Abstract
Objective
The PTPN22 rs2476601 polymorphism is associated with rheumatoid arthritis (RA); nonetheless, the association is weaker or absent in some southern European populations. The aim of the study was to evaluate the association between the PTPN22 rs2476601 polymorphism and RA in Italian subjects and to compare our results with those of other European countries, carrying out a meta-analysis of European data.
Methods
A total of 396 RA cases and 477 controls, all of Italic ancestry, were genotyped for PTPN22 rs2476601 polymorphism. Patients were tested for autoantibodies positivity. The meta-analysis was performed on 23 selected studies.
Results
The PTPN22 T1858 allele was significantly more frequent in RA patients compared to controls (5.7% vs. 3.7%, p = 0.045). No clear relationship arose with the autoantibodies tested. The 1858T allele frequency in Italian RA patients was lower than the one described in northern European populations and similar to the frequency found in Spain, Turkey, Greece, Tunisia. A clear-cut North-South gradient arose from the analysis.
Conclusions
The PTPN22 T1858 allele is associated with RA in the Italian population. A North-South gradient of the allele frequency seems to exist in Europe, with a lower prevalence of the mutation in the Mediterranean area.
Introduction
Genetic factors are thought to be responsible for up to 50–60% of the rheumatoid arthritis (RA) liability [1]. The minor allele (T) at 1858C>T (rs2476601) single-nucleotide polymorphism (SNP) in the protein tyrosine phosphatase non-receptor type 22 (PTPN22, gene map locus 1p13) gene has been extensively associated with susceptibility to various autoimmune diseases [2]. The rs2476601 determines a R620W substitution resulting in a gain-of-function form of the enzyme Lyp (encoded by the PTPN22 gene), thus leading to a stronger suppression of early T cell activation process [3]. The B cell compartment seems altered by this SNP as well [4]. The association of the PTPN22 1858C>T SNP with RA is well established among many populations all over the world, especially in anti-CCP (cyclic citrullinated peptides) antibodies positive RA patients [5]. However, a weaker or a complete lack of association has been reported in some southern European populations [6]–[9]. A review of the literature suggests a lower frequency of the T1858 allele in RA patients of the Mediterranean area. According to Orozco et al., there was no association of PTPN22 1858C>T SNP with early RA in Spain [6]. In Turkey, Sahin et al. found no association of the 1858C>T SNP with RA [7]. Moreover, a lack of association has also been reported in a Tunisian and in a Greek population [8], [9].
Although the role of PTPN22 rs2476601 SNP in autoimmunity and its association with RA are undoubted, as recently confirmed by genome-wide association analyses [10], it is important to take into account geographical and anthropological differences when performing genetic epidemiology studies.
To date, no data are available regarding the possible association between PTPN22 1858C>T polymorphism and rheumatoid arthritis in patients of Italic ancestry. The aims of the study were: 1. To evaluate the PTPN22 rs2476601 SNP distribution in an Italian cohort; 2. To define, by means of a systematic review and meta-analysis, the association between the PTPN22 1858C>T polymorphism distribution in Europe and rheumatoid arthritis.
Methods
Ethical statement
The ethical approval for the study was obtained from the Catholic University of the Sacred Heart Ethical Committee. All subjects gave their written informed consent on the analysis of the PTPN22 gene polymorphism and autoantibodies testing.
Case-control study
Population and setting
Cases were recruited from the Division of Rheumatology of the Catholic University of the Sacred Heart of Rome. Patients fulfilled at least four of the American College of Rheumatology criteria for RA [11]. When looking back at the database with characteristics of each patient, all satisfied the 2010 ACR criteria as well [12]. The controls sample includes healthy subjects matched for age, sex and geographical origin with case subjects.
In order to calculate the sample size, the following parameters were used: power 80%, level of confidence 95%, estimated frequency of PTPN22 T1858 allele in controls of 8.9% and of 15.1% in RA patients (based on the mean value of the European studies). Sample size was estimated to be: 462 cases and 462 controls. Patients were recruited consecutively between January 2008 and December 2010 from the Division of Rheumatology of the Catholic University of the Sacred Heart of Rome.
All patients' sera were tested for the presence of anti-CCP, IgM RF (rheumatoid factor) and IgA RF autoantibodies (ELISA method, Axis-Shield Diagnostics, Dundee, UK for anti-CCP and Orgentec diagnostika, Mainz, Germany for IgM and IgA RFs).
Genotyping
Genomic DNA was isolated from whole blood through FlexiGene DNA kit (Qiagen, Valencia, CA) according to the manufacturer instructions.
The PTPN22 1858C>T SNP was determined by the restriction fragment length polymorphism-polymerase chain reaction (PCR) based method as previously described [13], in all the patients and controls. Briefly, oligonucleotides 5′-TCACCAGCTTCCTCAACCACA-3′ and 5′-GATAATGTTGCTTCAACGGAATTT-3′ were used as primers for PTPN22 1858C>T SNP. The C→T transition at codon 620 creates in the T1858 allele a restriction site for XcmI (New England Biolabs, Beverly, MA. USA). The product of PCR was digested with XcmI at 37°C for 3 hours and each digestion was resolved on 3% agarose gel (Figure S1). Repeated typing was performed in 10% of patient samples, with identical results in all cases.
Meta-analysis
Identification of eligible studies and data extraction
The electronic medical databases used for the search were Pubmed, Embase and the Cochrane Library. In the research, we used the keywords: “arthritis”, “rheumatoid”, “PTPN22”, “polymorphism” applying the following algorithm: (PTPN22 OR “protein tyrosine phosphatase non-receptor type 22” OR Lyp) AND (rheumatoid AND/OR arthritis) AND (polymorphism OR SNP). The identification of eligible studies was carried out from 2000 until December 2010 and it was not restricted to English language. Studies references were also analyzed to find any study not available from the electronic databases. A study was included in the systematic extraction of the data if: 1. it was published before 2011; 2. it was about European patients with rheumatoid arthritis; 3. PTPN22 1858C>T SNP was evaluated and genotypes data were clearly expressed; 4 it was a case-control study; 5. it was not a transmission disequilibrium test in which family members were studied. Data related to the PTPN22 1858C>T SNP in RA patients and controls groups were extracted to perform the meta-analysis. Data extraction and quality assessment, according to a score sheet available for observational studies [14], were performed independently by two different investigators.
Statistical analysis
Data regarding the PTPN22 1858C>T SNP in our RA patients and controls groups were checked for deviation from Hardy-Weinberg equilibrium (χ2 test). Descriptive statistics was performed using frequencies and percentages. The association between alleles and genotypes with RA was investigated applying Exact Fisher's Test, and calculating Odds Ratio (OR) with 95% Confidence Interval (95%CI). Statistical analysis was performed with SPSS 19.0 software for Windows.
Three different meta-analyses were carried out using StatDirect statistical software Version2.7.8. The first one evaluated the association of PTPN22 1858C>T SNP with RA considering all published studies, present study included; the second one presented data excluding the Italian population; the third one considered the studies with a quality score ≥11, that corresponds to the median quality score.
Forest-plots graphs were produced in order to estimate the pooled association between the PTPN22 1858C>T SNP and RA. The Cochran's Q test was performed to evaluate studies heterogeneity, thus using the random effect model when the test highlighted differences between studies and the fixed effect model when no significant differences were shown. Publication bias was quantified by inspection of funnel plot and computation of Egger and Begg test probability values [15]. Significance threshold was set at p<0.05 (2-tailed) for all analyses.
Results
PTPN22 1858C>T SNP in Italy
The studied sample was composed of 396 RA patients and 477 controls (the power of the study was 75% for the cases). The genotype distribution of the PTPN22 1858C>T SNP was in Hardy-Weinberg equilibrium in both groups. Seventy-nine percent of the RA patients were female, 67% were positive for anti-CCP antibodies, 50% were positive for IgM RF (rheumatoid factor), and 34% were positive for IgA RF. Moreover, 74% were positive for at least one of the autoantibodies tested.
In our center, the analysis of RA patients and controls, all of Italic ancestry, showed a PTPN22 T1858 allele frequency of 5.7% in patients compared to 3.7% in controls (OR = 1.58; 95%CI = (1.01–2.49); p = 0.045) (Table 1). The frequency of positivity for anti-CCP antibodies tended to be higher in RA patients carrying the T allele (79.5%) compared to subjects with C/C genotype (65.6%; p = 0.09). No difference was detected in the percentage of IgA and IgM RFs positivity between RA patients carrying the T allele and patients with C/C genotype (data not shown).
Table 1. Analysis of the association of PTPN22 1858C>T SNP with RA patients and healthy subjects.
| Genotype | ||||||||
| Group | C/C | C/T | T/T | T Allele | HWE (p)1 | OR (95%CIs)2 | Allelic p 3 | |
| Females | RA patients (n = 313) (mean age: 55.7±13.7) | 276 (91.6%) | 36 (11.5%) | 1 (0.3%) | 38 (6.1%) | 1.00 | 1.56 (0.95–2.55) | 0.074 |
| Healthy controls (n = 377) (mean age: 55.5±14.1) | 348 (92.3%) | 28 (7.4%) | 1 (0.3%) | 30 (4.0%) | 0.45 | |||
| Males | RA patients (n = 83) (mean age: 59.7±12.2) | 76 (91.6%) | 7 (8.4%) | 0 (0.0%) | 7 (4.2%) | 1.00 | 1.72 (0.54–5.52) | 0.359 |
| Healthy controls (n = 100) (mean age: 59.4±12.0) | 95 (95.0%) | 5 (5.0%) | 0 (0.0%) | 5 (2.5%) | 1.00 | |||
| All | RA patients (n = 396) (mean age: 56.6±13.4) | 352 (88.9%) | 43 (10.9%) | 1 (0.2%) | 45 (5.7%) | 1.00 | 1.58 (1.01–2.49) | 0.045 |
| Healthy controls (n = 477) (mean age: 56.3±14.0) | 443 (92.9%) | 33 (6.9%) | 1 (0.2%) | 35 (3.7%) | 0.48 | |||
HWE: Hardy-Weinberg equilibrium.
Odds ratios expressed as carriers of T1858 allele vs. non-carriers considering healthy controls as control group.
Fisher's exact test of odds ratios.
Meta-analysis of the reported studies regarding the PTPN22 1858C>T SNP in Europe
The association of the PTPN22 1858C>T SNP with RA in Europe was found in 23 studies [5], [7], [8], [16]–[35] (see PRISMA Checklist S1 and Figure S2). The characteristics of each study are shown in Table 2.
Table 2. Characteristics of the selected studies (n = 24) concerning the association between PTPN22 1858C>T SNP and RA in Europe.
| Study | Ref. | Year | Country | Quality | RA C/T-T/T | Controls C/T-T/T | RA total | Controls total |
| Seldin et al. | 16 | 2005 | Finland | 10 | 372 | 406 | 1030 | 1400 |
| Plenge et al. | 17 | 2005 | Sweden | 11.5 | 432 | 203 | 1513 | 874 |
| Hinks et al. | 18 | 2005 | UK | 11 | 289 | 114 | 886 | 595 |
| Steer et al. | 19 | 2005 | UK | 10 | 84 | 62 | 302 | 374 |
| Wesoly et al. | 20 | 2005 | Netherlands | 10 | 93 | 155 | 416 | 891 |
| Zhernakova et al. | 21 | 2005 | Netherlands | 10 | 42 | 88 | 151 | 528 |
| Orozco et al. | 22 | 2005 | Spain | 11.5 | 163 | 146 | 826 | 1036 |
| Johansson et al. | 5 | 2006 | Sweden | 12 | 35 | 71 | 89 | 360 |
| Harrison et al. | 23 | 2006 | UK | 10 | 179 | 109 | 686 | 566 |
| Pierer et al. | 24 | 2006 | Germany | 11.5 | 148 | 67 | 390 | 349 |
| Viken et al. | 25 | 2007 | Norway | 10.5 | 264 | 119 | 861 | 557 |
| Lie et al. | 26 | 2007 | Norway | 12.5 | 75 | 119 | 221 | 555 |
| Kokkonen et al. | 27 | 2007 | Sweden | 12 | 166 | 209 | 504 | 970 |
| Majorczyk et al. | 28 | 2007 | Poland | 11 | 61 | 118 | 173 | 543 |
| Wesoly et al. | 29 | 2007 | Netherlands | 10.5 | 183 | 55 | 661 | 284 |
| Eike et al. | 30 | 2008 | Norway | 11 | 213 | 191 | 686 | 952 |
| Farago et al. | 31 | 2008 | Hungary | 11 | 158 | 24 | 399 | 107 |
| Starck et al. | 32 | 2009 | Slovakia | 12.5 | 158 | 63 | 514 | 302 |
| Sahin et al. | 7 | 2009 | Turkey | 10.5 | 11 | 9 | 167 | 177 |
| Chabchoub et al. | 8 | 2009 | Tunisia | 11 | 7 | 12 | 150 | 236 |
| Sfar et al. | 33 | 2009 | Tunisia | 11 | 33 | 2 | 133 | 100 |
| Morgan et al. | 34 | 2009 | UK | 10 | 1324 | 705 | 4789 | 3630 |
| Majorczyk et al. | 35 | 2010 | Poland | 11 | 259 | 118 | 371 | 543 |
| Present study | - | 2011 | Italy | - | 44 | 34 | 396 | 477 |
In the first meta-analysis a significant and positive association between the PTPN22 1858C>T SNP and RA was found: pooled OR = 1.79 with 95%CI = (1.60–2.01) (Figure 1). The Cochran's Q test established the presence of heterogeneity (χ2 = 79.42, df = 23, p<0.001), therefore a random effect model was applied.
Figure 1. Forest plot of the first meta-analysis.
Forest plot of published studies in relation to the first meta-analysis (24 studies). The association of the PTPN22 1858C>T SNP with RA was evaluated through the Odds ratios measures. The random effect model was used.
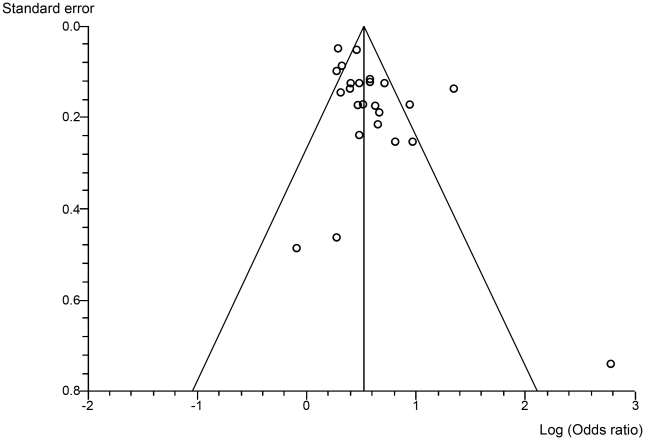
The funnel plot (Figure 2) did not show any publication bias, in accordance with the bias tests: Begg-Mazumdar: Kendall τ = −0.05, p = 0.76; Egger: bias = −0.55, p = 0.46.
Figure 2. Funnel plot.
Funnel plot of published studies in relation to the first meta-analysis (24 studies).
There was not a significant difference between the first meta-analysis and the second one: combined OR = 1.80 with 95%CI = (1.61–2.02) using random effect estimate (Cochran's Q test: χ2 = 79.42, df = 22, p<0.001).
The studies with a quality score ≥11 were 14. The relationship between PTPN22 1858C>T SNP and RA resulted even stronger: OR = 2.01, 95%CI = (1.67–2.43) using random effect model (Cochran's Q test: χ2 = 61.14, df = 13, p<0.001).
When looking at the T allele frequency in RA patients and controls, we noticed a North-South gradient with higher values in Finland, Germany, Hungary, and lower values in Spain, Italy, Tunisia, Greece and Turkey (Figure 3 [5], [7]–[9], [16]–[37]).
Figure 3. Geographical distribution in Europe.
Geographical distribution of the T allele frequency at PTPN22 rs2476601 SNP in European RA patients (red) and healthy controls (black) [5], [7]–[9], [16]–[37]. The ‘*’ symbol refers to countries in which a statistical significant different distribution of the PTPN22 T1858 allele among patients and controls was noted. In Tunisia the two existing articles regarding PTPN22 show opposite findings.
Discussion
The association between the T1858 allele at rs2476601 in the PTPN22 gene and RA has been documented in several cohorts, from the USA as well from Europe [38], [39], though it seems to be less relevant in other continents [40]. When considering the strength of the allele association in the analysis of the European consortium, the relationship arose quite clear; however regarding single countries, data are less clear-cut. As reported in Figure 3, a North-South gradient seems to be present in the distribution of the T1858 allele in both RA patients and controls, as previously remarked by Gregersen et al. in some European populations [38].
Furthermore, it is also noteworthy that while in Germany, the frequency of the T1858 allele was significantly higher in RA patients (21.3%) compared to controls (10.0%; with an OR of 2.43) and the association was present irrespective of the presence or absence of anti-CCP and RF [41], in France the European Consortium Group provided evidence for an association of T1858 allele only with RF positive cases but not with RF negative RA patients [36]. In Spain, there was no association with early RA but the association was significant with the anti-CCP positive RA [6].
Genetic differences within European populations have been once more underlined by a recent work of Rodríguez-Rodríguez et al. [42]. The authors described the association of another PTPN22 SNP, the R263Q, with RA in six different Caucasian populations. The 1858C>T SNP was also investigated using mostly previously published data. The T allele of the 1858C>T SNP showed an inhomogeneous distribution among the populations taken into account, with a prevalence of 10.5% in RA patients and 6.8% in controls in Spain, compared to 16.1% and 10.6% respectively in the other countries (Norway, UK, The Netherlands, Germany, New Zealand).
Our data revealed a higher frequency of the T1858 allele in RA Italian patients compared to the controls cohort. On the other hand, the frequency in controls was lower than that observed in France or in Germany and similar to Turkish, Greek and Tunisian populations.
Interestingly, Mediterranean populations are genetically linked by a common history of migrations, like the abiding one of Saracens and Moors. In fact a recent work, estimating the medieval North African contribution over Mediterranean countries through the analysis of the Y chromosome short tandem repeats, suggested a general correlation between historical and genetic data of Iberia, Sicily, Turkey and North Africa [43].
No relationship arose between the C/T-T/T genotypes presence and auto-antibodies positivity. The demonstration of a gain-of-function conferred by the T1858 allele in suppressing TCR (T cell receptor) function in T cells and BCR (B cell receptor) function in B cells raises new hypotheses on the role of tyrosine phosphatases. The T1858 allele might increase the threshold for a persistent activation of both autoreactive T and B cells thus leading to a more defined autoimmune subset of RA [4]. In our study, the trend for an association between the rs2476601 SNP and the positivity of anti-CCP seems to move towards this direction, though the only conclusion we can formulate with the data at hand is the geographical issue.
In conclusion, the geographical distribution of SNPs in the world, linked to different population origins, should be taken into account in studies regarding genetic associations. Given that specific therapies directed toward Lyp will be available in the near future for various autoimmune diseases [44], there could be clinical-therapeutic consequences as well, and, on these grounds, the approach based on PTPN22 might be different from North to South.
Supporting Information
Electrophoresis gel. Photo of the electrophoresis gel showing intact vs. cleaved PCR-amplified fragments from patients and controls, non-mutated, heterozygous or homozygous for the C>T substitution. A: intact PCR-amplified fragment; B: cleaved fragment from DNA of a patient non-mutated for the PTPN22 rs2476601 SNP; C: cleaved fragment from DNA of a patient heterozygous for the SNP; D: cleaved fragment from DNA of a patient homozygous for the SNP; E, F and G: same as for B, C and D, but from the DNA of a control subject.
(TIF)
PRISMA flow diagram. PRISMA 2009 flow diagram regarding the article selection.
(DOC)
PRISMA Checklist. PRISMA 2009 checklist regarding meta-analysis data and their position in the manuscript.
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by a grant from Schering-Plough and by Fondazione ASRALES ONLUS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.MacGregor AJ, Snieder H, Rigby AS, Koskenvuo M, Kaprio J, et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 2000;43:30–37. doi: 10.1002/1529-0131(200001)43:1<30::AID-ANR5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 2.Viken MK, Amundsen SS, Kvien TK, Boberg KM, Gilboe IM, et al. Association analysis of the 1858C>T polymorphism in the PTPN22 gene in juvenile idiopathic arthritis and other autoimmune diseases. Genes Immun. 2005;6:271–273. doi: 10.1038/sj.gene.6364178. [DOI] [PubMed] [Google Scholar]
- 3.Vang T, Congia M, Macis MD, Musumeci L, Orru V, et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of function variant. Nat Genet. 2005;37:1317–1319. doi: 10.1038/ng1673. [DOI] [PubMed] [Google Scholar]
- 4.Rieck M, Arechiga A, Onengut-Gumuscu S, Greenbaum C, Concannon P, et al. Genetic variation in PTPN22 corresponds to altered function of T and B lymphocytes. J Immunol. 2007;179:4704–4710. doi: 10.4049/jimmunol.179.7.4704. [DOI] [PubMed] [Google Scholar]
- 5.Johansson M, Alestig L, Hallmans G, Rantapaa-Dahlqvist S. PTPN22 polymorphism and anti-cyclic citrullinated peptide antibodies in combination strongly predicts future onset of rheumatoid arthritis and has a specificity of 100% for the disease. Arthritis Res Ther. 2006;8:R19. doi: 10.1186/ar1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orozco G, Pascual-Salcedo D, López-Nevot MA, Cobo T, Cabezón A, et al. Auto-antibodies, HLA and PTPN22: susceptibility markers for rheumatoid arthritis. Rheumatology. 2008;47:138–141. doi: 10.1093/rheumatology/kem343. [DOI] [PubMed] [Google Scholar]
- 7.Sahin N, Gunduz F, Inanc N, Direskeneli H, Saruhan-Direskeneli G. No association of PTPN22 gene polymorphism with rheumatoid arthritis in Turkey. Rheumatol Int. 2009;30:81–83. doi: 10.1007/s00296-009-0919-2. [DOI] [PubMed] [Google Scholar]
- 8.Chabchoub G, Teixiera EP, Maalej A, Ben Hamad M, Bahloul Z, et al. The R620W polymorphism of the protein tyrosine phosphatase 22 gene in autoimmune thyroid diseases and rheumatoid arthritis in the Tunisian population. Ann Hum Biol. 2009;36:342–349. doi: 10.1080/03014460902817968. [DOI] [PubMed] [Google Scholar]
- 9.Plant D, Flynn E, Mbarek H, Dieudé P, Cornelis F, et al. Investigation of potential non-HLA rheumatoid arthritis susceptibility loci in a European cohort increases the evidence for nine markers. Ann Rheum Dis. 2010;691:548–553. doi: 10.1136/ard.2009.121020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steer S, Abkevich V, Gutin A, Cordell HJ, Gendall KL, et al. Genomic DNA pooling for whole-genome association scans in complex disease: empirical demonstration of efficacy in rheumatoid arthritis. Genes Immun. 2006;8:57–68. doi: 10.1038/sj.gene.6364359. [DOI] [PubMed] [Google Scholar]
- 11.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 12.Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, et al. Rheumatoid Arthritis Classification Criteria: An American College of Rheumatology/European League Against Rheumatism Collaborative Initiative. Arthritis and Rheum. 2010;62:2569–2581. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- 13.Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2006;36:337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 14.La Torre G, Chiaradia G, Gianfagna F, De Laurentis A, Boccia S, et al. Quality assessment in meta-analysis. Ital J Publ Health. 2006;3:44–50. [Google Scholar]
- 15.Sutton AJ, Jones DR, Abrams KR, Sheldon TA, Song F. Publication bias. In: Sutton AJ, Jones DR, Abrams KR, Sheldon TA, Song F, editors. Methods for Meta-Analysis in Medical Research. Chichester, UK: John Wiley & Sons; 2000. pp. 109–132. [Google Scholar]
- 16.Seldin MF, Shigeta R, Laiho K, Li H, Saila H, et al. Finnish case-control and family studies support PTPN22 R620W polymorphism as a risk factor in rheumatoid arthritis, but suggest only minimal or no effect in juvenile idiopathic arthritis. Genes Immun. 2005;6:720–722. doi: 10.1038/sj.gene.6364255. [DOI] [PubMed] [Google Scholar]
- 17.Plenge RM, Padyukov L, Remmers EF, Purcell S, Lee AT, et al. Replication of putative candidate-gene associations with rheumatoid arthritis in >4,000 samples from North America and Sweden: association of susceptibility with PTPN22, CTLA4, and PADI4. Am J Hum Genet. 2005;77:1044–1060. doi: 10.1086/498651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinks A, Barton A, John S, Bruce I, Hawkins C, et al. Association between the PTPN22 gene and rheumatoid arthritis and juvenile idiopathic arthritis in a UK population: further support that PTPN22 is an autoimmunity gene. Arthritis Rheum. 2005;52:1694–1699. doi: 10.1002/art.21049. [DOI] [PubMed] [Google Scholar]
- 19.Steer S, Lad B, Grumley JA, Kingsley GH, Fisher SA. Association of R620W in a protein tyrosine phosphatase gene with a high risk of rheumatoid arthritis in a British population: evidence for an early onset/disease severity effect. Arthritis Rheum. 2005;52:358–360. doi: 10.1002/art.20737. [DOI] [PubMed] [Google Scholar]
- 20.Wesoly J, van der Helm-van Mil AH, Toes RE, Chokkalingam AP, Carlton VE, et al. Association of the PTPN22 C1858T single-nucleotide polymorphism with rheumatoid arthritis phenotypes in an inception cohort. Arthritis Rheum. 2005;52:2948–2950. doi: 10.1002/art.21294. [DOI] [PubMed] [Google Scholar]
- 21.Zhernakova A, Eerligh P, Wijmenga C, Barrera P, Roep BO, et al. Differential association of the PTPN22 coding variant with autoimmune diseases in a Dutch population. Genes Immun. 2005;6:459–461. doi: 10.1038/sj.gene.6364220. [DOI] [PubMed] [Google Scholar]
- 22.Orozco G, Sanchez E, Gonzalez-Gay MA, López-Nevot MA, Torres B, et al. Association of a functional single-nucleotide polymorphism of PTPN22, encoding lymphoid protein phosphatase, with rheumatoid arthritis and systemic lupus erythematosus. Arthritis Rheum. 2005;52:219–224. doi: 10.1002/art.20771. [DOI] [PubMed] [Google Scholar]
- 23.Harrison P, Pointon JJ, Farrar C, Brown MA, Wordsworth BP. Effects of PTPN22 C1858T polymorphism on susceptibility and clinical characteristics of British Caucasian rheumatoid arthritis patients. Rheumatology. 2006;45:1009–1011. doi: 10.1093/rheumatology/kei250. [DOI] [PubMed] [Google Scholar]
- 24.Pierer M, Kaltenhäuser S, Arnold S, Wahle M, Baerwald C, et al. Association of PTPN22 1858 single-nucleotide polymorphism with rheumatoid arthritis in a German cohort: higher frequency of the risk allele in male compared to female patients. Arthritis Res Ther. 2006;8:R75. doi: 10.1186/ar1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viken MK, Olsson M, Flåm ST, Førre O, Kvien TK, et al. The PTPN22 promoter polymorphism -1123G>C association cannot be distinguished from the 1858C>T association in a Norwegian rheumatoid arthritis material. Tissue Antigens. 2007;70:190–197. doi: 10.1111/j.1399-0039.2007.00871.x. [DOI] [PubMed] [Google Scholar]
- 26.Lie BA, Viken MK, Odegård S, van der Heijde D, Landewé R, et al. Associations between the PTPN22 1858C->T polymorphism and radiographic joint destruction in patients with rheumatoid arthritis: results from a 10-year longitudinal study. Ann Rheum Dis. 2007;66:1604–1609. doi: 10.1136/ard.2006.067892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kokkonen H, Johansson M, Innala L, Jidell E, Rantapää-Dahlqvist S. The PTPN22 1858C/T polymorphism is associated with anti-cyclic citrullinated peptide antibody-positive early rheumatoid arthritis in northern Sweden. Arthritis Res Ther. 2007;9:R56. doi: 10.1186/ar2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majorczyk E, Jasek M, Płoski R, Wagner M, Kosior A, et al. Association of PTPN22 single nucleotide polymorphism with rheumatoid arthritis but not with allergic asthma. Eur J Hum Genet. 2007;15:1043–1048. doi: 10.1038/sj.ejhg.5201879. [DOI] [PubMed] [Google Scholar]
- 29.Wesoly J, Hu X, Thabet MM, Chang M, Uh H, et al. The 620W allele is the PTPN22 genetic variant conferring susceptibility to RA in a Dutch population. Rheumatology. 2007;46:617–621. doi: 10.1093/rheumatology/kel381. [DOI] [PubMed] [Google Scholar]
- 30.Eike MC, Nordang GB, Karlsen TH, Boberg KM, Vatn MH, et al. The FCRL3 -169T>C polymorphism is associated with rheumatoid arthritis and shows suggestive evidence of involvement with juvenile idiopathic arthritis in a Scandinavian panel of autoimmune diseases. Ann Rheum Dis. 2008;67:1287–1291. doi: 10.1136/ard.2007.077826. [DOI] [PubMed] [Google Scholar]
- 31.Farago B, Talian GC, Komlosi K, Nagy G, Berki T, et al. Protein tyrosine phosphatase gene C1858T allele confers risk for rheumatoid arthritis in Hungarian subjects. Rheumatol Int. 2009;29:793–796. doi: 10.1007/s00296-008-0771-9. [DOI] [PubMed] [Google Scholar]
- 32.Starck K, Rovenský J, Blažičková S, Grosse-Wilde H, Ferencik S, et al. Association of common polymorphisms in known susceptibility genes with rheumatoid arthritis in a Slovak population using osteoarthritis patients as controls. Arthritis Res Ther. 2009;11:R70. doi: 10.1186/ar2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sfar I, Dhaouadi T, Habibi I, Abdelmoula L, Makhlouf M, et al. Functional polymorphisms of PTPN22 and FCGR genes in Tunisian patients with rheumatoid arthritis. Archs Inst Pasteur Tunis. 2009;86:1–4. [PubMed] [Google Scholar]
- 34.Morgan AW, Thomson W, Martin SG, Carter AM, et al. Yorkshire Early Arthritis Register. HLA–DRB1 Shared Epitope Alleles, PTPN22, and Smoking in Determining Susceptibility to Autoantibody-Positive and Autoantibody-Negative Rheumatoid Arthritis in a Large UK Caucasian Population. Arthritis Rheum. 2009;60:2565–2576. doi: 10.1002/art.24752. [DOI] [PubMed] [Google Scholar]
- 35.Majorczyk E, Pawlik A, Kuśnierczyk P. PTPN22 1858C>T polymorphism is strongly associated with rheumatoid arthritis but not with a response to methotrexate therapy. Int Immunopharmacol. 2010;10:1626–1629. doi: 10.1016/j.intimp.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 36.Dieudé P, Garnier S, Michou L, Petit-Teixeira E, Glikmans E, et al. Rheumatoid arthritis seropositive for the rheumatoid factor is linked to the protein tyrosine phosphatase nonreceptor 22-620W allele. Arthritis Res Ther. 2005;7:R1200–1207. doi: 10.1186/ar1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dieudé P, Guedj M, Wipff J, Avouac J, Hachulla E, et al. The PTPN22 620W allele confers susceptibility to systemic sclerosis. Arthris Rheum. 2008;58:2183–2188. doi: 10.1002/art.23601. [DOI] [PubMed] [Google Scholar]
- 38.Gregersen PK, Lee HS, Batliwalla F, Begovich AB. PTPN22: Setting threshold for autoimmunity. Semin Immunol. 2006;18:214–223. doi: 10.1016/j.smim.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 39.Coenen MJ, Gregersen PK. Rheumatoid arthritis: a view of the current genetic landscape. Genes Immun. 2008;10:101–111. doi: 10.1038/gene.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kochi Y, Suzuki A, Yamada R, Yamamoto K. Ethnogenetic heterogeneity of rheumatoid arthritis-implications for pathogenesis. Nat Rev Rheumatol. 2010;6:290–295. doi: 10.1038/nrrheum.2010.23. [DOI] [PubMed] [Google Scholar]
- 41.Burkhardt H, Hüffmeier U, Spriewald B, Böhm B, Rau R, et al. Association between protein tyrosine phosphatase 22 variant R620W in conjunction with the HLA-DRB1 shared epitope and humoral autoimmunity to an immunodominant epitope of cartilage-specific type II collagen in early rheumatoid arthritis. Arthritis Rheum. 2006;54:82–89. doi: 10.1002/art.21498. [DOI] [PubMed] [Google Scholar]
- 42.Rodríguez-Rodríguez L, Wan Taib WR, Topless R, Steer S, González-Escribano MF, et al. The PTPN22 R263Q Polymorphism Is a Risk Factor for Rheumatoid Arthritis in Caucasian Case–Control Samples. Arthritis Rheum. 2011;63:365–372. doi: 10.1002/art.30145. [DOI] [PubMed] [Google Scholar]
- 43.Capelli C, Onofri V, Brisighelli F, Boschi I, Scarnicci F, et al. Moors and Saracens in Europe: estimating the medieval North African male legacy in southern Europe. Eur J Hum Genet. 2009;17:848–852. doi: 10.1038/ejhg.2008.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie Y, Liu Y, Gong G, Rinderspacher A, Deng SX, et al. Discovery of a novel submicromolar inhibitor of the lymphoid specific tyrosine phosphatase. Bioorg Med Chem Lett. 2008;18:2840–2844. doi: 10.1016/j.bmcl.2008.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Electrophoresis gel. Photo of the electrophoresis gel showing intact vs. cleaved PCR-amplified fragments from patients and controls, non-mutated, heterozygous or homozygous for the C>T substitution. A: intact PCR-amplified fragment; B: cleaved fragment from DNA of a patient non-mutated for the PTPN22 rs2476601 SNP; C: cleaved fragment from DNA of a patient heterozygous for the SNP; D: cleaved fragment from DNA of a patient homozygous for the SNP; E, F and G: same as for B, C and D, but from the DNA of a control subject.
(TIF)
PRISMA flow diagram. PRISMA 2009 flow diagram regarding the article selection.
(DOC)
PRISMA Checklist. PRISMA 2009 checklist regarding meta-analysis data and their position in the manuscript.
(DOC)