Abstract
Emerging plant pathogens have largely been a consequence of the movement of pathogens to new geographic regions. Another documented mechanism for the emergence of plant pathogens is hybridization between individuals of different species or subspecies, which may allow rapid evolution and adaptation to new hosts or environments. Hybrid plant pathogens have traditionally been difficult to detect or confirm, but the increasing ease of cloning and sequencing PCR products now makes the identification of species that consistently have genes or alleles with phylogenetically divergent origins relatively straightforward. We investigated the genetic origin of Phytophthora andina, an increasingly common pathogen of Andean crops Solanum betaceum, S. muricatum, S. quitoense, and several wild Solanum spp. It has been hypothesized that P. andina is a hybrid between the potato late blight pathogen P. infestans and another Phytophthora species. We tested this hypothesis by cloning four nuclear loci to obtain haplotypes and using these loci to infer the phylogenetic relationships of P. andina to P. infestans and other related species. Sequencing of cloned PCR products in every case revealed two distinct haplotypes for each locus in P. andina, such that each isolate had one allele derived from a P. infestans parent and a second divergent allele derived from an unknown species that is closely related but distinct from P. infestans, P. mirabilis, and P. ipomoeae. To the best of our knowledge, the unknown parent has not yet been collected. We also observed sequence polymorphism among P. andina isolates at three of the four loci, many of which segregate between previously described P. andina clonal lineages. These results provide strong support that P. andina emerged via hybridization between P. infestans and another unknown Phytophthora species also belonging to Phytophthora clade 1c.
Introduction
Emerging plant pathogens threaten natural ecosystems, food security, and commercial interests. Major mechanisms underlying plant pathogen emergence include host range expansion and host jumps [1], [2]. Recently, these events have largely been the result of migration or movement of pathogens or hosts into new geographic regions [3], [4], [5]. Another mechanism is hybridization between species or individuals [6]. Known hybrid plant pathogens include the alder pathogen Phytophthora alni [7], the poplar rust Melampsora×columbiana [8], the crucifer pathogen Verticillium longisporum [9], the onion pathogen Botrytis allii [10], [11], and Heterobasidion forest pathogens [12], [13]. Hybridization and introgression are also hypothesized to be behind the continued epidemic of Dutch elm disease in Europe [14]. Hybridization between a recently introduced exotic pathogen and a resident pathogen may allow rapid evolution and adaptation to new hosts or environments [14], [15], [16], [17], because hybridization introduces genetic variation that has already been “tested by selection” in the resident parental species [18]. The continuing global movement of plant pathogens may be creating opportunities for new and virulent hybrid pathogens to arise [15], [19].
Hybrid plant pathogens have traditionally been difficult to detect or confirm and have generally been investigated for their unusual morphology, pathogenicity, or other phenotypic characters and subsequently identified as hybrids [15], [19]. Modern molecular techniques are currently the gold standard for identifying hybrid pathogens, in particular the sequencing of nuclear loci for which genealogies can be constructed and ancestral and derived states inferred. Based on DNA sequences, hybrids have been identified when sampled individuals consistently have genes or alleles with phylogenetically divergent origins. In diploids or polyploids one may observe that alleles at any one locus are from divergent origins. However, in the case of introgression, when hybrid offspring are not sterile and can backcross to one or the other parental species or strains, the hybridization event may be more difficult to detect if limited DNA sequences are available. Modern molecular methods and especially whole genome sequencing will likely identify additional ‘atypical’ plant pathogens as being hybrids or as having introgressed genes from past hybridization events.
The oomycete pathogen Phytophthora infestans is one of the most widely known emerging plant pathogens. It initially emerged in the early 1840s in the United States and Europe and rapidly spread across potato-growing regions, leading to the Irish potato famine. It causes an aggressive disease of potato and tomato, and is still considered a major threat to global food security [20]. In the 1950s, a diverse and sexual population of P. infestans was found in the Toluca Valley of central Mexico, on commercial potatoes and then wild relatives of potato, leading to the conventional wisdom that this devastating pathogen evolved in association with the diverse tuber-bearing Solanum plant community in the central highlands of Mexico [21], [22]. This scenario is supported by the presence of two closely related species, P. mirabilis and P. ipomoeae, also found in the Toluca Valley [23], [24]. However, the center of origin and primary center of diversity of potatoes is in the Andean highlands of South America, thus a competing hypothesis is that the Andean highlands are the center of origin of P. infestans. This scenario is supported by a genealogical analysis of P. infestans using two mitochondrial DNA loci and one nuclear locus that showed old lineages of the pathogen in the Andes and not Mexico [25]. One of the arguments for an Andean origin of P. infestans has also been that the closest known relative of P. infestans, P. andina (formerly known as P. infestans sensu lato), is morphologically indistinguishable from P. infestans and is found only in the Andean highlands [25], [26]. Furthermore, several apparent lineages of P. infestans-like pathogens, all now classified as P. andina, has led to the suggestion that the Andes are a hotspot of Phytophthora diversification [25].
Phytophthora andina was originally discovered when a broader range of blighted Solanum species, particularly non-tuber-bearing species, were sampled in Ecuador [26], [27], [28]. These isolates were quickly identified as being genetically distinct from P. infestans despite their shared morphology. Specifically, they had new RFLP fingerprints (EC-2 and EC-3) and some EC-2 isolates had a distinct mtDNA haplotype, designated Ic [26], [28]. There are currently three distinct clonal lineages within P. andina, defined by RFLP fingerprint (also readily distinguished by AFLP), mitochondrial DNA haplotype, and mating type [26], [29]. Initially these lineages were referred to as P. infestans sensu lato, but recently they were all reclassified as P. andina Adler & Flier, sp. nov. [29]. Due to the genetic differences among the P. andina lineages, this species description is controversial [30]. Host use by P. infestans and P. andina in Ecuador overlap minimally, with P. infestans found infecting S. tuberosum (potato), S. lycopersicum (tomato), and close relatives (Solanum sections Petota, Lycopersicon, and Juglandifolium), and P. andina primarily infecting S. betaceum (section Pachyphylla), S. muricatum (section Basarthrum), S. quitoense (section Lasiocarpa), S. hispidum (section Torva), and species in the section Anarrhichomenum [29], [31]. Both species have been isolated from S. muricatum, S. quitoense, and S. ochranthum [26], [29]. Genetic variation within P. andina may be correlated with host use, suggesting the possibility of host specialization by P. andina lineages in the field [26], [29], [31].
P. infestans and P. andina share identical or nearly identical ITS sequences [29], [32], which is the traditional molecular marker used in species definition in oomycetes and fungi. P. mirabilis and P. ipomoeae also have identical or nearly identical ITS sequences to P. infestans [23]. These four closely related species, plus P. phaseoli, make up Phytophthora clade 1c [33], [34], [35]. Direct sequencing of nuclear genes in P. andina produced identical sequences in all P. andina isolates examined [29], [32], but also revealed high levels of heterozygosity with several of these sites differentiating P. infestans from P. mirabilis sequences [29], [32], [35]. Based on the observed heterozygous sites, it was hypothesized that P. andina may be a hybrid between P. infestans and P. mirabilis [32] or between P. infestans and another unspecified parent [29], [35], but the question was not investigated further. Resolution of the ancestry of P. andina, particularly whether it is of hybrid origin, is necessary for accurate interpretation of its population structure, evolution, and genetics. Here, we investigate the evolutionary history of P. andina and determine whether P. andina is in fact a hybrid of P. infestans and another species by cloning four nuclear loci to obtain haplotypes to infer the phylogenetic relationships of these alleles in relation to P. infestans and related species. Because of the considerable methodological and analytical challenges posed by both the large (∼240 Mb) and highly repetitive (∼74%) P. infestans genome [36] and the phasing of haplotypes in short-read, high throughput sequencing approaches, our work relied on traditional PCR cloning of coding sequences.
Results
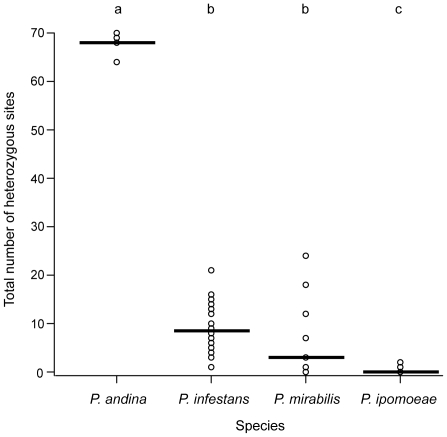
Every P. andina isolate was heterozygous at each of the four loci sequenced, as evidenced by double peaks in chromatograms from direct sequencing of PCR products. The total number of heterozygous sites summed across the four sequenced loci was significantly higher in P. andina compared to P. infestans, P. ipomoeae, and P. mirabilis (Figure 1; P<0.0001 for each comparison with P. andina by Tukey HSD). On average, P. andina isolates had greater than seven times more heterozygous sites than the other three species (Figure 1). Heterozygosity for indels was also observed in both regions of ypt1, btub and PITG11126, such that chromatograms showed overlapping PCR products of different lengths. Heterozygosity was observed in P. infestans, P. mirabilis, and in one isolate of P. ipomoeae, but with many fewer heterozygous sites per locus. When maximum likelihood gene trees were constructed using genotypes, P. andina could not be distinguished from P. infestans (Figure S1).
Figure 1. Total number of heterozygous sites across four nuclear loci sequenced in each isolate by species.
Lines represent mean values for each species and circles represent values of individual isolates (circles are overlapping). Lowercase letters above graph indicate significance, such that significantly different means (P<0.05) by Tukey's HSD are shown by different letters. The number of heterozygous sites observed in P. andina isolates was at least two to three times higher than isolates from the other species and P-values of comparisons with P. andina were less than 0.0001.
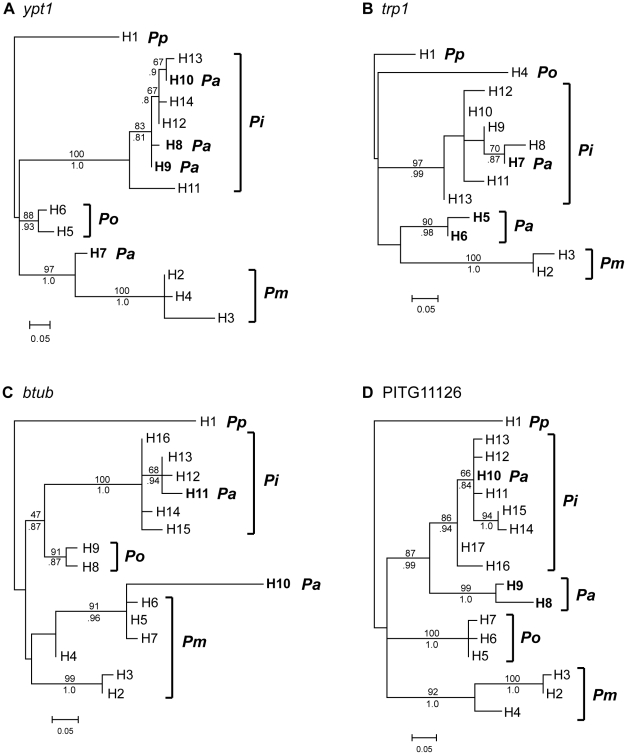
Sequencing of cloned PCR products in every case revealed two distinct haplotypes for P. andina isolates (Table 1). For btub, trp1, and PITG11126, one haplotype was identical to the most common P. infestans haplotype, found in isolates from the Andes, the United States, Mexico, and the United Kingdom (Table S1). For ypt1, P. andina isolates had one of two P. infestans haplotypes (H9 or H10) differing by 2 bp, with the exception of EC_3678, which had a P. infestans-like haplotype (H8) that differed from H9 at one nucleotide site (Table S2A). The second haplotype in each isolate was more or less distantly related to P. infestans depending on the locus (Figure 2). There were two versions of the non-P. infestans haplotypes for trp1 and PITG11126, which differed by 1 and 5 bp, respectively (Table S2). Much of the observed variation within P. andina segregates between the three P. andina lineages (Table 2).
Table 1. P. andina isolates and haplotypes of each locus sequenced.
| Haplotypesa | |||||||
| Isolate | Origin | Host | mtDNAb | ypt1 | trp1 | btub | PITG11126 |
| EC 3163 | Ecuador | Anarrichomenum group | Ic | H7/H9 | H5/H7 | H10/H11 | H8/H10 |
| EC 3189 | Ecuador | Anarrichomenum group | Ic | H7/H10 | H5/H7 | H10/H11 | H8/H10 |
| EC 3399 | Ecuador | Anarrichomenum group | Ia | H7/H10 | H5/H7 | H10/H11 | H8/H10 |
| EC 3510 | Ecuador | S. betaceum | Ia | H7/H9 | H6/H7 | H10/H11 | H9/H10 |
| EC 3540 | Ecuador | Anarrichomenum group | Ic | H7/H9 | H5/H7 | H10/H11 | H8/H10 |
| EC 3561 | Ecuador | S. quitoense | Ia | H7/H10 | H5/H7 | H10/H11 | H8/H10 |
| EC 3563 | Ecuador | S. quitoense | Ia | H7/H10 | H5/H7 | H10/H11 | H8/H10 |
| EC 3655 | Ecuador | S. hispidum | Ic | H7/H9 | H5/H7 | H10/H11 | H8/H10 |
| EC 3678 | Ecuador | Anarrichomenum group | Ic | H7/H8 | H5/H7 | H10/H11 | H8/H10 |
| EC 3780 | Ecuador | S. hispidum | Ic | H7/H9 | H5/H7 | H10/H11 | H8/H10 |
| EC 3818 | Ecuador | Anarrichomenum group | Ia | H7/H10 | H5/H7 | H10/H11 | H8/H10 |
| EC 3821 | Ecuador | Anarrichomenum group | Ia | H7/H10 | H5/H7 | H10/H11 | H8/H10 |
| EC 3836 | Ecuador | S. betaceum | Ia | H7/H9 | H6/H7 | H10/H11 | H9/H10 |
| EC 3860 | Ecuador | Torva group | Ic | H7/H9 | H5/H7 | H10/H11 | H8/H10 |
| EC 3864 | Ecuador | Torva group | Ic | H7/H9 | H5/H7 | H10/H11 | H8/H10 |
| POX 102 | Peru | S. betaceum | Ic | H7/H9 | H5/H7 | H10/H11 | H8/H10 |
| POX 103 | Peru | S. betaceum | Ic | H7/H9 | H5/H7 | H10/H11 | H8/H10 |
Figure 2. Maximum likelihood trees of haplotypes for each locus sequenced.
Loci are A. ypt1, B. trp1, C. btub, and D. PITG11126, sequenced in P. andina (Pa) and four other closely related species (Pi: P. infestans; Pm: P. mirabilis; Po: P. ipomoeae; and Pp: P. phaseoli). The haplotype designation is shown for each branch tip, corresponding to Tables 1, 2, and S1, S2, S3, S4, S5, S6, S7, S8. P. andina haplotypes are bolded. Trees have been rooted with P. phaseoli. Bootstrap support values obtained by maximum likelihood are shown above branches and Bayesian posterior probabilities are shown below branches. Values are not shown for branches that had less then 80% support/probability by both methods.
Table 2. Summary of sequence variation among P. andina lineages.
| P. andina lineagea | |||||
| Locus | Variable allele | Segregating sites | EC2 Ia | EC2 Ic | EC3 Ia |
| ypt1 | P. infestans | 2b | H10 | H8c, H9, H10c | H9 |
| trp1 | Non-P. infestans | 1 | H5 | H5 | H6 |
| PITG11126 | Non-P. infestans | 5 | H8 | H8 | H9 |
P. andina lineage is given as the RG57 genotype followed by the mtDNA haplotype.
Two differences between H9 and H10. H8 had one difference from H9 and three from H10.
One EC2 Ic isolate had P. infestans haplotype H10 and one had a P. infestans-like haplotype H8.
The non-P. infestans P. andina haplotypes (hereafter Pa-unknown) for ypt1, btub, and trp1, were related to P. mirabilis, but clearly distinct. Different possible phylogenetic relationships among Pa-unknown, P. infestans, P. mirabilis, and P. ipomoeae were statistically tested using the approximately unbiased (AU) test and Shimodaira–Hasegawa (SH) tests (Table S9). All trees were essentially star phylogenies for trp1, thus the AU test failed and no trees were rejected by the SH test. For ypt1, trees that did not contain a derived {Pa-unknown, P. mirabilis} clade had low P values by the AU and SH tests (0.05<P<0.1), but no trees were rejected at the P = 0.05 level. For btub, trees with the complex {Pa-unknown, P. mirabilis} clade had high P values. But one tree with monophyletic P. mirabilis as sister species to Pa-unknown was also not rejected, as well as two trees in which P. infestans and P. ipomoeae formed a derived clade (P>0.1 by the AU test, 0.05<P<0.1 by the SH test). Species relationships were qualitatively different for the PITG11126 locus. The Pa-unknown haplotype was more closely related to P. infestans than P. mirabilis (Fig. 2D). Unlike the other loci, sites in PITG11126 that differed between P. infestans and P. phaseoli, P. mirabilis, and P. ipomoeae were in the P. infestans state in P. andina (Table S2D). The AU test rejected all trees that did not include a derived {Pa-unknown, P. infestans} clade or a derived {Pa-unknown, P. ipomoeae} clade.
Discussion
We tested the hypothesis that Phytophthora andina is a hybrid pathogen and found that it is a hybrid between P. infestans and an unknown species that is closely related but distinct from P. mirabilis and P. ipomoeae. Cloning and sequencing of four nuclear loci clearly shows that P. andina isolates have one allele derived from a P. infestans parent and a second divergent allele from the unknown species. The P. infestans haplotypes found in P. andina appear to be common worldwide, found in North and South America, Europe, and Asia. Because P. andina has a different host range than P. infestans and the three P. andina lineages may have different host ranges themselves [26], [29], [31], it is probable that hybridization led to host range expansion or shifts.
Host shifts are likely to require several rapid genetic changes, but P. andina may have been in a unique position to undergo the necessary changes and rapidly adapt to new hosts. First, hybridization may facilitate adaptation to a new environment by rapidly introducing genetic variation, and not random variation but rather a complement of alleles that have been subjected to selection in the parental species [18]. Second, P. infestans has a genome structure which may have contributed to its ability to rapidly evolve virulence to resistant potato varieties in the near absence of sexual reproduction [36]. In particular, P. infestans has a very large genome with expanded repeat-rich gene-sparse regions where pathogenicity effectors, genes involved in virulence and host range, are primarily located. Comparisons to the genome sequences of two distantly related Phytophthora species show considerable expansion of effector genes in P. infestans and suggest that the repeat-rich gene-sparse regions are highly dynamic, exhibiting gene duplications and gene loss by tandem duplication, non-allelic homologous recombination, and pseudogenization. P. ipomoeae, P. mirabilis, and P. phaseoli have similar genome structures to P. infestans and comparisons among these species show greater gene copy number variation and presence/absence polymorphisms in the repeat-rich regions compared to the gene-dense repeat-poor regions where core orthologs are found [37]. The repeat-rich regions are also enriched in genes induced during infection. P. infestans is also known to exhibit aneuploidy, particularly in the clonal lineages that dominate much of its current geographic distribution [38], [39], [40]. Thus, P. andina had a potential mechanism for rapid change in its genic and allelic composition following hybridization. Different evolutionary paths taken by hybrid progeny could also explain the genetic variation observed within P. andina.
We cloned parental haplotypes at different frequencies from P. andina isolates for several loci, and while it is possible that P. andina is tetraploid or aneuploid and that the haplotypes are actually present in P. andina at different frequencies, it is more likely that there is a bias in the efficiency of the primers between haplotypes. The primers were designed from P. infestans and may contain mismatches with the sequences of the other Phytophthora clade 1c species. Cloned recombinant haplotypes are likely to be chimeras from PCR error, as PCR conditions were not optimized to reduce these sorts of errors [9], [41]. Illumina sequence reads from P. andina graciously shared with us [S. Kamoun personal communication, [37]] were examined, but these data could not be analyzed because depth of coverage was not sufficient to call heterozygous sites with high confidence [42] and the short reads were problematic for determining haplotype phase. More extensive deep sequencing may elucidate the genome composition of P. andina, particularly using sequencing technologies that generate longer read lengths. Genome-wide analysis would also allow for examination of alternate hypotheses for the observed pattern of phylogenetically distinct alleles at each locus, including mechanisms such as gene duplication or horizontal gene transfer.
The hybrid P. alni, a lethal pathogen of alder, is another example of hybridization between closely related Phytophthora species resulting in a host range expansion or shift [7], [43], [44]. Additional examples include hybrid Phytophthora pathogens causing disease on loquat trees in Peru and Taiwan [45], [46] and in ornamental nurseries where exotic pathogens are brought together under artificial conditions [47], [48], [49]. Host range expansions by Phytophthora hybrids have been documented for both these naturally occurring hybrids and for hybrids created in the laboratory [6]. P. infestans and P. mirabilis are outcrossers, occur in sympatry on different hosts in the Toluca Valley of central Mexico, and are thought to have evolved by sympatric speciation via host shifts [21], thus the potential for interspecific mating between these species has been investigated. Population genetic analysis suggested some gene flow between P. infestans and P. mirabilis populations [21], [50]. However, initial crosses between P. infestans and P. mirabilis produced hybrid offspring that were largely unable to infect either host group and had poor viability [51]. Nevertheless, a recent cross between a P. infestans isolate (virulent on potato and tomato) and a P. mirabilis isolate (virulent on Mirabilis jalapa), both from central Mexico, produced F1 and F2 progeny that were pathogenic on tomato and one F2 isolate had an expanded host range, able to infect all parental hosts [52]. Interestingly, the ability to infect tomato segregated as a dominant single locus trait in this cross. Sexual crosses have also been attempted between P. infestans and P. andina [53]. A limited number of viable progeny were obtained, but further crosses with these progeny were not successful. Here we examined only four nuclear loci, yet we observed both parental haplotypes at each locus for each isolate, which suggests that these P. andina lineages were not the result of backcrosses.
Reproductive barriers between closely related species are usually stronger when the species occur in sympatry than when the species have evolved in allopatry [54]. There is not yet strong evidence for this pattern specifically for fungal or oomycete pathogens, in part because the native distributions of many of these pathogens are not well known. Essentially, it is not clear where pathogens evolved and therefore whether sister species evolved in sympatry or allopatry. On the other hand, host shift speciation may also occur without intrinsic reproductive barriers when pathogens must sexually reproduce on their host [1]. It has nevertheless been hypothesized that Phytophthora hybrids are offspring of two exotic species or of an exotic and resident species [6], [48]. If this pattern does hold true for Phytophthora, it would suggest that at least one of the P. andina parent species is introduced and did not co-evolve with the Andean Solanum host community [52].
Synthetic hybridization experiments have been used to recreate hybrids, to a certain extent, observed in the wild in order to validate the hybrid origin of species (e.g. [18], [55], [56]). For P. andina, one of the hybrid parents remains unknown and so these experiments remain to be conducted, pending the collection and identification of the unknown parent. However, locating this species could be challenging. Disease epidemics caused by P. mirabilis and P. ipomoeae are infrequent and incidence of infection is low (N. J. Grünwald, personal observation). This would probably also be true of other relatives of P. infestans that infect wild and patchy host populations. The unknown species suggests that there is undiscovered diversity in Phytophthora clade 1c that may be found in the Andes, although the evolutionary origin of this species in relationship to P. infestans and its Mexican sister species remains unclear.
Phytophthora diseases are currently being managed with fungicides or, preferably, resistant plant varieties when available. Global movement and interspecific hybridization of plant pathogens multiply the considerable challenges already faced by crop breeding programs and growers trying to manage disease. The global movement of plant pathogens may increase the risk of formation of novel hybrid Phytophthora pathogens if hybridization is more likely between previously allopatric species brought together by migration events. Understanding the ecological and genetic processes that result in hybrid pathogens with novel host ranges or virulence, as appears to be the case for P. andina, should suggest conditions under which special vigilance and increased monitoring for emerging pathogens is warranted.
Materials and Methods
Isolates
Isolates or genomic DNA of clade 1c species were kindly provided by several researchers (Table S1). P. andina was distinguished from P. infestans based on the host from which isolates were collected, an apparently complementary mating system, AFLP markers, and sequence differences at in Ras intron 1 gene [29]. Isolates were received as genomic DNA or maintained on Rye A agar [57]. Total genomic DNA was extracted from mycelium grown in pea broth (P. infestans and P. andina) or clarified V8 (other species) using the FastDNA SPIN kit (MP Biomedicals LLC, Solon, OH).
mtDNA RFLP
Mitochondrial DNA haplotype sensu Griffith and Shaw [58] was determined for each P. andina isolate by amplifying and digesting the P2 and P4 regions as described by Griffith and Shaw.
Nuclear gene sequencing
The following genes known to contain variation within and among Phytophthora species were chosen for sequencing: the Ras gene ypt1 [25], [59]; trp1, btub, [33], [35], [60], and an additional gene that also exhibited variation within and among 1c species in preliminary sequencing (PITG11126, [61]). Primers for ypt1 were from Gomez-Alpizar et al. [25]. These amplify a fragment of the 5′ untranslated region of the gene (intron1, IR) and a larger downstream fragment including both exons and introns (RAS). These two fragments were concatenated for analysis. Primers for the other genes were designed from the P. infestans genome sequence [36] (Table 3). All isolates were directly sequenced from the PCR product. For each locus, two to six P. andina isolates were selected for cloning of the PCR product to obtain haplotypes (Table 3). For ypt1, 6 isolates were additionally cloned across the entire region to obtain haplotypes across both amplified fragments. Several P. infestans and P. mirabilis isolates with heterozygous sites were also cloned to obtain haplotypes. When a chromatogram indicated that the isolate was heterozygous for an indel at a sequenced locus, the preliminary sequence was determined using the sequence obtained from each primer up to the indel (i.e. sequence was inferred from one strand). Then, isolates representing each inferred genotype were cloned to obtain haplotypes and confirm the genotype inferred from direct sequencing. Specific cloning and sequencing methods and protocols differed among the laboratories (Fry, Grünwald, Restrepo) where they were performed and are available upon request (see also [60], [62]).
Table 3. Loci sequenced and P. andina isolates cloned.
| Locus | Lengtha | Pa isolates cloned | Primers | Ta b |
| ypt1 (IR) | 227 | EC 3399, EC 3561, EC 3563, EC 3818, EC 3821, EC 3189 | IRF – TTGCAGCACAACCCAAGACG; IRR – TGCACGTACTATTCGGGGTTC | 61C |
| ypt1 (RAS) | 544 | EC 3163, EC 3399, EC 3563, EC 3821 | RASF – CGTGTCTGCTTCTCCGTTTCG; RASR – CCAGGCTTTCGGCAAATTCC | 61C |
| ypt1 (IRRAS) | 987 | EC 3163, EC 3510, EC 3563, EC 3655, EC 3818, POX 102 | IRF – TTGCAGCACAACCCAAGACG; RASR – CCAGGCTTTCGGCAAATTCC | 61C |
| trp1 | 814 | EC 3818, POX 102 | F3 – GGGTAACATCCTGGAGGAGA; R3 - TCGTACTTGACCACGTCTGC | 63C |
| beta-tubulin | 1592 | EC 3836, POX 102 | F1 – GTCCGAATTCTCCTCAGAGC; F2 – CGCTATCGGTACACCAGCTT; F3b – ACCATAACGAAGGGAAAGG; R1 – GATGCCAAGCCACTAACCTC; R2 – CCTCATTGTCCAGGCACAT | 58C |
| PITG11126 | 788 | EC 3163, EC 3510, POX 102 | F1 – GGGGACTTCGCTGTTTGTTA; R1 – ATGTTCATGTACGGCTGACG | 59C |
Length of the multiple sequence alignment across all sequenced isolates.
Optimal annealing temperature of primers for PCR based on experience in the Grünwald lab.
The number of heterozygous sites was summed across the four sequenced loci for each isolate for which sequence was obtained for all loci. This total per isolate was used to examine differences in the number of heterozygous sites among species using analysis of variance, implemented in R 2.11.1 for Mac OS. Post-hoc multiple comparisons were conducted using Tukey's HSD.
Haplotype inference
More than two haplotypes were often obtained from cloning P. andina isolates (Tables S3, S4, S5, S6, S7, S8). Haplotypes that were common across isolates were inferred to be the non-recombinant (parental) haplotypes. Other haplotypes cloned from P. andina were recombinants of the two parental haplotypes and were not included in the analyzed data sets. For some loci and P. andina isolates, the inferred parental haplotypes were cloned at unequal frequencies (Tables S3 and S6, S7, S8).
Haplotypes for each P. infestans isolate were inferred from genotypes using PHASE v2.1 [63], [64]. Selected isolates were cloned to confirm inferred haplotypes. When the cloned sequences did not match the inferred haplotypes because the genotype was a combination of three alleles (btub in two Colombian isolates), all three alleles were included in the data set. When the inferred haplotypes were recovered by cloning, but additional recombinant haplotypes were also cloned, the recombinant haplotypes were not included in the analyzed sequences. All haplotypes included in the analysis were submitted to Genbank (Accession numbers JF919525–JF919609). Recombinant haplotypes are provided as supporting data.
Phylogenetic methods
Sequences were aligned using ClustalW [65]. Sequence alignments were collapsed to unique haplotypes, removing invariable sites and indels using Map in SNAP WORKBENCH [66], [67]. Jmodeltest [68] was used to estimate a nucleotide substitution model using maximum-likelihood trees estimated for each model and model selection by AIC.
Maximum likelihood (ML) gene trees were inferred using PhyML [69], implemented in Geneious 5.0.2 (Biomatters Ltd.), using the substitution model selected by jmodeltest (HKY for trp1, GTR for ypt1, btub, and PITG11126). The transition/transversion ratio, proportion of invariable sites, and gamma distribution parameter were estimated from the data in PhyML using 6 rate categories. Data sets were bootstrapped using 500 samples.
Gene trees were also inferred using MrBayes [70], implemented in Geneious 5.0.2. The same nucleotide substitution model was used as for PhyML. MCMC used 4 heated chains of 1.1×106 steps sampled every 200 steps. Posterior trees were summarized excluding the initial 500 trees as burn-in. The default priors were used.
The approximately unbiased (AU) test of Shimodaira [71] and Shimodaira–Hasegawa (SH) test [72] was used to test among tree topologies using the program CONSEL [73]. We tested 15 topologies for each locus, in which the phylogenetic relationships among P. infestans, P. ipomoeae, P. mirabilis, and the non-P. infestans parent of P. andina (Pa-unknown) were varied. All trees were rooted with P. phaseoli. Site likelihoods were estimated in PhyML as described above with the exception that the topology was set to the input tree and not optimized. Three additional trees were tested for btub, for which ML and Bayesian trees showed P. mirabilis forming two clades with Pa-unknown embedded in one of these clades. Monophyly of P. mirabilis was forced in 15 trees while three additional trees tested the relative relationship of the inferred complex {P. mirabilis, Pa-unknown} clade to P. infestans and P. ipomoeae.
Supporting Information
Maximum likelihood trees of genotypes for each locus sequenced in P. andina and four other closely related species. Loci are A. ypt1, B. trp1, C. btub, and D. PITG11126. Genotypes are shown as combinations of haplotypes. Bootstrap support values obtained by maximum likelihood are shown above branches.
(TIF)
Isolates and haplotype designations for each locus sequenced.
(DOCX)
Variable sites at each sequenced locus: A. ypt1 , B. trp1 , C. btub , and D. PITG11126. For each locus, the consensus sequence across clade 1c species is shown and identity to this sequence indicated with a dot. Haplotype numbers for each locus correspond with those in Tables 1, 2, and S1, and Figure 2. Site numbers indicate position in multispecies alignment. Indels are not included; see Tables S3, S4, S5, S6, S7, S8 for indels that are heterozygous in P. andina. Sites with shared nucleotides between the non-P. infestans haplotype in P. andina and P. mirabilis, P. ipomoeae, or P. infestans are shown in bold.
(DOCX)
P. andina ypt1 haplotypes obtained from cloning for the full region (IR through RAS). Italicized sites are between sequenced regions and were not included in the analysis.
(DOCX)
P. andina haplotypes obtained from cloning just the IR region, including those shown in Table S3.
(DOCX)
P. andina haplotypes obtained from cloning just the RAS region, including those shown in Table S3.
(DOCX)
P. andina trp1 haplotypes obtained from cloning.
(DOCX)
P. andina btub haplotypes obtained from cloning.
(DOCX)
P. andina PITG11126 haplotypes obtained from cloning.
(DOCX)
Results of AU and SH tests of tree topologies for ypt1 , btub , and PITG11126.
(DOCX)
Acknowledgments
We thank Valerie Fieland, Karan Fairchild, Kim Henslee, and Caroline Press for excellent technical support. Mention of trade names or commercial products in this manuscript are solely for the purpose of providing specific information and do not imply recommendation or endorsement.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Financial support was provided by the United States Department of Agriculture–Agricultural Research Service (USDA-ARS) CRIS 5358-22000-034-00. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Giraud T, Gladieux P, Gavrilets S. Linking the emergence of fungal plant diseases with ecological speciation. Trends in Ecology & Evolution. 2010;25:387–395. doi: 10.1016/j.tree.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stukenbrock EH, McDonald BA. The origins of plant pathogens in agro-ecosystems. Annual Review of Phytopathology. 2008;46:75–100. doi: 10.1146/annurev.phyto.010708.154114. [DOI] [PubMed] [Google Scholar]
- 3.Anderson PK, Cunningham AA, Patel NG, Morales FJ, Epstein PR, et al. Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends in Ecology & Evolution. 2004;19:535–544. doi: 10.1016/j.tree.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 4.Brasier CM. The biosecurity threat to the UK and global environment from international trade in plants. Plant Pathology. 2008;57:792–808. [Google Scholar]
- 5.Brown JKM, Hovmoller MS. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science. 2002;297:537–541. doi: 10.1126/science.1072678. [DOI] [PubMed] [Google Scholar]
- 6.Ersek T, Nagy ZA. Species hybrids in the genus Phytophthora with emphasis on the alder pathogen Phytophthora alni: a review. European Journal of Plant Pathology. 2008;122 [Google Scholar]
- 7.Brasier CM, Cooke DEL, Duncan JM. Origin of a new Phytophthora pathogen through interspecific hybridization. Proceedings of the National Academy of Sciences, USA. 1999;96:5878–5883. doi: 10.1073/pnas.96.10.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newcombe G, Stirling B, McDonald S, Bradshaw HD., Jr Melampsora×columbiana, a natural hybrid of M. medusae and M. occidentalis. Mycological Research. 2000;104:261–274. [Google Scholar]
- 9.Inderbitzin P, Davis RM, Bostock RM, Subbarao KV. The ascomycete Verticillium longisporum is a hybrid and a plant pathogen with an expanded host range. PLoS ONE. 2011;6:e18260. doi: 10.1371/journal.pone.0018260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nielsen K, Yohalem DS. Origin of a polypoloid Botrytis pathogen through interspecific hybridization between Botrytis aclada and B. byssoidea. Mycologia. 2001;93:1064–1071. [Google Scholar]
- 11.Staats M, van Baarlen P, van Kan JAL. Molecular phylogeny of the plant pathogenic genus Botrytis and the evolution of host specificity. Mol Biol Evol. 2005;22:333–346. doi: 10.1093/molbev/msi020. [DOI] [PubMed] [Google Scholar]
- 12.Garbelotto M, Ratcliff A, Bruns TD, Cobb FW, Otrosina WJ. Use of taxon-specific competitive-priming PCR to study host specificity, hybridization, and intergroup gene flow in intersterility groups of Heterobasidion annosum. Phytopathology. 1996;86:543–551. [Google Scholar]
- 13.Gonthier P, Nicolotti G, Linzer R, Guglielmo F, Garbelotto M. Invasion of European pine stands by a North American forest pathogen and its hybridization with a native interfertile taxon. Molecular Ecology. 2007;16:1389–1400. doi: 10.1111/j.1365-294X.2007.03250.x. [DOI] [PubMed] [Google Scholar]
- 14.Brasier CM. Rapid evolution of introduced plant pathogens via interspecific hybridization. BioScience. 2001;51:123–133. [Google Scholar]
- 15.Brasier CM. The rise of the hybrid fungi. Nature. 2000;405:134–135. doi: 10.1038/35012193. [DOI] [PubMed] [Google Scholar]
- 16.Giraud T, Refregier G, Le Gac M, de Vienne DM, Hood ME. Speciation in fungi. Fungal Genetics and Biology. 2008;45:791–802. doi: 10.1016/j.fgb.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Schardl CL, Craven KD. Interspecific hybridization in plant-associated fungi and oomycetes: a review. Molecular Ecology. 2003;12:2861–2873. doi: 10.1046/j.1365-294x.2003.01965.x. [DOI] [PubMed] [Google Scholar]
- 18.Reiseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, et al. Major ecological transitions in wild sunflowers facilitated by hybridization. Science. 2003;301:1211–1216. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- 19.Yan S, Liu H, Mohr TJ, Jenrette J, Chiodini R, et al. Role of recombination in the evolution of the model plant pathogen Pseudomonas syringae pv. tomato DC3000, a very atypical tomato strain. Applied and Environmental Microbiology. 2008;74:3171–3181. doi: 10.1128/AEM.00180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pennisi E. Armed and Dangerous. Science. 2010;327:804–805. doi: 10.1126/science.327.5967.804. [DOI] [PubMed] [Google Scholar]
- 21.Grünwald NJ, Flier WG. The biology of Phytophthora infestans at its center of origin. Annual Review of Phytopathology. 2005;43:171–190. doi: 10.1146/annurev.phyto.43.040204.135906. [DOI] [PubMed] [Google Scholar]
- 22.Grünwald NJ, Flier WG, Sturbaum AK, Garay-Serrano E, van den Bosch TBM, et al. Population Structure of Phytophthora infestans in the Toluca Valley Region of Central Mexico. Phytopathology. 2001;91:882–890. doi: 10.1094/PHYTO.2001.91.9.882. [DOI] [PubMed] [Google Scholar]
- 23.Flier WG, Grünwald NJ, Kroon LPNM, van den Bosch TBM, Garay-Serrano E, et al. Phytophthora ipomoeae sp. nov., a new homothallic species causing leaf blight on Ipomoea longipedunculata in the Toluca Valley of central Mexico. Mycological Research. 2002;106:848–856. [Google Scholar]
- 24.Galindo J, Hohl HR. Phytophthora mirabilis, a new species of Phytophthora. Sydowia. 1985;38:87–96. [Google Scholar]
- 25.Gomez-Alpizar L, Carbone I, Ristaino JB. An Andean origin of Phytophthora infestans inferred from mitochondrial and nuclear gene genealogies. Proceedings of the National Academy of Sciences, USA. 2007;104:3306–3311. doi: 10.1073/pnas.0611479104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adler NE, Erselius LJ, Chacon MG, Flier WG, Ordonez ME, et al. Genetic diversity of Phytophthora infestans sensu lato in Ecuador provides new insight into the origin of this important plant pathogen. Phytopathology. 2004;94:154–162. doi: 10.1094/PHYTO.2004.94.2.154. [DOI] [PubMed] [Google Scholar]
- 27.Oyarzun PJ, Ordonez ME, Forbes GA. First report of Phytophthora infestans A2 mating type in Ecuador. Plant Disease. 1997;81:311. doi: 10.1094/PDIS.1997.81.3.311C. [DOI] [PubMed] [Google Scholar]
- 28.Ordonez ME, Hohl HR, Velasco JA, Ramon MP, Oyarzun PJ, et al. A novel population of Phytophthora, similar to P. infestans, attacks wild Solanum species in Ecuador. Phytopathology. 2000;90:197–202. doi: 10.1094/PHYTO.2000.90.2.197. [DOI] [PubMed] [Google Scholar]
- 29.Oliva RF, Kroon LPNM, Chacon G, Flier WG, Ristaino JB, et al. Phytophthora andina sp. nov., a newly identified heterothallic pathogen of solanaceous hosts in the Andean highlands. Plant Pathology. 2010;59:613–625. [Google Scholar]
- 30.Cárdenas M, Tabima J, Fry WE, Grünwald NJ, Bernal A, et al. Defining species boundaries in the genus Phytophthora: The case of Phytophthora andina. Plant Pathology. 2011 In press. [Google Scholar]
- 31.Oliva RF, Chacon G, Cooke DEL, Lees AK, Forbes GA. Is Phytophthora infestans a good taxonomist? Host recognition and co-evolution in the Phytophthora/Solanum interaction. Acta Horticulturae. 2007;745:465–471. [Google Scholar]
- 32.Gomez-Alpizar L, Hu C-H, Oliva R, Forbes G, Ristaino JB. Phylogenetic relationships of Phytophthora andina, a new species from the highlands of Ecuador that is closely related to the Irish potato famine pathogen Phytophthora infestans. Mycologia. 2008;100:590–602. doi: 10.3852/07-074r1. [DOI] [PubMed] [Google Scholar]
- 33.Blair JE, Coffey MD, Park SY, Geiser DM, Kang S. A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genetics and Biology. 2008;45:266–277. doi: 10.1016/j.fgb.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 34.Cooke DEL, Drenth A, Duncan JM, Wagels G, Brasier CM. A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genetics and Biology. 2000;30:17–32. doi: 10.1006/fgbi.2000.1202. [DOI] [PubMed] [Google Scholar]
- 35.Kroon LP, Bakker FT, van den Bosch GB, Bonants PJ, Flier WG. Phylogenetic analysis of Phytophthora species based on mitochondrial and nuclear DNA sequences. Fungal Genetics and Biology. 2004;41:766–782. doi: 10.1016/j.fgb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 37.Raffaele S, Farrer RA, Cano LM, Studholme DJ, MacLean D, et al. Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science. 2010;330:1540–1543. doi: 10.1126/science.1193070. [DOI] [PubMed] [Google Scholar]
- 38.Catal M, King L, Tumbalam P, Wiriyajitsomboon P, Kirk WW, et al. Heterokaryotic nuclear conditions and a heterogeneous nuclear population are observed by flow cytometry in Phytophthora infestans. Cytometry Part A. 2010;77A:769–775. doi: 10.1002/cyto.a.20888. [DOI] [PubMed] [Google Scholar]
- 39.Tooley PW, Therrien CD. Cytophotometric determination of the nuclear DNA content of 23 Mexican and 18 non-Mexican isolates of Phytophthora infestans. Experimental Mycology. 1987;11:19–26. [Google Scholar]
- 40.van der Lee T, Testa A, Robold A, van 't Klooster J, Govers F. High-density genetic linkage maps of Phytophthora infestans reveal trisomic progeny and chromosomal rearrangements. Genetics. 2004;167:1643–1661. doi: 10.1534/genetics.104.029652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beser J, Hagblom P, Fernandez V. Frequent in vitro recombination in internal transcribed spacers 1 and 2 during genotyping of Pneumocystis jirovecii. Journal of Clinical Microbiology. 2007;45:881–886. doi: 10.1128/JCM.02245-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, et al. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nature Review Genetics. 2011;12:499–510. doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- 43.Ioos R, Andrieux A, Marcais B, Frey P. Genetic characterization of the natural hybrid species Phytophthora alni as inferred from nuclear and mitochondrial DNA analyses. Fungal Genetics and Biology. 2006;43:511–529. doi: 10.1016/j.fgb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 44.Brasier CM, Kirk SA, Delcan J, Cooke DEL, Jung T, et al. Phytophthora alni sp. nov. and its variants: designation of emerging heteroploid hybrid pathogens spreading on Alnus trees. Mycological Research. 2004;108:1172–1184. doi: 10.1017/s0953756204001005. [DOI] [PubMed] [Google Scholar]
- 45.Hurtado-Gonzales OP, Aragon-Caballero LM, Flores-Torres JG, Man In'T Veld W, Lamour KH. Molecular comparison of natural hybrids of Phytophthora nicotianae and P. cactorum infecting loquat trees in Peru and Taiwan. Mycologia. 2009;1010:496–502. doi: 10.3852/08-079. [DOI] [PubMed] [Google Scholar]
- 46.Man In'T Veld WA. First report of natural hybrids of Phytophthora nicotianae and P. cactorum on loquat in Taiwan. Plant Disease. 2001;85:98. doi: 10.1094/PDIS.2001.85.1.98B. [DOI] [PubMed] [Google Scholar]
- 47.Bonants PJM, Hagenaar-de Weerdt M, Man In'T Veld WA, Baayen RP. Molecular characterization of natural hybrids of Phytophthora nicotianae and P. cactorum. Phytopathology. 2000;90:867–874. doi: 10.1094/PHYTO.2000.90.8.867. [DOI] [PubMed] [Google Scholar]
- 48.Man In'T Veld WA, de Cock AWAM, Summerbell RC. Natural hybrids of resident and introduced Phytophthora species proliferating on multiple new hosts. European Journal of Plant Pathology. 2007;117:25–33. [Google Scholar]
- 49.Man In'T Veld WA, Veenbaas-Rijks WJ, Ilieva E, de Cock AWAM, Bonants PJM, et al. Natural hybrids of Phytophthora nicotianae and P. cactorum demonstrated by isozyme analysis and random amplified polymorphic DNA. Phytopathology. 1998;88:922–929. doi: 10.1094/PHYTO.1998.88.9.922. [DOI] [PubMed] [Google Scholar]
- 50.Flier WG, Grünwald NJ, Kroon LPNM, Sturbaum AK, van den Bosch TBM, et al. The population structure of Phytophthora infestans from the Toluca Valley of Central Mexico suggests genetic differentiation between populations from cultivated potato and wild Solanum spp. Phytopathology. 2003;93:382–390. doi: 10.1094/PHYTO.2003.93.4.382. [DOI] [PubMed] [Google Scholar]
- 51.Goodwin SB, Legard DE, Smart CD, Levy M, Fry WE. Gene flow analysis of molecular markers confirms that Phytophthora mirabilis and P. infestans are separate species. Mycologia. 1999;91:796–810. [Google Scholar]
- 52.Kroon LPNM. PhD Thesis: The genus Phytophthora; phylogeny, speciation and host specificity. Wageningen, Netherlands: Wageningen University; 2010. 184 [Google Scholar]
- 53.Oliva Perez RF. PhD Thesis: Occurance of sympatric Phytophthora species in the highland of Ecuador. Zurich, Switzerland: Swiss Federal Institute of Technology; 2009. [Google Scholar]
- 54.Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer Associates, Inc; 2004. [Google Scholar]
- 55.Rieseberg LH, Sinervo B, Linder CR, Ungerer MC, Arias DM. Role of gene interactions in hybrid speciation: Evidence from ancient and experimental hybrids. Science. 1996;272:741–745. doi: 10.1126/science.272.5262.741. [DOI] [PubMed] [Google Scholar]
- 56.Mavarez J, Salazar CA, Bermingham E, Salcedo C, Jiggins CD, et al. Speciation by hybridization in Heliconius butterflies. Nature. 2006;441:868–871. doi: 10.1038/nature04738. [DOI] [PubMed] [Google Scholar]
- 57.Caten CE, Jinks JL. Spontaneous variability of single isolates of Phytophthora infestans. I. Cultural variation. Canadian Journal of Botany/Revue Canadienne de Botanique. 1968;46:329–348. [Google Scholar]
- 58.Griffith GW, Shaw DS. Polymorphisms in Phytophthora infestans: Four mitochondrial haplotypes are detected after PCR amplification of DNA from pure cultures or from host lesions. Applied and Environmental Microbiology. 1998;64:4007–4014. doi: 10.1128/aem.64.10.4007-4014.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Y, Roxby R. Characterization of a Phytophthora infestans gene involved in vesicle transport. Gene. 1996;181:89–94. doi: 10.1016/s0378-1119(96)00469-6. [DOI] [PubMed] [Google Scholar]
- 60.Goss EM, Carbone I, Grünwald NJ. Ancient isolation and independent evolution of the three clonal lineages of the exotic sudden oak death pathogen Phytophthora ramorum. Molecular Ecology. 2009;18:1161–1174. doi: 10.1111/j.1365-294X.2009.04089.x. [DOI] [PubMed] [Google Scholar]
- 61.Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 62.Cardenas M, Grajales A, Sierra R, Rojas A, Gonzalez-Almario A, et al. Genetic diversity of Phytophthora infestans in the Northern Andean region. BMC Genetics. 2011;12:23. doi: 10.1186/1471-2156-12-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction. American Journal of Human Genetics. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. American Journal of Human Genetics. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aylor DL, Price EW, Carbone I. SNAP: Combine and Map modules for multilocus population genetic analysis. Bioinformatics. 2006;22:1399–1401. doi: 10.1093/bioinformatics/btl136. [DOI] [PubMed] [Google Scholar]
- 67.Price EW, Carbone I. SNAP: workbench management tool for evolutionary population genetic analysis. Bioinformatics. 2005;21:402–404. doi: 10.1093/bioinformatics/bti003. [DOI] [PubMed] [Google Scholar]
- 68.Posada D. jModelTest: Phylogenetic model averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 69.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 70.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 71.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Systematic Biology. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- 72.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Molecular Biology and Evolution. 1999;16:1114–1116. [Google Scholar]
- 73.Shimodaira H, Hasegawa M. CONSEL: For assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maximum likelihood trees of genotypes for each locus sequenced in P. andina and four other closely related species. Loci are A. ypt1, B. trp1, C. btub, and D. PITG11126. Genotypes are shown as combinations of haplotypes. Bootstrap support values obtained by maximum likelihood are shown above branches.
(TIF)
Isolates and haplotype designations for each locus sequenced.
(DOCX)
Variable sites at each sequenced locus: A. ypt1 , B. trp1 , C. btub , and D. PITG11126. For each locus, the consensus sequence across clade 1c species is shown and identity to this sequence indicated with a dot. Haplotype numbers for each locus correspond with those in Tables 1, 2, and S1, and Figure 2. Site numbers indicate position in multispecies alignment. Indels are not included; see Tables S3, S4, S5, S6, S7, S8 for indels that are heterozygous in P. andina. Sites with shared nucleotides between the non-P. infestans haplotype in P. andina and P. mirabilis, P. ipomoeae, or P. infestans are shown in bold.
(DOCX)
P. andina ypt1 haplotypes obtained from cloning for the full region (IR through RAS). Italicized sites are between sequenced regions and were not included in the analysis.
(DOCX)
P. andina haplotypes obtained from cloning just the IR region, including those shown in Table S3.
(DOCX)
P. andina haplotypes obtained from cloning just the RAS region, including those shown in Table S3.
(DOCX)
P. andina trp1 haplotypes obtained from cloning.
(DOCX)
P. andina btub haplotypes obtained from cloning.
(DOCX)
P. andina PITG11126 haplotypes obtained from cloning.
(DOCX)
Results of AU and SH tests of tree topologies for ypt1 , btub , and PITG11126.
(DOCX)