Abstract
Skin protects the body from the environment and is an important component of the innate and adaptive immune systems. Atopic dermatitis and contact dermatitis are among the most frequent inflammatory skin diseases and are both determined by multigenic predisposition, environmental factors, and aberrant immune response. Peptidoglycan Recognition Proteins (Pglyrps) are expressed in the skin and we report here that they modulate sensitivity to experimentally-induced atopic dermatitis and contact dermatitis. Pglyrp3 −/− and Pglyrp4 −/− mice (but not Pglyrp2 −/− mice) develop more severe oxazolone-induced atopic dermatitis than wild type (WT) mice. The common mechanism underlying this increased sensitivity of Pglyrp3 −/− and Pglyrp4 −/− mice to atopic dermatitis is reduced recruitment of Treg cells to the skin and enhanced production and activation Th17 cells in Pglyrp3 −/− and Pglyrp4 −/− mice, which results in more severe inflammation and keratinocyte proliferation. This mechanism is supported by decreased inflammation in Pglyrp3 −/− mice following in vivo induction of Treg cells by vitamin D or after neutralization of IL-17. By contrast, Pglyrp1 −/− mice develop less severe oxazolone-induced atopic dermatitis and also oxazolone-induced contact dermatitis than WT mice. Thus, Pglyrp3 and Pglyrp4 limit over-activation of Th17 cells by promoting accumulation of Treg cells at the site of chronic inflammation, which protects the skin from exaggerated inflammatory response to cell activators and allergens, whereas Pglyrp1 has an opposite pro-inflammatory effect in the skin.
Introduction
Skin protects the body from the environment and is the largest organ in mammals. Besides forming a mechanical barrier, skin is an important component of the innate and adaptive immune systems rich in anti-microbial peptides and antigen-sensing cells, and it maintains the proper homeostatic balance between pro- and anti-inflammatory responses. Atopic dermatitis and contact dermatitis are among the most frequent inflammatory skin diseases, both determined by multigenic predisposition, environmental factors, and aberrant immune response. Atopic dermatitis has a prevalence of 15–30% in children and 2–10% in adults, involves loss of barrier function of the skin and type I hypersensitivity to environmental allergens, and is manifested by pruritic erythematosus skin eruptions and increased IgE response, often with aggravating bacterial infections [1]–[3]. Allergic contact dermatitis (contact hypersensitivity) has a prevalence of 2–20% and involves type IV (delayed-type) hypersensitivity to environmental allergens, manifested by erythematosus skin infiltrations with inflammatory cells [2], [4], [5].
Peptidoglycan Recognition Proteins (PGRPs or Pglyrps) are a family of innate immunity proteins expressed in the skin. PGRPs are conserved from insects to mammals, recognize bacterial peptidoglycan, and function in antibacterial immunity. Mammals have four PGRPs, Pglyrp1, Pglyrp2, Pglyrp3, and Pglyrp4, which were initially named PGRP-S, PGRP-L, PGRP-Iα, and PGRP-Iβ, respectively [6], [7]. Three PGRPs, Pglyrp1, Pglyrp3, and Pglyrp4 are directly bactericidal [8]–[11], whereas Pglyrp2 is an N-acetylmuramoyl-L-alanine amidase that hydrolyzes peptidoglycan [12], [13]. Pglyrp1 is highly expressed in PMN's granules and to a much lower extent in other cells [7], [14], [15]. Pglyrp2 is constitutively expressed in the liver and secreted into blood, and its expression is induced in keratinocytes and other epithelial cells [12], [13], [16]–[20]. Pglyrp3 and Pglyrp4 have the highest expression in the skin and are also expressed in the salivary glands, throat, tongue, esophagus, stomach, intestine, and eyes [8], [21], [22]. Similar to Pglyrp1 and Pglyrp2, Pglyrp3 and Pglyrp4 are secreted and their expression in these tissues is both constitutive and inducible.
We hypothesized that PGRPs play a role in the development atopic dermatitis and contact dermatitis because of (a) the prominent expression of PGRPs in the skin, (b) the location of Pglyrp3 and Pglyrp4 genes in the epidermal differentiation gene cluster in the psoriasis sensitivity psors4 locus, (c) coordinated expression of Pglyrp3 and Pglyrp4 with other genes in the psors4 locus, (d) previous evidence of genetic association of Pglyrp3 and Pglyrp4 variants with psoriasis, which is another genetically- and environmentally-determined skin disease [23], [24], and (e) the ability of mammalian PGRPs to protect mice against experimental colitis [22] and to modulate the development of experimental arthritis [25]. Here we tested this hypothesis using PGRP-deficient mice and mouse models of chemically-induced atopic dermatitis and contact dermatitis.
Our results show that Pglyrp3 −/− and Pglyrp4 −/− mice (but not Pglyrp2 −/− mice) are more sensitive to the development of experimental atopic dermatitis than wild type (WT) mice. The common mechanism underlying this increased sensitivity of Pglyrp3 −/− and Pglyrp4 −/− mice is reduced recruitment of Treg cells to the skin and enhanced production and activation Th17 cells in Pglyrp3 −/− and Pglyrp4 −/− mice, which results in more severe inflammation and keratinocyte proliferation. By contrast, Pglyrp1 −/− mice are less sensitive than WT mice to both experimental atopic dermatitis and contact dermatitis. Thus, Pglyrp3 and Pglyrp4 limit over-activation of Th17 cells by promoting accumulation of Treg cells at the site of chronic inflammation, which protects the skin from exaggerated inflammatory response to cell activators and allergens. By contrast, Pglyrp1 has an opposite pro-inflammatory effect in the skin.
Results
Pglyrp3 −/− and Pglyrp4 −/− mice have enhanced inflammatory response in the oxazolone model of atopic dermatitis
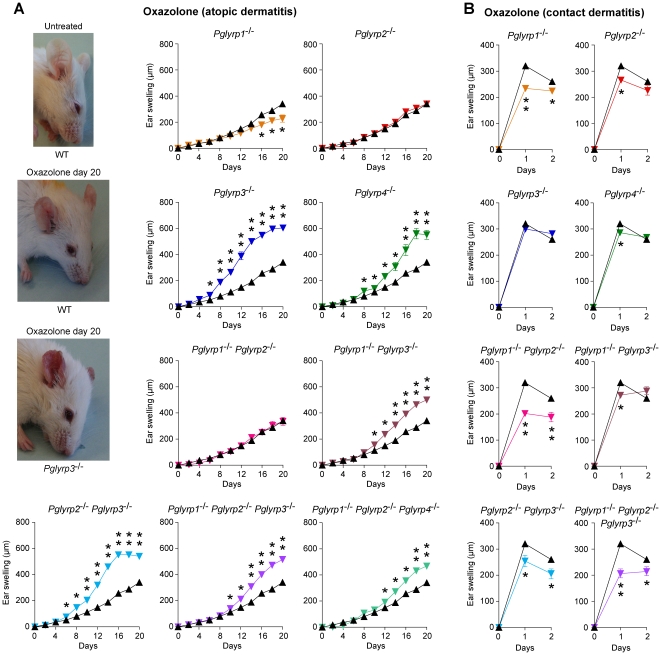
Repeated epicutaneous sensitization with oxazolone is an established mouse model of atopic dermatitis [3], [26]. Initial sensitization of mice with oxazolone through abdominal skin, followed by 6-day rest and application of oxazolone to the ears every other day for 20 days induced in WT BALB/c mice progressive moderate inflammation manifested by some redness and swelling (Figure 1A). Similar application of oxazolone to Pglyrp3 −/− or Pglyrp4 −/− mice induced significantly enhanced inflammation, manifested first by increased redness and significantly increased swelling, accompanied by scaling (Figure 1A). This enhanced response was unique for Pglyrp3 −/− and Pglyrp4 −/− mice, because it was not observed in Pglyrp1 −/−, Pglyrp2 −/−, and Pglyrp1 −/− Pglyrp2 −/− mice, and was dominant, because it was still observed in Pglyrp1 −/− Pglyrp3 −/− and Pglyrp2 −/− Pglyrp3 −/− double-knockout mice and in Pglyrp1 −/− Pglyrp2 −/− Pglyrp3 −/− and Pglyrp1 −/− Pglyrp2 −/− Pglyrp4 −/− triple-knockout mice (Figure 1A). Deletion of Pglyrp1 had the opposite effect – Pglyrp1 −/− single knockout mice showed reduced ear swelling on days 16 through 20 of oxazolone application (Figure 1A). These results indicate that in WT mice both Pglyrp3 and Pglyrp4 have a protective effect against severe atopic dermatitis-like inflammation, whereas Pglyrp1 has an enhancing proinflammatory effect and Pglyrp2 has little effect on the response to oxazolone.
Figure 1. Pglyrp3 −/− and Pglyrp4 −/− mice have enhanced response in the oxazolone model of atopic dermatitis and Pglyrp1 −/− and other Pglyrp −/− mice have reduced response in the oxazolone model of contact dermatitis.
(A) Atopic dermatitis model: sensitization followed by 10 applications of oxazolone to the ears every other day induces mild inflammation (left) and mild ear swelling (right) in WT mice (black triangles) and severe inflammation with increased redness, scaling, and extensive ear swelling in Pglyrp3 −/− and Pglyrp4 −/− mice (color triangles). (B) Contact dermatitis model: sensitization followed by a single application of oxazolone to the ears induces strong ear swelling in WT mice (black triangles) and reduced ear swelling in Pglyrp1 −/−, Pglyrp2 −/−, and Pglyrp4 −/− mice (color triangles). Means ± SEM (SEM were often smaller than the symbols in this and other figures); N = 9–17 mice/group; significance of differences between Pglyrp −/− and WT mice: *, P<0.02; **, P<0.0001.
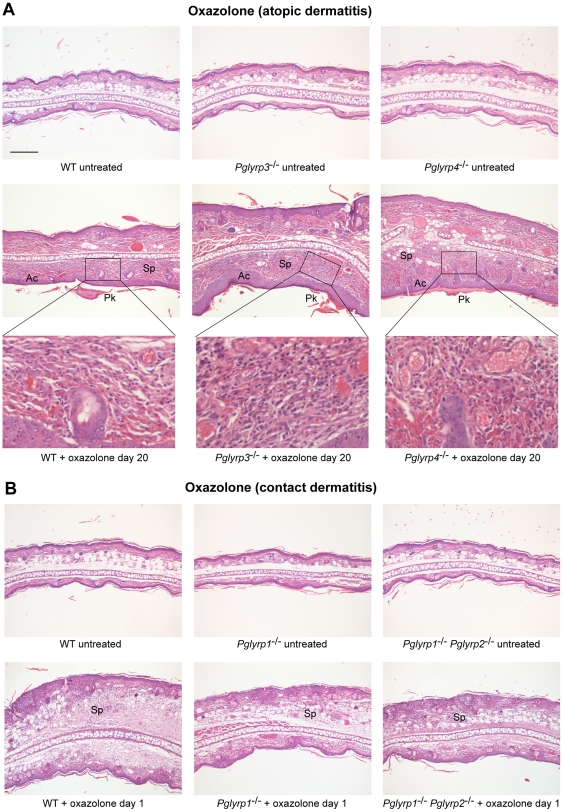
To determine the pathologic basis of higher sensitivity of Pglyrp3 −/− and Pglyrp4 −/− mice to oxazolone, we compared the histology of oxazolone-induced skin lesions in WT and Pglyrp-deficient mice. Ears in untreated WT mice have one- to two-cell thick epidermis and few-cell thick subepidermal layer with blood vessels, sebaceous glands, hair follicles, muscle bundles, and central fat and connective tissue layer, with total thickness of approximately 200 µm. Histology of all untreated Pglyrp-deficient mice was similar to WT mice (Figure 2). Sensitization and 10 oxazolone applications to the ears (every other day) induced strong inflammatory response that was very severe in Pglyrp3 −/− mice and Pglyrp4 −/− mice. Cross-sections of the oxazolone-treated ears revealed severe acanthosis (thickening of the epidermis due to proliferation of keratinocytes), parakeratosis (retention of keratinocytes' nuclei in stratum corneum), and marked thickening of the sub-epidermal layer with spongiosis (intercellular edema) and dense cellular infiltrates (composed primarily of mononuclear cells and some polymorphonuclear cells) that were all highly prominent in Pglyrp3 −/− mice and Pglyrp4 −/− mice (Figure 2A) and in all other double and triple knockout mice deficient in Pglyrp3 (not shown). All these changes are highly characteristic of atopic dermatitis lesions. These mice did not develop rete pegs (downward papillary projections of epidermis), which are characteristic of psoriasis, but not atopic dermatitis. WT mice (Figure 2A), Pglyrp1 −/− mice, Pglyrp2 −/− mice, and Pglyrp1 −/− Pglyrp2 −/− mice (not shown) all showed much less severe acanthosis, parakeratosis, edema, and cell infiltrations, and less thickening of the subepidermal layer.
Figure 2. Ear histology in WT and Pglyrp-deficient mice in atopic dermatitis and contact dermatitis models of skin inflammation.
(A) Oxazolone model of atopic dermatitis: sensitization and 10 applications of oxazolone to the ears every other day induced acanthosis (Ac), parakeratosis (Pk), marked thickening of the sub-epidermal layer with spongiosis (Sp) and dense cellular infiltrates of primarily mononuclear and some polymorphonuclear cells (high magnification insets), that were all highly prominent in Pglyrp3 −/− mice and Pglyrp4 −/− mice and much less severe in WT mice. (B) Oxazolone model of contact dermatitis: sensitization and a single application of oxazolone to the ears induced strong inflammatory response in WT mice with marked spongiosis of the sub-epidermal layer (Sp) and cellular infiltrates of epidermal and sub-epidermal layers, composed of mononuclear and polymorphonuclear cells; Pglyrp1 −/− and Pglyrp1 −/− Pglyrp2 −/− mice still had cellular infiltrates, but had substantially reduced swelling, compared to WT mice, mostly due to reduced edema. H&E stained cross-sections; bar = 200 µm for all panels, except high magnification insets (the magnified areas are shown by rectangles).
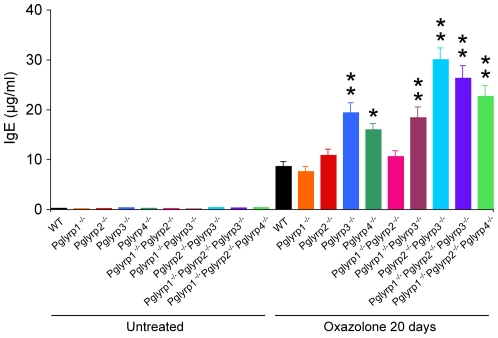
Inflammatory response to oxazolone was accompanied by a marked increase in the concentration of serum IgE, consistent with oxazolone inflammation being a model of atopic dermatitis (which is type I hypersensitivity associated with increased IgE production). The serum IgE levels in Pglyrp3 −/− mice and Pglyrp4 −/− mice, as well as Pglyrp1 −/− Pglyrp3 −/− and Pglyrp2 −/− Pglyrp3 −/− double-knockout mice and in Pglyrp1 −/− Pglyrp2 −/− Pglyrp3 −/− and Pglyrp1 −/− Pglyrp2 −/− Pglyrp4 −/− triple-knockout mice were all significantly higher than in WT and in Pglyrp1 −/−, Pglyrp2 −/−, and Pglyrp1 −/− Pglyrp2 −/− mice (Figure 3). Pglyrp2 also played a minor role, because Pglyrp2 −/− Pglyrp3 −/− double-knockout mice and Pglyrp1 −/− Pglyrp2 −/− Pglyrp3 −/− and Pglyrp1 −/− Pglyrp2 −/− Pglyrp4 −/− triple-knockout mice had higher serum IgE levels than Pglyrp3 −/− and Pglyrp4 −/− single-knockout mice.
Figure 3. Pglyrp3 −/− and Pglyrp4 −/− mice have higher serum IgE levels than WT mice in the oxazolone model of atopic dermatitis.
Sensitization followed by 10 applications of oxazolone to the ears every other day induced higher level of serum IgE in Pglyrp3 −/− and Pglyrp4 −/− mice than in WT, Pglyrp1 −/− or Pglyrp2 −/− mice; means ± SEM of 8–20 mice/group; *, P<0.002; **, P<0.0001, Pglyrp −/− versus WT.
These results demonstrate that deletion of Pglyrp3 or Pglyrp4 highly predisposes mice to atopic dermatitis-like lesions in response to oxazolone, and thus in WT mice Pglyrp3 or Pglyrp4 protect the skin from excessive inflammation in the oxazolone model of atopic dermatitis.
Pglyrp-deficient mice have reduced response in the oxazolone model of contact dermatitis
Single epicutaneous sensitization with oxazolone followed by a single epicutaneous challenge in a different area is an established mouse model of allergic contact dermatitis (contact hypersensitivity) due to type IV hypersensitivity [27], [28]. Sensitization of mice with oxazolone through abdominal skin, followed by 6-day rest and a single application of oxazolone to the ears induced on the next day a strong inflammatory response in WT BALB/c mice, manifested by redness and swelling (Figure 1B). Similar application of oxazolone to Pglyrp-deficient mice induced significantly lower inflammation, manifested by significantly reduced ear swelling compared to WT mice, especially in Pglyrp1 −/− mice, and to some extent in Pglyrp2 −/− and Pglyrp4 −/− mice. The reduced responsiveness was dominant, because it was still observed in Pglyrp1 −/− Pglyrp2 −/− (most affected), Pglyrp1 −/− Pglyrp3 −/−, and Pglyrp2 −/− Pglyrp3 −/− double-knockout mice and in Pglyrp1 −/− Pglyrp2 −/− Pglyrp3 −/− triple-knockout mice (Figure 1B).
To determine the pathologic basis of this lower response to oxazolone in the contact dermatitis model in Pglyrp-deficient mice, we compared the ear histology in oxazolone-treated WT and Pglyrp-deficient mice. Sensitization and single oxazolone application to the ears induced strong inflammatory response in WT mice manifested by marked spongiosis of the sub-epidermal layer with cellular infiltrates of epidermal and sub-epidermal layers with mononuclear and polymorphonuclear cells (but no acanthosis, parakeratosis, rete pegs, or scaling, which histologically differentiates contact dermatitis from atopic dermatitis and psoriasis) (Figure 2B). Pglyrp1 −/− and Pglyrp1 −/− Pglyrp2 −/− mice still had cellular infiltrates, but had substantially reduced swelling, compared to WT mice, mostly due to reduced edema (Figure 2B). These results indicate that in WT mice Pglyrp1 (and also to some extent Pglyrp2 and Pglyrp4) has an enhancing proinflammatory role in this model of contact dermatitis.
Thus, altogether our results indicate that individual PGRPs have selective and distinct effects on skin inflammation. In WT mice Pglyrp3 and Pglyrp4 offer protection in the oxazolone atopic dermatitis model, Pglyrp1 has a pro-inflammatory effect both atopic and contact dermatitis models, and Pglyrp2 has little effect on the atopic dermatitis and has a pro-inflammatory effect in the contact dermatitis model of skin inflammation.
Expression of PGRPs in inflamed ears
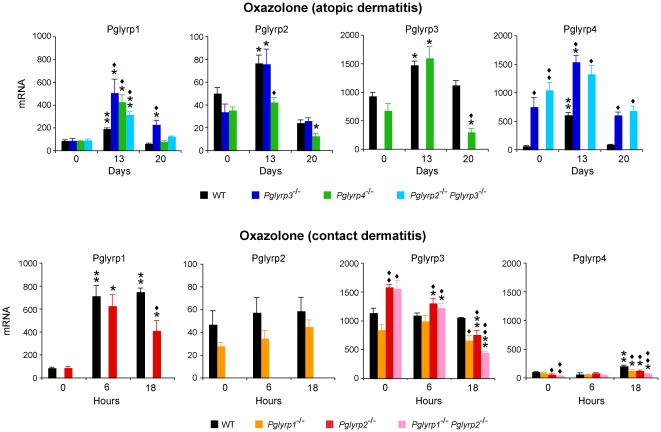
To gain further insight how individual PGRPs influence sensitivity to both models of skin inflammation, we compared expression of all PGRPs in the ears in untreated and oxazolone-treated mice. In both atopic and contact dermatitis models treatment with oxazolone induced increased Pglyrp1 mRNA expression in the ears in all strains of mice (except Pglyrp1 −/− mice, in which Pglyrp1 gene is deleted) that was significantly higher than in untreated mice (Figure 4). The expression of Pglyrp1 was significantly higher in all Pglyrp-deficient mice− than in WT mice early (day 13) in the atopic dermatitis model, which correlates with their higher inflammatory response. This increase in Pglyrp1 mRNA in the ears is likely due to increased infiltration with PMNs, because PMNs highly express Pglyrp1 and expression of Pglyrp1 is not inducible in any of the cell types previously studied, including lymphocytes, monocytes, macrophages, and keratinocytes [8], [14], [15]. The expression of Pglyrp2 was only modestly increased early in the atopic dermatitis model and returned to the untreated level later in this model, and was not significantly changed at any time point in the contact dermatitis model, consistent with the little effect of Pglyrp2 in both atopic and contact dermatitis models (Figure 1). Pglyrp1 and Pglyrp2 were also constitutively expressed in the cervical lymph nodes at a similar level as in the ears, but following oxazolone treatment their expression in the cervical lymph nodes did not significantly change (data not shown), despite extensive stimulation and expansion of immune cells in the draining lymph nodes, which increased in diameter from <0.5 mm in untreated mice to >3 mm after 20 days of oxazolone treatment.
Figure 4. Pglyrp1, Pglyrp2, Pglyrp3, and Pglyrp4 expression is increased in the affected skin in the atopic dermatitis and contact dermatitis models of inflammation.
The amounts of each PGRP mRNA in WT mice and in the indicated Pglyrp-deficient mice sensitized and treated with oxazolone every other day (atopic dermatitis model) or sensitized and treated with oxazolone at 0 hrs only (contact dermatitis model) were measured by qRT-PCR. The results are means of 3–4 mice ± SEM; *, P<0.04; **, P<0.001, treated versus untreated; ♦, P<0.04; ♦♦, P<0.001, Pglyrp −/− versus WT.
Pglyrp3 has high constitutive expression in untreated skin [8], [21]. Pglyrp3 mRNA expression initially increased and later decreased in the atopic dermatitis model, and decreased in the contact dermatitis model. Pglyrp4 has lower constitutive expression in untreated skin than Pglyrp3. Expression of Pglyrp4 mRNA was highly increased in the atopic dermatitis model and was the highest in Pglyrp2 −/− Pglyrp3 −/− and Pglyrp3 −/− mice, suggesting a possibility of compensatory expression of Pglyrp4 in mice deficient in Pglyrp3 or changes in the regulation of transcription or stability of Pglyrp4 mRNA, since Pglyrp3 and Pglyrp4 genes are tightly linked in the psoriasis sensitivity locus on chromosome 3 in mice and their expression is correlated with the expression of keratinocytes differentiation genes [21]. To further investigate which cells express Pglyrp3 and Pglyrp4 in the untreated and oxazolone-treated skin, we analyzed the expression of Pglyrp3 and Pglyrp4 proteins by immunohistochemistry. Pglyrp3 and Pglyrp4 were expressed in epidermal keratinocytes in untreated ears, and in oxazolone-treated ears they were primarily expressed in the upper layers of differentiated keratinocytes (Figure 5). Pglyrp3 and Pglyrp4 expression was not induced in other cell types, such as infiltrating inflammatory cells in oxazolone-treated ears. Moreover, Pglyrp3 and Pglyrp4 were not expressed in the cervical lymph nodes in untreated mice and their expression there was not induced following oxazolone treatment (data not shown). These results indicate that Pglyrp3 and Pglyrp4 are primarily expressed in keratinocytes, but not in immune or inflammatory cells, and thus these results raise the possibility that Pglyrp3 and Pglyrp4 exert their protective effects in the oxazolone-induced atopic dermatitis by modulating the function of keratinocytes, which are known to produce many pro-inflammatory cytokines and chemokines.
Figure 5. Pglyrp3 and Pglyrp4 are expressed primarily in the differentiated epidermal cells in both untreated and oxazolone-treated skin in the atopic dermatitis model.
Transverse sections of the ears from either untreated or oxazolone-treated mice (sensitization followed by 10 applications of oxazolone to the ears every other day) were stained by an immunoperoxidase method with either anti-Pglyrp3 or anti-Pglyrp4 antibodies or IgG (negative control); bar = 50 µm.
Pglyrp3 −/− and Pglyrp4 −/− mice have increased Th17 cells and Th17 responses in the skin
To determine the cellular basis for the differences in the inflammatory responses in Pglyrp-deficient mice, we next determined the types of inflammatory cells in the ears in both models of skin inflammation, and then we determined which cell types significantly differed in Pglyrp-deficient mice compared to WT mice. This was first accomplished by measuring the amounts of mRNA for several marker genes characteristic of various immune and inflammatory cell types in the untreated and the affected ears. To determine which marker genes (and thus cell types) are increased or decreased in Pglyrp-deficient mice compared to WT mice, we calculated how many times higher or lower they were induced in Pglyrp-deficient mice than in WT mice (fold induction in Pglyrp-deficient mice/fold induction in WT mice).
Prolonged treatment with oxazolone in the atopic dermatitis model induced increases in mRNA of several cell types in WT mice, and especially mature B cells, CD8+ T cells, monocytes, PMNs, and mast cells, as well as changes associated with de-differentiation, proliferation, and activation of keratinocytes, which are all expected changes consistent with the atopic dermatitis model (Figure 6A and Figure S1). Atopic dermatitis-sensitive mice (Pglyrp3 −/−, Pglyrp4 −/−, and Pglyrp2 −/− Pglyrp3 −/− mice) had increased expression of genes characteristic of B cells and T cells. Also increased was Rorγt mRNA, which is preferentially expressed in Th17 cells [29] (Figure 6A and Figure S1).
Figure 6. Pglyrp3 −/− and Pglyrp4 −/− mice have increased Th17 cells in the affected skin in the atopic dermatitis model, whereas Pglyrp1 −/− and Pglyrp2 −/− mice have most cell types decreased (except B cells and Treg cells) in the contact dermatitis model.
Expression of a panel of marker genes characteristic of various inflammatory cell types in the ears of mice after (A) sensitization and 10 applications of oxazolone to the ears every other day, or (B) after sensitization and 6 hrs after a single application of oxazolone to the ears, measured by qRT-PCR is shown. For WT mice (left panels in A and B), the ratio of the amount of mRNA in oxazolone-treated to untreated mice for each gene (fold induction by oxazolone) is shown; for Pglyrp −/− mice (right panels in A and B), the results are the ratios of fold induction of each gene by oxazolone in Pglyrp −/− mice to fold induction of each gene by oxazolone in WT mice (which represents the fold difference in the response to oxazolone in Pglyrp −/− versus WT mice). The results are means of 3 arrays from 4–5 mice/group in heat map format. The means ± SEM bar graphs for these results are shown in Figures S1 and S2.
Short-term treatment with oxazolone in the contact dermatitis model in WT mice induced increases primarily in monocytes and PMNs at 6 hrs after challenge (Figure 6B and Figure S2) and later in T cells (18 hrs, not shown). Contact dermatitis-resistant mice (Pglyrp1 −/−, Pglyrp2 −/−, and Pglyrp1 −/− Pglyrp2 −/− mice) had initially (6 hrs after challenge) increased B cells and Treg cells and decreased other cell-types, compared to WT mice (Figure 6B and Figure S2). Later (18 hrs after challenge) the cell markers in Pglyrp2 −/− and Pglyrp1 −/− Pglyrp2 −/− mice were similar (most cells) or increased (monocytes), compared to WT mice (not shown), consistent with comparable cell infiltrates seen in all mice on tissue sections on day 1 (Figure 2B).
To further define the cell types responsible for differential sensitivity of Pglyrp-deficient mice to atopic dermatitis and contact dermatitis, we measured the expression of an extended panel of cytokines, chemokines, and other marker genes characteristic of Th1, Th2, Th17, Treg, NK, and other cell types to determine which of these genes were differentially induced in the affected skin in Pglyrp-deficient mice, compared to WT mice. We included these cell types, because in addition to Th1 and Th2 cells, Th17 cells and other cell types may also be involved in these skin diseases.
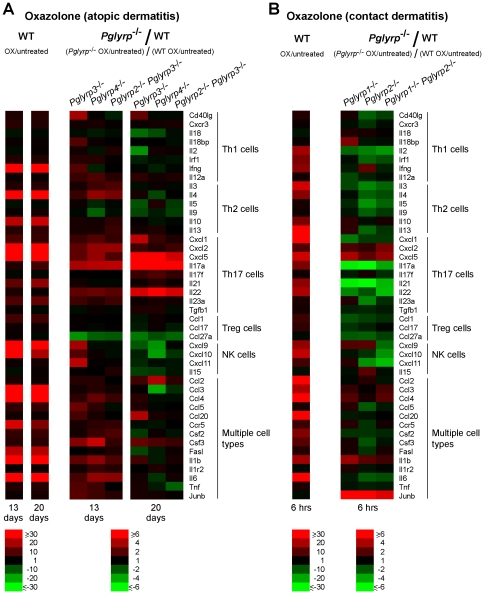
Prolonged epicutaneous sensitization with oxazolone (20 days), which induced atopic dermatitis-like skin inflammation, was accompanied in WT mice by high activation (more than 15-fold) of 14 out of 44 studied genes in the affected skin. These genes were Ifng (Th1), Il4 and Il10 (Th2), Cxcl2, Cxcl5, and Il21 (Th17), Cxcl9 and Cxcl10 (NK cells), and Ccl3, Ccl4, Ccr5, Fasl, Il1b, and Il6 (several cell types) (Figure 7A and Figures S3 and S4). Atopic dermatitis-sensitive mice initially (day 13) had several genes activated higher than in WT mice, characteristic of several cell types, including Th2 and Th17 (Figure 7A and Figure S3). Later (day 20), atopic dermatitis-sensitive mice (Pglyrp3 −/−, Pglyrp4 −/−, and Pglyrp2 −/− Pglyrp3 −/− mice) had four genes characteristic of Th17 cells (Cxcl1, Cxcl5, IL17a, IL22) and one gene characteristic of several cell types (Ccl2) activated more than three-fold higher in Pglyrp-deficient than in WT mice (Figure 7A and Figure S4).
Figure 7. Th17 gene expression profile is preferentially induced in the atopic dermatitis model in Pglyrp3 −/− and Pglyrp4 −/− mice, whereas expression of most immune genes is reduced in the contact dermatitis model in Pglyrp −/− mice.
Expression of a panel of cytokines, chemokines, and other marker genes characteristic of Th1, Th2, Th17, Treg, NK, and other cell types in the ears of mice was measured by qRT-PCR. (A) After sensitization and 10 applications of oxazolone to the ears every other day the expression of several Th17 marker genes was higher in Pglyrp3 −/− and Pglyrp4 −/− mice than in WT mice. (B) After sensitization and a single application of oxazolone to the ears the expression of most of immune marker genes was lower in Pglyrp −/− mice than in WT mice. For WT mice (left panels in A and B), the ratio of the amount of mRNA in oxazolone-treated to untreated mice for each gene (fold induction by oxazolone) is shown; for Pglyrp −/− mice (right panels in A and B), the results are the ratios of fold induction of each gene by oxazolone in Pglyrp −/− mice to fold induction of each gene by oxazolone in WT mice (which represents the fold difference in the response to oxazolone in Pglyrp −/− versus WT mice). The results are means of 3 arrays from 4–5 mice/group in heat map format. The means ± SEM bar graphs for these results are shown in Figures S3, S4, and S5.
A single oxazolone challenge in sensitized WT mice (in the contact dermatitis model) also induced many genes characteristic of several cell types, and the early (6 hrs) activation of these genes in Pglyrp-deficient mice was mostly reduced, compared to WT mice (Figure 7B and Figure S5). These results are consistent with lower clinical responses of Pglyrp-deficient mice to a single oxazolone challenge in the contact dermatitis model (Figures 1B and 2B).
The above results indicate that the atopic dermatitis-sensitive Pglyrp-deficient mice have increased activity of Th17 cells in the affected skin, compared to WT mice. To further investigate the role Th17 cells (and other Th cell types) in increased sensitivity of Pglyrp-deficient mice in atopic dermatitis model, we used flow cytometry to directly measure Th cell types in the ears, draining lymph nodes, and the spleen.
Untreated ears in WT and Pglyrp −/− mice had <400 CD4+ cells/ear, whereas after sensitization and 20 days of oxazolone treatment the numbers of CD4+ cells/ear increased >50 times to ∼18,000–19,000/ear in WT and Pglyrp3 −/− mice (Figure 8A). Regarding Th cell subpopulations, oxazolone treatment for 13 days induced significantly higher numbers of Th2 cells (CD4+IL-4+) in the affected ears in Pglyrp3 −/− mice compared to WT mice, whereas oxazolone treatment for 20 days induced significantly higher numbers of Th17 cells (CD4+IL-17+) in the affected ears in Pglyrp3 −/− mice compared to WT mice (Figure 8A, B and C). Thus, on day 20 in Pglyrp3 −/− mice the numbers of Th17 cells in the ears increased from undetectable (<10/ear) to ∼650 Th17 cells/ear, 3.5 times higher than in WT mice (Figure 8A). Virtually all detectable IL-17+ cells in the oxazolone-treated ears were CD4+ (Th17 cells) and there were very few (<50/ear, not shown) other IL-17+ cells in the inflamed skin (such as CD8+, γ/δ T cells, or NKT cells), and therefore the observed increases in the numbers IL-17+ cells mostly represent increases in Th17 cells (CD4+IL-17+). There was no significant difference in the numbers of Th1 (CD4+IFN-γ+) and Th2 (CD4+IL-4+) cells in the ears of WT and Pglyrp3 −/− mice on day 20 (Figure 8C). Oxazolone-treated mice had substantially swollen cervical lymph nodes (>3 mm in diameter, compared to <0.5 mm in untreated mice), where on day 13 the numbers of Th2 cells and on day 20 the numbers of all Th cell types were significantly higher in Pglyrp3 −/− mice compared to WT mice (Figure 8B and C).
Figure 8. Pglyrp3 −/− mice have high numbers Th17 and low numbers of Treg cells in the affected skin in the oxazolone model of atopic dermatitis.
(A) Numbers of CD4+ cells and Th17 cells in the ears or (B–D) percentages of Th1, Th2, Th17, and Treg cells in the ears, cervical lymph nodes, and spleen in sensitized WT and Pglyrp3 −/− mice on days 13 or 20 of ear treatment with oxazolone, measured by flow cytometry; means ± SEM of 5–9 mice/group (*, P<0.05; **, P<0.005; Pglyrp3 −/− versus WT) or representative dot plots are shown. (E) Expression of receptors for chemokines that attract Treg cells in cervical lymph nodes of sensitized WT and Pglyrp3 −/− mice on day 20 of ear treatment with oxazolone measured by qRT-PCR; amounts of mRNA are shown as means ± SEM of 3 arrays from 5 mice/group.
These results indicate initial (day 13) preferential activation of Th2 cells in the affected ears and draining lymph nodes in Pglyrp3 −/− mice compared to WT mice, consistent with B-cell-dependence of atopic dermatitis model. However, continued treatment with oxazolone (20 days) showed a switch to preferential infiltration of the affected ears with Th17 cells in Pglyrp3 −/− mice compared to WT mice (Figure 8), consistent with our mRNA gene expression data (Figures 6 and 7 and Figures S1, S3, and S4).
IL-17 is required for enhanced response to oxazolone in Pglyrp3 −/− mice
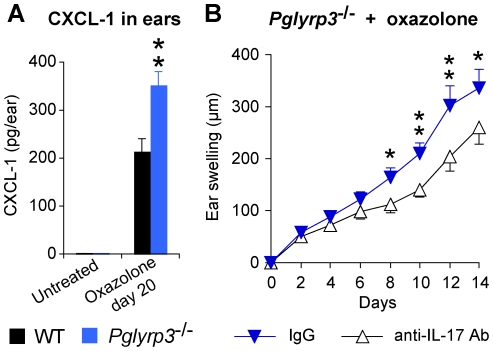
To further study the role of IL-17 (Th17 cytokine) in high sensitivity of Pglyrp3 −/− mice to oxazolone-induced atopic dermatitis, we determined the protein levels of an IL-17-induced chemokine, CXCL-1, in the ears of WT and Pglyrp3 −/− mice. CXCL-1 was undetectable (<7 pg/ear) in the ears of untreated mice, and after sensitization and 20 days of skin treatment with oxazolone, the amount of CXCL-1 increased to >350 pg/ear in Pglyrp3 −/− mice, the level that was significantly higher than in WT mice (Figure 9A).
Figure 9. IL-17 is required for enhanced response to oxazolone in Pglyrp3 −/− mice.
(A) The level of IL-17-induced chemokine, CXCL-1, is higher in the ears of Pglyrp3 −/− mice than WT mice after sensitization and application of oxazolone for 20 days. (B) Ear swelling in Pglyrp3 −/− mice sensitized and treated 7 times with oxazolone every other day and also treated with neutralizing anti-IL-17 mAbs is lower than in Pglyrp3 −/−mice similarly treated with oxazolone and isotype control IgG. Means ± SEM; N = 6 mice/group; significance of differences between Pglyrp3 −/− and WT mice (A) or IgG control and anti-IL-17 mAbs-treated mice (B): *, P<0.05; **, P<0.005.
To determine whether IL-17 is required for the high sensitivity of Pglyrp3 −/− mice to atopic dermatitis, we compared the severity of ear inflammation in oxazolone-treated Pglyrp3 −/− mice in which IL-17 activity was inhibited with neutralizing anti-IL-17 mAb. In vivo neutralization of IL-17 activity in Pglyrp3 −/− mice in the oxazolone-induced atopic dermatitis significantly reduced ear inflammation, compared to mice treated with an isotype control IgG (Figure 9B). These results demonstrate that IL-17 is required for full manifestation of severe skin inflammation in Pglyrp3 −/− mice in the atopic dermatitis model.
Pglyrp3 −/− and Pglyrp4 −/− mice have decreased numbers of Treg cells in the skin
Because WT mice were able to limit skin inflammation in the atopic dermatitis model more effectively than Pglyrp3 −/− and Pglyrp4 −/− mice, we then tested whether this difference is due to impaired generation or function of regulatory T cells (CD4+FoxP3+ Treg) in Pglyrp-deficient mice.
In the atopic dermatitis model WT mice efficiently recruited Treg cells into the affected skin, as evidenced by an increase in FoxP3-expressing Treg cells in the affected skin shown both by the qRT-PCR (Figure 6A and Figure S1) and by flow cytometry, in which high numbers of CD4+FoxP3+ Treg cells were found in the affected skin in WT mice (Figure 8B–D). By contrast, atopic dermatitis-sensitive Pglyrp-deficient mice (Pglyrp3 −/−, Pglyrp4 −/− and Pglyrp2 −/− Pglyrp3 −/− mice) all had lower expression of FoxP3 mRNA in the affected ears compared to WT mice (Figure 6A and Figure S1). Pglyrp3 −/− mice in the atopic dermatitis model also had significantly lower numbers of CD4+FoxP3+ Treg cells in the affected skin compared to WT mice measured by flow cytometry (Figure 8B–D). These results suggest impaired recruitment and/or maintenance of Treg cells in the inflamed skin in Pglyrp3 −/− and Pglyrp4 −/− mice.
To further investigate whether Pglyrp3 −/− mice have less efficient generation of induced Treg cells in lymphoid tissues in general or less efficient recruitment and/or maintenance of these cells in the inflamed skin, we compared the numbers of Treg cells in the draining cervical lymph nodes and in the spleen of WT and Pglyrp3 −/− mice treated with oxazolone. Oxazolone-treated Pglyrp3 −/− mice on day 13 had lower numbers of Treg cells than WT mice in cervical lymph nodes and spleen (Figure 8B). However, at the peak of inflammation (day 20) Pglyrp3 −/− mice had similar numbers of Treg cells in the draining lymph nodes and spleen compared to WT mice (Figure 8C). These results indicate that Pglyrp3 −/− mice can eventually generate sufficient numbers of induced Treg cells in lymphoid organs and suggest a possible reduced migration and retention of Treg cells in the affected skin.
There could be at least two reasons for this less efficient recruitment of Treg cells to the skin in Pglyrp3 −/− mice: insufficient production of Treg-attracting chemokines in the skin, and/or insufficient expression of receptors for these chemokines in Treg cells in Pglyrp3 −/− mice. Our results show lower expression of mRNA for Treg-attracting chemokines, CCL-27 (both on days 13 and 20) and CCL-17 (on day 20), in the ears of oxazolone-treated Pglyrp3 −/− mice compared to WT mice (Figure 7A and Figures S3 and S5), indicating insufficient production of Treg-attracting chemokines in the skin in Pglyrp3 −/− mice. To investigate the second of the above-mentioned possibilities, we determined whether Treg cells in the draining cervical lymph nodes in Pglyrp3 −/− mice had sufficient expression of receptors for Treg-attracting chemokines (Ccr4, Ccr8, and Ccr10). The expression of mRNA for Ccr4, Ccr8, and Ccr10 in the draining cervical lymph nodes in oxazolone-treated Pglyrp3 −/− mice and WT mice was similar (Figure 8E). These results support the conclusion that Treg cells in the draining lymph nodes in oxazolone-treated Pglyrp3 −/− mice have sufficient expression of receptors for Treg-attracting chemokines, but that these Treg cells are not recruited to the inflamed skin, likely because of the insufficient production of Treg-attracting chemokines in the skin (Figure 7A and Figures S3 and S4). Our results thus indicate that Pglyrp3 promotes efficient population of the skin with Treg cells in oxazolone-induced atopic dermatitis.
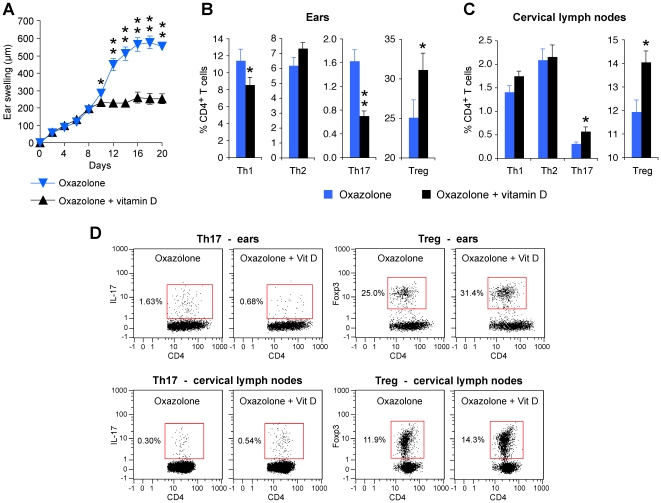
Induction of Treg cells in Pglyrp3 −/− mice reduces Th17 cells and sensitivity to atopic dermatitis
To further investigate the role of Treg cells in high sensitivity of Pglyrp3 −/− mice to atopic dermatitis, we induced generation of Treg cells by application of vitamin D to the skin (which is known to induce Treg cells [30], [31]) together with the sensitizing allergen, oxazolone. Vitamin D applied to the ears of Pglyrp3 −/− mice together with oxazolone significantly reduced ear swelling compared to Pglyrp3 −/− mice similarly treated with oxazolone alone (Figure 10A). Vitamin D applied to the ears (together with oxazolone) also significantly increased the percentages of Treg cells both in the ears and cervical lymph nodes (as expected [30], [31]), and, moreover, it significantly reduced the percentages of Th17 cells in the ears compared to the ears treated with oxazolone alone, measured on day 20 by flow cytometry (Figure 10B–D). Thus, increasing the numbers of Treg cells in the affected skin in Pglyrp3 −/− mice to the numbers found in WT mice could revert the inflammatory phenotype of Pglyrp3 −/− mice to the less inflammatory phenotype characteristic of WT mice. These results further demonstrate the critical role of Treg cells in preventing high levels of Th17 cells in the skin and excessive inflammation in the oxazolone model of atopic dermatitis. In summary, our results indicate that in WT mice Pglyrp3 and Pglyrp4 promote efficient population of the skin with Treg cells in the experimental model of atopic dermatitis.
Figure 10. Induction of Treg cells by vitamin D reduces the inflammatory response to oxazolone and decreases the numbers of Th17 cells in the skin of Pglyrp3 −/− mice.
(A) Vitamin D applied to the skin of Pglyrp3 −/− mice together with oxazolone reduces ear swelling compared to Pglyrp3 −/− mice similarly treated with oxazolone alone. (B–D) Vitamin D applied to the skin of Pglyrp3 −/− mice together with oxazolone increases the percentages of Treg cells in the ears and cervical lymph nodes and reduces the percentages of Th17 cells in the ears compared to the application of oxazolone alone, measured on day 20 by flow cytometry. Means ± SEM of 8 mice/group (A–C; *, P<0.05; **, P<0.005; oxazolone versus oxazolone + vitamin D) or representative dot plots (D) are shown.
Discussion
Skin diseases such as atopic dermatitis and contact dermatitis involve complex interactions of many cell types. Atopic dermatitis is thought to have Th2 bias [2], [26], but recent findings also show involvement of Th17 cells [32], [33]. The initially identified in vivo role of Th17 cells was promoting some autoimmune diseases and recruitment of PMNs to the sites of inflammation [34]–[37]. However, Th17 cells have many other functions – they play a role in inflammatory bowel diseases, skin diseases, asthma, graft rejection, atherosclerosis, periodontal disease, and arthritis [32], [33], [38]–[40]. We extend these findings by showing that Th17 cells exacerbate skin inflammation in experimental model of atopic dermatitis in a PGRP-dependent manner.
We demonstrate here that Pglyrp3 −/− and Pglyrp4 −/− mice develop more severe oxazolone-induced atopic dermatitis than WT mice. By contrast, Pglyrp1 −/− mice develop less severe oxazolone-induced atopic dermatitis and also less severe contact dermatitis than WT mice. Thus, individual PGRPs play distinct roles in these two models of skin diseases: in WT mice Pglyrp3 and Pglyrp4 protect mice from the development of experimental atopic dermatitis, whereas Pglyrp1 enhances the development of both atopic and contact dermatitis and Pglyrp2 has less effect on both disease models.
The common mechanism underlying these protective effects of PGRPs is decreased recruitment and activity of Treg cells and enhanced production and activation of Th17 cells in the affected skin in Pglyrp3 −/− and Pglyrp4 −/− mice, which results in more severe inflammation and keratinocyte proliferation. Thus, in WT mice Pglyrp3 and Pglyrp4 promote recruitment and retention of Treg cells in the inflamed skin, which limits over-activation of Th17 cells and protects the skin from exaggerated inflammatory response to allergens.
Our results do show prominent activation of Th2 cells in the oxazolone model of atopic dermatitis and prominent production of IgE, characteristic of Th2 bias in atopic diseases. In our model, however, the main difference at the peak of inflammation between WT and atopic dermatitis-sensitive Pglyrp-deficient mice, which determines the enhanced inflammatory responses and higher IgE production in Pglyrp-deficient mice, is the overactivation of Th17 cells and reduced numbers of Treg cells in the atopic dermatitis-sensitive mice. Our results indicate that initial Th2 response changes with time to Th17 response and that Th17 cells play an important role in enhancing inflammation and production of IgE in atopic dermatitis. Our results thus further extend recent findings of the enhancing role of Th17 cells in B cell maturation and differentiation [41]–[43]. Therefore, our results indicate that Pglyrp3 and Pglyrp4 are involved in controlling multiple functions of Treg and Th17 cells in the skin in atopic dermatitis.
Contact dermatitis, which is a skin model of type IV hypersensitivity, is usually considered to be mediated by Th1 cells. However, this is an oversimplification, because recent findings show involvement of multiple cell types [2], [4], [5], [44]. Our results show that Pglyrp1 (and other Pglyrps to a lesser extent) promotes Th1 responses, because Pglyrp1 −/− mice (and also Pglyrp2 −/− and Pglyrp4 −/− mice) have reduced inflammation in oxazolone-induced contact dermatitis. This shift of balance to Th1 cells is likely beneficial for the desirable anti-microbial responses and may taper exaggerated inflammatory responses in the skin to allergens. Note that Pglyrp1 is mainly delivered to the sites of inflammation by PMNs, which are usually recruited to fight infections.
Our results show that PGRPs, a family of innate immunity proteins, influence the functions of both innate and adaptive immune cells with an outcome of enhancing the activity of Treg cells and inhibiting the activity of Th17 cells. Pglyrp3 −/− and Pglyrp4 −/− mice have decreased numbers of Treg cells and increased numbers of Th17 cells in the inflamed skin, compared to WT mice. Allergens and proinflammatory stimuli (such as oxazolone) in WT mice initially induce vigorous cytokine and chemokine production. However, upon chronic exposure, WT mice recruit and maintain large numbers of Treg cells in the inflamed skin and are able to limit the proinflammatory response by reducing the number of proinflammatory genes that are activated and reducing the level of their activation. By contrast, Pglyrp3 −/− and Pglyrp4 −/− mice have fewer Treg cells and higher numbers of Th17 cells in the affected skin and are unable to limit inflammatory responses. This T cell imbalance in Treg/Th17 cells in Pglyrp3 −/− and Pglyrp4 −/− mice could come from reduced recruitment of Treg cells and increased recruitment of Th17 cells to the affected skin, and/or from enhanced local differentiation of T cells into Th17 cells (including conversion of Treg cells into Th17 cells) under the influence of locally produced chemokines and cytokines. T cell populations are dynamic and have considerable plasticity based on local cytokine milieu, as, for example, Treg cells can differentiate into Th17 under the influence of locally-produced proinflammatory cytokines [45], [46].
The enhanced recruitment and differentiation of Th17 cells is supported by higher production of Th17 cell promoting cytokines (IL-17, IL-22, IL-23) in the inflamed skin in Pglyrp3 −/− and Pglyrp4 −/− mice. Decreased recruitment of Treg cells to the inflamed skin is supported by the presence of adequate numbers of Treg cells and adequate expression of receptors for Treg-attracting chemokines on these Treg cells in the draining lymph nodes and spleen in oxazolone-treated Pglyrp3 −/− mice, but lower numbers in the skin than in WT mice. These results suggest efficient generation of induced Treg cells in lymphoid organs but defective recruitment to the inflamed skin. This mechanism is further supported by decreased production of Treg cell-attracting chemokines (CCL1, CCL17, CCL27) in the skin of Pglyrp3 −/− and Pglyrp4 −/− mice. Thus both increased recruitment and generation of Th17 cells and decreased recruitment and retention of Treg cells in the skin are likely responsible for increased inflammation in Pglyrp3 −/− and Pglyrp4 −/− mice.
Pglyrp3 and Pglyrp4 are primarily expressed in keratinocytes and other epithelial cells, but not in immune cells and stimulation of immune cells does not induce expression of Pglyrp3 and Pglyrp4 [7], [8], [18], [20], [21]. Our results extend these findings by showing expression of Pglyrp3 and Pglyrp4 in differentiated keratinocytes in untreated and oxazolone-treated skin, and no expression of Pglyrp3 and Pglyrp4 in the inflammatory cells infiltrating the skin and in the draining lymph nodes, which primarily contain resting and activated lymphocytes and antigen-presenting cells that migrated from the inflamed skin. Thus, Pglyrp3 and Pglyrp4 most likely exert their anti-inflammatory effect in the skin through their expression in keratinocytes. Keratinocytes are an important local source of chemokines and cytokines, and activation of keratinocytes by proinflammatory stimuli also leads to increased expression of Pglyrp3 and Pglyrp4 in the skin, which correlates with the ability of WT mice to reduce chronic inflammation in the skin. By contrast, increased Pglyrp1 expression in the inflamed skin likely comes from the influx of PMNs, because PMNs express high amounts of Pglyrp1 in their granules and, unlike other PGRPs, Pglyrp1 expression is not increased by proinflammatory stimuli in epithelial cells, including keratinocytes [7], [8], [14], [15]. Thus, the effects of PGRPs in the inflamed skin are likely exerted through a change in the local production of chemokines and cytokines in the skin, which modulates the recruitment and activity of these Treg and Th17 cells. In Pglyrp3 −/− and Pglyrp4 −/− mice reduced numbers of Treg cells allow dominating expansion of Th17 cells, which can increase inflammatory responses in the atopic dermatitis model, but may reduce Th1-mediated response in the contact dermatitis model by shifting T cell differentiation into Th17 cells instead of Th1 cells. Thus in WT mice, compared to Pglyrp3 −/− and Pglyrp4 −/− mice, the immune balance is shifted towards Th1 cells, which are protective against microbial infections (rather than Th17 cells) and towards Treg cells that control detrimental inflammation induced by proinflammatory chemicals and allergens. Our results suggest that defects in Pglyrp3 and Pglyrp4 genes could be predisposing to atopic dermatitis through the aforementioned shifts in immune homeostasis.
Materials and Methods
Ethics statement
All experiments on mice were performed according to the guidelines and approved by the Indiana University School of Medicine–Northwest Institutional Animal Care and Use Committee (approval number IUSM-NW-16).
Mice
We generated Pglyrp1 −/−, Pglyrp2 −/−, Pglyrp3 −/−, and Pglyrp4 −/− mice as described previously [15], [22], [25]. We generated Pglyrp1 −/− Pglyrp2 −/−, Pglyrp1 −/− Pglyrp3 −/−, and Pglyrp2 −/− Pglyrp3 −/− double knockout mice and Pglyrp1 −/− Pglyrp2 −/− Pglyrp3 −/− and Pglyrp1 −/− Pglyrp2 −/− Pglyrp4 −/− triple knockout mice by breeding single and double knockout mice (all on BALB/c background) and screening for homozygous deletion of each Pglyrp gene by PCR analysis of genomic DNA as previously described [15], [22], [25]. The lack of expression of the Pglyrp genes was confirmed by qRT-PCR in mRNA from the ears. Double and triple homozygous Pglyrp knockout mice were viable and fertile, bred normally, and yielded the expected male∶female ratios and similar litter size as the wild type and heterozygous mice. They had similar weight as the WT and single Pglyrp knockout mice and developed normally with no obvious defects. Their major internal organs had normal macroscopic appearance, and normal histological appearance on hematoxylin/eosin-stained sections.
All mice used in experiments were 8–10 week-old and on BALB/c background. The original colony founder WT BALB/c breeder mice were obtained from Harlan-Sprague-Dawley. All knockout mice were backcrossed to the same WT BALB/c mice from our breeding colony, and all WT and knockout mice were bred and kept under conventional pathogen-free conditions in the same room in our facility to minimize the influence of differences in the environment. For each experiment, mice from several different cages and breeder pairs were used. The BALB/c background of Pglyrp-deficient mice and their negative status for all common viral and bacterial pathogens and parasites were confirmed as previously described [22].
Oxazolone atopic dermatitis and contact dermatitis models
To induce atopic dermatitis female mice were first sensitized with 10 µl of 5% oxazolone (in 80% acetone, 20% olive oil) applied to the abdomen (after removing hair with Nair cream); 6 days later applications of 30 µl of 0.1% oxazolone (in 80% acetone, 20% olive oil) to each ear (15 µl to each side) were started (day 0) and continued every other day through day 18 [26]. In some experiments to induce Treg cells [30], [31] 3 µM vitamin D (1α,25-dihydroxyvitamin D3 from Sigma) was added to the oxazolone solution and used for the initial sensitization and applications to the ears. Ear thickness was measured each time before oxazolone application with Digimatic Micrometer (Mitutoyo, Japan) under constant pressure at the lowest setting. Ear swelling was determined by subtracting the untreated ear thickness. The significance of differences in ear swelling was determined using t-test.
To induce contact dermatitis female mice were first sensitized with 50 µl of 2% oxazolone (Sigma, in 80% acetone, 20% olive oil) applied to the abdomen (after removing hair with Nair cream) and 5 µl applied to each paw. The contact dermatitis reaction was then elicited 6 days later with a single application of 20 µl of 1% oxazolone (in 80% acetone, 20% olive oil) to each ear (10 µl to each side) [27], [28]. Ear thickness was measured as described above before and 24 and 48 hrs after oxazolone application, and ear swelling was determined by subtracting the untreated ear thickness. The significance of differences in ear swelling was determined using t-test.
Histology and immunohistochemistry
For histological analysis ears were fixed in Bouin's fixative, postfixed in 70% ethanol, and embedded in paraffin, and 5 µm cross-sections were stained with hematoxylin/eosin, and evaluated microscopically. For immunohistochemistry, antibodies to mouse Pglyrp3 and Pglyrp4 were obtained by immunizing rabbits with peptides corresponding to the following amino acids: CLVPQHSEIPKKA for Pglyrp3 (exon 5), and CWENPQTDQVSEG for Pglyrp4 (exon 2), coupled to KLH, followed by affinity purification on SulfoLink gel (Pierce) with corresponding peptides linked through the N-terminal Cys, elution with Tris-glycine buffer, pH 2.5, and dialysis against PBS, pH 7.2. A rabbit IgG antibody to a different peptide, which did not react with mouse Pglyrp3 and Pglyrp4, prepared and purified by the same method, was used as a negative control. Paraffin 5 µm cross-sections of were stained by the immunoperoxidase method as previously described [8], including standard deparaffinization, re-hydration, quenching of endogenous peroxidase by 30 min incubation in 0.3% H2O2, and incubation with 0.5 µg/ml of anti-Pglyrp3 or Pglyrp4 antibodies or control IgG overnight, followed by biotinylated second Ab and Vectastain Elite ABC kit (Vector) with DAB as a substrate (which generates brown reaction product) and counterstaining with hematoxylin (blue).
RNA and quantitative real-time reverse transcription PCR (qRT-PCR)
RNA was isolated from either the entire untreated or treated ears or lymph nodes using the TRIZOL method (InVitrogen), followed by digestion with RNase-free DNase (Qiagen) and purification on RNeasy spin columns using RNeasy Minikit (Qiagen). Quantitative reverse transcription real-time PCR (qRT-PCR) was used to quantify the amounts of mRNA in the ears or lymph nodes using custom RT2 Profiler PCR Arrays designed by us and manufactured by Qiagen/SA Biosciences, as previously described [22], [25]. The arrays typically included 30 to 44 assay genes, 5 housekeeping genes and reverse transcription efficiency and DNA contamination controls. All primer sets were from Qiagen/SA Biosciences, except the following primers designed by us: Pglyrp1, exons 1 and 2 primers, GTGGTGATCTCACACACAGC and GTGTGGTCACCCTTGATGTT; Pglyrp2, exons 3 and 4 primers, ACCAGGATGTGCGCAAGTGGGAT and AGTGACCCAGTGTAGTTGCCCA; and Pglyrp4, exons 4 and 5 primers, CGACCAGGGCTACAAGAA and CCAGGCAGTCTTCACTTTTC. cDNA was synthesized from 2 µg of RNA using RT2 PCR Array First Strand Kit (Qiagen/SA Biosciences) and the arrays were performed according to the manufacturer instructions using Qiagen/SA Biosciences Master Mix. The lists of genes are provided in the figures. The experiments were performed on RNA pooled from 4–5 mice/group and repeated 3 times usually with another set of 4–5 mice/group (usually total of 8–10 mice per treatment).
For each gene, ΔCt was calculated using the same threshold (0.2) for all genes and Ct≤35 considered as no expression, followed by normalization to 5 housekeeping genes (Hsp90ab1, Gusb, Hprt1, Gapdh, and Actb) included in each array, followed by calculation of ΔΔCt for each gene from two arrays: ΔΔCt = ΔCt1−ΔCt2, where ΔCt1 is the oxazolone treated mice and ΔCT2 is the untreated mice, using the program provided by Qiagen/SA Biosciences. This calculation gives the fold increase in expression of each gene in the treated mice versus untreated mice per µg RNA. The genomic DNA contamination controls, reverse transcription controls, and positive PCR controls were included in each array and were all passed. Additional control to assure amplification from RNA, but not from possible contaminating DNA included parallel reaction sets from which reverse transcriptase was omitted, and which showed no amplification. To compare baseline gene expression in untreated mice, ΔCT1 was from untreated PGRP-deficient mice and ΔCT2 was from untreated WT mice.
The results were reported as mean fold increases after oxazolone treatment (treated/untreated) for WT mice, or ratios of fold increases in Pglyrp-deficient to WT mice, calculated as follows: [(Pglyrp −/− treated)/(Pglyrp −/− untreated)]/[(WT treated)/(WT untreated)] and presented as heat maps or bar graphs. The latter fold differences (ratios) of >1 or <1 reflect higher or lower expression levels of the genes (respectively) in Pglyrp-deficient than in WT mice. Heat maps were generated using Java TreeView after converting <1 ratios to negative fold difference using the formula: (−1)/ratio. In some bar graph figures, <1 ratios were also converted to negative fold difference using the formula: (−1)/ratio. The significance of differences in gene activation between groups of mice was determined using the two-sample one-tailed t-test, and typically the differences of >2 fold were significant at P<0.05.
Expression of mRNA for PGRPs and some chemokine receptors was similarly measured by qRT-PCR using Qiagen/SA Biosciences First Strand Kit (with random primers) and amplification for 40 cycles with Qiagen/SA Biosciences SYBR Green Master Mix, and calculated using comparative cycle threshold method with 5 housekeeping genes (Hsp90ab1, Gusb, Hprt1, Gapdh, and Actb) as controls.
Isolation of cells and flow cytometry
Mouse ears were placed in RPMI-1640 with 3 mg/ml of Dispase II (Roche), separated into dorsal and ventral halves and scored on the dermal side with a scalpel. The tissue was digested for 8 hrs at 37°C in 5% CO2. Dermis was then separated from the epidermis and epidermis was further digested with 0.25% trypsin in RPMI-1640 for 10 min at 37°C. Cells were washed twice with RPMI-1640 with 5% fetal bovine serum (FBS) and incubated for 20 hrs in the same medium at 37°C in 5% CO2. Cells were then strained through a 40 µm filter and resuspended at 2.0×107 cells/ml in RPMI-1640 with 5% FBS. Single cells from cervical lymph nodes and spleen were obtained by passing the tissue through a 40 µm filter, red blood cells were removed from the spleen cells with a lysis buffer (Biolegend), and cells were suspended at 2.0×107 cells/ml in RPMI-1640 with 5% FBS.
1×106 cells were stained with CD4-APC (clone RM4-5, Biolegend) antibody for 20 min at 4°C. CD4-stained cells were then stained for Foxp3-PE (clone FJK-16s, eBioscience) or for cytokines IFN-γ-PE (clone XMG1.2), IL4-PE (clone 11B11) and IL-17-PE (clone TC11-18H10.1) with antibodies from Biolegend, used at 0.2 mg/1×106 cells according to Biolegend protocols using Biolegend buffers. Prior to staining for cytokines, CD4-APC stained cells were activated with TPA (12-O-Tetradecanoylphorbol 13-acetate, 25 ng/ml) and ionomycin (250 ng/ml) in the presence of the Golgi inhibitor, monensin, for 4 hrs at 37°C in 5% CO2. Cells were analyzed by flow cytometry using MACSQuant (Miltenyi) cytometer. Foxp3, IFN-γ, IL-4 and IL-17 positive cells were measured within the CD4+ gate.
Neutralization of IL-17 and ELISA
IL-17 was neutralized by intravenous injections of anti-IL-17 mAb (specific for IL-17A and not reactive with IL-1F, rat clone 50104, endotoxin-free from R&D Systems) 100 µg at sensitization and 50 µg on days 0, 3, 6, 9, 12, and 15 of oxazolone treatment. Control mice were similarly treated with isotype control rat IgG2aκ mAb (clone 16-4321, endotoxin-free from eBioscience). The amount of CXCL-1 in ears was measured by ELISA as previously described [25], after each ear was homogenized with Polytron in 0.5 ml of PBS (without Ca/Mg) with 1 mM EDTA, 1 mM PMSF, 1∶100 dilution of protease inhibitors (Sigma P1860), and 0.5% Triton X-100, followed by sonication and centrifugation.
Supporting Information
Pglyrp3 −/− and Pglyrp4 −/− mice have increased Th17 cells in the affected skin in the oxazolone atopic dermatitis model. Expression of a panel of marker genes characteristic of various inflammatory cell types in the ears of mice after sensitization and 10 applications of oxazolone to the ears every other day measured by qRT-PCR (day 20) is shown. For WT mice (top panel), the ratio of the amount of mRNA in oxazolone-treated to untreated mice for each gene (fold induction by oxazolone) is shown; for Pglyrp −/− mice, the results are the ratios of fold induction of each gene by oxazolone in Pglyrp −/− mice to fold induction of each gene by oxazolone in WT mice (which represents the fold difference in the response to oxazolone in Pglyrp −/− versus WT mice). The results are means ± SEM of 3 arrays from 4–5 mice/group and are shown as heat maps in Figure 6A in the main article.
(PDF)
In the oxazolone contact dermatitis model in Pglyrp1 −/− and Pglyrp2 −/− mice most cell types are decreased in the affected skin, but Treg cells and B cells are increased. Expression of a panel of marker genes characteristic of various inflammatory cell types in the ears of mice after sensitization and 6 hrs after single application of oxazolone to the ears measured by qRT-PCR is shown. For WT mice (top panel), the ratio of the amount of mRNA in oxazolone-treated to untreated mice for each gene (fold induction by oxazolone) is shown; for Pglyrp −/− mice, the results are the ratios of fold induction of each gene by oxazolone in Pglyrp −/− mice to fold induction of each gene by oxazolone in WT mice (which represents the fold difference in the response to oxazolone in Pglyrp −/− versus WT mice). The results are means ± SEM of 3 arrays from 4–5 mice/group and are shown as heat maps in Figure 6B in the main article.
(PDF)
Multiple inflammatory and immune genes are induced at an early stage of oxazolone model of atopic dermatitis. Expression of a panel of cytokines, chemokines, and other marker genes characteristic of Th1, Th2, Th17, Treg, NK, and other cell types in the ears of mice after sensitization and 7 applications of oxazolone to the ears every other day measured by qRT-PCR (day 13). For WT mice (top panel), the ratio of the amount of mRNA in oxazolone-treated to untreated mice for each gene (fold induction by oxazolone) is shown; for Pglyrp3 −/− or Pglyrp4 −/− mice, the results are the ratios of fold induction of each gene by oxazolone in Pglyrp −/− mice to fold induction of each gene by oxazolone in WT mice (which represents the fold difference in the response to oxazolone in Pglyrp −/− versus WT mice). The results are means ± SEM of 3 arrays from 4–5 mice/group and are shown as heat maps in Figure 7A in the main article.
(PDF)
Th17 gene expression profile is preferentially induced in the oxazolone model of atopic dermatitis in Pglyrp3 −/− and Pglyrp4 −/− mice. Expression of a panel of cytokines, chemokines, and other marker genes characteristic of Th1, Th2, Th17, Treg, NK, and other cell types in the ears of mice after sensitization and 10 applications of oxazolone to the ears every other day shows higher induction of several Th17 marker genes in Pglyrp3 −/− and Pglyrp4 −/− compared to WT mice measured by qRT-PCR. For WT mice (top panel), the ratio of the amount of mRNA in oxazolone-treated to untreated mice for each gene (fold induction by oxazolone) is shown; for Pglyrp −/− mice, the results are the ratios of fold induction of each gene by oxazolone in Pglyrp −/− mice to fold induction of each gene by oxazolone in WT mice (which represents the fold difference in the response to oxazolone in Pglyrp −/− versus WT mice). The results are means ± SEM of 3–4 arrays from 4–5 mice/group and are shown as heat maps in Figure 7A in the main article.
(PDF)
Expression of most immune genes is reduced in the oxazolone model of contact dermatitis in Pglyrp −/−mice. Expression of a panel of cytokines, chemokines, and other marker genes characteristic of Th1, Th2, Th17, Treg, NK, and other cell types in the ears of mice after sensitization and a single application of oxazolone to the ears shows lower induction of immune marker genes in Pglyrp −/− mice compared to WT mice measured by qRT-PCR. For WT mice (top panel), the ratio of the amount of mRNA in oxazolone-treated to untreated mice for each gene (fold induction by oxazolone) is shown; for Pglyrp −/− mice, the results are the ratios of fold induction of each gene by oxazolone in Pglyrp −/− mice to fold induction of each gene by oxazolone in WT mice (which represents the fold difference in the response to oxazolone in Pglyrp −/− versus WT mice, negative numbers show lower gene induction in Pglyrp −/− than in WT mice). The results are means ± SEM of 3–4 arrays from 4–5 mice/group and are shown as heat maps in Figure 7B in the main article.
(PDF)
Acknowledgments
We are grateful to Patrick Bankston for help in interpreting histology slides, and Robert Rukavina, Julie Cook, Panida Girddonfag, and Tiffany Caluag for maintaining and breeding our mice.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by United States Public Health Services Grants AI028797 and AI073290 from the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.De Benedetto A, Agnihothri R, McGirt LY, Bankova LG, Beck LA. Atopic dermatitis: a disease caused by innate immune defects? J Invest Dermatol. 2009;129:14–30. doi: 10.1038/jid.2008.259. [DOI] [PubMed] [Google Scholar]
- 2.Fonacier LS, Dreskin SC, Leung DY. Allergic skin diseases. J Allergy Clin Immunol. 2010;125(Suppl 2):S138–149. doi: 10.1016/j.jaci.2009.05.039. [DOI] [PubMed] [Google Scholar]
- 3.Jin H, He R, Oyoshi M, Geha RS. Animal models of atopic dermatitis. J Invest Dermatol. 2009;129:31–40. doi: 10.1038/jid.2008.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang B, Feliciani C, Freed I, Cai Q, Sauder DN. Insights into molecular mechanisms of contact hypersensitivity gained from gene knockout studies. J Leukoc Biol. 2001;70:185–191. [PubMed] [Google Scholar]
- 5.Martin SF, Jakob T. From innate to adaptive immune responses in contact hypersensitivity. Curr Opin Allergy Clin Immunol. 2008;8:289–293. doi: 10.1097/ACI.0b013e3283088cf9. [DOI] [PubMed] [Google Scholar]
- 6.Kang D, Liu G, Lundstrom A, Gelius E, Steiner H. A peptidoglycan recognition protein in innate immunity conserved from insects to humans. Proc Natl Acad Sci USA. 1998;95:10078–10082. doi: 10.1073/pnas.95.17.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu C, Xu Z, Gupta D, Dziarski R. Peptidoglycan recognition proteins: a novel family of four human innate immunity pattern recognition molecules. J Biol Chem. 2001;276:34686–34694. doi: 10.1074/jbc.M105566200. [DOI] [PubMed] [Google Scholar]
- 8.Lu X, Wang M, Qi J, Wang H, Li X, et al. Peptidoglycan recognition proteins are a new class of human bactericidal proteins. J Biol Chem. 2006;281:5895–5907. doi: 10.1074/jbc.M511631200. [DOI] [PubMed] [Google Scholar]
- 9.Tydell CC, Yuan J, Tran P, Selsted ME. Bovine peptidoglycan recognition protein-S: antimicrobial activity, localization, secretion, and binding properties. J Immunol. 2006;176:1154–1162. doi: 10.4049/jimmunol.176.2.1154. [DOI] [PubMed] [Google Scholar]
- 10.Wang M, Liu L-H, Wang S, Li X, Lu X, et al. Human peptidoglycan recognition proteins require zinc to kill both Gram-positive and Gram-negative bacteria and are synergistic with antibacterial peptides. J Immunol. 2007;178:3116–3125. doi: 10.4049/jimmunol.178.5.3116. [DOI] [PubMed] [Google Scholar]
- 11.Kashyap DR, Wang M, Liu L-H, Boons G-J, Gupta D, Dziarski R. Peptidoglycan recognition proteins kill bacteria by activating protein-sensing two-component systems. Nature Med. 2011;17:676–683. doi: 10.1038/nm.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gelius E, Persson C, Karlsson J, Steiner H. A mammalian peptidoglycan recognition protein with N-acetylmuramoyl-L-alanine amidase activity. Biochem Biophys Res Commun. 2003;306:988–994. doi: 10.1016/s0006-291x(03)01096-9. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z-M, Li X, Cocklin RR, Wang M, Wang M, et al. Human peptidoglycan recognition protein-L is an N-acetylmuramoyl-L-alanine amidase. J Biol Chem. 2003;278:49044–49052. doi: 10.1074/jbc.M307758200. [DOI] [PubMed] [Google Scholar]
- 14.Liu C, Gelius E, Liu G, Steiner H, Dziarski R. Mammalian peptidoglycan recognition protein binds peptidoglycan with high affinity, is expressed in neutrophils, and inhibits bacterial growth. J Biol Chem. 2000;275:24490–24499. doi: 10.1074/jbc.M001239200. [DOI] [PubMed] [Google Scholar]
- 15.Dziarski R, Platt KA, Gelius E, Steiner H, Gupta D. Defect in neutrophil killing and increased susceptibility to infection with non-pathogenic Gram-positive bacteria in peptidoglycan recognition protein-S (PGRP-S)-deficient mice. Blood. 2003;102:689–697. doi: 10.1182/blood-2002-12-3853. [DOI] [PubMed] [Google Scholar]
- 16.Lo D, Tynan W, Dickerson J, Mendy J, Chang HW, et al. Peptidoglycan recognition protein expression in mouse Peyer's Patch follicle associated epithelium suggests functional specialization. Cell Immunol. 2003;224:8–16. doi: 10.1016/s0008-8749(03)00155-2. [DOI] [PubMed] [Google Scholar]
- 17.Xu M, Wang Z, Locksley RM. Innate immune responses in peptidoglycan recognition protein L-deficient mice. Mol Cell Biol. 2004;24:7949–7957. doi: 10.1128/MCB.24.18.7949-7957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Gupta D, Li X, Dziarski R. Peptidoglycan recognition protein 2 (N-acetylmuramoyl-L-Ala amidase) is induced in keratinocytes by bacteria through the p38 kinase pathway. Infect Immun. 2005;73:7216–7225. doi: 10.1128/IAI.73.11.7216-7225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, van der Fits L, Voerman JS, Melief M-J, Laman JD, et al. Identification of serum N-acetylmuramoyl-L-alanine amidase as liver peptidoglycan recognition protein 2. Biochim Biophys Acta. 2005;1752:34–46. doi: 10.1016/j.bbapap.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Wang S, Wang H, Gupta D. Differential expression of peptidoglycan recognition protein 2 in the skin and liver requires different transcription factors. J Biol Chem. 2006;281:20738–20748. doi: 10.1074/jbc.M601017200. [DOI] [PubMed] [Google Scholar]
- 21.Mathur P, Murray B, Crowell T, Gardner H, Allaire N, et al. Murine peptidoglycan recognition proteins PglyrpIα and PglyrpIβ are encoded in the epidermal differentiation complex and are expressed in epidermal and hematopoietic tissues. Genomics. 2004;83:1151–1163. doi: 10.1016/j.ygeno.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Saha S, Jing X, Park SY, Wang S, Li X, et al. Peptidoglycan recognition proteins protect mice from experimental colitis by promoting normal gut flora and preventing induction of interferon-γ. Cell Host Microbe. 2010;8:147–162. doi: 10.1016/j.chom.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun C, Mathur P, Dupuis J, Tizard R, Ticho B, et al. Peptidoglycan recognition proteins Pglyrp3 and Pglyrp4 are encoded from the epidermal differentiation complex and are candidate genes for the Psors4 locus on chromosome 1q21. Hum Genet. 2006;119:113–125. doi: 10.1007/s00439-005-0115-8. [DOI] [PubMed] [Google Scholar]
- 24.Kainu K, Kivinen K, Zucchelli M, Suomela S, Kere J, et al. Association of psoriasis to PGLYRP and SPRR genes at PSORS4 locus on 1q shows heterogeneity between Finnish, Swedish and Irish families. Exp Dermatol. 2009;18:109–115. doi: 10.1111/j.1600-0625.2008.00769.x. [DOI] [PubMed] [Google Scholar]
- 25.Saha S, Qi J, Wang S, Wang M, Li X, et al. PGLYRP-2 and Nod2 are both required for peptidoglycan-induced arthritis and local inflammation. Cell Host Microbe. 2009;5:137–150. doi: 10.1016/j.chom.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Man MQ, Hatano Y, Lee SH, Man M, Chang S, et al. Characterization of a hapten-induced, murine model with multiple features of atopic dermatitis: structural, immunologic, and biochemical changes following single versus multiple oxazolone challenges. J Invest Dermatol. 2008;128:79–86. doi: 10.1038/sj.jid.5701011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lange-Asschenfeldt B, Weninger W, Velasco P, Kyriakides TR, von Andrian UH, et al. Increased and prolonged inflammation and angiogenesis in delayed-type hypersensitivity reactions elicited in the skin of thrombospondin-2-deficient mice. Blood. 2002;99:538–545. doi: 10.1182/blood.v99.2.538. [DOI] [PubMed] [Google Scholar]
- 28.Kunstfeld R, Hirakawa S, Hong YK, Schacht V, Lange-Asschenfeldt B, et al. Induction of cutaneous delayed-type hypersensitivity reactions in VEGF-A transgenic mice results in chronic skin inflammation associated with persistent lymphatic hyperplasia. Blood. 2004;104:1048–1057. doi: 10.1182/blood-2003-08-2964. [DOI] [PubMed] [Google Scholar]
- 29.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, et al. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 30.Gorman S, Kuritzky LA, Judge MA, Dixon KM, McGlade JP, et al. Topically applied 1,25-dihydroxyvitamin D3 enhances the suppressive activity of CD4+CD25+ cells in the draining lymph nodes. J Immunol. 2007;179:6273–6283. doi: 10.4049/jimmunol.179.9.6273. [DOI] [PubMed] [Google Scholar]
- 31.Ghoreishi M, Bach P, Obst J, Komba M, Fleet JC, et al. Expansion of antigen-specific regulatory T cells with the topical vitamin D analog calcipotriol. J Immunol. 2009;182:6071–6078. doi: 10.4049/jimmunol.0804064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koga C, Kabashima K, Shiraishi N, Kobayashi M, Tokura Y. Possible pathogenic role of Th17 cells for atopic dermatitis. J Invest Dermatol. 2008;128:2625–2630. doi: 10.1038/jid.2008.111. [DOI] [PubMed] [Google Scholar]
- 33.Oyoshi MK, Murphy GF, Geha RS. Filaggrin-deficient mice exhibit TH17-dominated skin inflammation and permissiveness to epicutaneous sensitization with protein antigen. J Allergy Clin Immunol. 2009;124:485–493. doi: 10.1016/j.jaci.2009.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye P, Rodriguez FH, Kanaly S, Stocking KL, Schurr J, et al. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J Exp Med. 2001;194:519–527. doi: 10.1084/jem.194.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 36.Murphy CA, Langrish CL, Chen Y, Blumenschein W, McClanahan T, et al. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J Exp Med. 2003;198:1951–1957. doi: 10.1084/jem.20030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, et al. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 39.Miossec P, Korn T, Kuchroo VK. Interleukin-17 and type 17 helper T cells. N Engl J Med. 2009;361:888–898. doi: 10.1056/NEJMra0707449. [DOI] [PubMed] [Google Scholar]
- 40.Alcorn JF, Crowe CR, Kolls JK. TH17 cells in asthma and COPD. Annu Rev Physiol. 2010;72:495–516. doi: 10.1146/annurev-physiol-021909-135926. [DOI] [PubMed] [Google Scholar]
- 41.Hsu HC, Yang P, Wang J, Wu Q, Myers R, et al. Interleukin 17-producing T helper cells and interleukin 17 orchestrate autoreactive germinal center development in autoimmune BXD2 mice. Nat Immunol. 2008;9:166–175. doi: 10.1038/ni1552. [DOI] [PubMed] [Google Scholar]
- 42.Mitsdoerffer M, Lee Y, Jäger A, Kim HJ, Korn T, et al. Proinflammatory T helper type 17 cells are effective B-cell helpers. Proc Natl Acad Sci USA. 2010;107:14292–14297. doi: 10.1073/pnas.1009234107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Leary JG, Goodarzi M, Drayton DL, von Andrian UH. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat Immunol. 2006;7:507–516. doi: 10.1038/ni1332. [DOI] [PubMed] [Google Scholar]
- 45.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 46.Campbell DJ, Koch MA. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat Rev Immunol. 2011;11:119–130. doi: 10.1038/nri2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pglyrp3 −/− and Pglyrp4 −/− mice have increased Th17 cells in the affected skin in the oxazolone atopic dermatitis model. Expression of a panel of marker genes characteristic of various inflammatory cell types in the ears of mice after sensitization and 10 applications of oxazolone to the ears every other day measured by qRT-PCR (day 20) is shown. For WT mice (top panel), the ratio of the amount of mRNA in oxazolone-treated to untreated mice for each gene (fold induction by oxazolone) is shown; for Pglyrp −/− mice, the results are the ratios of fold induction of each gene by oxazolone in Pglyrp −/− mice to fold induction of each gene by oxazolone in WT mice (which represents the fold difference in the response to oxazolone in Pglyrp −/− versus WT mice). The results are means ± SEM of 3 arrays from 4–5 mice/group and are shown as heat maps in Figure 6A in the main article.
(PDF)
In the oxazolone contact dermatitis model in Pglyrp1 −/− and Pglyrp2 −/− mice most cell types are decreased in the affected skin, but Treg cells and B cells are increased. Expression of a panel of marker genes characteristic of various inflammatory cell types in the ears of mice after sensitization and 6 hrs after single application of oxazolone to the ears measured by qRT-PCR is shown. For WT mice (top panel), the ratio of the amount of mRNA in oxazolone-treated to untreated mice for each gene (fold induction by oxazolone) is shown; for Pglyrp −/− mice, the results are the ratios of fold induction of each gene by oxazolone in Pglyrp −/− mice to fold induction of each gene by oxazolone in WT mice (which represents the fold difference in the response to oxazolone in Pglyrp −/− versus WT mice). The results are means ± SEM of 3 arrays from 4–5 mice/group and are shown as heat maps in Figure 6B in the main article.
(PDF)
Multiple inflammatory and immune genes are induced at an early stage of oxazolone model of atopic dermatitis. Expression of a panel of cytokines, chemokines, and other marker genes characteristic of Th1, Th2, Th17, Treg, NK, and other cell types in the ears of mice after sensitization and 7 applications of oxazolone to the ears every other day measured by qRT-PCR (day 13). For WT mice (top panel), the ratio of the amount of mRNA in oxazolone-treated to untreated mice for each gene (fold induction by oxazolone) is shown; for Pglyrp3 −/− or Pglyrp4 −/− mice, the results are the ratios of fold induction of each gene by oxazolone in Pglyrp −/− mice to fold induction of each gene by oxazolone in WT mice (which represents the fold difference in the response to oxazolone in Pglyrp −/− versus WT mice). The results are means ± SEM of 3 arrays from 4–5 mice/group and are shown as heat maps in Figure 7A in the main article.
(PDF)
Th17 gene expression profile is preferentially induced in the oxazolone model of atopic dermatitis in Pglyrp3 −/− and Pglyrp4 −/− mice. Expression of a panel of cytokines, chemokines, and other marker genes characteristic of Th1, Th2, Th17, Treg, NK, and other cell types in the ears of mice after sensitization and 10 applications of oxazolone to the ears every other day shows higher induction of several Th17 marker genes in Pglyrp3 −/− and Pglyrp4 −/− compared to WT mice measured by qRT-PCR. For WT mice (top panel), the ratio of the amount of mRNA in oxazolone-treated to untreated mice for each gene (fold induction by oxazolone) is shown; for Pglyrp −/− mice, the results are the ratios of fold induction of each gene by oxazolone in Pglyrp −/− mice to fold induction of each gene by oxazolone in WT mice (which represents the fold difference in the response to oxazolone in Pglyrp −/− versus WT mice). The results are means ± SEM of 3–4 arrays from 4–5 mice/group and are shown as heat maps in Figure 7A in the main article.
(PDF)
Expression of most immune genes is reduced in the oxazolone model of contact dermatitis in Pglyrp −/−mice. Expression of a panel of cytokines, chemokines, and other marker genes characteristic of Th1, Th2, Th17, Treg, NK, and other cell types in the ears of mice after sensitization and a single application of oxazolone to the ears shows lower induction of immune marker genes in Pglyrp −/− mice compared to WT mice measured by qRT-PCR. For WT mice (top panel), the ratio of the amount of mRNA in oxazolone-treated to untreated mice for each gene (fold induction by oxazolone) is shown; for Pglyrp −/− mice, the results are the ratios of fold induction of each gene by oxazolone in Pglyrp −/− mice to fold induction of each gene by oxazolone in WT mice (which represents the fold difference in the response to oxazolone in Pglyrp −/− versus WT mice, negative numbers show lower gene induction in Pglyrp −/− than in WT mice). The results are means ± SEM of 3–4 arrays from 4–5 mice/group and are shown as heat maps in Figure 7B in the main article.
(PDF)