype (F1,20 = 9.19, P = 0.007) on the number of NREMS bouts; TLR4 had fewer NREMS bouts per h (5.6 NREMS bouts per h) than WT mice (7.2 NREMS bouts per h).
Figure 2.
Measures of the primary waking bout beginning at the onset of dark in baseline conditions. (A) Amount of time spent awake during the first 3 h of the dark phase of the LD12:12 cycle; (B) latency to the onset of consolidated sleep (i.e., 1 consecutive minute of sleep); and (C) latency to one accumulated hour of sleep are shown for wild-type (gray) and TLR4 KO (white) mice allowed to sleep spontaneously in an LD12:12 cycle. *sP < 0.05 vs. wild-type, Student t.
Consolidation of both wakefulness and NREMS during baseline sleep was unaffected by TLR4-deficiency (data not shown). Bout duration, averaged in 3-h bins across the light/dark cycle of NREMS and wake did not differ between genotypes. The reduction in time spent in REMS in TLR4 KO mice relative to wild-type was paralleled by a change in the frequency of occurrence of REMS episodes. ANOVA performed for the average number of REMS bouts per h across 3-h bins during baseline yielded a significant effect of genotype (F1,20 = 5.50,P = 0.029). The number of bouts per hour was less in TLR4 KO mice (2.1 ± 0.1) than wild-type mice (2.3 ± 0.1). REMS bout duration was equivalent in TLR4 KO (0.92 ± 0.04 min) and wild-type (0.99 ± 0.04 min) mice. Thus, a reduction in the frequency, but not the duration, of REMS bouts explains the reduction of total time spent in REMS observed in TLR4 KO mice.
TLR4 KO Effect Opposes that of Sleep Restriction on Sleep States
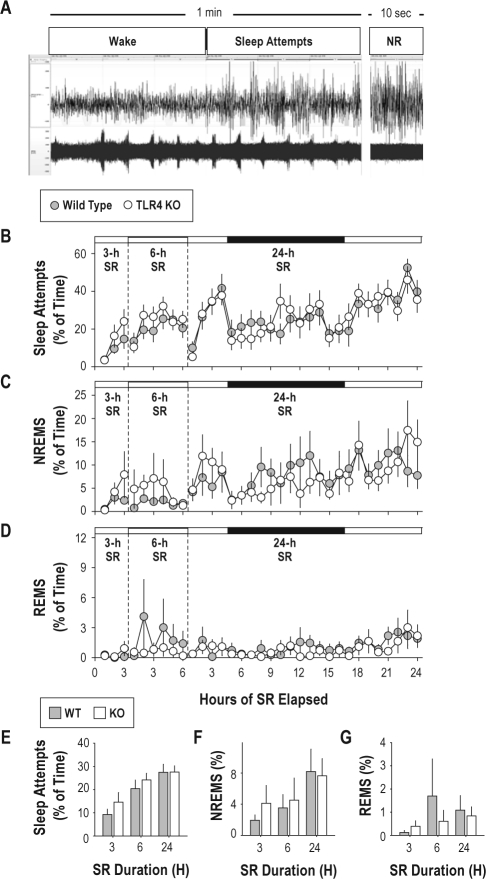
Both short-term (3-h or 6-h) and long-term (24-h) sleep restriction reduced, but did not abolish, the occurrence of sleep (Figure 3). At the beginning of sleep restriction, animals spent nearly 100% of time awake. However, sleep attempts became more frequent with increasing time spent in sleep restriction. Sleep attempts were episodes of high-amplitude EEG and low EMG activity alternating with low-amplitude EEG and high EMG activity at intervals of < 10 sec. These awakenings occurred at 0.2 Hz (once every 5 sec) due to the awakening effect of the rotating bar in the base of the cage (Figure 3A). ANOVA on data from the 24-h sleep restriction indicated a significant effect of time on the percentage of epochs scored as sleep attempts (F23,460 = 5.77, P < 0.001); the occurrence of sleep attempts was more frequent over time spent in sleep restriction (Figure 3B). Despite the nearly continuous somatosensory stimulation provided by the automated sleep system (0.2 Hz; once with each half-rotation of the bar), episodes of NREMS (Figure 3C), and rarely REMS (Figure 3D), occurred during sleep restriction. Thus, while sleep was grossly disrupted during sleep restriction, deprivation was not complete. The amount of time spent in sleep attempts, NREMS, and REMS during sleep restriction were unaffected by genotype (Figure 3E-G).
Figure 3.
Effect of automated sleep restriction procedure on sleep states. (A): EEG (upper) and EMG (lower) traces from a wild-type mouse subjected to sleep restriction. The left trace contains 1-min of continuously recorded potentials. The first 30 sec was classified as wake and the remainder as sleep attempts. The right trace contains EEG/EMG data from a 10-sec epoch of NREMS (NR) from the same animal at a later time. (B-D): Sleep states in wild type (gray symbols) and TLR4 KO (white symbols) mice during 3, 6, and 24-h sleep restriction. Data from the 3 sleep restriction sessions are separated by vertical dashed lines. Time spent in sleep attempts (B), NREMS (C), and REMS (D) are plotted as a percentage of each h. Time spent in darkness (h 5-16 of 24-h sleep restriction only) is indicated by the black bars at the top of each graph in B-D. (E-G): Percent of time spent in sleep attempts (E), NREMS (F), and REMS (G) averaged across the entirety of the 3, 6, and 24-h sleep restriction sessions.
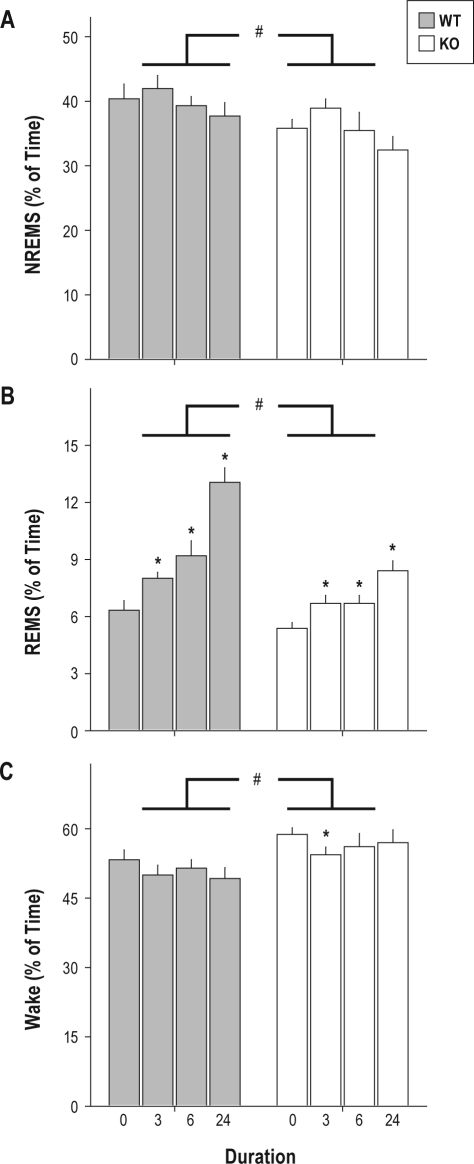
Sleep restriction had significant effects on a number of sleep parameters collected in the 6-h interval immediately subsequent to termination of sleep restriction (Table 1). Percent of time spent in NREMS (Figure 4A), REMS (Figure 4B), and wake (Figure 4C) all exhibited main effects of sleep restriction (F3,60 ≥ 3.38, P ≤ 0.023). Time spent in NREMS and REMS increased after sleep restriction, whereas time spent awake decreased. The numbers of bouts of both NREMS and REMS, the duration of REMS bouts, and NREMS EEG delta power were elevated after sleep restriction relative to baseline (Table 1). Wake bout duration was significantly affected by sleep restriction and was reduced by 38% in wild-type mice and 29% in TLR4 KO mice after 24-h sleep restriction relative to baseline (albeit not significantly with Bonferroni correction; Table 1). With the exception of NREMS EEG delta power and NREMS bout duration, all of the variables significantly affected by sleep restriction were affected in the opposite direction in TLR4 KO mice relative to wild-type mice. For instance, while the percent of time spent in REMS was significantly increased by approximately 2-fold after 24-h sleep restriction relative to baseline in wild-type mice, TLR4 KO mice exhibited 28% less REMS than wild-type mice on average across all sleep restriction durations (Figure 4B).
Table 1.
Effects of sleep restriction on state consolidation and NREMS EEG delta power
| Genotype | Wake Bout Duration (Minutes) |
Main Effects |
||||
|---|---|---|---|---|---|---|
| 0-h SR | 3-h SR | 6-h SR | 24-h SR | Genotype1 | Sleep Restriction2 | |
| Wild Type | 13 ± 2 | 13 ± 2 | 14 ± 2 | 8 ± 1 | F = 5.61, P = 0.028 | F = 4.75, P = 0.005 |
| TLR4 KO | 17 ± 2 | 16 ± 1 | 19 ± 2 | 12 ± 2 | ||
| Genotype | Number of Wake Bouts |
Main Effects |
||||
| 0-h SR | 3-h SR | 6-h SR | 24-h SR | Genotype | Sleep Restriction | |
| Wild Type | 26 ± 2 | 31 ± 4 | 32 ± 3 | 30 ± 3 | F = 4.98, P = 0.041 | N.S. |
| TLR4 KO | 22 ± 1 | 23 ± 2 | 26 ± 2 | 27 ± 3 | ||
| Genotype | REMS Bout Duration (Minutes) |
Main Effects |
||||
| 0-h SR | 3-h SR | 6-h SR | 24-h SR | Genotype | Sleep Restriction | |
| Wild Type | 1.1 ± 0.1 | 1.4 ± 0.1 | 1.5 ± 0.1 | 1.7 ± 0.1 | F = 6.52, P = 0.019 | F = 9.42, P < 0.001 |
| TLR4 KO | 1.1 ± 0.1 | 1.2 ± 0.1 | 1.3 ± 0.1† | 1.5 ± 0.1† | ||
| Genotype | Number of REMS Bouts |
Main Effects |
||||
| 0-h SR | 3-h SR | 6-h SR | 24-h SR | Genotype | Sleep Restriction | |
| Wild Type | 17 ± 1 | 18 ± 1 | 21 ± 2 | 22 ± 3 | F = 4.57, P = 0.045 | F = 10.89, P < 0.001 |
| TLR4 KO | 15 ± 1 | 16 ± 1 | 16 ± 3 | 20 ± 3 | ||
| Genotype | NREMS Bout Duration (Minutes) |
Main Effects |
||||
| 0-h SR | 3-h SR | 6-h SR | 24-h SR | Genotype | Sleep Restriction | |
| Wild Type | 2.7 ± 0.2 | 2.5 ± 0.2 | 2.1 ± 0.1 | 2.6 ± 0.2 | N.S. | F = 5.07, P = 0.003 |
| TLR4 KO | 2.8 ± 0.2 | 2.9 ± 0.2 | 2.2 ± 0.2† | 2.5 ± 0.1 | ||
| Genotype | Number of NREMS Bouts |
Main Effects |
||||
| 0-h SR | 3-h SR | 6-h SR | 24-h SR | Genotype | Sleep Restriction | |
| Wild Type | 51 ± 5 | 56 ± 5 | 59 ± 3 | 48 ± 3 | F = 11.47, P = 0.003 | F = 4.42, P = 0.007 |
| TLR4 KO | 39 ± 3 | 43 ± 2* | 44 ± 3* | 39 ± 2 | ||
| Genotype | NREMS EEG Delta Power (μV2/Hz) |
Main Effects |
||||
| 0-h SR | 3-h SR | 6-h SR | 24-h SR | Genotype | Sleep Restriction | |
| Wild Type | 240 ± 47 | 424 ± 98 | 454 ± 108 | 378 ± 104 | N.S. | F = 4.11, P = 0.010 |
| TLR4 KO | 374 ± 93 | 689 ± 190 | 593 ± 106 | 622 ± 144† | ||
main effect of genotype; degrees of freedom = 1,20.
main effect of sleep restriction (SR); degrees of freedom = 3,60.
significantly different from wild type, same sleep restriction condition, Student t with Bonferroni correction.
significantly different from 0 h sleep restriction within genotype, Student t with Bonferroni correction.
Figure 4.
Effects of 3, 6, or 24-h sleep restriction on sleep state timing. (A) NREMS, (B) REMS, and (C) wake as a percentage of time during the 6-h interval immediately subsequent to termination of sleep restriction. Duration ‘0’ refers to data from the analogous interval in the baseline recording session. ANOVAs on all 3 variables yielded significant effects of sleep restriction (F ≥ 3.38, P ≤ 0.023). *P < 0.05 vs. same genotype, baseline control, Student t with Bonferroni correction. #P < 0.05, ANOVA main effect of genotype (F ’ 6.64, P ≤ 0.018).
TLR4 KO Attenuates the Reduction in Monocyte Cell Counts Induced by 24-h Sleep Restriction
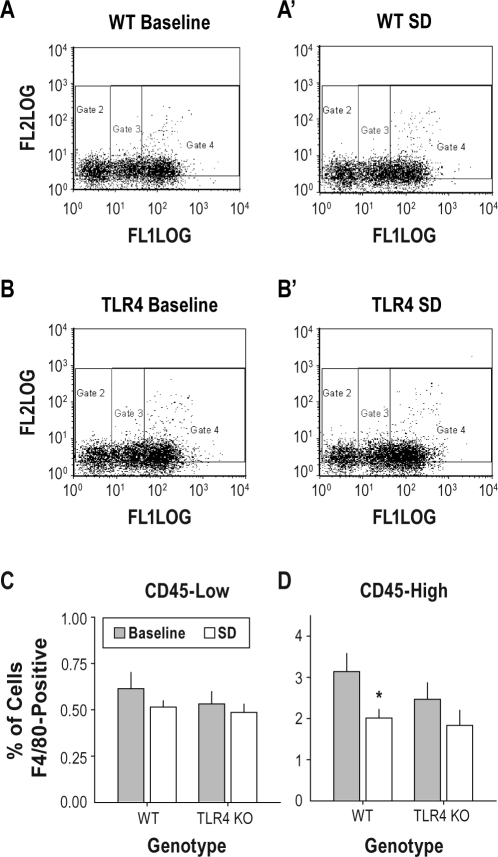
TLR4 KO mice and wild type controls were euthanized either immediately after 24-h sleep restriction or as time of day controls. Flow cytometry on dissociated brains indicated that 3.3% ± 0.5% of cells in TLR4 KO mice and 4.1% ± 0.5% of cells in wild-type mice were F4/80-positive under baseline conditions. F4/80-positive cells were further divided into CD45-high and CD45-low cell groups (Figure 5) to distinguish infiltrating monocytes (CD45-high cells) and resident microglia (CD45-low cells26) for statistical analyses. In the CD45-low population, ANOVA with treatment (sleep restriction vs. spontaneous sleep) and genotype (TLR4 KO vs. wild-type) as grouping factors yielded no significant main effects or interactions (all P > 0.20; Figure 5C). In the CD45-high population, ANOVA yielded a significant main effect of treatment (F1,18 = 6.09, P = 0.024). Sleep restriction caused a reduction in the percentage of brain cells classified as F4/80-positive, CD45-high. The reduction in the number of F4/80-positive, CD45-high cells due to sleep restriction was statistically significant in wild-type mice (36% reduction) but not in the KO (26% reduction; Figure 5D).
Figure 5.
Effects of 24-h sleep restriction on flow cytometric measures of monocyte cell surface antigens. Dissociated brains were incubated with fluorochrome conjugated anti-F4/80 and anti-CD45 immunoglobulins. The fluorochrome conjugated to the F4/80 antibody fluoresces at 580 nM (labeled “FL2 log” in A and B panels). The fluorochrome conjugated to the CD45 antibody fluoresces at 520 nM (labeled “FL1 log” in A and B panels). (A, B) Flow cytometric data from dissociated brains of individual wild-type mice (A = baseline control; A′ = 24-h sleep restriction) and TLR4 KO (B = baseline control; B′ = 24-h sleep restriction) are shown. Gates 2, 3, and 4 divide F4/80 positive cells into CD45-low, medium and high classes of cells, respectively. (C, D) Group mean F4/80 positive cell counts in the CD45-low (C) and CD45-high (D) classes. *P < 0.05 vs. same genotype, vs. baseline control, Student t with Bonferroni correction.
TLR4 Knockout Does Not Attenuate Effects of 24-h Sleep Restriction on Gene Expression in the Brain
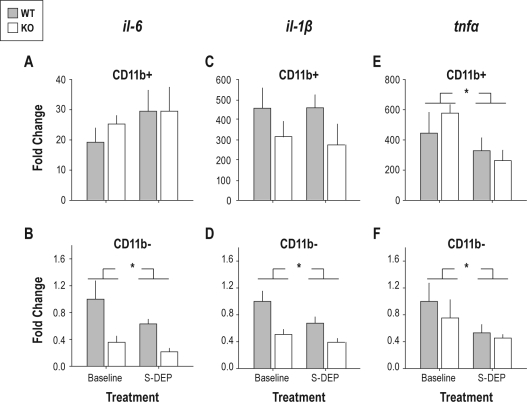
We used an antibody-based segregation procedure, followed by real-time reverse transcriptase-polymerase chain reaction (RT-PCR), to measure the enrichment of transcripts in CD11b-positive vs. negative brain cell populations, and to assess the effects of TLR4-deficiency and 24-h sleep restriction on these transcripts. cd11b expression at the mRNA level was enriched by well over 2000-fold in CD11b-positive cells relative to CD11b negative cells, irrespective of genotype and sleep restriction condition (Table 2). This high level of enrichment verifies the stringency of CD11b antibody-based isolation of the CD11b-positive population. Transcripts tlr4 (measured only in wild-type cell pools), c-fos and p2y12 were enriched in CD11b-positive cells (Table 2) but unaffected by 24-h sleep restriction or genotype (data not shown). npy and pbr transcripts were enriched in CD11b-negative cells (Table 2) but unaffected by treatment or genotype. Transcripts encoding 3 cytokines known to regulate sleep, il-1β, il-6 and tnf, were highly enriched in CD11b-positive cells relative to CD11b-negative cells (Table 2). Il-6 was affected by 24-h sleep restriction in CD11b negative cells only (Figure 6B). Despite relative expression levels less than 50% of wild-type in CD11b-negative cells from TLR4-deficient mice, genotype did not have a significant effect on il-6 expression in CD11b-negative cells (F1,18 = 3.88, P = 0.065). Il-1β was the only transcript to be significantly affected by genotype. Levels were significantly lower in CD11b-negative cells from TLR4 KO mice relative to CD11b-negative cells from wild-type mice (Figure 6D). Additionally,Il-1β levels were attenuated by 24-h sleep restriction in the CD11b-negative cell pool irrespective of genotype; genotype × sleep restriction interaction was not significant (Figure 6D). Tnf was unaffected by genotype within either cell pool. Tnf was suppressed by 24-h sleep restriction in both cell pools (Figure 6E, F) irrespective of genotype.
Table 2.
Target gene enrichment in CD11b-positive vs. CD11b-negative cell pools from mouse brain
| Target Gene | Target Gene CT |
18S CT |
Target Gene Fold Difference (Positive/Negative) | ||||
|---|---|---|---|---|---|---|---|
| Mean ± SEM |
ANOVA (df = 1.42) | Mean ± SEM |
ANOVA (df = 1.42) | ||||
| CD11b ± | CD11b- | CD11b ± | CD11b- | ||||
| CD11b | 21.1 ± 0.3 | 32.2 ± 0.4 | F = 327.1, P < 0.001 | 13.2 ± 0.2 | 12.9 ± 0.3 | NS | 2622.41 |
| c-fos | 20.6 ± 0.3 | 23.8 ± 0.5 | F = 32.1, P < 0.001 | 14.5 ± 0.2 | 14.2 ± 0.3 | NS | 11.18 |
| IL-1b | 23.5 ± 0.3 | 30.8 ± 1.5 | F = 22.2, P < 0.001 | 14.1 ± 0.2 | 13.9 ± 0.3 | NS | 633.00 |
| IL-6 | 27.9 ± 0.3 | 33.9 ± 0.5 | F = 92.4, P < 0.001 | 14.4 ± 0.2 | 14.2 ± 0.3 | NS | 52.56 |
| NpY | 35.6 ± 0.3 | 28.9 ± 0.5 | F = 134.9, P < 0.001 | 14.3 ± 0.2 | 14.0 ± 0.3 | NS | 0.01 |
| P2Y12 | 20.0 ± 0.3 | 30.1 ± 0.5 | F = 332.8, P < 0.001 | 14.1 ± 0.2 | 13.9 ± 0.3 | NS | 1286.78 |
| Pbr | 30.1 ± 0.2 | 29 ± 0.3 | F = 8.17, P = 0.007 | 14.0 ± 0.2 | 13.8 ± 0.3 | NS | 0.56 |
| Tlr4* | 26.3 ± 0.5* | 29.3 ± 0.5* | F = 15.6, P < 0.001* | 14.5 ± 0.3* | 14.0 ± 0.4* | NS* | 9.47 |
| TNFa | 23.3 ± 0.2 | 32.5 ± 0.6 | F = 183.4, P < 0.001 | 14.2 ± 0.2 | 14.0 ± 0.3 | NS | 696.22 |
Values less than one indicate enrichment in CD11b-negative cell pools relative to CD11b-positive cell pools.
Data are from wild type brains only. All other transcripts were assays in both genotypes.
Figure 6.
Effects of TLR4 KO and sleep restriction on cytokine gene expression in CD11b-positive and CD11b-negative cell pools. For each transcript, data are normalized to the mean of the values from the baseline control wild-type group in the CD11b-negative cell pool. (A, B) Effects of sleep restriction and TLR4 KO on expression of il-6 in (A) CD11b-positive and (B) CD11b-negative cell pools. (C, D) Effects of sleep restriction and TLR4 KO on expression of il-1β in (C) CD11b-positive and (D) CD11b-negative cell pools. (E, F) Effects of sleep restriction and TLR4 on expression of tnf in (E) CD11b-positive and (F) CD11b-negative cell pools. *P < 0.05, main effect for sleep restriction vs. baseline control, ANOVA.
DISCUSSION
We have shown that TLR4-deficiency causes subtle modifications in the timing of sleep and wakefulness in mice. We have additionally shown that CD45-high monocytes in the brain, a population in which TLR4 is enriched by 10-fold relative to other cell types, decrease in number as a consequence of 24-h sleep restriction, and that this effect of sleep restriction is attenuated in the absence of TLR4.When TLR4 senses the presence of the bacterial cell wall component lipopolysaccharide or endogenous ligands (see below), it initiates an intracellular signaling cascade, which then increases release of pro-inflammatory cytokines (TNF, IL-6, and IL-1β) and other pro-inflammatory agents, such as nitric oxide, cyclooxygenase, prostaglandin E2, and adenosine triphosphate,12 all of which regulate the timing of sleep/wake cycles. The current data document the importance of TLR4 and the intracellular signaling pathways that it activates in the regulation of sleep.
Under baseline conditions, the NREMS/wake distribution was modulated by TLR4 genotype only at the onset of dark, the time at which the primary daily waking bout occurs. This bout of wakefulness was larger in TLR4 KO mice than in wild-type mice. Additionally, the decrease in wakefulness, relative to baseline, that occurred in wild type mice after sleep restriction was attenuated in TLR4 KO mice. Sleep restriction sessions ended 8 hours into the light phase of the LD cycle and post-sleep restriction data were collected during the last 4 h of the light phase and the first 2 hours of the dark phase. The balance of sleep and wake at this time of day reflects the antagonistic interactions of a circadian waking signal and a homeostatic sleep drive.29 Either of these factors might be modulated by TLR4-deficiency; the data presented here do not discriminate between the two. However, we favor the hypothesis that the homeostatic sleep drive, rather than the circadian clock, is modulated by genotype. There is considerable evidence that cell types and molecular components of innate immune signaling pathways participate in homeostatic sleep regulation (see introduction), and scant evidence that they are necessary for circadian clock output signaling. The sleep phenotype of TLR4 KO mice at the EEG level was modest: over the entire 24-h baseline period, there was no genotype effect on NREMS or wake as a percentage of time. Therefore, TLR4 is not absolutely necessary for the detection of, or reaction to, sleep loss. At the biochemical level, TLR4 regulates expression of pro-inflammatory cytokines. Despite the expectation that the dynamics of cytokine expression would be modified in the absence of TLR4, they were not, with the exception of levels of IL-1β, which was modestly reduced in CD11b-negative cell pools from TLR4 KO mice relative to wild type mice (Figure 6D). The effects of sleep restriction on the expression of tnf in CD11b-positive and CD11b-negative cell pools and on il-6 and il-1β in CD11b-negative cells were intact in TLR4 KO mice. These observations do not nullify the possibility that innate immune effector pathways are necessary for regulation of cytokine expression by sleep/wake changes. Like TLR4, TLR2 detects both bacterial ligands (lipoteichoic acid, peptidoglycan12) and endogenous ligands associated with cellular damage and stress (the heat shock protein HSP6030). TLR2 also activates an intracellular signaling cascade similar to the one activated by TLR412. The EEG changes that occurred in TLR4 KO mice in reaction to sleep restriction may have been mediated in part by TLR2 signaling.
The significant reduction in F4/80-positive, CD45-high monocyte counts that occurred in wild type mice in response to sleep restriction was absent in TLR4-deficient mice. Since the number of F4/80-positive, CD45-high monocyte counts decreased (nonsignificantly) in TLR4-deficient mice, we predict that a larger sample size would reveal a significant, but blunted, decrease in TLR4-deficient animals. Therefore, the reduction in F4/80-positive, CD45-high monocyte counts, like the EEG response to sleep restriction, is probably driven by redundant innate immune pathways including stimulation of more than one TLR.
Some effects of short-term sleep restriction (3- or 6-h duration) were not detected after 24-h sleep restriction in this study. At the biochemical level, the reduction in cd11b expression and simultaneous increase in c-fos expression, which we observed elsewhere after 3-h sleep restriction,1 were not seen in mice euthanized after 24-h sleep restriction relative to TOD controls in the current study. The failure of 24-h sleep restriction to modify these transcripts is likely a consequence of the failure of the automated sleep restriction system to maintain wakefulness over the entirety of the sleep restriction session. During the last 8 hours of the 24-h sleep restriction, mice spent a significant portion of the time in either NREMS or sleep attempts (46% in wild-type, 47% in TLR4 KO mice; Figure 3). Thus, although NREMS was fragmented, it occurred for much of the time. By contrast, animals spent virtually no time in REMS during this 8-h period (1.7% in wild type, 1.4% in TLR4 KO mice) as well as the preceding 16 hours of sleep restriction, and sustained a high REMS drive as a consequence. The differential efficacy of 24-h sleep restriction in suppressing REMS is apparent in the EEG data collected after 24-h sleep restriction. The strongest effect of 24-h sleep restriction at the EEG level was in REM sleep parameters (Table 1); in wild-type mice, REMS bout duration increased by more than 50% and the amount of time spent in REMS more than doubled. NREMS parameters, by contrast, were more robustly affected by short-term sleep restriction (3-h or 6-h sleep restriction, Table 1). The response of mice to 24-h sleep restriction resembles that of rats subjected to 4-days of sleep restriction (“sleep deprivation” per the authors of that study) by the automated disk-over-water method,31 during which a significant amount of NREMS occurs. Subsequent to 4-day SDEP, rats exhibit very robust increases in REMS and little to no recovery of high-amplitude NREMS. It is likely that the mice in the current study and the rats studied by Rechtschaffen et al.31 were able to discharge NREMS need through NREMS episodes and/or “sleep attempts” during sleep restriction. To the extent that the biochemical changes we observed after 24-h sleep restriction reflect changes in sleep need, they are likely to reflect increased REMS need rather than increased NREMS need.
The flow cytometric data, together with the previous observation that cd11b mRNA is reduced by short-term SDEP, support the contention that sleep loss reduces monocyte infiltration into the brain. These measures are admittedly indirect measures. The CD45-high, F4/80 positive cell count in particular, while thought to reflect newly infiltrated monocytes26 in the non-diseased brain, is sensitive to the threshold for inclusion chosen by the investigator and may reflect either the total number of cells present, or the density of the cell surface antigen on individual cells (either of which would be evidence of an effect of sleep loss on monocyte function).
The real time PCR data presented here demonstrate that TLR4 and the cytokines IL-1β, IL-6, and TNF were all enriched in CD11b-positive, presumed microglial26 cells of the brain parenchyma. Since IL-1β,32 IL-6,33 TNF,34 and, as shown here, TLR4, are all regulators of sleep, their enrichment in CD11b-positive cells alone is reason to think that cells of the monocyte lineage regulate sleep. However, the EEG and biochemical phenotypes of TLR4-deficient mice are not necessarily due to deficits in microglial function. Our data demonstrate a ten-fold enrichment for TLR4 mRNA in the CD11b-positive cell pool of the brain, but TLR4 immunoreactivity is nonetheless detectable on neurons, including enteric sensory neurons that feed into the vagus nerve.35 The time of day at which a sleep phenotype is apparent in TLR4 KO mice is also the time of day at which animals begin to feed intensively (i.e., dark onset).36 Therefore, given that the vagus nerve mediates effects of gut contents on sleep/wake cycles at the EEG level,37 the sleep phenotype that we observed in baseline and after sleep restriction may be due, in part, to TLR4-deficiency in vagal or other afferents to the CNS.
The current report adds to a body of literature linking immunity in general, and innate immune cells and signaling systems in particular, to sleep regulation. This relationship appears to be widespread in a phylogenetic sense. In Drosophila, knockout of the innate immune transcriptional regulatory protein NFκB-Relish (an ortholog of the mammalian TLR4 target, NF-κB) modifies the effects of immune challenge on sleep.38,39 In zebrafish embryos, a large scale screen found that immunomodulatory molecules had profound effects on behaviorally-defined sleep.40 Many details of the relationship between innate immune functions and sleep remain to be understood. But the fact that monocytes, unlike neurons, are amenable to genetic manipulations and transplantation,41,42 renders this a promising route for therapeutic applications in the area of sleep disorders.
Future Directions
TLR4 is one of more than ten mammalian Toll-like receptors, all of which serve as important components of innate immunity by detecting and initiating responses to pathogen- or danger-associated molecular patterns.28 In interpreting the modest sleep phenotype of TLR4 KO mice, it is worth considering the mutually supportive roles of these receptors in detecting and reacting to pathogens.43 Interactions among the receptors may play an analogous role in sleep regulation. An assessment of sleep/wake regulation in TLR2/4 doubly-deficient mice, or mice deficient for Myd88, a protein necessary for the intracellular signaling cascade triggered by all cell-surface TLRs,28 might reveal a more robust sleep phenotype. And whether TLR-mediated changes in sleep and wakefulness are in reaction to changes in gut bacterial load36 (possibly conveyed by vagal afferents to the brainstem)37 or endogenous “danger-associated” ligands in the brain remains to be addressed. Whether the effects of TLR4-deficiency on sleep are mediated by changes in the inflammatory responses of brain monocytes remains uncertain; cytokine mRNAs are regulated by TLR4 activation, but they were not upregulated by 24-h sleep restriction in wild-type or TLR4 animals. This observation and the failure of NREMS EEG variables to be robustly affected by 24-h sleep restriction are likely indications that NREMS need did not increase during 24-h sleep restriction. Total sleep deprivation, even if it can only be achieved over a shorter term, may reveal more information about the dependence of biochemical changes induced by sleep loss on TLR4 signaling. Experimental measurement of the discrete contributions of the circadian clock and sleep homeostat to the sleep phenotype of TLR4 KO mice (possibly with a forced desynchrony protocol44) will also be informative in identifying the root cause of the sleep phenotype observed here.
A question unrelated to the function of TLR4 in sleep regulation is whether the effect of sleep loss on CD11b mRNA1 and monocyte cell counts (shown here) are caused by a reduced rate of monocyte infiltration into the brain or changes in the biochemical profiles of resident monocyte-derived cells. Resident microglia in the adult brain are likely to be derived from brain parenchymal progenitors that are present from early development, rather than from continuously infiltrating monocytes.45 In the non-diseased adult brain, infiltration of monocytes derived from bone marrow progenitors is an infrequent occurrence.42,45 A direct measure of monocyte infiltration, such as monitoring the rate of infiltration of bone marrow-derived, genetically tagged cells,42 will be necessary to determine which population or populations of cerebral monocytes are affected by sleep loss.
DISCLOSURE STATEMENT
This was not an industry supported study. The authors have indicated no financial conflicts of interest.
ACKNOWLEDGMENTS
These experiments were supported by a Washington State University, Spokane Faculty Seed grant and a Washington State University New Faculty Seed grant. Erik Naylor, an employee of Pinnacle Technologies, Inc., fact-checked the subsections of the methods section in which the in vivo data collection and automated sleep restriction technologies are described. The authors thank him for doing so.
REFERENCES
- 1.Wisor JP, Schmidt MA, Clegern WC. Sleep. 2011. Evidence for neuroinflammatory and microglial changes in the cerebral response to sleep loss; pp. 261–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frey DJ, Fleshner M, Wright KP., Jr The effects of 40 hours of total sleep deprivation on inflammatory markers in healthy young adults. Brain Behav Immun. 2007;21:1050–7. doi: 10.1016/j.bbi.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Imeri L, Opp MR. How (and why) the immune system makes us sleep. Nat Rev Neurosci. 2009;10:199–210. doi: 10.1038/nrn2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krueger JM, Obal FJ, Fang J, Kubota T, Taishi P. The role of cytokines in physiological sleep regulation. Ann N Y Acad Sci. 2001;933:211–21. doi: 10.1111/j.1749-6632.2001.tb05826.x. [DOI] [PubMed] [Google Scholar]
- 5.Matsui T, Svensson CI, Hirata Y, Mizobata K, Hua XY, Yaksh TL. Release of prostaglandin E(2) and nitric oxide from spinal microglia is dependent on activation of p38 mitogen-activated protein kinase. Anesth Analg. 2010;111:554–60. doi: 10.1213/ANE.0b013e3181e3a2a2. [DOI] [PubMed] [Google Scholar]
- 6.Wisor JP, Clegern WC. Quantification of short-term slow wave sleep homeostasis and its disruption by minocycline in the laboratory mouse. Neurosci Lett. 2011;490:165–9. doi: 10.1016/j.neulet.2010.11.034. [DOI] [PubMed] [Google Scholar]
- 7.Nonaka K, Nakazawa Y, Kotorii T. Effects of antibiotics, minocycline and ampicillin, on human sleep. Brain Res. 1983;288:253–9. doi: 10.1016/0006-8993(83)90101-4. [DOI] [PubMed] [Google Scholar]
- 8.Gyoneva S, Orr AG, Traynelis SF. Differential regulation of microglial motility by ATP/ADP and adenosine. Parkinsonism Relat Disord. 2009;15(Suppl 3):S195–9. doi: 10.1016/S1353-8020(09)70813-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haynes SE, Hollopeter G, Yang G, et al. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat Neurosci. 2006;9:1512–9. doi: 10.1038/nn1805. [DOI] [PubMed] [Google Scholar]
- 10.Benington JH, Kodali SK, Heller HC. Stimulation of A1 adenosine receptors mimics the electroencephalographic effects of sleep deprivation. Brain Res. 1995;692:79–85. doi: 10.1016/0006-8993(95)00590-m. [DOI] [PubMed] [Google Scholar]
- 11.Krueger JM, Taishi P, De A, et al. ATP and the purine type 2 X7 receptor affect sleep. J Appl Physiol. 2010;109:1318–27. doi: 10.1152/japplphysiol.00586.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy K, Travers P, Walport M. Innate immunity. In: Janeway CA, Travers P, Walport M, Shlomchik MJ, editors. Immunobiology the immune system in health and disease. 6th ed. New York: Garland Science; 2005. pp. 36–97. [Google Scholar]
- 13.Cirelli C, Tononi G. Gene expression in the brain across the sleep-waking cycle. Brain Res. 2000;885:303–21. doi: 10.1016/s0006-8993(00)03008-0. [DOI] [PubMed] [Google Scholar]
- 14.Terao A, Steininger TL, Hyder K, et al. Differential increase in the expression of heat shock protein family members during sleep deprivation and during sleep. Neuroscience. 2003;116:187–200. doi: 10.1016/s0306-4522(02)00695-4. [DOI] [PubMed] [Google Scholar]
- 15.Pawlyk AC, Ferber M, Shah A, Pack AI, Naidoo N. Proteomic analysis of the effects and interactions of sleep deprivation and aging in mouse cerebral cortex. J Neurochem. 2007;103:2301–13. doi: 10.1111/j.1471-4159.2007.04949.x. [DOI] [PubMed] [Google Scholar]
- 16.Lehnardt S, Schott E, Trimbuch T, et al. A vicious cycle involving release of heat shock protein 60 from injured cells and activation of toll-like receptor 4 mediates neurodegeneration in the CNS. J Neurosci. 2008;28:2320–31. doi: 10.1523/JNEUROSCI.4760-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutchinson MR, Bland ST, Johnson KW, Rice KC, Maier SF, Watkins LR. Opioid-induced glial activation: mechanisms of activation and implications for opioid analgesia, dependence, and reward. ScientificWorldJournal. 2007;7:98–111. doi: 10.1100/tsw.2007.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weinstein JR, Swarts S, Bishop C, Hanisch UK, Moller T. Lipopolysaccharide is a frequent and significant contaminant in microglia-activating factors. Glia. 2008;56:16–26. doi: 10.1002/glia.20585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsan MF, Gao B. Endogenous ligands of Toll-like receptors. J Leukoc Biol. 2004;76:514–9. doi: 10.1189/jlb.0304127. [DOI] [PubMed] [Google Scholar]
- 20.Rivest S. Molecular insights on the cerebral innate immune system. Brain Behav Immun. 2003;17:13–9. doi: 10.1016/s0889-1591(02)00055-7. [DOI] [PubMed] [Google Scholar]
- 21.Hanisch UK. Microglia as a source and target of cytokines. Glia. 2002;40:140–55. doi: 10.1002/glia.10161. [DOI] [PubMed] [Google Scholar]
- 22.Institute of Laboratory Animal Resources, National Research Council. Guide for Care and Use of Laboratory Animals. Washington, DC: National Academy Press; 1996. [Google Scholar]
- 23.Sasmono RT, Hume DA. The biology of macrophages. In: Kaufmann SH, Medzhitov R, Gordon S, editors. The innate immune response to infection. Washington, DC: American Society for Microbiology Press; 2004. pp. 71–94. [Google Scholar]
- 24.Tambuyzer BR, Ponsaerts P, Nouwen EJ. Microglia: gatekeepers of central nervous system immunology. J Leukoc Biol. 2008:352–70. doi: 10.1189/jlb.0608385. 352–70. [DOI] [PubMed] [Google Scholar]
- 25.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 26.Sedgwick JD, Schwender S, Imrich H, Dorries R, Butcher GW, ter Meulen V. Isolation and direct characterization of resident microglial cells from the normal and inflamed central nervous system. Proc Natl Acad Sci U S A. 1991;88:7438–42. doi: 10.1073/pnas.88.16.7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shankaran M, Marino ME, Busch R, et al. Measurement of brain microglial proliferation rates in vivo in response to neuroinflammatory stimuli: application to drug discovery. J Neurosci Res. 2007;85:2374–84. doi: 10.1002/jnr.21389. [DOI] [PubMed] [Google Scholar]
- 28.Abbas AK, Lichtman AH. Cellular and molecular immunology. Philadelphia: Saunders; 2003. Innate immunity; pp. 275–97. [Google Scholar]
- 29.Edgar DM, Dement WC, Fuller CA. Effect of SCN lesions on sleep in squirrel monkeys: evidence for opponent processes in sleep-wake regulation. J Neurosci. 1993;13:1065–79. doi: 10.1523/JNEUROSCI.13-03-01065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Graaf R, Kloppenburg G, Kitslaar PJ, Bruggeman CA, Stassen F. Human heat shock protein 60 stimulates vascular smooth muscle cell proliferation through Toll-like receptors 2 and 4. Microbes Infect. 2006;8:1859–65. doi: 10.1016/j.micinf.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 31.Rechtschaffen A, Bergmann BM, Gilliland MA, Bauer K. Effects of method, duration, and sleep stage on rebounds from sleep deprivation in the rat. Sleep. 1999;22:11–31. doi: 10.1093/sleep/22.1.11. [DOI] [PubMed] [Google Scholar]
- 32.Baker FC, Shah S, Stewart D, et al. Interleukin 1beta enhances non-rapid eye movement sleep and increases c-Fos protein expression in the median preoptic nucleus of the hypothalamus. Am J Physiol Regul Integr Comp Physiol. 2005;288:R998–R1005. doi: 10.1152/ajpregu.00615.2004. [DOI] [PubMed] [Google Scholar]
- 33.Morrow JD, Opp MR. Sleep-wake behavior and responses of interleukin-6-deficient mice to sleep deprivation. Brain Behav Immun. 2005;19:28–39. doi: 10.1016/j.bbi.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Kapas L, Bohnet SG, Traynor TR, et al. Spontaneous and influenza virus-induced sleep are altered in TNF-alpha double-receptor deficient mice. J Appl Physiol. 2008;105:1187–98. doi: 10.1152/japplphysiol.90388.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barajon I, Serrao G, Arnaboldi F, et al. Toll-like receptors 3, 4, and 7 are expressed in the enteric nervous system and dorsal root ganglia. J Histochem Cytochem. 2009;57:1013–23. doi: 10.1369/jhc.2009.953539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Everson CA, Toth LA. Systemic bacterial invasion induced by sleep deprivation. Am J Physiol Regul Integr Comp Physiol. 2000;278:R905–16. doi: 10.1152/ajpregu.2000.278.4.R905. [DOI] [PubMed] [Google Scholar]
- 37.Hansen MK, Kapas L, Fang J, Krueger JM. Cafeteria diet-induced sleep is blocked by subdiaphragmatic vagotomy in rats. Am J Physiol. 1998;274:R168–74. doi: 10.1152/ajpregu.1998.274.1.R168. [DOI] [PubMed] [Google Scholar]
- 38.Kuo TH, Pike DH, Beizaeipour Z, Williams JA. Sleep triggered by an immune response in Drosophila is regulated by the circadian clock and requires the NFkappaB Relish. BMC Neurosci. 2010;11:17. doi: 10.1186/1471-2202-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams JA, Sathyanarayanan S, Hendricks JC, Sehgal A. Interaction between sleep and the immune response in Drosophila: a role for the NFkappaB relish. Sleep. 2007;30:389–400. doi: 10.1093/sleep/30.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rihel J, Prober DA, Arvanites A, et al. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science. 2010;327:348–51. doi: 10.1126/science.1183090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kimura Y, Zhou L, Miwa T, Song WC. Genetic and therapeutic targeting of properdin in mice prevents complement-mediated tissue injury. J Clin Invest. 2010;120:3545–54. doi: 10.1172/JCI41782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lebson L, Nash K, Kamath S, et al. Trafficking CD11b-positive blood cells deliver therapeutic genes to the brain of amyloid-depositing transgenic mice. J Neurosci. 2010;30:9651–8. doi: 10.1523/JNEUROSCI.0329-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Underhill DM, Ozinsky A. Toll-like receptors: key mediators of microbe detection. Curr Opin Immunol. 2002;14:103–10. doi: 10.1016/s0952-7915(01)00304-1. [DOI] [PubMed] [Google Scholar]
- 44.Yasenkov R, Deboer T. Circadian regulation of sleep and the sleep EEG under constant sleep pressure in the rat. Sleep. 2010;33:631–41. doi: 10.1093/sleep/33.5.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ginhoux F, Greter M, Leboeuf M, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–4. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]