Abstract
Background
Here we report specific activities of all seven naturally occurring LinA variants towards three different isomers, α, γ and δ, of a priority persistent pollutant, hexachlorocyclohexane (HCH). Sequence-structure-function differences contributing to the differences in their stereospecificity for α-, γ-, and δ-HCH and enantiospecificity for (+)- and (−)-α -HCH are also discussed.
Methodology/Principal Findings
Enzyme kinetic studies were performed with purified LinA variants. Models of LinA2B90A A110T, A111C, A110T/A111C and LinA1B90A were constructed using the FoldX computer algorithm. Turnover rates (min−1) showed that the LinAs exhibited differential substrate affinity amongst the four HCH isomers tested. α-HCH was found to be the most preferred substrate by all LinA's, followed by the γ and then δ isomer.
Conclusions/Significance
The kinetic observations suggest that LinA-γ1-7 is the best variant for developing an enzyme-based bioremediation technology for HCH. The majority of the sequence variation in the various linA genes that have been isolated is not neutral, but alters the enantio- and stereoselectivity of the encoded proteins.
Introduction
Hexachlorocyclohexane (HCH) consists of four main isomers (α-, β-, γ- and δ-HCH), all of which are highly toxic to vertebrates and one of which (γ-HCH; lindane) is a potent insecticide [1]. Toxicity concerns have led to deregistration of lindane in many countries, and large dumps of unused HCH now pose major environmental hazards [2], [3], [4]. Bacterial strains have evolved to degrade HCH, with the initial steps in their degradation of the α-, γ- and δ- isomers catalyzed by the enzyme LinA [2]. The crystal structure of LinA shows that it shares a structural fold and an Asp-His catalytic dyad with enzymes of the scytalone dehydratase family [5], [6]. LinA catalyses E2 elimination reactions at biaxial H-Cl pairs of atoms in α-, γ- and δ- HCH [2], [7], [8]. Seven naturally occurring LinA variants have been identified (Table 1), which differ in as many as 10% of their residues. However, little is known of their functional differences other than that LinA1B90A and LinA2B90A preferentially catalyze degradation of the (+)-α-HCH and (−)-α-HCH enantiomers, respectively [2], [9]. Here we report the specific activities of all seven previously cloned linA gene variants towards α-, γ- and δ-HCH and analyse how their sequence differences contribute to differences in their activities.
Table 1. Turnover rates (min−1) of the published LinA variants towards α-, γ- and δ-HCH, along with the amino acid differences amongst thema.
| Residue variations | Turnover number (min−1) | ||||||||||||||||||||||||||||
| 20 | 23 | 35 | 68 | 71 | 96 | 110 | 111 | 113 | 115 | 126 | 129 | 131 | 133 | 145 | 149 | 150 | 151 | 152 | 153 | 154 | 155 | 156 | α | γ | α/γ | δ | |||
| Group 1 | A | LinA2B90A | K | A | I | F | C | L | A | A | F | D | F | R | A | T | A | I | H | F | A | P | S | G | A | 3815±3 | 733±13 | 5.2 | 174±4 |
| LinAγ1-7 | . | . | . | . | . | . | T | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 3001±30 | 11794±526 | 0.2 | 284±32 | ||
| LinAbITRC-5 | . | . | . | . | . | . | T | C | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | 2306±29 | 2778±180 | 0.8 | 246±3 | ||
| LinANM05 | . | . | . | . | . | . | T | C | . | . | . | . | . | . | . | . | . | L | . | . | T | . | . | 11267±27 | 889±107 | 12.6 | 10±1 | ||
| b | LinADS3-1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | L | L | Q | K | . | . | . | 7884±14 | 5376±359 | 1.4 | 214±35 | |
| Group 2 | a | LinAaITRC-5 | . | G | V | Y | T | C | . | . | Y | N | . | L | G | M | V | A | . | . | . | . | . | – | – | 965±0 | 65±15 | 14.8 | 49±1 |
| b | LinA1B90A | Q | G | . | Y | T | C | . | . | Y | N | L | L | G | M | . | A | L | L | Q | K | . | – | – | 418±4 | 89±7 | 4.6 | 10±1 | |
LinA2B90A derives from Sphingobium indicum B90A and is identical to LinA from Sphinogobium sps. strains UT26, Sp+, DS2, γ16-1, BHC-A and Rhodanobacter lindaniclasticus; LinAγ1-7 is from Sphingomonas sp. γ1-7; LinAbITRC-5 is from Pseudomonas aeruginosa ITRC-5; LinANM05 comes from Sphingomonas sp. NM05; LinADS3-1 is from Sphingomonas sp. DS3-1 and is identical to LinA of Sphingobium sp. α1-2; LinAaITRC-5 is from Pseudomonas aeruginosa ITRC-5; LinA1B90A is from Sphingobium indicum B90A and is identical to LinA of Sphingomonas sp. α4-2 [2].
Materials and Methods
Codon optimized linA genes for expression of all the variants in E. coli were synthesized by Geneart AG, Regensburg Germany (GenBank accession numbers HM447244–HM447250). The synthetic linA genes were PCR amplified with attB1, attB2 and attB2-R2 primers (Table S1) and the amplicons were then cloned into pDONR201 and transferred to pDEST17 using the BP and LR reactions, respectively, following the manufacturers' instructions (Invitrogen, CA). The host E. coli BL21-AI™ (Invitrogen) cells co-expressed chaperones from the plasmid pGro7 (Takara, Japan).
The bacterial clones were cultured in 100 ml of LB at 28°C [10]. When the culture reached an OD600 of 0.5, L-(+)-arabinose was added to a final concentration of 2 g/L. Cells were harvested after overnight incubation, washed with 10 mM imidazole buffer (pH 7.5) and disrupted by 1× bugbuster (Novagen, Darmstadt). The lysate was centrifuged at 16,000 g for 20 min and the supernatant was used to purify his-tagged proteins using NTA-Ni2+ agarose (Qiagen, GmbH) following the manufacturers' instructions. The purified protein was quantified using Nanodrop (Thermo Scientific, DE). The purified enzyme was stored in storage buffer (pH 7.5) containing 1 mM 2-mercaptoethanol and 10% glycerol at a concentration of about 1 mg/ml at 4°C.
Enzyme assays were performed within 3 days of purification. In this period no measurable loss of enzyme activity was observed (data not shown). LinA activity was assayed by estimating the depletion of HCH isomers using gas chromatography. The assay reaction was initiated by the addition of enzyme to a reaction mixture (500 µl) containing 1.7 µM of the respective HCH isomer in 1× Tris glycine buffer (25 mM Tris, 192 mM glycine pH 8.3) at 22°C and was stopped by the addition of 0.3% (v/v) formic acid (final concentration). The incubation times for reaction mixtures containing α- and γ-HCH as substrates were 30 sec and 1 min, respectively, and those for δ-HCH assays were 2 or 5 min, depending on the activity of the enzyme. The samples were extracted in an equal volume of hexane by vortexing for 5 min and quantitatively analyzed on a GC equipped with a BPX-50 capillary column (30 m by 0.25 mm by 0.32 µm; SGE Analytical) and an electron capture detector. The temperature program was isothermal at 100°C for 5 min, followed by an increase to 200°C at 20°C/min, and the carrier gas (He) flow rate was 3.0 ml/min.
Models of LinA2B90A A110T, A111C, A110T/A111C and LinA1B90A were constructed using the FoldX computer algorithm (http://foldx.crg.es/), which permits the construction of low-energy models and calculation of interaction energies contributing to the stability of proteins [11], [12]. These mutations were modeled using the available crystal structure of LinA2B90A (PDB ID: 3A76). Prior to mutagenesis, the RepairPDB option of FoldX was used to optimize the total energy of the protein, which involved identifying and repairing residues with bad torsion angles and van der Waals clashes. Mutagenesis was subsequently performed using the BuildModel option of FoldX. The effects of the mutations on protein stability were obtained from the output file. The geometries of HCH isomers used in the docking analyses were taken from Brittain et al. [7]. The substrates were docked by superimposing them on the conformations of the transition states as docked in that work and with reference to the nearby histidine.
Results and Discussion
Histidine-tagged LinA proteins were heterologously expressed in E. coli co-expressing the chaperone GroEL and purified to homogeneity by affinity chromatography as described by Brittain et al. [7]. Specific activities of the various proteins with α-, γ- and δ-HCH were measured using gas chromatography with electron capture detection. The seven variants fall into two main groups according to sequence differences at positions 20, 23, 35, 68, 71, 96, 113, 115, 126, 129, 131 and 133 and the first and larger group has higher activities for all isomers than those of the second group (Table 1). However there is also functionally important variation in two sections of the sequence within these groups: 110 and 111, where A–T and A–C differences co-occur, and at the C-terminus (149–156), where most sequences are either I-H-F-A-P-S-G-A or A-L-L-Q-K-S, or minor variants thereof. Several specific comparisons show the effects of these variable regions on isomer specific activities. Firstly, LinA2B90A and LinAγ1-7, which differ only in the A110T/A111C pair, show radically different α-HCH:γ-HCH isomer specificities (5.2 cf. 0.25). Secondly, differences in the 149–154 region between LinAγ1-7, LinAbITRC-5 and LinANM05 also have a large effect on this ratio (0.25 cf. 14.8 and 12.6, respectively). Thirdly, differences in the 149–156 region between LinA2B90A and LinADS3-1 also have large effects on activity, with LinADS3-1 being more active for all isomers, but particularly for γ-HCH (7 fold, cf. 2 fold for α-HCH). Interestingly, this C-terminal variation is thought to result from insertion of the IS6100 transposable element [2], [13].
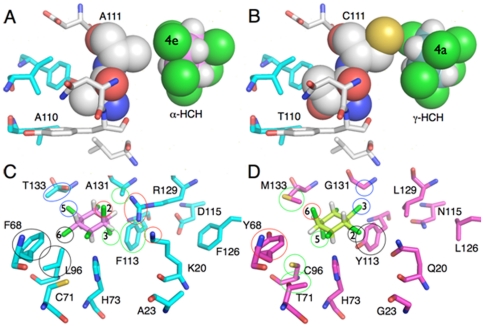
To investigate how the 110/111 sequence differences change the isomer specificity, the FoldX force-field [12] was used to create models of LinAγ1-7 and LinA1B90A from the crystal structure of LinA2B90A (PDB ID: 3A76) [5] and to estimate the stabilizing/destabilizing effects of these mutations (similar analysis could not be carried out for the C-terminal differences because the structure of that region is not well resolved). Fig. 1 shows that the reversal in α-HCH: γ-HCH isomer specificities (5.2 cf. 0.25) on changing from A110/A111 in LinA2B90A to T110/C111 in LinAγ1-7 is a result of the A111C mutation, which is opposite the 3-position of the HCH ring, the only position where α- and γ-HCH differ. This mutation will provide closer, more favourable contact with γ-HCH but generate some level of steric clash with α-HCH. However, this A111C change is predicted to be highly destabilizing (ΔΔG 0.75 kcal/monomer), whereas A110T appears to provide a compensating, stabilizing effect (ΔΔG -1.28 kcal/monomer) by extending into a hydrophobic cavity at the trimer interface. These mutations thus provide a clear example of the role of stabilizing mutations (A110T) in allowing function-changing mutations (A111C), which would otherwise result in aggregation of the protein, to be tolerated [14].
Figure 1. Effect of mutation and sequence differences in the LinA enzymes.
The effect of the A111C mutation is shown in panels A and B, illustrating that the cysteine residue will clash with the equatorial chlorine at the 4-position of (+)-α-HCH (4e), but provides complementary contacts with γ-HCH when the chlorine at the 4-position is axial (4a). Panels C and D show the effects of the sequence differences between LinA2B90A and LinA1B90A on enantioselectivity, with (+)- and (−)-α-HCH docked in each active site. The important sequence differences in the LinA enzymes and structural differences in the α-HCH enantiomers are circled.
All seven variants show the same broad isomer preferences as described previously for the LinAUT26 enzyme (identical sequence to LinA2B90A above), i.e. high activity towards α- and γ-HCH and less activity towards δ-HCH [2], [8], [15], [16]. Quantitatively, however, there are large differences among the seven variants in their absolute and relative activities (Table 1): α-HCH turnover varies from 418 to 11267 min−1, γ-HCH turnover varies from 65 to 11795 min−1 and the ratio (α∶γ) varies from 14.8 to 0.25. Similarly -HCH activities show over 200 fold variation in absolute terms, with values ranging from ∼2 to 75% of the corresponding γ-HCH value in relative terms.
Suar et al. [9] have previously determined the enantioselectivity of certain LinA variants for the (+) and (−) enantiomers of α-HCH, finding LinA1B90A and LinA2B90Aδ have strong preferences for (+)- and (−)-α-HCH, respectively. LinA1B90A and LinA2B90A are typical of the two main sequence groups in Table 1 and we can now use the structure described by Okai et al. [5] to analyse how the 18 amino acid differences between these sequences may contribute to the enantioselectivity differences. To do this we docked the two enantiomers in the active site based on the transition state geometry of the elimination reaction [5]. The most noticeable difference between the structures is that the positive charge on R129, which is catalytically essential and provides stabilising interactions with the leaving group in the transition state for LinA2B90A [5], is absent in LinA1B90A, with L129 unable to fulfil this role. The positively charged side chain of K20 is also found in this region in LinA2B90A and may fulfil a similar, if less critical, role that also cannot be replicated by Q20 in LinA1B90A. These findings are consistent with the leaving group changing sides from the 2-position in LinA2 B90A to the 6-position in LinA1B90A. Three other mutations in the second shell most likely compensate for these mutations (A23G, D115N, F126L). Other sequence differences account for changes in the position of the axial groups in the ring: F68Y provides a H-bonding group at the site of the axial leaving group in the 6-position, which will stabilize the transition state; the C71T/L96C/T133M mutations reshape the active site to account for the equatorial chlorine in the 5-position of (−)-α-HCH being axial in (+)-α-HCH; and similarly, the F113Y difference fills the space formed from the axial chlorine at the 3-position of (−)-α-HCH changing to equatorial in (+)-α-HCH. P153K might be involved in leaving group stabilization to fulfil an analogous role to that which K20 and R129 perform in LinA2 B90A.
Our analysis suggests that a majority of the sequence variation in the various linA genes that have been isolated is not neutral, but alters the enantio- and stereoselectivity of the encoded proteins. Significantly, some organisms, for example Sphingobium indicum B90A and Pseudomonas aeruginosa ITRC5, have at least two copies of the genes, in each case one from each of the two major sequence groups, thus providing those organisms with enhanced substrate ranges, covering α- and HCH as well as (+)-α- and (−)-α-HCH. Notably, linA has a different codon bias and G+C content than other lin genes encoding subsequent steps in the HCH degradation pathway [2], suggesting that it may be recently acquired. It will be interesting to monitor ongoing evolution of the lin system, and in particular LinA, to see whether this recently emerged pathway continues to adapt to the challenges of life in soils polluted with HCH.
Supporting Information
lin A specific primers used in the current study to clone lin A variants into gateway vector pDONR201 (Invitrogen).
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Indo-Australian Biotechnology Fund from the Department of Education Science and Technology (DEST), Australia and the Department of Biotechnology (DBT), India. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Slade RE. The gamma isomer of hexachlorocyclohexane (gammexane). An insecticide with outstanding properties. Chem Ind. 1945;40:314–319. [Google Scholar]
- 2.Lal R, Pandey G, Sharma P, Kumari K, Malhotra S, et al. Biochemistry of microbial degradation of hexachlorocyclohexane and prospects for bioremediation. Microbiol Mol Biol Rev. 2010;74:58–80. doi: 10.1128/MMBR.00029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phillips TM, Lee H, Trevors JT, Seech AG. Full-scale in situ bioremediation of hexachlorocyclohexane-contaminated soil. J Chem Technol Biotechnol. 2006;81:289–298. [Google Scholar]
- 4.Rubinos DA, Villasuso R, Muniategui S, Barral MT, Diaz Fierros F. Using the landfarming technique to remediate soils contaminated with hexachlorocyclohexane isomers. Water Air Soil Pollut. 2007;181:385–399. [Google Scholar]
- 5.Okai M, Kubota K, Fukuda M, Nagata Y, Nagata K, et al. Crystal structure of γ-hexachlorocyclohexane dehydrochlorinase LinA from Sphingobium japonicum UT26. J Mol Biol. 2010;403:260–269. doi: 10.1016/j.jmb.2010.08.043. [DOI] [PubMed] [Google Scholar]
- 6.Nagata Y, Mori K, Takagi M, Murzin AG, Damborsky J. Identification of protein fold and catalytic residues of γ-hexachlorocyclohexane dehydrochlorinase LinA. Proteins. 2001;45:471–477. doi: 10.1002/prot.10007. [DOI] [PubMed] [Google Scholar]
- 7.Brittain DRB, Pandey R, Kumari K, Sharma P, Pandey G, et al. Competing SN2 and E2 reaction pathways for hexachlorocyclohexane degradation in the gas phase, solution and enzymes. Chem Commun. 2011;47:976–978. doi: 10.1039/c0cc02925d. [DOI] [PubMed] [Google Scholar]
- 8.Trantirek L, Hynkova K, Nagata Y, Murzin A, Ansorgova A, et al. Reaction mechanism and stereochemistry of γ-hexachlorocyclohexane dehydrochlorinase LinA. J Biol Chem. 2001;276:7734–7740. doi: 10.1074/jbc.M007452200. [DOI] [PubMed] [Google Scholar]
- 9.Suar M, Hauser A, Poiger T, Buser HR, Muller MD, et al. Enantioselective transformation of alpha-hexachlorocyclohexane by the dehydrochlorinases LinA1 and LinA2 from the soil bacterium Sphingomonas paucimobilis B90A. Appl Environ Microbiol. 2005;71:8514–8518. doi: 10.1128/AEM.71.12.8514-8518.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 11.Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 12.Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, et al. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33(Suppl 2):W382–388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lal R, Dogra C, Malhotra S, Sharma P, Pal R. Diversity, distribution and divergence of lin genes in hexachlorocyclohexane-degrading sphingomonads. Trends Biotechnol. 2006;24:121–130. doi: 10.1016/j.tibtech.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature. 2006;444:929–932. doi: 10.1038/nature05385. [DOI] [PubMed] [Google Scholar]
- 15.Nagata Y, Miyauchi K, Takagi M. Complete analysis of genes and enzymes for γ-hexachlorocyclohexane degradation in Sphingomonas paucimobilis UT26. J Ind Microbiol Biotechnol. 1999;23:380–390. doi: 10.1038/sj.jim.2900736. [DOI] [PubMed] [Google Scholar]
- 16.Nagata Y, Hatta T, Imai R, Kimbara K, Fukuda M, et al. Purification and characterization of γ-hexachlorocyclohexane (γ-HCH) dehydrochlorinase (LinA) from Pseudomonas paucimobilis. Biosci Biotech Biochem. 1993;59:1582–1583. doi: 10.1271/bbb.57.703. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lin A specific primers used in the current study to clone lin A variants into gateway vector pDONR201 (Invitrogen).
(DOC)