Abstract
Previous expression quantitative trait loci (eQTL) studies have performed genetic association studies for gene expression, but most of these studies examined lymphoblastoid cell lines from non-diseased individuals. We examined the genetics of gene expression in a relevant disease tissue from chronic obstructive pulmonary disease (COPD) patients to identify functional effects of known susceptibility genes and to find novel disease genes. By combining gene expression profiling on induced sputum samples from 131 COPD cases from the ECLIPSE Study with genomewide single nucleotide polymorphism (SNP) data, we found 4315 significant cis-eQTL SNP-probe set associations (3309 unique SNPs). The 3309 SNPs were tested for association with COPD in a genomewide association study (GWAS) dataset, which included 2940 COPD cases and 1380 controls. Adjusting for 3309 tests (p<1.5e-5), the two SNPs which were significantly associated with COPD were located in two separate genes in a known COPD locus on chromosome 15: CHRNA5 and IREB2. Detailed analysis of chromosome 15 demonstrated additional eQTLs for IREB2 mapping to that gene. eQTL SNPs for CHRNA5 mapped to multiple linkage disequilibrium (LD) bins. The eQTLs for IREB2 and CHRNA5 were not in LD. Seventy-four additional eQTL SNPs were associated with COPD at p<0.01. These were genotyped in two COPD populations, finding replicated associations with a SNP in PSORS1C1, in the HLA-C region on chromosome 6. Integrative analysis of GWAS and gene expression data from relevant tissue from diseased subjects has located potential functional variants in two known COPD genes and has identified a novel COPD susceptibility locus.
Introduction
Gene expression levels in humans are highly heritable [1], [2]. Multiple published studies have examined the associations between single nucleotide polymorphism (SNP) variation and microarray gene expression measurements to identify expression Quantitative Trait Loci (eQTLs ), single nucleotide polymorphisms (SNPs) that influence gene expression [3]–[5]. However, most of the published studies have examined gene expression in lymphoblastoid cell lines (LCL) from unphenotyped individuals [3], [4], though a recent paper has described eQTLs in peripheral blood CD4+ lymphocytes of patients with asthma [6]. Integrative genomic analyses can provide functional information regarding significant SNPs found through genomewide association studies (GWAS) or identify the key genes within a locus identified through GWAS. For example, genome-wide expression profiling in LCL from children with asthma [5] was used to localize ORMDL3 (ORM1-like 3 (S. cerevisiae) [MIM 610075]) as the likely gene for childhood asthma in the multi-gene chromosome 17q21 locus found through GWAS [7]. However, this study did not determine whether the eQTLs identified were relevant in primary human tissues in asthma. Integrative genomics studies can also be used to implicate novel genes for complex traits, such as the association between MMP20 (matrix metallopeptidase 20 [MIM 604629]) and age related decline in kidney function [8].
Chronic obstructive pulmonary disease (COPD [MIM 606963]), which includes emphysema and chronic bronchitis, is a complex disease with genetic and environmental influences [9]. COPD is a major source of morbidity and mortality in the U.S. and worldwide [10]. Previous GWAS have identified three susceptibility loci for COPD, including HHIP (hedgehog interacting protein [MIM 606178]), FAM13A (family with sequence similarity 13, member A [MIM 613299]), and a multi-gene locus on chromosome 15q25 containing candidate genes CHRNA5 (cholinergic receptor, nicotinic, alpha 5 [MIM 118505]), CHRNA3 (MIM 118503), and IREB2 (iron-responsive element binding protein 2 [MIM 147582]) [11]–[13]. Cough and phlegm production is common among COPD patients, and sputum samples may provide a non-invasive window into pathobiologic processes in the lungs of COPD patients. Therefore, we integrated GWAS data with microarray gene expression profiles from induced sputum samples from well-characterized COPD subjects participating in the Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points (ECLIPSE) Study [14]. We addressed two hypotheses: (1) eQTL analysis will improve understanding of previously known COPD susceptibility loci, such as chromosome 15q25; and (2) eQTL SNPs can be used to identify novel COPD susceptibility genes. Limiting the search to functional eQTL SNPs can reduce the multiple testing burden found in traditional GWAS. Although eQTL studies have now been performed in several human tissues besides blood, our study represents one of the first integrative genomics analyses performed in affected patients in order to gain insights into a common disease.
Methods
Ethics Statement
Study subjects provided written informed consent, and all studies were approved by the Institutional Review Boards at Partners Healthcare and all participating centers.
ECLIPSE Study
ECLIPSE was a three year observational study conducted at 46 centers in 12 countries [14]. ECLIPSE recruited 2083 COPD subjects ages 40–75 with a smoking history of at least 10 pack-years (cigarettes smoked per day multiplied by years smoked, divided by 20 to convert to packs), 332 control smokers with at least 10 pack-years smoking history and normal lung function, and 237 non-smoking controls [15]. COPD was defined by GOLD stage 2 or greater (FEV1/FVC<0.7 with FEV1<80% predicted) [10]. Genome-wide SNP genotyping was performed on all ECLIPSE subjects using the Illumina HumanHap550 BeadChip. GWAS analysis included 1736 cases COPD cases and 175 controls [11]. Sputum induction was performed on a subset of COPD cases at 14 sites, using a standard protocol [16]. RNA was extracted from sputum cell pellets using TRIzol and amplified with the Nugen Ovation RNA Amplification kit. Gene expression profiling was performed on RNA extracted from sputum samples of 145 COPD cases (all ex-smokers) using the Affymetrix Human U133 Plus2 array [17]. MIAME-compliant array data are available in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo), accession GSE22148. Only Caucasian subjects were included in this analysis.
Other GWAS Populations
Subjects from two additional COPD case-control studies were merged with the ECLIPSE subjects in the combined GWAS analysis and the GWAS meta-analysis [11]. COPD cases and control smokers were Caucasians recruited in Bergen, Norway [18], [19]. Cases were defined by GOLD stage 2 or greater COPD; smoking controls had normal lung function. Both cases and controls had smoking history of at least 2.5 pack-years. GWAS included 838 cases and 791 controls, genotyped using the Illumina HumanHap550 BeadChip [12].
The National Emphysema Treatment Trial (NETT) cases have FEV1≤45% predicted and emphysema on chest CT scan [20], [21]. Thus, NETT cases have COPD severity of GOLD Stage 3 or greater. All NETT Genetics Ancillary Study subjects are former smokers; only white subjects are included in this analysis. The Normative Aging Study (NAS) is a cohort study of initially healthy men followed by the Boston VA [22]. To define a control group for comparison to NETT cases, we selected Caucasian subjects meeting the following criteria: FEV1>80% predicted, FEV1/FVC>90% predicted, at least 10 pack-years of smoking, and an adequate DNA sample [23]. Genomewide SNP genotyping has been performed in the NETT-NAS study (366 cases, 414 controls) using the Illumina 610-Quad BeadChip [11].
Replication Populations
The International COPD Genetics Network (ICGN) was a family-based study of COPD at ten centers in North America and Europe [18], [24]. Probands were ages 45–65 with post-bronchodilator FEV1<60% predicted, FEV1/VC<90% predicted, a smoking history of at least 5 pack-years, and at least one sibling with ≥5 pack-year smoking history. Genotyping was performed on Caucasian subjects only (Table 1).
Table 1. Characteristics of Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points (ECLIPSE) study subjects in the integrative genomics analysis as well as subjects from the International COPD Genetics Network (ICGN) and the Genetic Epidemiology of COPD study (COPDGene) included in follow-up analyses.
| ECLIPSE | ICGN | COPDGene | |||
| Cases | Probands | Relatives | Cases | Controls | |
| N | 131 | 983 | 1876 | 496 | 498 |
| Male sex | 87 (66.4%) | 580 (59.0%) | 970 (51.7%) | 244 (49.2%) | 251 (50.4%) |
| Age | 64.9 (±5.5) | 58.4 (±5.4) | 57.9 (±9.5) | 64.7 (±8.1) | 60.3 (±8.6) |
| Pack-years of smoking | 46.8 (±28.3) | 52.7 (±29.5) | 39.2 (±25.3) | 54.8 (±26.8) | 38.8 (±21.0) |
| Current smoker | 0 | 320 (32.6%) | 956 (51.0%) | 149 (30.0%) | 168 (33.7%) |
| Post-bronchodilator FEV1, % predicted | 49.3 (±15.3) | 35.5 (±13.1) | 83.3 (±26.0) | 48.8 (±18.4) | 98.0 (±11.3) |
| Post-bronchodilator FEV1/FVC | 0.43 (±0.12) | 0.37 (±0.12) | 0.64 (±0.14) | 0.48 (±0.13) | 0.78 (±0.05) |
Values are presented as mean (±SD) or N (%).
FEV1 = forced expiratory volume in 1 second.
FVC = forced vital capacity.
The Genetic Epidemiology of COPD Study (COPDGene) enrolled COPD cases and control smokers at 21 clinical centers throughout the United States [25]. Subjects are 45–80 years old and have a smoking history of at least 10 pack-years. This analysis included the first 994 non-Hispanic white case and control subjects enrolled in COPDGene (Table 1). In these samples, a set of 75 ancestry informative markers has been previously genotyped and did not show evidence of population stratification [11].
SNP genotyping in ICGN and COPDGene SNPs was done using the iPLEX Gold assay on the Sequenom (San Diego, CA) MassARRAY system [26] or the TaqMan 5′ exonuclease assay (Applied Biosystems, Foster City, CA) [27].
Statistical Analysis
A total of 145 COPD subjects had sputum samples with gene expression data available; two arrays failed quality control. Of the remaining 143 subjects, 131 had corresponding genomewide SNP data and phenotype data. The Affymetrix HG-U133 Plus 2 array contains 54,675 probe sets. After filtering out 17,420 probe sets which were not annotated with a specific gene symbol in the hgu133plus2.db R/Bioconductor database or which mapped to the X or Y chromosomes, 37,255 probe sets remained. Microarray preprocessing used the robust multiarray average method and quantile normalization [28], implemented in Bioconductor. QC of microarrays was performed using the Bioconductor package affyQCReport; QC results are available in the Data S1 and in Figure S1. QC of genomewide SNP data in ECLIPSE has been reported [11]. SNPs with minor allele frequency <0.05 in the 131 ECLIPSE cases were additionally excluded.
In the integrative analysis, each expression probe set was mapped to its corresponding gene and all genotyped SNPs were identified within 50 kb of the transcription start site (TSS). General linear models were used to detect cis-acting associations between probe set expression levels and SNP genotypes, adjusted for age, gender, and the first six genetic ancestry principal components derived from the genotype data on all ECLIPSE COPD cases [29]. False discovery rate adjusted p-value<0.05 defined statistical significance. eQTL analysis utilized the GGTools Bioconductor package [30].
Each significant cis-eQTL SNP was then tested for association with COPD in the combined GWAS dataset from ECLIPSE, Norway, and NETT-NAS [11]. The published combined GWAS analysis was a mega-analysis of individual-level genotype data, using logistic regression, adjusted for age, pack-years of smoking and principal components for genetic ancestry. In the published meta-analysis, stratified logistic regression was performed within each case-control study and results were combined using Z-scores for weighting by the inverse variance. SNPs associated with COPD at p<0.01 in either the combined GWAS analysis (mega-analysis) or the GWAS meta-analysis were genotyped for replication in ICGN and COPDGene. In the COPDGene study, case-control data were analyzed with linear regression models, adjusted for age, sex, and pack-years of smoking, using PLINK version 1.0.7 [31]. Family-based ICGN data were analyzed in PBAT version 3.6.1, adjusted for age, sex, and pack-years of smoking [32].
We also tested for eQTL SNPs influencing the expression of genes in previously identified COPD loci. On chromosome 15q25, we defined a region starting 50 kb centromeric from IREB2 extending 50 kb telomeric from CHRNB4 (approx. 300 kb total) and tested all genotyped SNPs within this region for association with expression levels of probe sets for six genes: IREB2, AGPHD1, PSMA4, CHRNA5, CHRNA3, and CHRNB4. For the other two COPD loci, we expanded the cis-eQTL analysis to all SNPs with 200 kb of the TSS of the genes HHIP and FAM13A.
Results
Sputum eQTL Analysis
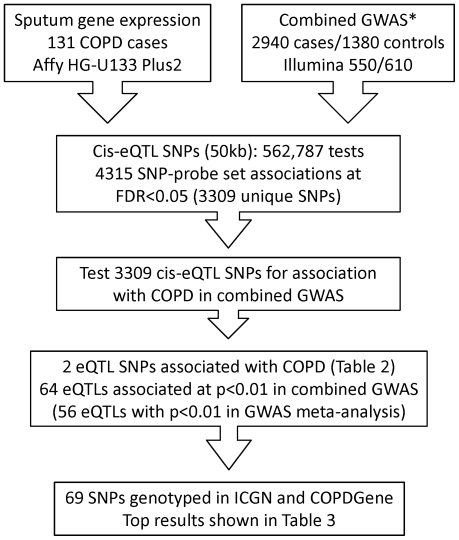
Characteristics of the 131 ECLIPSE COPD subjects in the eQTL analysis are shown in Table 1. On average, COPD subjects had a heavy smoking history and severely impaired lung function, similar to the full set of ECLIPSE GWAS cases [11]. The data analysis is outlined in Figure 1. Combining the gene expression data with genomewide SNP data and limiting analysis to potential cis-acting SNPs (within 50 kb of TSS) yielded 562,787 SNP-probe set association tests. Of these, 4315 SNP-probe set associations were significant at FDR-adjusted p<0.05 (corresponding to unadjusted p = 3.8e-4), representing 3309 unique SNPs and 1399 unique probe sets, covering 1086 genes (Table S1).
Figure 1. Overview of integrative genomics data analysis.
*Combined genomewide association study (GWAS) = Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points (ECLIPSE), Bergen Norway, and National Emphysema Treatment Trial (NETT)-Normative Aging Study (NAS) [11]. COPD = chronic obstructive pulmonary disease. eQTL = expression quantitative trait locus. FDR = false discovery rate. ICGN = International COPD Genetics Network. SNP = single nucleotide polymorphism.
The top eQTL was for SNP rs104664 within the gene FAM118A (family with sequence similarity 118, member A). This SNP was found to be highly associated with FAM118A expression (Affymetrix Human 1.0 ST Exon array) in human osteoblasts [33], suggesting cross-tissue generalizability of this eQTL association. Other significant eQTL associations observed in sputum included CHURC1 (churchill domain containing 1 [MIM 608577]), HLA-DQB1 (major histocompatibility complex, class II, DQ beta 1 [MIM 604305]) and HLA-DQA1 (MIM 146880), all of which were previously been found in other tissues, such as LCL [4], [34] and brain [35], according to a search of the GTEx (Genotype-Tissue Expression) eQTL Browser (http://www.ncbi.nlm.nih.gov/gtex, accessed 11/30/2010).
Sputum eQTLs associated with COPD
We queried the 3309 significant cis-eQTL SNPs in the combined GWAS dataset including ECLIPSE, Norway, and NETT-NAS subjects [11]. Using a strict Bonferroni correction, there were two cis-eQTL SNPs significantly associated with COPD at p<0.05/3309 = 1.5e-5 (Table 2). These two SNPs on chromosome 15q25 are located in CHRNA5 and IREB2, genes with known COPD associations.[12], [36] At a nominal threshold of p<0.01, there were 64 cis-eQTL SNPs associated with COPD (Table S2). There were 56 eQTL SNPs associated with COPD at p<0.01 in the meta-analysis of the ECLIPSE, Norway, NETT-NAS GWAS studies (Table S3), as opposed to the combined analysis of individual-level genotype data. Merging the 64 SNPs from the combined GWAS analysis and the 56 SNPs from the GWAS meta-analysis left 76 unique SNPs, which were brought to replication analysis.
Table 2. Sputum cis-eQTL SNPs significantly associated with COPD in the combined COPD GWAS.
| Chrom | location | SNP | Affy probe | Gene | eQTL pvalue | FDR-adjusted pvalue | GWAS COPD ORa | GWAS COPD pvaluea | GWAS pvalue (adjusted)b |
| 15 | 76532762 | rs2656069 | 1555476_at | IREB2 | 0.00030 | 0.042 | 0.75 | 6.8E-06 | 0.023 |
| 15 | 76681394 | rs1051730 | 206533_at | CHRNA5 | 0.00015 | 0.026 | 1.29 | 2.8E-06 | 0.0093 |
Combined COPD genomewide association study (GWAS) according to Cho et al. [11].
Bonferroni correction for 3309 sputum expression quantitative trait locus (eQTL) single nucleotide polymorphisms (SNPs) tested.
Replication Studies
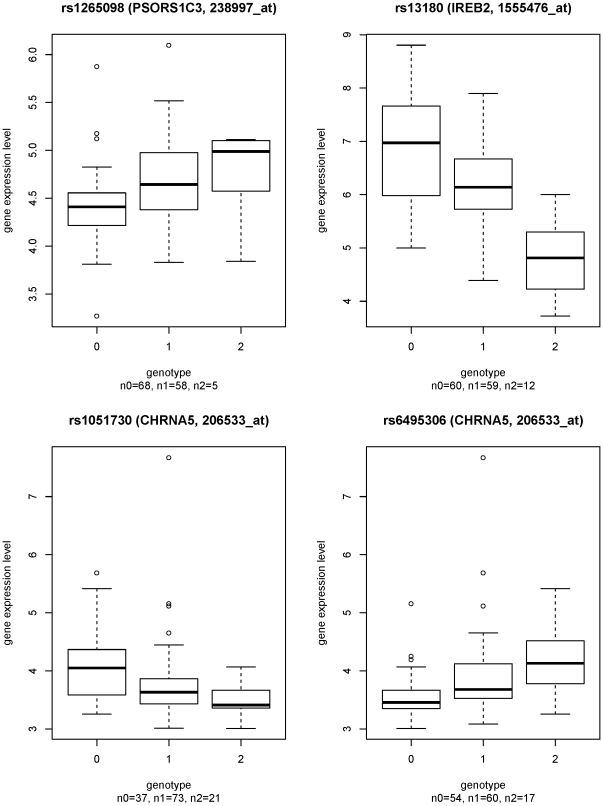
Characteristics of the ICGN and COPDGene subjects in the replication analysis are reported in Table 1. The two SNPs in Table 2 were analyzed in previous reports [12], [36] and were not retested. Of the remaining 74 SNPs, 69 were successfully genotyped in ICGN and COPDGene. Screening in the larger ICGN study found 8 SNPs with p<0.1 (Table 3). Of these, only one had p<0.1 in COPDGene. SNP rs1265098 was significantly associated with COPD in ICGN and had a trend for significance in COPDGene. The effect direction for rs1265098 was consistent in ICGN, COPDGene, and the combined GWAS; the minor allele was associated with increased COPD risk in all three studies. SNP rs1265098 maps to the gene PSORS1C1 (psoriasis susceptibility 1 candidate 1 [MIM 613525]) on chromosome 6, yet is associated with transcript levels of the neighboring gene PSORS1C3 (p = 8.2e-05, FDR-adjusted p = 0.016) (Figure 2).
Table 3. Genetic association analysis of sputum expression quantitative trait locus (eQTL) single nucleotide polymorphisms (SNPs) with COPD susceptibility.
| SNP | Chrom | BP | Gene | Effect on COPD risk in ICGN | ICGN pvalue | COPDGene OR | COPDGene pvalue | GWAS ORa | GWAS pvaluea |
| rs1999261 | 6 | 6515106 | INTERGENIC | Increase | 0.042 | 0.94 | 0.53 | 1.15 | 0.0080 |
| rs1265098 | 6 | 31214156 | PSORS1C1 | Increase | 0.024 | 1.20 | 0.098 | 1.18 | 0.0065 |
| rs4750277 | 10 | 12954527 | INTERGENIC | Increase | 0.0062 | 0.97 | 0.80 | 0.83 | 0.0097 |
| rs1025607 | 12 | 94884637 | AMDHD1 | Increase | 0.011 | 1.12 | 0.26 | a | a |
| rs2347279 | 18 | 2528545 | METTL4 | Decrease | 0.097 | 1.01 | 0.91 | 0.83 | 0.0025 |
| rs1878553 | 18 | 2560155 | NDC80 | Decrease | 0.030 | 1.03 | 0.83 | 0.83 | 0.0028 |
| rs4803481 | 19 | 46758396 | INTERGENIC | Decrease | 0.055 | 0.97 | 0.81 | a | a |
| rs2302188 | 19 | 46777713 | CEACAM21 | Decrease | 0.090 | 0.97 | 0.77 | 0.85 | 0.0075 |
SNPs with p-value<0.1 in the International COPD Genetics Network (ICGN) are shown.
Odds Ratio and p-value from combined genomewide association study (GWAS) analysis [11] are shown, except as noted:
rs1025607 p = 0.0089 in GWAS meta-analysis.
rs4803481 p = 0.0043 in GWAS meta-analysis.
Figure 2. Boxplots of sputum gene expression levels stratified by genotype in 131 Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points (ECLIPSE) subjects with chronic obstructive pulmonary disease.
a) rs1265098 - PSORS1C3 (238997_at), p = 8.2e-5. b) rs13180 - IREB2 (1555476_at), p = 6.7e-9. c) rs1051730 - CHRNA5 (206533_at), p = 2.2e-4; LD bin 1 (see Table 4). d) rs6495306 - CHRNA5 (206533_at), p = 9.9e-6; LD bin 3 (see Table 4).
Sputum eQTLs in COPD Candidate Loci
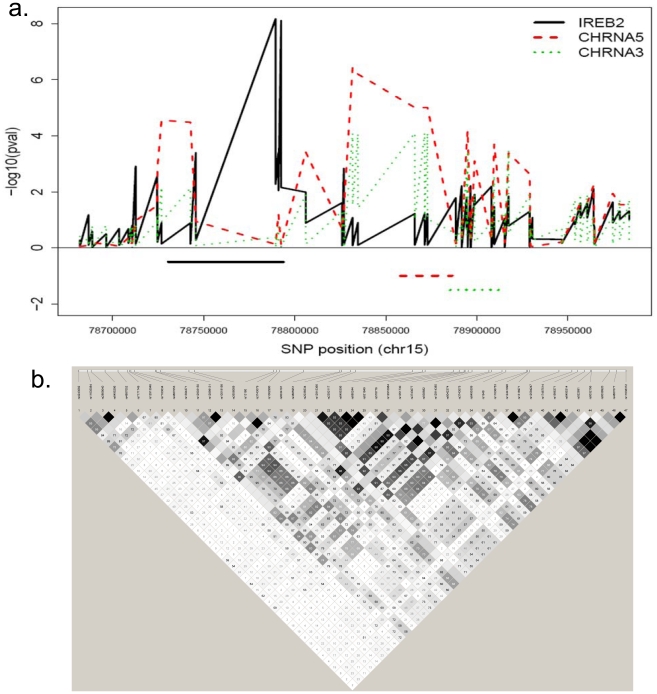
Previous GWAS have identified three loci associated with COPD susceptibility: HHIP on chromosome 4q31 [12], [13], FAM13A on chromosome 4q22 [11], and a region on chromosome 15q25 encompassing candidate genes CHRNA5, CHRNA3 and IREB2, among others [12], [36]. On chromosome 15q25, cis-eQTL associations for IREB2 mapped to that gene (Figure 3a). Genetic regulation of CHRNA5 was more complex. Previous studies have demonstrated cis-acting effects of multiple SNPs on CHRNA5 expression. Saccone et al. defined 4 LD bins surrounding CHRNA5 with varying associations with cigarette smoking, lung cancer, and COPD [37]. Bins 1–3 were represented in our dataset, tagged by SNPs rs1051730, rs938682, and rs6495306, respectively (Table 4). SNPs in bins 1 and 3 were associated with CHRNA5 expression in sputum (Figure 2), as has been demonstrated in brain [38] and lung tissue [39]. SNPs in bin 2 were not eQTLs for CHRNA5. We added additional SNPs to these bins, based on strong LD with tag SNPs in the larger ECLIPSE GWAS dataset. We also identified 3 sets of SNPs (3a, b, c in Table 4) with cis-eQTL associations for CHRNA5 and moderate LD with SNPs in bin 3 (r2 0.57–0.76). SNPs in bins 1 and 2, but not bin 3, showed evidence of association with COPD in the combined GWAS dataset, though they were not genomewide significant (bin 1: rs1051730, p = 2.8e-6; bin 2: rs938682, p = 5.6e-5; bin 3: rs6495306, p = 0.2). These results suggest that the COPD-associated SNP rs1051730 (bin 1) may influence phenotype by its effect on gene expression, while COPD-associated SNPs in bin 2 (tagged by rs938682) may exert their effect through other mechanisms. SNPs in bin 3, although eQTLs, were not associated with COPD risk.
Figure 3. Detailed analysis of the chromosome 15q25 chronic obstructive pulmonary disease (COPD) locus.
a) Association between single nucleotide polymorphisms (SNPs) in the chromosome 15q25 COPD locus and expression levels of IREB2 (1555476_at), CHRNA5 (206533_at) and CHRNA3 (211587_x_at) in sputum samples from 131 Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points (ECLIPSE) subjects. SNP rs numbers are listed in Table 4. b) Linkage disequilibrium r2 values between SNPs in the chromosome 15q25 COPD locus (listed in Table 4) in 131 ECLIPSE subjects.
Table 4. Single nucleotide polymorphism (SNP) associations with expression of IREB2 (1555476_at) and CHRNA5 (206533_at) in induced sputum samples from COPD subjects.
| SNP | Position | IREB2 eQTL pvalue | CHRNA5 eQTL pvalue | LD bin | r2 with locus tag | r2 with rs13180 |
| rs4243082 | 78682076 | 0.88 | 0.89 | |||
| rs11635084 | 78686823 | 0.066 | 0.82 | |||
| rs2869040 | 78688834 | 0.26 | 0.68 | |||
| rs6495296 | 78696492 | 0.32 | 0.71 | |||
| rs4887052 | 78703631 | 0.26 | 0.88 | |||
| rs7171749 | 78708715 | 0.41 | 0.44 | |||
| rs12913946 | 78709937 | 0.30 | 0.72 | |||
| rs7183034 | 78710766 | 0.16 | 0.66 | |||
| rs4887053 | 78712699 | 0.0012 | 0.11 | |||
| rs1394371 | 78724469 | 0.0028 | 0.031 | |||
| rs12903150 | 78724645 | 0.014 | 0.0014 | |||
| rs12899131 | 78726885 | 0.71 | 2.8E-05 | 3a | 0.57 | <0.01 |
| rs10519198 | 78742754 | 0.73 | 3.3E-05 | 3a | 0.57 | <0.01 |
| rs2656069 | 78745707 | 4.0E-04 | 0.11 | IREB2 | 0.44 | |
| rs13180 | 78789488 | 6.7E-09 | 0.74 | IREB2 | ||
| rs3743079 | 78791061 | 3.9E-04 | 0.063 | IREB2 | 0.36 | |
| rs1062980 | 78792527 | 7.7E-09 | 0.78 | IREB2 | 0.99 | |
| rs8034191 | 78806023 | 0.010 | 3.8E-04 | 1 | 0.90 | |
| rs3885951 | 78825917 | 0.025 | 0.20 | |||
| rs2036534 | 78826948 | 0.0015 | 0.043 | 2 | 0.79 | |
| rs12915366 | 78831753 | 0.83 | 3.7E-07 | 3 | 0.80 | |
| rs2292117 | 78834689 | 0.80 | 6.4E-07 | 3 | 0.82 | |
| rs6495306 | 78865893 | 0.79 | 9.9E-06 | 3 | <0.01 | |
| rs680244 | 78871288 | 0.79 | 9.9E-06 | 3 | 0.99 | |
| rs621849 | 78872861 | 0.79 | 9.9E-06 | 3 | 0.99 | |
| rs578776 | 78888400 | 0.016 | 0.70 | 2 | 0.72 | |
| rs12910984 | 78891627 | 0.0061 | 0.45 | 2 | 0.99 | |
| rs1051730 | 78894339 | 0.016 | 2.2E-04 | 1 | 0.19 | |
| rs3743077 | 78894896 | 0.93 | 6.7E-05 | 3 | 0.98 | |
| rs938682 | 78896547 | 0.0061 | 0.45 | 2 | 0.26 | |
| rs12914385 | 78898723 | 0.014 | 8.0E-04 | 1 | 0.81 | |
| rs8042374 | 78908032 | 0.0064 | 0.53 | 2 | 0.98 | |
| rs3743075 | 78909452 | 0.76 | 2.0E-04 | 3b | 0.76 | 0.01 |
| rs6495309 | 78915245 | 0.088 | 0.64 | 2 | 0.90 | |
| rs1948 | 78917399 | 0.16 | 4.2E-04 | 3c | 0.60 | 0.03 |
| rs11636753 | 78928946 | 0.91 | 0.0023 | |||
| rs12441998 | 78929372 | 0.14 | 0.85 | |||
| rs1316971 | 78930510 | 0.083 | 0.89 | |||
| rs12594247 | 78946633 | 0.63 | 0.62 | |||
| rs17487514 | 78953785 | 0.10 | 0.068 | |||
| rs1996371 | 78956806 | 0.025 | 0.025 | |||
| rs6495314 | 78960529 | 0.025 | 0.025 | |||
| rs922691 | 78963994 | 0.14 | 0.0060 | |||
| rs8032156 | 78964498 | 0.51 | 0.66 | |||
| rs8038920 | 78974545 | 0.059 | 0.011 | |||
| rs4887077 | 78978364 | 0.049 | 0.029 | |||
| rs11638372 | 78983559 | 0.049 | 0.029 |
Linkage disequilibrium (LD) bins for CHRNA5 associations are defined as in Saccone et al. [37]. SNPs tags for LD bins are shown in bold. Additional SNPs added to bins based on LD in ECLIPSE genomewide association study dataset are shown in italics.
SNPs in IREB2 were both cis-eQTLs for that gene (Table 4, Figure 2) and were associated with COPD in the combined GWAS (rs13180, p = 5.0e-7). Even though some of the significant eQTL SNPs for CHRNA5 mapped to IREB2 (bin 3a), SNPs in all 3 bins were not in LD with the IREB2 eQTL SNPs (Figure 3b). No SNPs were significantly associated with AGPHD1, PSMA4, or CHRNB4 gene expression. For the other two COPD GWAS loci, HHIP and FAM13A, we found no significant cis-eQTL SNPs within 50 kb, so we expanded the assessment of cis-eQTLs to all SNPs within 200 kb of the TSS of each gene. There were no significant cis-eQTLs within 200 kb of either HHIP or FAM13A.
Discussion
In a cohort of well-characterized COPD subjects, we integrated genomewide SNP and gene expression data derived from induced sputum, a biologically-relevant tissue in COPD, to identify a set of eQTL SNPs affecting gene expression levels. The SNPs were then tested for association with the clinical phenotype of COPD; gene expression was not tested for association with disease status in this set of COPD cases only. Using the eQTL results, we implicated two distinct COPD susceptibility genes in a previously identified region of chromosome 15q25. Additionally, we provide evidence for a potential novel COPD susceptibility locus in the HLA region on chromosome 6.
The initial GWAS in COPD found significant associations on chromosome 15q25, with SNPs in the genes CHRNA3 and CHRNA5, encoding two subunits of the nicotinic acetylcholine receptor [12]. This region has also been associated with lung cancer, peripheral arterial disease, and smoking behavior [37], [40]–[43], so it is not clear whether these genes have a direct effect on COPD susceptibility, or their effects are at least partially influenced through cigarette smoking, the major environmental risk factor for COPD [44], [45]. In terms of genetic regulation of expression of the chromosome 15q25 genes, we found similar eQTL associations with CHRNA5 expression in induced sputum as has been found in brain [38] and lung tissue [39]. We found additional sputum eQTL SNPs for CHRNA5 in moderate LD with previously defined eQTLs. The previous papers on brain and lung tissue gene expression did not report testing IREB2, a gene previously associated with COPD [11], [36]. The specific IREB2 SNPs associated in GWAS (rs13180) [11] and in a candidate gene analysis of differentially expressed genes (rs2656069) [36] were in only moderate LD (r2 = 0.44) with each other, implying independent effects on IREB2 expression. The IREB2 and CHRNA5 eQTL SNPs were not in LD with each other, suggesting the presence of at least two COPD susceptibility genes on chromosome 15q25. Previous studies have similarly used eQTL analyses to add functional information about genes identified through GWAS, including studies of asthma [7], celiac disease [46], and Crohn's disease [47]. However, these prior studies have examined gene expression in blood cells, and not primary disease tissues.
However, we did not finding significant cis-eQTL SNPs for two other known COPD loci, HHIP and FAM13A. The associated SNPs found through GWAS may exert their effects on phenotype via other mechanisms besides influencing gene expression. Alternatively, the GWAS SNPs may actually be eQTLs acting in other tissues besides sputum, such as alveolar or bronchial epithelial cells, which were not assessed in our study.
Besides improving understanding of the COPD susceptibility locus on chromosome 15q25, we identified a potential novel COPD locus on chromosome 6. The SNP maps to gene PSORS1C1, but it is associated with expression levels of the neighboring gene PSORS1C3. Variants in PSORS1C3 have been reported to be associated with psoriasis [48], an immune-mediated skin disease. PSORS1C3 is located in the major histocompatability (MHC) region, and subsequent papers have found that the associations with psoriasis may be due to variants in HLA-C (MIM 142840) [49], [50]. Interestingly, one study has reported an epidemiologic association between psoriasis and COPD [51], and cigarette smoking is a risk factor for psoriasis as well [52]. Although there are no reports of HLA-C associations with COPD, alleles of other MHC class I genes, HLA-A and HLA-B, have been associated with COPD [53], [54]. The locus encompassing PSORS1C1/3 and HLA-C will require additional replication studies and functional validation to confirm its role in COPD susceptibility.
Prior studies have also used eQTL analyses to identify novel genes for complex traits, including age related decline in kidney function [8] and body mass index [55]. In contrast to our study, these papers first found gene transcripts correlated with the phenotype, then tested SNPs in/near these genes for association with expression levels. We performed the cis-eQTL analysis as the initial step, then tested the eQTL SNPs for phenotype association. This limits multiple testing compared to a GWAS, enriching for eQTL SNPs which may be more likely to be associated with disease [56].
This study has several limitations. The sample size of 131 subjects, though adequate for gene expression analyses, may be underpowered to detect all potential eQTL associations. Therefore, we limited the cis-acting analysis to SNPs within 50 kb from the gene, to limit the multiple testing burden. Based on RNA sequencing data, Pickrell et al. estimate that 90% of eQTL SNPs are within 15 kb of a gene [57]. Previous papers have used a 50 kb limit to define cis-acting eQTLs [6]. Using this method, we were able to replicate published eQTL associations from other tissues and were able to identify a set of significant eQTL SNPs to carry forward for COPD association studies. However, our method would be unable to detect cis-eQTLs located >50 kb from the TSS, such as a SNP in an upstream enhancer or in the 3′ UTR of a large gene. Due to the sample size, we limited our investigation to cis-acting eQTL SNPs, as a full search for trans-acting regulatory SNPs greatly increases the number of tests performed. The literature suggests that sample sizes under 200 subjects may be inadequate to find true trans-eQTLs [58].
Several groups have compared eQTLs in different tissues from the same individual, finding both overlapping and tissue-specific eQTLs [59]–[61]. Multiple tissues are known to be important in COPD biology, including large and small airways, lung parenchyma and immune cells. By only surveying sputum, we may have missed significant eQTLs for COPD genes that are expressed in other tissues. Multiple cell types may be present in sputum, yet neutrophils have been shown to be the predominant cell type in the sputum samples from COPD subjects in ECLIPSE [16]. Despite these limitations, sputum is a clinically important tissue in COPD and is more accessible for genomic and biomarkers studies than lung tissue. Studying diseased individuals may be advantageous to identify eQTL SNPs for potential disease genes, which may only be expressed, or may be expressed at higher levels, in patients compared to healthy controls.
In conclusion, we combined genomewide SNP genotyping with genomewide expression profiling from a relevant tissue in well-characterized subjects with a common chronic disease. Using this strategy, we were able to gain insights into the functional role of SNPs previously associated through GWAS, as well as identify a potential novel disease susceptibility gene which would have been missed using standard GWAS analysis. Previous eQTL studies have provided important information about genetic control of human gene expression. Integrative genomics studies in relevant tissue from well-phenotyped individuals, as we have performed, will be required to apply this knowledge to human disease.
Supporting Information
Principal components plot of RMA expression values, demonstrating lack of batch effects based on hybridization dates or other systematic effects.
(TIF)
Microarray Quality Control.
(DOC)
The top cis-expression quantitative trait loci (eQTLs) in sputum samples from 131 ECLIPSE COPD subjects. Single nucleotide polymorphism (SNP)-probe set associations with FDR-adjusted p<0.05 are shown.
(XLS)
Cis-expression quantitative trait locus (eQTL) single nucleotide polymorphisms (SNPs) from sputum samples from 131 ECLIPSE COPD subjects associated with COPD case-control status in combined ECLIPSE, NETT-NAS, and Norway GWAS (Cho et al. 2010). SNPs associated at p<0.01 are shown.
(DOC)
Cis-expression quantitative trait locus (eQTL) single nucleotide polymorphisms (SNPs) from sputum samples from 131 ECLIPSE COPD subjects associated with COPD case-control status in a meta-analysis of ECLIPSE, NETT-NAS, and Norway GWAS (Cho et al. 2010). SNPs with combined p<0.01 are shown.
(DOC)
Acknowledgments
ECLIPSE Steering Committee: Harvey Coxson (Canada), Lisa Edwards (GlaxoSmithKline, USA), Katharine Knobil (Co-chair, GlaxoSmithKline, UK), David Lomas (UK), William MacNee (UK), Edwin Silverman (USA), Ruth Tal-Singer (GlaxoSmithKline, USA), Jørgen Vestbo (Co-chair, Denmark), Julie Yates (GlaxoSmithKline, USA).
ECLIPSE Scientific Committee: Alvar Agusti (Spain), Peter Calverley (UK), Bartolome Celli (USA), Courtney Crim (GlaxoSmithKline, USA), Bruce Miller (GlaxoSmithKline, UK), William MacNee (Chair, UK), Stephen Rennard (USA), Ruth Tal-Singer (GlaxoSmithKline, USA), Emiel Wouters (The Netherlands), Julie Yates (GlaxoSmithKline, USA).
ECLIPSE Investigators: Bulgaria: Yavor Ivanov, Pleven; Kosta Kostov, Sofia. Canada: Jean Bourbeau, Montreal, Que Mark Fitzgerald, Vancouver, BC; Paul Hernandez, Halifax, NS; Kieran Killian, Hamilton, On; Robert Levy, Vancouver, BC; Francois Maltais, Montreal, Que; Denis O'Donnell, Kingston, On. Czech Republic: Jan Krepelka, Praha. Denmark: Jørgen Vestbo, Hvidovre. The Netherlands: Emiel Wouters, Horn-Maastricht. New Zealand: Dean Quinn, Wellington. Norway: Per Bakke, Bergen. Slovenia: Mitja Kosnik, Golnik. Spain: Alvar Agusti, Jaume Sauleda, Palma de Mallorca. Ukraine: Yuri Feschenko, Kiev; Vladamir Gavrisyuk, Kiev; Lyudmila Yashina, Kiev; Nadezhda Monogarova, Donetsk. United Kingdom: Peter Calverley, Liverpool; David Lomas, Cambridge; William MacNee, Edinburgh; David Singh, Manchester; Jadwiga Wedzicha, London. United States of America: Antonio Anzueto, San Antonio, TX; Sidney Braman, Providence, RI; Richard Casaburi, Torrance CA; Bart Celli, Boston, MA; Glenn Giessel, Richmond, VA; Mark Gotfried, Phoenix, AZ; Gary Greenwald, Rancho Mirage, CA; Nicola Hanania, Houston, TX; Don Mahler, Lebanon, NH; Barry Make, Denver, CO; Stephen Rennard, Omaha, NE; Carolyn Rochester, New Haven, CT; Paul Scanlon, Rochester, MN; Dan Schuller, Omaha, NE; Frank Sciurba, Pittsburgh, PA; Amir Sharafkhaneh, Houston, TX; Thomas Siler, St. Charles, MO, Edwin Silverman, Boston, MA; Adam Wanner, Miami, FL; Robert Wise, Baltimore, MD; Richard ZuWallack, Hartford, CT.
Footnotes
Competing Interests: The authors have the following competing interests: Dr. Riley, Dr. Anderson, Dr. Tal-Singer, and Dr. Fox are full-time employees of GlaxoSmithKline. The International COPD Genetics Network and The ECLIPSE Study were funded by GlaxoSmithKline. Dr. Singh has received lectures fees, support for conference attendance, advisory board fees and research grants from a range of pharmaceutical companies including GSK, Chiesi Pharmaceuticals, AstraZeneca, Pfizer, CIPLA, Novartis, Forest, MSD, Boehringer and Allmiral. Dr. Lomas has received an educational grant, fees for speaking and acts as a consultant for GSK. Dr. Rennard has consulted for Able Associates, Adelphi Research, APT Pharma/Britnall, Aradigm, AstraZeneca, Boehringer-Ingelheim, Chiesi, CommonHealth, Consult Complete, and COPDForum, and has served on advisory boards for Almirall, Novartis, Nycomed, and Pfizer. Dr. Rennard has received lecture fees from American Thoracic Society, AstraZeneca, Boehringer-Ingelheim, California Allergy Society, and Creative Educational Concept, and has received industry-sponsored grants from AstraZeneca, Biomarck, Centocor, Mpex, and Nabi. Dr. Silverman has received grant support for two studies of COPD genetics and consulting fees from GlaxoSmithKline, and has received consulting fees from Astra-Zeneca. Dr. Silverman has also received honoraria from Wyeth, Bayer, GlaxoSmithKline, and Astra-Zeneca. Dr. Qiu, Dr. Cho, Dr. Bakke, Dr. Gulsvik, Dr. Litonjua, Dr. Crapo, Dr. Beaty, Dr. Celli, and Dr. Hersh do not report any conflicts of interest related to this manuscript. There are no patents, products in development or marketed products to declare. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: This work was supported by the National Institutes of Health grants K08HL080242 (Dr. Hersh), R01HL094635 (Dr. Hersh), R01HL084323 (Dr. Silverman), P01HL083069 (Dr. Silverman), and a grant from the Alpha-1 Foundation. The ECLIPSE Study is funded by GlaxoSmithKline (SCO104960, NCT00292552). The National Emphysema Treatment Trial was supported by the National Heart, Lung, and Blood Institute (N01HR76101, N01HR76102, N01HR76103, N01HR76104, N01HR76105, N01HR76106, N01HR76107, N01HR76108, N01HR76109, N01HR76110, N01HR76111, N01HR76112, N01HR76113, N01HR76114, N01HR76115, N01HR76116, N01HR76118, N01HR76119), Centers for Medicare and Medicaid Services, and the Agency for Healthcare Research and Quality. The Normative Aging Study is supported by the Cooperative Studies Program/Epidemiology Research and Information Center of the US Department of Veterans Affairs and is a component of the Massachusetts Veterans Epidemiology Research and Information Center (MAVERIC), Boston, Massachusetts. The Genetic Epidemiology of COPD Study (COPDGene) is supported by National Institutes of Health grants U01HL089856 (Dr. Silverman) and U01HL089897 (Dr. Crapo). The International COPD Genetics Network was supported by GlaxoSmithKline. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 2.Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen KY, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 3.Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, et al. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 6.Murphy A, Chu JH, Xu M, Carey VJ, Lazarus R, et al. Mapping of numerous disease-associated expression polymorphisms in primary peripheral blood CD4+ lymphocytes. Hum Mol Genet. 2010;19:4745–4757. doi: 10.1093/hmg/ddq392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 8.Wheeler HE, Metter EJ, Tanaka T, Absher D, Higgins J, et al. Sequential use of transcriptional profiling, expression quantitative trait mapping, and gene association implicates MMP20 in human kidney aging. PLoS Genet. 2009;5:e1000685. doi: 10.1371/journal.pgen.1000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hersh CP, DeMeo DL, Silverman EK. Chronic Obstructive Pulmonary Disease. In: Silverman EK, Shapiro SD, Lomas DA, Weiss ST, editors. Respiratory Genetics. New York: Hodder Arnold; 2005. pp. 253–296. [Google Scholar]
- 10.Rabe KF, Hurd S, Anzueto A, Barnes PJ, Buist SA, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007;176:532–555. doi: 10.1164/rccm.200703-456SO. [DOI] [PubMed] [Google Scholar]
- 11.Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42:200–202. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilk JB, Chen TH, Gottlieb DJ, Walter RE, Nagle MW, et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;5:e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vestbo J, Anderson W, Coxson HO, Crim C, Dawber F, et al. Evaluation of COPD Longitudinally to Identify Predictive Surrogate End-points (ECLIPSE). Eur Respir J. 2008;31:869–873. doi: 10.1183/09031936.00111707. [DOI] [PubMed] [Google Scholar]
- 15.Lomas DA, Silverman EK, Edwards LD, Miller BE, Coxson HO, et al. Evaluation of serum CC-16 as a biomarker for COPD in the ECLIPSE cohort. Thorax. 2008;63:1058–1063. doi: 10.1136/thx.2008.102574. [DOI] [PubMed] [Google Scholar]
- 16.Singh D, Edwards L, Tal-Singer R, Rennard S. Sputum neutrophils as a biomarker in COPD: findings from the ECLIPSE study. Respir Res. 2010;11:77. doi: 10.1186/1465-9921-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh D, Fox SM, Tal-Singer R, Plumb J, Bates S, et al. Induced sputum genes associated with spirometric and radiological disease severity in COPD ex-smokers. Thorax. 2011;66:489–495. doi: 10.1136/thx.2010.153767. [DOI] [PubMed] [Google Scholar]
- 18.Zhu G, Warren L, Aponte J, Gulsvik A, Bakke P, et al. The SERPINE2 gene is associated with chronic obstructive pulmonary disease in two large populations. Am J Respir Crit Care Med. 2007;176:167–173. doi: 10.1164/rccm.200611-1723OC. [DOI] [PubMed] [Google Scholar]
- 19.Brogger J, Steen VM, Eiken HG, Gulsvik A, Bakke P. Genetic association between COPD and polymorphisms in TNF, ADRB2 and EPHX1. Eur Respir J. 2006;27:682–688. doi: 10.1183/09031936.06.00057005. [DOI] [PubMed] [Google Scholar]
- 20.The National Emphysema Treatment Trial Research Group. Rationale and design of The National Emphysema Treatment Trial: a prospective randomized trial of lung volume reduction surgery. Chest. 1999;116:1750–1761. doi: 10.1378/chest.116.6.1750. [DOI] [PubMed] [Google Scholar]
- 21.Fishman A, Martinez F, Naunheim K, Piantadosi S, Wise R, et al. A randomized trial comparing lung-volume-reduction surgery with medical therapy for severe emphysema. N Engl J Med. 2003;348:2059–2073. doi: 10.1056/NEJMoa030287. [DOI] [PubMed] [Google Scholar]
- 22.Bell B, Rose CL, Damon H. The Normative Aging Study: an interdisciplinary and longitudinal study of health and aging. Aging Hum Dev. 1972;3:5–17. [Google Scholar]
- 23.Hersh CP, Pillai SG, Zhu G, Lomas DA, Bakke P, et al. Multistudy fine mapping of chromosome 2q identifies XRCC5 as a chronic obstructive pulmonary disease susceptibility gene. Am J Respir Crit Care Med. 2010;182:605–613. doi: 10.1164/rccm.200910-1586OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel BD, Coxson HO, Pillai SG, Agusti AG, Calverley PM, et al. Airway wall thickening and emphysema show independent familial aggregation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008;178:500–505. doi: 10.1164/rccm.200801-059OC. [DOI] [PubMed] [Google Scholar]
- 25.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, et al. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Storm N, Darnhofer-Patel B, van den Boom D, Rodi CP. MALDI-TOF mass spectrometry-based SNP genotyping. Methods Mol Biol. 2003;212:241–262. doi: 10.1385/1-59259-327-5:241. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 28.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 29.Wan ES, Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, et al. Genome-Wide Association Analysis of Body Mass in Chronic Obstructive Pulmonary Disease. Am J Respir Cell Mol Biol. 2010 doi: 10.1165/rcmb.2010-0294OC. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carey VJ, Davis AR, Lawrence MF, Gentleman R, Raby BA. Data structures and algorithms for analysis of genetics of gene expression with Bioconductor: GGtools 3.x. Bioinformatics. 2009;25:1447–1448. doi: 10.1093/bioinformatics/btp169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet. 2004;74:367–369. doi: 10.1086/381563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwan T, Grundberg E, Koka V, Ge B, Lam KC, et al. Tissue effect on genetic control of transcript isoform variation. PLoS Genet. 2009;5:e1000608. doi: 10.1371/journal.pgen.1000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeMeo DL, Mariani T, Bhattacharya S, Srisuma S, Lange C, et al. Integration of genomic and genetic approaches implicates IREB2 as a COPD susceptibility gene. Am J Hum Genet. 2009;85:493–502. doi: 10.1016/j.ajhg.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saccone NL, Culverhouse RC, Schwantes-An TH, Cannon DS, Chen X, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010;6:e1001053. doi: 10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang JC, Cruchaga C, Saccone NL, Bertelsen S, Liu P, et al. Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Hum Mol Genet. 2009;18:3125–3135. doi: 10.1093/hmg/ddp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Falvella FS, Galvan A, Frullanti E, Spinola M, Calabro E, et al. Transcription deregulation at the 15q25 locus in association with lung adenocarcinoma risk. Clin Cancer Res. 2009;15:1837–1842. doi: 10.1158/1078-0432.CCR-08-2107. [DOI] [PubMed] [Google Scholar]
- 40.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 42.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caporaso N, Gu F, Chatterjee N, Sheng-Chih J, Yu K, et al. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS One. 2009;4:e4653. doi: 10.1371/journal.pone.0004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lambrechts D, Buysschaert I, Zanen P, Coolen J, Lays N, et al. The 15q24/25 susceptibility variant for lung cancer and chronic obstructive pulmonary disease is associated with emphysema. Am J Respir Crit Care Med. 2010;181:486–493. doi: 10.1164/rccm.200909-1364OC. [DOI] [PubMed] [Google Scholar]
- 45.Wang J, Spitz MR, Amos CI, Wilkinson AV, Wu X, et al. Mediating effects of smoking and chronic obstructive pulmonary disease on the relation between the CHRNA5-A3 genetic locus and lung cancer risk. Cancer. 2010;116:3458–3462. doi: 10.1002/cncr.25085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dubois PC, Trynka G, Franke L, Hunt KA, Romanos J, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fransen K, Visschedijk MC, van Sommeren S, Fu JY, Franke L, et al. Analysis of SNPs with an effect on gene expression identifies UBE2L3 and BCL3 as potential new risk genes for Crohn's disease. Hum Mol Genet. 2010;19:3482–3488. doi: 10.1093/hmg/ddq264. [DOI] [PubMed] [Google Scholar]
- 48.Chang YT, Chou CT, Shiao YM, Lin MW, Yu CW, et al. Psoriasis vulgaris in Chinese individuals is associated with PSORS1C3 and CDSN genes. Br J Dermatol. 2006;155:663–669. doi: 10.1111/j.1365-2133.2006.07420.x. [DOI] [PubMed] [Google Scholar]
- 49.Holm SJ, Sanchez F, Carlen LM, Mallbris L, Stahle M, et al. HLA-Cw*0602 associates more strongly to psoriasis in the Swedish population than variants of the novel 6p21.3 gene PSORS1C3. Acta Derm Venereol. 2005;85:2–8. doi: 10.1080/00015550410023527. [DOI] [PubMed] [Google Scholar]
- 50.Nair RP, Stuart PE, Nistor I, Hiremagalore R, Chia NV, et al. Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am J Hum Genet. 2006;78:827–851. doi: 10.1086/503821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dreiher J, Weitzman D, Shapiro J, Davidovici B, Cohen AD. Psoriasis and chronic obstructive pulmonary disease: a case-control study. Br J Dermatol. 2008;159:956–960. doi: 10.1111/j.1365-2133.2008.08749.x. [DOI] [PubMed] [Google Scholar]
- 52.Setty AR, Curhan G, Choi HK. Smoking and the risk of psoriasis in women: Nurses' Health Study II. Am J Med. 2007;120:953–959. doi: 10.1016/j.amjmed.2007.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kauffmann F, Kleisbauer JP, Cambon-De-Mouzon A, Mercier P, Constans J, et al. Genetic markers in chronic air-flow limitation. A genetic epidemiologic study. Am Rev Respir Dis. 1983;127:263–269. doi: 10.1164/arrd.1983.127.3.263. [DOI] [PubMed] [Google Scholar]
- 54.Anagnostopoulou U, Toumbis M, Konstantopoulos K, Kotsovoulou-Fouskaki V, Zervas J. HLA-A and -B antigens in chronic bronchitis. J Clin Epidemiol. 1993;46:1413–1416. doi: 10.1016/0895-4356(93)90141-m. [DOI] [PubMed] [Google Scholar]
- 55.Naukkarinen J, Surakka I, Pietilainen KH, Rissanen A, Salomaa V, et al. Use of genome-wide expression data to mine the “Gray Zone” of GWA studies leads to novel candidate obesity genes. PLoS Genet. 2010;6:e1000976. doi: 10.1371/journal.pgen.1000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Franke L, Jansen RC. eQTL analysis in humans. Methods Mol Biol. 2009;573:311–328. doi: 10.1007/978-1-60761-247-6_17. [DOI] [PubMed] [Google Scholar]
- 59.Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 60.Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, et al. Tissue-Specific Genetic Control of Splicing: Implications for the Study of Complex Traits. PLoS Biology. 2008;6:e1. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Principal components plot of RMA expression values, demonstrating lack of batch effects based on hybridization dates or other systematic effects.
(TIF)
Microarray Quality Control.
(DOC)
The top cis-expression quantitative trait loci (eQTLs) in sputum samples from 131 ECLIPSE COPD subjects. Single nucleotide polymorphism (SNP)-probe set associations with FDR-adjusted p<0.05 are shown.
(XLS)
Cis-expression quantitative trait locus (eQTL) single nucleotide polymorphisms (SNPs) from sputum samples from 131 ECLIPSE COPD subjects associated with COPD case-control status in combined ECLIPSE, NETT-NAS, and Norway GWAS (Cho et al. 2010). SNPs associated at p<0.01 are shown.
(DOC)
Cis-expression quantitative trait locus (eQTL) single nucleotide polymorphisms (SNPs) from sputum samples from 131 ECLIPSE COPD subjects associated with COPD case-control status in a meta-analysis of ECLIPSE, NETT-NAS, and Norway GWAS (Cho et al. 2010). SNPs with combined p<0.01 are shown.
(DOC)