Abstract
Chromatin boundary elements serve as cis-acting regulatory DNA signals required to protect genes from the effects of the neighboring heterochromatin. In the yeast genome, boundary elements act by establishing barriers for heterochromatin spreading and are sufficient to protect a reporter gene from transcriptional silencing when inserted between the silencer and the reporter gene. Here we dissected functional topography of silencers and boundary elements within circular minichromosomes in Saccharomyces cerevisiae. We found that both HML-E and HML-I silencers can efficiently repress the URA3 reporter on a multi-copy yeast minichromosome and we further showed that two distinct heterochromatin boundary elements STAR and TEF2-UASrpg are able to limit the heterochromatin spreading in circular minichromosomes. In surprising contrast to what had been observed in the yeast genome, we found that in minichromosomes the heterochromatin boundary elements inhibit silencing of the reporter gene even when just one boundary element is positioned at the distal end of the URA3 reporter or upstream of the silencer elements. Thus the STAR and TEF2-UASrpg boundary elements inhibit chromatin silencing through an antisilencing activity independently of their position or orientation in S. cerevisiae minichromosomes rather than by creating a position-specific barrier as seen in the genome. We propose that the circular DNA topology facilitates interactions between the boundary and silencing elements in the minichromosomes.
Introduction
The DNA in the nuclei of eukaryotic cells is packed into nucleosomes, establishing the primary level of chromatin packing. The DNA transcription, replication, recombination, and repair processes occur in the context of DNA packed into nucleosome arrays [1], [2]. The structural-functional relationship between chromatin packing and DNA transcription is manifested by segregation of nuclear chromatin into the open and active euchromatin and the condensed and repressed heterochromatin [3], [4]. The question how the transcriptionally active euchromatin is functionally separated from the inactive heterochromatin has been of considerable interest.
Previous research has focused on identifying cis-acting genetic elements termed “boundary elements” that demarcate the heterochromatin from the euchromatin [5], [6]. Such DNA elements were identified in evolutionary diverse organisms ranging from yeast to humans [7]–[9]. The heterochromatin boundary elements establish boundaries of chromatin domains by limiting the spread of silencing signals to the adjoining regions [10]. These elements are especially important when transcriptionally active genes are surrounded by condensed heterochromatin as they stop the incursion of silencing signals from the surrounding regions thereby protecting the genes from position-dependent variegation.
Similar to positional effects in higher eukaryotes, in the budding yeast Saccharomyces cerevisiae the telomeres and the silent mating-type loci (HML and HMR), all represent well-defined heterochromatin domains, where genes are transcriptionally silent. The transcriptionally silent copies of the mating-type genes are located at the HML and HMR silent loci near telomeres. The HML and HMR silent loci are flanked by “Essential” (E) and “Important” (I) silencer elements [11]–[14]. The cis-acting E and I elements are necessary and sufficient for initiating and mediating silencing in an orientation-dependent manner by interacting with a large number of trans-acting factors to repress transcription [11], [13], [15], [16]. Transcriptional repression at HM loci is a gene-nonspecific event and the silencers can repress any reporter gene [17], [18].
The silent chromatin structure does not extend indefinitely and is restricted within the HM loci and telomeres by the heterochromatin boundary elements that block silencer-mediated repression of the gene in mating-type loci as well as shield repressive positional effects of telomeric heterochromatin [7], [19]–[21]. It has been suggested that the HML-I silencer may itself establish a heterochromatin boundary by organizing heterochromatin in a uni-directional manner within the HML locus [22]. Furthermore, a tRNA gene surrounding the HMR locus in S. cerevisiae has been shown to have barrier activity and restrict the spread of silencing from the HMR locus [7], [8]. In addition, two special heterochromatin boundary elements, STAR and TEF2-UASrpg, were shown to have boundary activity in S. cerevisiae genome [19], [20].
In S. cerevisiae, the chromosomal ends contain the X and/or Y' subtelomeric repeat elements abutting the telomeric repetitive DNA. Sequences within these X and Y' subtelomeric repeats block silencing, exhibiting heterochromatin barrier activity and thus are named subtelomeric anti-silencing regions (STAR). The STAR boundary element has also been shown to counteract the silencer-driven repression of reporter genes at the HML locus in the genome, when interposed between the silencer and the reporter gene without transcriptional activation of the reporter [20], [23].
The TEF2-UASrpg located on Chromosome II in S. cerevisiae is an example of heterochromatin boundary element. It was identified by the silencer-blocking assay by positioning the boundary element between the silencer and the reporter gene to test its ability to counteract the silencing mechanism. The TEF2 gene encodes the translational elongation factor-1alpha and the upstream activation sequence of TEF2 is able to block the silencing activity and the spread of heterochromatin without transcriptional activation. In the genome, when the TEF2-UASrpg was placed at the HML locus it was able to resist transcriptional silencing of native or reporter genes in a position and orientation dependent manner [19], [24].
Distinct models have been suggested for chromatin boundary formation. In one model, boundary elements act by creating nucleosomal gaps and establishing barriers for example when placed between the silencer and the regulated gene but not upstream of the silencer or downstream of the gene [24], [25], [26]. In the other model, the boundary element could form loops reaching out to and inhibiting silencers. Within this model the boundary elements may act independently of their position versus the silencer element and are assigned to have a desilencing or anti-silencing rather than barrier activity [27]–[31]. The exact molecular mechanism may vary between distinct boundary elements and different organisms and still remains largely unknown.
In order to understand the molecular mechanism of barrier formation by STAR and TEF2-UASrpg, two distinct heterochromatin boundary elements, we turned to the yeast minichromosome system. Yeast minichromosomes are multi-copy circular plasmids that assemble into chromatin in-vivo [32] and have been used to study nucleosome positioning, chromatin remodeling, and interaction of trans-acting factors with cis-acting elements [33]–[36].
To dissect the topographic relationship of the silencers and boundary elements on a minichromosome, we generated a number of minichromosomal constructs containing different combinations of the “E” and “I” silencers from the HML locus with STAR and TEF2-UASrpg heterochromatin boundary elements. The URA3 has been used as the reporter gene and the TRP1 as the selection marker in all the minichromosome constructs examined. Identification of whether the URA3 reporter gene is ON/OFF has been tested using 5-FOA biochemical selection screen in addition to growth on Uracil-deficient media.
We report here that the URA3 reporter gene was efficiently silenced by the E and/or I silencers in the absence of heterochromatin boundary elements and the URA3 reporter was de-repressed in the presence of STAR and TEF2-UASrpg elements in circular minichromosomes similar to previous studies in the yeast genome [19], [20]. However, our findings showed that the STAR and TEF2-UASrpg elements exhibit an antisilencing rather than boundary activity in S. cerevisiae minichromosomes. We propose that the topology of circular minichromosomes may help to bypass the strict positional requirements of chromatin boundaries that operate in linear chromosomes.
Results
Characterization of URA3-based reporter minichromosomes
The minichromosome constructs generated were tested for their functionality upon transformation into trp1- and ura3- deficient yeast strains. The dependence upon TRP1 selection marker has been used as a control for all experiments and the URA3 served as a reporter gene for transcriptional silencing in this study (Figure 1A). Cell growth on 5-FOA [37] and inability to grow on URA- media [38] indicates that the URA3 is repressed and therefore the cells are 5-FOA resistant, allowing us to identify whether the URA3 reporter is silenced in the presence of silencer elements and expressed in the presence of boundary elements. Recent reports point out to a potential problem with 5-FOA screenings, due to metabolic changes caused by the 5-FOA and suggest to check the reporter gene expression for epigenetic mechanisms or heterochromatic silencing studies [39], [40]. However, in our study we have also directly assessed the URA3 reporter gene activity in medium lacking uracil for analysis of URA3 expression in the presence or absence of silencers and boundary elements independently of the 5-FOA assays.
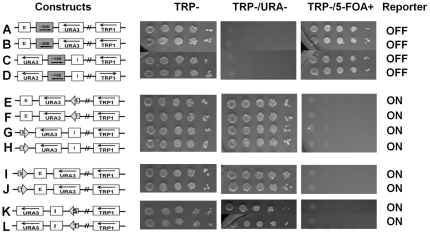
Figure 1. Schematic representation of the reporter, silencer, and boundary minichromosomal constructs.
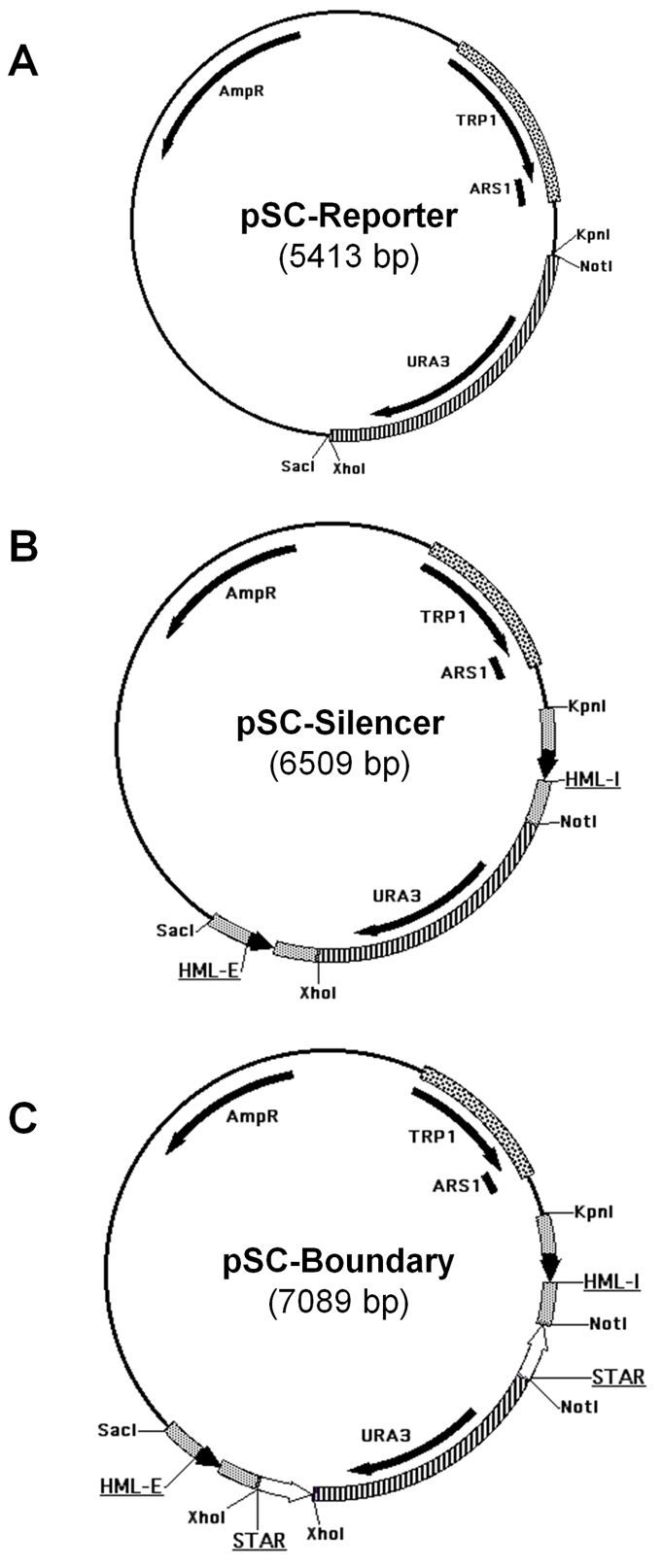
A: Physical map of the Reporter construct containing URA3 reporter gene, TRP1-ARS1 for selection and propagation in S. cerevisiae, and AmpR for modifications in E. coli. B: Physical map of the Silencer construct containing the HML - E and I silencer elements with flanking sequences on either side of the URA3 reporter gene. C: Physical map of one of the Boundary constructs. This example contains the STAR boundary element. Other boundary constructs may contain either TEF2-UASrpg or STAR element or both positioned either downstream or upstream of the silencing elements in different orientations (see schemes in Figures 2– 6).
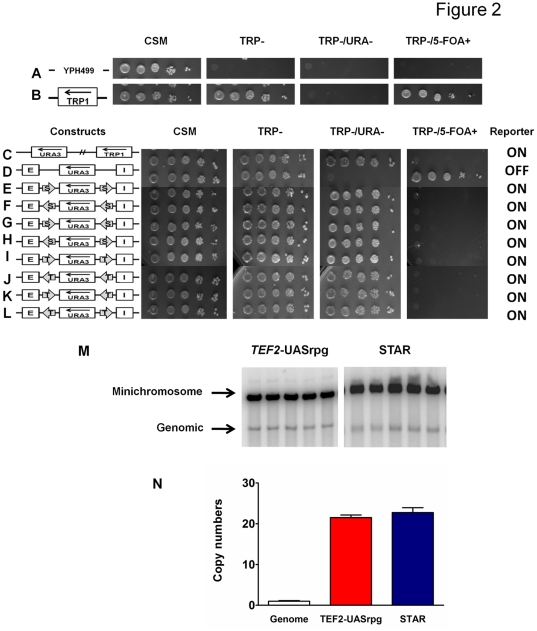
We checked the S. cerevisiae strain YPH499 (a-cells) for its genotype. These cells were only able to grow on complete synthetic media (CSM) without any dropouts, but could not grow on any selective media such as TRP− or TRP−/URA− or TRP−/5-FOA+ media (Figure 2A). In the presence of the TRP1 marker, the cells were able to grow on CSM, TRP- and TRP-/5-FOA+ selective media being 5-FOA resistant, but were unable to grow on TRP-/URA- due to the absence of URA3 gene product (Figure 2B). In the presence of both TRP1 marker and URA3 reporter, the cells were able to grow on CSM, TRP- and TRP-/URA- selective media, but were unable to grow on TRP-/5-FOA+ plates due to the presence of a functional URA3 gene product the cells exhibited sensitivity to 5-FOA (Figure 2C). Efficient ten-fold serial dilutions were established to cover the range of selection from ∼2×107 cells/ml to ∼2×103 cells/ml in the spotting assays. The copy numbers of the multi-copy S. cerevisiae minichromosomes were tested throughout this work and were found to be constant for different minichromosome constructs. The numbers of minichromosomes were determined to be ∼20 copies compared to the genomic copy by southern hybridization with specific TRP1-ARS1 probe and (Figure 2M) and quantified using the ImageQuant software (Figure 2N).
Figure 2. Boundary elements STAR and TEF2-UASrpg block the activity of the E and I silencers irrespective of their orientations.
A: Strain YPH499 (trp-, ura-) can grow on CSM, but is unable to grow on selective media conditions [TRP-; TRP-/URA-; TRP-/5-FOA+]. B: Minichromosome containing the TRP1 marker gene is able to grow on CSM; TRP-; unable to grown on TRP-/URA- (due to lack of URA3) and is 5-FOA resistant. C: Minichromosome containing both the TRP1 marker and the URA3 reporter genes is able to grow on CSM; TRP-; TRP-/URA- and is 5-FOA sensitive (due to the presence of the functional URA3 product). D: Minichromosome containing the HML-E and I silencer elements is able to silence the URA3 reporter gene and the cells are able to grow on CSM; TRP-; unable to grown on TRP-/URA- (due to lack of URA3) and is 5-FOA resistant. E–L: Two boundary elements (shown by arrows) STAR (S) and TEF2-UASrpg (T) were examined in different orientations in the presence of both the HML – E and I silencer elements. The URA3 reporter gene expression status (on or off) was assayed by the growth phenotypes of S. cerevisiae cells containing various minichromosome constructs tested in different selective media by serial dilutions. M: Southern blot of linearized minichromosomal and genomic DNA probed with radiolabeled TRP1-ARS1 containing fragment. Five independent clones transformed with minichromosome construct containing TEF2-UASrpg (left panel) and STAR (right panel). N: Graphic representation of minichromosome copy numbers quantified by scanning of the Southern blots (such as shown in Figure 2M) and normalized to the genomic TRP1-ARS1 signal. Error bars represent Standard Deviations.
Establishment of silencing in circular multi-copy minichromosomes
In order to study heterochromatin barrier function we had to establish robust silencing in the S. cerevisiae minichromosome system. Earlier reports indicate that silencing of a gene placed between the two silencer elements, HML – E and I, in the yeast genome is uniformly high and does not depend on the chromosomal context beyond the silenced locus [15], [22], [41].
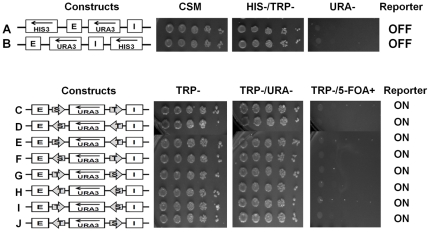
The silencer minichromosome construct (Figure 1B) has been designed to study the effects of both the HML “E” and the “I” silencer elements on silencing the expression of the URA3 reporter gene. The differences in growth phenotypes and the silencing of the URA3 reporter gene were studied in the absence (Figure 2C) or presence of both the HML-E and the HML-I silencers placed on either side of the URA3 reporter gene to reproduce the genomic topography as closely as possible (see schemes in Fig. 2D). Serial dilutions of the strains constructed were spotted for assaying the silencing efficiency under different selective media conditions (Figure 2C and 2D). Both HML – E and I silencers were capable of silencing the expression of the URA3 reporter gene, so these cells were unable to grow on TRP-/URA- and are 5-FOA resistant (Figure 2D). Using serial dilution assay, we observed that URA3 gene was repressed strongly enough to mimic the URA- phenotype of the control plasmid indicating that the silencers repressed the reporter gene completely. In contrast, the TRP1 marker gene located upstream of the HML – I silencer was not silenced by either E or I silencer in the circular minichromosomes, apparently because both HML – E and I silencers have directionality in the minichromosomes as seen in the genome and only silence the expression of genes positioned in between the silencers in an orientation-dependent manner [15], [22], [41]. As an additional control we also placed HIS3 marker gene upstream of the HML – E silencer and downstream of the I silencer, and we found that neither E nor I silencer were able to silence the expression of the HIS3 gene (Figure 3A and 3B).
Figure 3. Combination of two different boundary elements blocks the silencing of the reporter from both E and I silencers.
A: Control showing that E silencer in the minichromosome has directionality similar to the genome and only silences URA3, but not the HIS3 gene placed upstream of the E silencer. These cells are able to grow on CSM and HIS-/TRP-, but are unable to grow on URA- media. B: Control showing that I silencer in the minichromosome also has directionality similar to the genome (like the E silencer) and only silences URA3, but not the HIS3 gene placed upstream of the I silencer. These cells are able to grow on CSM and HIS-/TRP-, but are unable to grow on URA- media. C–J: Combination of two BE - boundary elements (shown by arrows) STAR (S) and TEF2-UASrpg (T) were examined in different positions and orientations in the presence of both the E and I silencers. The URA3 reporter gene expression status (on or off) was assayed as in Figure 2.
STAR and TEF2-UASrpg inhibit silencing of the reporter on yeast minichromosomes irrespective of its orientation
We examined if the URA3 reporter gene would be protected from silencing by two heterochromatin boundary elements, STAR and TEF2-UASrpg that are able to counteract the silencing in an orientation-dependent manner in the yeast genome [19], [20]. Here, we found that either two STAR elements (Figure 1C, 2E–H) or two TEF2-UASrpg elements (Figure 2I–L) bracketing the URA3 reporter gene were able to inhibit the silencing by the HML-E and HML-I silencers in an orientation-independent manner in S. cerevisiae minichromosomes (Figure 2E–L). There were no significant differences observed in the efficiency of the cell growth on TRP-/URA- selective media and growth inhibition on TRP-/5-FOA+ media. Thus in yeast minichromosome system, unlike the genomic studies [19], [20], the two boundary elements STAR and TEF2-UASrpg can block both the HML-E and HML-I silencers in an orientation-independent manner.
Combination of STAR and TEF2-UASrpg boundary elements on minichromosomes
To determine if two identical boundary elements are required at both ends to protect the URA3 reporter from the silencing effects of the HML - E and I silencer elements we have placed STAR on one end and TEF2-UASrpg heterochromatin boundary element at the other end. We found that all possible combinations of STAR and TEF2-UASrpg boundary elements on either ends of the URA3 in different orientations were able to protect the reporter gene from being silenced (Figure 3C–J). Thus in the context of minichromosomes two identical boundary elements at either end of the reporter gene are not required to counteract HML-E and HML-I driven silencing, and a combination of STAR and TEF2-UASrpg boundary elements are able to protect the URA3 from being silenced.
A single boundary element is sufficient to block a single silencer
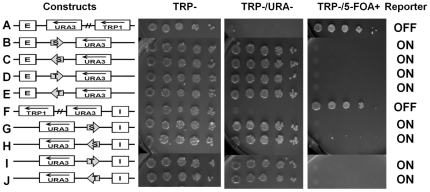
We asked if a single silencer either E or I is sufficient to silence the expression of the URA3 reporter gene in our minichromosome system. This was important to examine since due to limiting Sir protein concentrations (amount of silencing factors) in the yeast cells we expected a weaker silencing activity for the multi-copy minichromosomes than a single-copy silenced domain in the genome [42]. We first tested a minichromosome where only the HML–E silencer element has been inserted upstream of the URA3 reporter gene (Figure 4A). We found that the HML–E alone was capable of silencing the expression of the URA3 reporter gene, therefore these cells were unable to grow on TRP-/URA- plates and they are 5-FOA resistant due to the absence of the functional URA3 gene product (Figure 4A). It has been reported earlier that in the genome, the HML-E and the HML-I silencer elements were capable of silencing alone [19], [20], [43]. By series of dilutions, we have confirmed here that the HML-E silencer alone was able to silence the URA3 reporter gene on a minichromosome as efficiently as a pair of two silencers.
Figure 4. A single boundary element can counteract the silencing of the URA3 reporter by either E or I silencer.
Single BE (shown by arrows) either STAR (S) or TEF2-UASrpg (T) were examined in the presence of a single E silencer (A–E) or a single I silencer (F–J) in different orientations. The URA3 reporter gene expression status (on or off) was assayed as in Figure 2.
In another series of experiments we observed similar results with a single HML–I silencer element has been inserted downstream of the URA3 reporter gene (Figure 4F). Thus the minichromosome-borne E and I silencers are both functional and efficient in silencing URA3 reporter gene on multi-copy circular minichromosomes. Each single silencer, either E or I, is capable and sufficient to silence the reporter gene even in the absence of the other silencer in the minichromosome constructs.
Next, we examined if a single heterochromatin boundary element, either STAR or TEF2-UASrpg, was sufficient to block silencing imposed by HML-E or HML-I on circular minichromosomes. The boundary elements STAR or TEF2-UASrpg were positioned between either the HML-E or HML-I silencer elements and the URA3 reporter. As observed in the genome, we found that a single boundary element STAR or TEF2-UASrpg was sufficient in blocking the silencing imposed by a single silencer either HML-E or HML-I in any orientation (Figure 4B–E and G–J).
A single boundary is sufficient to protect URA3 from silencing in presence of two silencer elements in circular minichromosomes
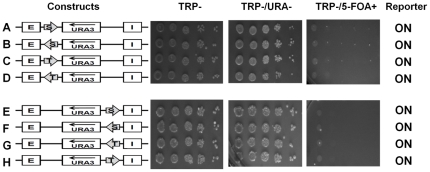
To determine if the STAR or TEF2-UASrpg elements behave as barriers in minichromosomes or they are able to overcome the silencing of both the E and I silencers in the presence of only one boundary element, we positioned a single boundary element downstream of either the E or the I silencer, leaving the other silencer upstream or downstream of the URA3 reporter intact (Figure 5A–D and E–H). We found that a single boundary element was able to overcome the silencing of both the E and I silencer elements on the URA3 reporter, even though the URA3 was protected only from one side and there was an equal opportunity for URA3 to be silenced by the other silencer. This finding is in striking contrast to the previous studies in linear chromosomes where the reporter gene had to be bracketed by two boundary elements to prevent the silencing in presence of both the silencers [19], [20].
Figure 5. A single boundary efficiently blocks the silencing of the URA3 reporter from both the E and I silencers.
Single BE (shown by arrows) either STAR (S) or TEF2-UASrpg (T) were examined in the presence of both the HML – E and I silencer elements, placing the BE between E and URA3 reporter (A–D) or between I and URA3 reporter (E–H) in different orientations and the reporter gene on/off was determined by the growth phenotypes of the yeast cells tested in different selective media.
STAR and TEF2-UASrpg sequences are specific in blocking of silencing
To confirm the specificity of the STAR and TEF2-UASrpg sequences in inhibiting silencing in the context of minichromosomes we used DNA sequences of similar length from the Leu2 ORF in different orientations replacing the ∼300 bp of STAR and ∼150 bp of TEF2-UASrpg sequences. The control sequences inserted either between the HML-E or the HML-I silencer and the URA3 reporter gene were unable to block the silencing mediated by the silencers and the URA3 reporter was completely repressed by either E or I silencer elements (Figure 6A–D). Thus, blocking of the silencing activity is specific for the STAR and TEF2-UASrpg DNA sequences in the minichromosomes, as random sequences of similar length to STAR or TEF2-UASrpg were unable to protect URA3 reporter from being silenced.
Figure 6. STAR or TEF2-UASrpg activity is sequence-specific and acts by imposing antisilencing.
A–D: Minichromosome constructs with ∼300 bp or ∼150 bp of Leu2 ORF sequences replacing the STAR or TEF2-UASrpg boundary element were positioned in between the E silencer or the I silencer and the URA3 reporter. The E or I silencers was capable of silencing the expression of the URA3 reporter gene. The cells were able to grow on TRP- and TRP-/5-FOA+ media being 5-FOA resistant, but unable to grow on TRP-/URA- selective media. E–H: Boundary elements positioned downstream of the silencer and the URA3 reporter gene are able to block the silencing of the URA3 reporter independent of its position unlike in the genome. These cells are able to grow on TRP- and TRP-/URA- media, but are unable to grow on TRP-/5-FOA+, as the URA3 gene is not repressed and exhibits sensitivity to 5-FOA. I–L: STAR and TEF2-UASrpg are positioned upstream of the HML-E and I silencer and the URA3 reporter gene. Unlike in the genome where the BE has to be positioned in between the reporter and the silencer, in minichromosomes the upstream BE is able to counteract silencing exhibiting antisilencing mechanism. These cells are able to grow on TRP- and TRP-/URA- media, but are unable to grow on TRP-/5-FOA+, as the URA3 gene is not repressed and exhibits sensitivity to 5-FOA.
STAR and TEF2-UASrpg boundary elements exhibit antisilencing activity in S. cerevisiae minichromosomes
To further dissect the mechanism of inhibition of silencing, we conducted a genetic test showing whether STAR and TEF2-UASrpg act as barriers or exhibit antisilencing activity in minichromosomes. The anti-silencing activity is defined as an ability of a boundary element to block the silencing independently of its position in relation to the silencer or the reporter distinct from the desilencing activity [28], [30]. We therefore positioned the STAR and TEF2-UASrpg elements downstream of the HML-E or the HML-I silencers (Figure 6E–H). We found that either STAR or TEF2-UASrpg was able to protect the URA3 from being silenced even though they were not interposed between the silencer and the reporter, but was instead placed downstream of the silencers (Figure 6E–H).
Unlike the position downstream of the silencer elements, at which there is a possibility of competition between the boundary and the silencer to either activate or repress URA3, we next placed the STAR and TEF2-UASrpg upstream of the HML-E or the HML-I silencers and the URA3 reporter gene where the silencer is in closer proximity to the reporter than the boundary. We found that in this setting the STAR and TEF2-UASrpg displayed a strong antisilencing activity (Figure 6I–L) as effectively as it had already been observed with positioning STAR and TEF2-UASrpg downstream of the silencers. Only one copy of either STAR or TEF2-UASrpg irrespective of its position in relation to the silencers or the reporter was sufficient to stop the silencing of the URA3 reporter. Thus, in contrast to linear chromosomes where the STAR or TEF2-UASrpg boundary elements show a position- and orientation-dependent barrier function [19], [20], in the S. cerevisiae minichromosomes these elements restrict silencing in a position and orientation independent manner and exhibit an antisilencing rather than barrier activity.
Discussion
The objective of this study is creating a minichromosome model system recapitulating functional and spatial relationships between genetic elements controlling heterochromatin in yeast and facilitating its topographic analysis. We conducted a detailed characterization of STAR and TEF2-UASrpg heterochromatin boundary elements in minichromosomes in S. cerevisiae to determine if these elements act as barriers separating an active locus from silenced locus or are they able to inhibit silencing where their topology is not essential. We found that S. cerevisiae can maintain and pack episomal DNA into chromatin and minichromosomes at a stable copy number and thus provide a robust model for studying relationships between the heterochromatin elements and gene regulation independently of the chromosomal context. Our newly established minichromosome system can be employed as a screen for testing other candidate barriers elements such as tRNA genes and positioned nucleosomes.
We used the HML-E and HML-I silencer elements for our study, since both the E and I silencers at the HML locus are equally capable in silencing, unlike the silencers at the HMR locus [11], [17] and as the HML silencers have been used in previous experiments with STAR and TEF2-UASrpg barriers [19], [20]. In the genomic HM loci the E and I silencers are ∼3.5 Kb apart and the silencing is known to work if that distance is increased only up to a certain extent (∼6–7 Kb), after which the silencing activity decreases [7], [17], [29], [44], [45] as it is limited by the silencing propagating factors such as Sir3 [45]. In this study, in the minichromosome context, the silencing-initiating HML– E and I elements are ∼2 Kb apart but with ∼20 copies of the minichromosome the total DNA length through which silencing is propagated in the minichromosomes exceeds ∼10-fold the effective spreading length limit between the HML- E and I silencer elements in the genome. We found that the URA3 reporter gene was completely repressed in all the ∼20 minichromosomal copies in the yeast cells, since expression of only one gene copy was sufficient for growth on URA- media as well as for inhibiting growth on 5-FOA. As the plasmid-borne silencers are less constrained than those in the genome, the silencers on minichromosomes can effectively silence the reporter genes and efficiently maintain a total length of ∼40 Kb silenced loci on minichromosomes. Similar to earlier reports, the silencing in the minichromosomes is much more robust and multi-fold higher than seen in the genome [46]. Furthermore, consistent with earlier reports [11], [47], stating that a single silencer element is capable of acting alone in the genome, we have shown for the first time that a single silencer element (either HML – E or I) is sufficient in silencing the URA3 reporter gene even in circular multi-copy minichromosomes. Thus we were able to construct a circular minichromosome model system where both HML- E and I silencers were functional and efficient in silencing the URA3 reporter gene in multi-copy minichromosomes in the yeast cells.
Surprisingly, in sharp contrast to the genome, where the STAR or TEF2-UASrpg are known to block the spreading of silencing acting as barriers, i.e. only when interposed between a silencer and the reporter gene [19], [20] with minichromosomes, we found that both STAR and TEF2-UASrpg were able to inhibit the silencing of URA3 irrespective of their orientations and positions in relation to the silencer or the reporter. In minichromosomes the STAR and TEF2-UASrpg exhibit position-independency and antisilencing activity where only one copy of either the STAR or TEF2-UASrpg is sufficient in inhibiting silencing of the URA3 reporter gene. Although the silencer elements exhibit efficient silencing of the reporter gene in a direction-dependent manner, similar to what is exhibited by silencer elements in the genome, we cannot rule out that the altered function of barrier elements on minichromosomes is (at least partially) due to the altered properties of the silencer.
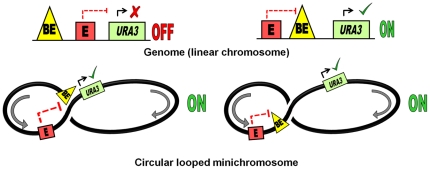
To explain the antisilencing mechanism observed in the circular minichromosomes, we propose that within a minichromosome, its circular topology would promote interactions between the boundary element and the silencer bypassing the topographical constraints (Figure 7). This would allow the boundary element to block silencing of the URA3 reporter gene by the E silencer, irrespective of the position of the boundary element in relation to the silencer or the reporter. Thus unlike in the genome, the STAR or TEF2-UASrpg elements in the S. cerevisiae minichromosomes are capable of inhibiting the silencing of the URA3 reporter gene by the HML-E and HML-I silencers in a position independent manner.
Figure 7. Model illustrating antisilencing activity of boundary element facilitated by minichromosome looping.
Top panels: Within the genome, the boundary element (BE) would block silencing when located between the silencer (E) and the URA3 gene but not upstream of the silencer. Bottom panels: In the minichromosomes, a close contact between the silencer (E) and the boundary element - BE (either STAR or TEF2-UASrpg) established by looping would prevent the silencing of the URA3 reporter by the silencer (E) irrespective of the position of the boundary element in relation to either the silencer or the URA3 reporter gene.
Why do boundary elements act so differently on a minichromosome as compared to the genome? In the genome, the silent loci are known to have an altered chromatin organization that, in addition to spreading of the silencing complex and histone deacetylation, also involves chromatin higher-order structural transitions [44], [48]–[50]. In the genome, the boundary elements need to be positioned between the silencer element and the promoter because otherwise they will be spatially hindered from the silenced region that forms a stable fold-back loop. Perhaps, this is not the case with the minichromosome where sliding of the interwound DNA helices of the covalently closed supercoiled DNA ring against each other “slithering” [51] facilitates DNA interaction at a distance and brings distal regulatory elements into close contact [52]. Furthermore, on a minichromosome associated with nucleosomes, the nucleosomal folding per se promotes contacts between the distal regulatory elements without supercoiling [53]. Looping of the telomeric heterochromatin has been shown to facilitate transcription by bringing Upstream Activating Sequences in a close contact with promoter [54] when the former was positioned downstream of the gene. We suggest that a facilitated looping may explain the more efficient functioning of the boundary elements in the minichromosome where they could find an easier access to the promoter (Figure 7) and out-compete silencers even when positioned downstream of the gene or upstream of the silencer. Based on the study of DNA minicircles excised from the yeast genome, it has been previously shown that DNA topology changes are associated with silencing [55]. Here we propose that DNA topology may regulate interactions between the boundary and silencing elements in the genome and its effect on silencing can be functionally dissected in the minichromosome system.
It is known that plasmid-borne silencers exhibit very strong silencing [46], similarly we show here that the STAR and TEF2-UASrpg may exhibit robust antisilencing in a minichromosomal environment. This “easy come – easy go” mode of silencing on a minichromosome implies that in this system the heterochromatin structure is relatively relaxed compared to the more rigid chromatin organization in the genome. The yeast minichromosome system that we have genetically characterized here is especially suited for isolating of the minichromosome in different functional states [32], [56] for subsequent ultrastructural analysis that may finally clarify the actual 3D chromatin organization of the silent and active minichromosomes.
Materials and Methods
Minichromosome Constructs
A) Reporter constructs:
The URA3 reporter minichromosome construct was generated by inserting ∼1.3 kb FspI DNA fragment from the YIp5 plasmid containing ∼800 bp URA3 reporter gene (GenBank accession number NC_001137.3, Chromosome V, 116167 to 116970). The URA3 reporter gene (FspI fragment) is cleaved from YIp5 plasmid and inserted at the multiple cloning site (SmaI) of the ALT (ARS1, lac-operator, TRP1) plasmid. All minichromosome shuttle vectors contain an autonomously replicating sequence - ARS1 (GenBank accession number NC_001136.10, Chromosome IV, 462354 to 463192), and a selectable marker - TRP1 gene for selection in S. cerevisiae [56] (GenBank accession number NC_001136.10, Chromosome IV, 461842 to 462516) and a pBR322 vector-derived sequence with an AmpR gene for selection and ColE1 origin for propagation in E. coli. The URA3 reporter is adjacent to the TRP1 marker gene and in the same orientation in all the minichromosomal constructs (Figure 1A).
B) Silencer constructs:
The silencer constructs were generated by inserting HML-E and HML-I silencer elements into the minichromosome backbone. An ∼500 bp fragment containing the “E silencer” with ∼200 bp upstream and downstream flanking sequences (GenBank accession number NC_001135.5; chromosome III, 10966 to 11499), were PCR amplified from the HML locus using genomic DNA and primers with unique SacI and XhoI restriction enzyme sites for integrating into the minichromosome (Table S1). Similarly an ∼500 bp fragment containing the “I silencer” with ∼200 bp upstream and downstream flanking sequences (GenBank accession number NC_001135.5, Chromosome III, 14364 to 14912), were PCR amplified from the HML locus using genomic DNA and primers with unique NotI and KpnI restriction enzyme sites (Table S1) for integrating into the minichromosome and were verified by DNA sequencing. The PCR products were cloned into pGEMT vectors. The silencer elements were excised from the pGEMT vectors and ligated into the minichromosome constructs. The SacI site in the minichromosome vector was too close to the XhoI site, this problem was overcome by adding a short DNA linker containing a SacI site. The HML - E and I silencer elements have the same directionality in the minichromosome as in the genome (Figure 1B). The control HIS3 gene (GenBank accession number NC_001147.6, Chromosome XV, 721946 to 722608) was PCR amplified from pRS413 vector (ATCC pRS series) and inserted at the SacI restriction site upstream of the E silencer or at the BamHI restriction site upstream of the I silencer. The HIS3 is in the same orientation as the TRP1 and the URA3 in the minichromosome constructs.
C) Boundary constructs:
The heterochromatin boundary element constructs were generated by inserting TEF2-UASrpg and STAR boundary elements in the minichromosome. The ∼150 bp TEF2-UASrpg (GenBank accession number NC_001134.8, Chromosome II, 477109 to 477257) and ∼300 bp STAR (GenBank accession number NC_001143.9, Chromosome XI, 70 to 345) were PCR amplified from genomic DNA using specific primer sets with unique restriction enzyme sites (Table S1) and verified by DNA sequencing. The PCR products were cloned into pGEMT vectors. The boundary elements were excised from the pGEMT vectors and ligated into the minichromosome constructs. The TEF2-UASrpg and STAR boundary elements has XhoI ends inserted in between the E-silencer and the URA3-reporter and has NotI ends inserted in between the I-silencer and URA3-reporter in the minichromosomes (Figure 1C). The STAR and TEF2-UASrpg were also positioned upstream of the E or the I silencer using SacI and BamHI restriction sites. The TEF2-UASrpg and the STAR boundary elements have been inserted in both orientations and in different positions in the various minichromosome constructs. The control sequences of ∼300 bp (similar to STAR in length) and the ∼150 bp (similar to TEF2-UASrpg in length) were PCR amplified from Leu2 ORF (GenBank accession number NC_001135.5) from pRS415 vector (ATCC pRS series) and inserted at the XhoI or NotI restriction sites between the E or I silencer and the URA3 reporter replacing STAR or TEF2-UASrpg boundary elements.
Yeast strains and media
All minichromosome constructs were transformed into E. coli DH5α competent cells and bacterial colonies were screened using restriction enzyme digests, PCR analysis and DNA sequencing. The minichromosome constructs isolated from bacteria were re-transformed into S. cerevisiae a-cells YPH499 strain (MATa, ade2–101°, his3-Δ200, leu2-Δ1, lys2–801a, trp1-Δ63, ura3–52) [57].
Yeast colonies grown on complete synthetic media lacking tryptophan (TRP-) were selected for all minichromosome constructs containing TRP1 marker gene in the construct backbone. The functionality of the regulatory elements in various minichromosome constructs and the expression of the URA3 reporter gene in the presence or absence of the HML-E and I silencer elements and STAR or TEF2-UASrpg boundary elements were determined using different selective media. The yeast colonies were grown on CSM (complete synthetic media, TRP- (lacking tryptophan), TRP-/URA- (lacking both tryptophan and uracil), TRP-/5-FOA+ (lacking tryptophan, but containing 5-fluro-orotic acid) and HIS- (lacking histidine).
Southern Hybridization
The minichromosome DNA integrity and copy number were examined by Southern blotting. DNA was purified, linearized with XmnI restriction enzyme digestion, subjected to electrophoretic separation on 1% agarose gel, and then transferred to Hybond-NX membrane (Amersham Biosciences), as per standard procedures [58]. The DNA was cross-linked to the membranes with UV light, and hybridized with TRP1-ARS1 specific minichromosome probe (∼1.4 kb EcoRI fragment) that was gel purified and random primer labeled with [α-32P] dATP. After hybridization and washing the membranes were exposed to imaging screen (Bio-Rad) and the signal intensities were analyzed using Typhoon 9400 Phosphoimager (Amersham Biosciences) and quantified by the ImageQuant 5.2 software (Molecular Dynamics). The genomic hybridization signal was normalized to the size of the genomic TRP1-ARS1 fragment recognized by the probe to determine the copy numbers.
Spotting Assay
All yeast strains containing different minichromosomal constructs were grown to mid-log phase (A600 of ∼1.0) in liquid TRP- media with 2% dextrose at 30°C with aeration by shaking at 250 RPM. The a-cells not containing any minichromosome construct were grown in CSM. The optical density of all yeast cultures were adjusted to absorbance 1 at 600 nm wavelength containing ∼2×107 cells/ml. Ten-fold serial dilution up to ∼2×103 cells/ml of each strain was made for the spotting assay to assess URA3 expression for assaying the silencing and insulating efficiency of the strains under different growth conditions [20], [46]. For each strain at least 6 independent transformants were verified by Southern blot analysis. Transformed cells from isolated colonies were inoculated and grown in TRP- liquid medium and spotted on to different selective media CSM, TRP-, TRP-/URA- (to check if 5-FOA resistance is due to silencing and not due to URA3 mutation), and TRP-/5-FOA+ [Toronto Research Chemicals]. Cells with repressed URA3 are able to form colonies in the presence of 5-FOA compound known to be toxic for cells expressing a functional URA3 gene [38]. The selective media plates were spotted with 5 µl cells per spot and grown for 2 days at 30°C prior to imaging the plates to study the differences in growth phenotypes.
Supporting Information
List of primers used in this study.
(DOCX)
Acknowledgments
We thank the Department of Biochemistry and Molecular Biology (Penn State, University Park) for equipment support, Drs. John Diller, Abid Kazi, Ralph Keil, and Laura Carrel for their technical advice, reagents and helpful discussions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Funding support was received from National Science Foundation grant MCB-1021681 to SAG and by the American Association of University Women International Doctoral Fellowship Award to SAC. This project was also funded, in part, by the Department of Biochemistry and Molecular Biology (Penn State, University Park) and a grant from the Pennsylvania Department of Health using Tobacco Settlement Funds. The Department specifically disclaims responsibility for any analyses, interpretations, or conclusions. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg R, Lorch Y. Chromatin rules. Nature Structural and Molecular Biology. 2007;14:986–988. [Google Scholar]
- 3.Rusche LN, Lynch PJ. Assembling heterochromatin in the appropriate places: A boost is needed. Journal of cellular physiology. 2009;219:525–528. doi: 10.1002/jcp.21749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oliver B, Parisi M, Clark D. Gene expression neighborhoods. Journal of biology. 2002;1:4. doi: 10.1186/1475-4924-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donze D, Kamakaka RT. Braking the silence: how heterochromatic gene repression is stopped in its tracks. Bioessays. 2002;24:344–349. doi: 10.1002/bies.10072. [DOI] [PubMed] [Google Scholar]
- 6.Giles KE, Gowher H, Ghirlando R, Jin C, Felsenfeld G. Chromatin Boundaries, Insulators, and Long-Range Interactions in the Nucleus. Cold Spring Harbor symposia on quantitative biology. 2010;75:006. doi: 10.1101/sqb.2010.75.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donze D, Kamakaka RT. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. Embo J. 2001;20:520–531. doi: 10.1093/emboj/20.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurudatta BV, Corces VG. Chromatin insulators: lessons from the fly. Briefings in functional genomics & proteomics. 2009;8:276–282. doi: 10.1093/bfgp/elp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 11.Mahoney DJ, Broach JR. The HML mating-type cassette of Saccharomyces cerevisiae is regulated by two separate but functionally equivalent silencers. Mol Cell Biol. 1989;9:4621–4630. doi: 10.1128/mcb.9.11.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brand AH, Breeden L, Abraham J, Sternglanz R, Nasmyth K. Characterization of a “silencer” in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell. 1985;41:41–48. doi: 10.1016/0092-8674(85)90059-5. [DOI] [PubMed] [Google Scholar]
- 13.Feldman JB, Hicks JB, Broach JR. Identification of sites required for repression of a silent mating type locus in yeast. J Mol Biol. 1984;178:815–834. doi: 10.1016/0022-2836(84)90313-9. [DOI] [PubMed] [Google Scholar]
- 14.Abraham J, Nasmyth KA, Strathern JN, Klar AJ, Hicks JB. Regulation of mating-type information in yeast. Negative control requiring sequences both 5′ and 3′ to the regulated region. J Mol Biol. 1984;176:307–331. doi: 10.1016/0022-2836(84)90492-3. [DOI] [PubMed] [Google Scholar]
- 15.Shei GJ, Broach JR. Yeast silencers can act as orientation-dependent gene inactivation centers that respond to environmental signals. Mol Cell Biol. 1995;15:3496–3506. doi: 10.1128/mcb.15.7.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahoney DJ, Marquardt R, Shei GJ, Rose AB, Broach JR. Mutations in the HML E silencer of Saccharomyces cerevisiae yield metastable inheritance of transcriptional repression. Genes & development. 1991;5:605–615. doi: 10.1101/gad.5.4.605. [DOI] [PubMed] [Google Scholar]
- 17.Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Annu Rev Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 18.Holmes SG, Broach JR. Silencers are required for inheritance of the repressed state in yeast. Genes & development. 1996;10:1021–1032. doi: 10.1101/gad.10.8.1021. [DOI] [PubMed] [Google Scholar]
- 19.Bi X, Broach JR. UASrpg can function as a heterochromatin boundary element in yeast. Genes Dev. 1999;13:1089–1101. doi: 10.1101/gad.13.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fourel G, Revardel E, Koering CE, Gilson E. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. Embo J. 1999;18:2522–2537. doi: 10.1093/emboj/18.9.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamakaka RT. Silencers and locus control regions: opposite sides of the same coin. Trends Biochem Sci. 1997;22:124–128. doi: 10.1016/s0968-0004(96)10074-8. [DOI] [PubMed] [Google Scholar]
- 22.Bi X, Braunstein M, Shei GJ, Broach JR. The yeast HML I silencer defines a heterochromatin domain boundary by directional establishment of silencing. Proc Natl Acad Sci U S A. 1999;96:11934–11939. doi: 10.1073/pnas.96.21.11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fourel G, Boscheron C, Revardel E, Lebrun E, Hu YF, et al. An activation-independent role of transcription factors in insulator function. EMBO Rep. 2001;2:124–132. doi: 10.1093/embo-reports/kve024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bi X, Broach JR. Chromosomal boundaries in S. cerevisiae. Curr Opin Genet Dev. 2001;11:199–204. doi: 10.1016/s0959-437x(00)00179-9. [DOI] [PubMed] [Google Scholar]
- 25.Fourel G, Magdinier F, Gilson E. Insulator dynamics and the setting of chromatin domains. BioEssays : news and reviews in molecular, cellular and developmental biology. 2004;26:523–532. doi: 10.1002/bies.20028. [DOI] [PubMed] [Google Scholar]
- 26.Bi X, Yu Q, Sandmeier JJ, Zou Y. Formation of boundaries of transcriptionally silent chromatin by nucleosome-excluding structures. Mol Cell Biol. 2004;24:2118–2131. doi: 10.1128/MCB.24.5.2118-2131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 2002;109:551–562. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- 28.Ishii K, Laemmli UK. Structural and dynamic functions establish chromatin domains. Mol Cell. 2003;11:237–248. doi: 10.1016/s1097-2765(03)00010-8. [DOI] [PubMed] [Google Scholar]
- 29.Kamakaka RT. Chromatin: a connection between loops and barriers? Curr Biol. 2002;12:R535–537. doi: 10.1016/s0960-9822(02)01032-1. [DOI] [PubMed] [Google Scholar]
- 30.Stover DM, Zehner ZE. Identification of a cis-acting DNA antisilencer element which modulates vimentin gene expression. Molecular and cellular biology. 1992;12:2230–2240. doi: 10.1128/mcb.12.5.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schedl P, Broach JR. Making good neighbors: the right fence for the right job. Nature structural biology. 2003;10:241–243. doi: 10.1038/nsb0403-241. [DOI] [PubMed] [Google Scholar]
- 32.Simpson RT, Ducker CE, Diller JD, Ruan C. Purification of native, defined chromatin segments. Methods Enzymol. 2004;375:158–170. doi: 10.1016/s0076-6879(03)75010-1. [DOI] [PubMed] [Google Scholar]
- 33.Simpson RT. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990;343:387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- 34.Patterton HG, Simpson RT. Nucleosomal location of the STE6 TATA box and Mat alpha 2p-mediated repression. Mol Cell Biol. 1994;14:4002–4010. doi: 10.1128/mcb.14.6.4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morse RH. Nucleosome disruption by transcription factor binding in yeast. Science. 1993;262:1563–1566. doi: 10.1126/science.8248805. [DOI] [PubMed] [Google Scholar]
- 36.Kim Y, Shen CH, Clark DJ. Purification and nucleosome mapping analysis of native yeast plasmid chromatin. Methods. 2004;33:59–67. doi: 10.1016/j.ymeth.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 37.Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods in enzymology. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 38.Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Molecular & general genetics : MGG. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 39.Rossmann MP, Luo W, Tsaponina O, Chabes A, Stillman B. A common telomeric gene silencing assay is affected by nucleotide metabolism. Molecular cell. 2011;42:127–136. doi: 10.1016/j.molcel.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takahashi YH, Schulze JM, Jackson J, Hentrich T, Seidel C, et al. Dot1 and Histone H3K79 Methylation in Natural Telomeric and HM Silencing. Molecular cell. 2011;42:118–126. doi: 10.1016/j.molcel.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bi X. Domains of gene silencing near the left end of chromosome III in Saccharomyces cerevisiae. Genetics. 2002;160:1401–1407. doi: 10.1093/genetics/160.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, et al. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes & development. 1996;10:1796–1811. doi: 10.1101/gad.10.14.1796. [DOI] [PubMed] [Google Scholar]
- 43.Rusche LN, Kirchmaier AL, Rine J. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2207–2222. doi: 10.1091/mbc.E02-03-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loo S, Rine J. Silencers and domains of generalized repression. Science. 1994;264:1768–1771. doi: 10.1126/science.8209257. [DOI] [PubMed] [Google Scholar]
- 45.Boscheron C, Maillet L, Marcand S, Tsai-Pflugfelder M, Gasser SM, et al. Cooperation at a distance between silencers and proto-silencers at the yeast HML locus. The EMBO journal. 1996;15:2184–2195. [PMC free article] [PubMed] [Google Scholar]
- 46.Lebrun E, Fourel G, Defossez PA, Gilson E. A methyltransferase targeting assay reveals silencer-telomere interactions in budding yeast. Mol Cell Biol. 2003;23:1498–1508. doi: 10.1128/MCB.23.5.1498-1508.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 48.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein K. SIR2 and SIR4 interactions differ in core extended telomeric heterochromatin in yeast. GenDevel. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 49.de Bruin D, Kantrow SM, Liberatore RA, Zakian VA. Telomere folding is required for the stable maintenance of telomere position effects in yeast. Mol Cell Biol. 2000;20:7991–8000. doi: 10.1128/mcb.20.21.7991-8000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valenzuela L, Dhillon N, Dubey RN, Gartenberg MR, Kamakaka RT. Long-range communication between the silencers of HMR. Mol Cell Biol. 2008;28:1924–1935. doi: 10.1128/MCB.01647-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang J, Schlick T, Vologodskii A. Dynamics of site juxtaposition in supercoiled DNA. Proc Natl Acad Sci U S A. 2001;98:968–973. doi: 10.1073/pnas.98.3.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bondarenko VA, Jiang YI, Studitsky VM. Rationally designed insulator-like elements can block enhancer action in vitro. Embo J. 2003;22:4728–4737. doi: 10.1093/emboj/cdg468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rubtsov MA, Polikanov YS, Bondarenko VA, Wang YH, Studitsky VM. Chromatin structure can strongly facilitate enhancer action over a distance. Proc Natl Acad Sci U S A. 2006;103:17690–17695. doi: 10.1073/pnas.0603819103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Bruin D, Zaman Z, Liberatore RA, Ptashne M. Telomere looping permits gene activation by a downstream UAS in yeast. Nature. 2001;409:109–113. doi: 10.1038/35051119. [DOI] [PubMed] [Google Scholar]
- 55.Ansari A, Gartenberg MR. Persistence of an alternate chromatin structure at silenced loci in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:343–348. doi: 10.1073/pnas.96.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ducker CE, Simpson RT. The organized chromatin domain of the repressed yeast a cell-specific gene STE6 contains two molecules of the corepressor Tup1p per nucleosome. Embo J. 2000;19:400–409. doi: 10.1093/emboj/19.3.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ausubel F. Current protocols in molecular biology. Brooklyn, NYPa., Media: J. Wiley & Sons Inc; 1999. pp. 2.9.1–2.9-20. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of primers used in this study.
(DOCX)