Abstract
Here we extend the ability to predict hydrodynamic coefficients and other solution properties of rigid macromolecular structures from atomic-level structures, implemented in the computer program HYDROPRO, to models with lower, residue-level resolution. Whereas in the former case there is one bead per nonhydrogen atom, the latter contains one bead per amino acid (or nucleotide) residue, thus allowing calculations when atomic resolution is not available or coarse-grained models are preferred. We parameterized the effective hydrodynamic radius of the elements in the atomic- and residue-level models using a very large set of experimental data for translational and rotational coefficients (intrinsic viscosity and radius of gyration) for >50 proteins. We also extended the calculations to very large proteins and macromolecular complexes, such as the whole 70S ribosome. We show that with proper parameterization, the two levels of resolution yield similar and rather good agreement with experimental data. The new version of HYDROPRO, in addition to considering various computational and modeling schemes, is far more efficient computationally and can be handled with the use of a graphical interface.
Introduction
As a result of the increasing proliferation of structural determinations, renewed interest in measuring hydrodynamic and other solution properties, and the development of new measurement techniques, the importance of predicting such properties from models with high or medium resolution has increased notably over the past decade. A relevant development for the prediction of properties of rigid, globular proteins from their atomic-level structures was the methodology implemented in our programs HYDROPRO (1) and HYDRONMR (2,3). These tools have been widely employed for the analysis of hydrodynamic coefficients and other solution properties.
In the HYDROPRO and HYDRONMR procedures, each Na nonhydrogen atom is replaced by one sphere in the so-called primary hydrodynamic model (PHM). This is the model whose properties are actually calculated. The radius of the sphere, a, has been estimated to be ∼3 Å. In this model, a number of such spheres are fully internal and not exposed to the solvent. Furthermore, the spheres overlap considerably with their neighbors. In principle, both circumstances prevent a conventional bead-model calculation (although we shall later consider this alternative), such as that used in the HYDRO++ program (4), which is intended for more open arrays of nonoverlapping or scarcely overlapping beads. Instead, HYDROPRO employs internally the shell-model strategy, in which the surface of the particle under consideration is represented by a shell consisting of Ns small, tangent, nonoverlapping minibeads of radius σ. Then, a bead-model calculation is done with the shell of minibeads for a series of decreasing values of σ, with a subsequent increase in Ns, and the results are extrapolated to . This shell-model strategy was first suggested by Bloomfield et al. (5, 6), used for detailed, atomic models of proteins for the first time by Teller et al. (7), and further developed and tested by our group (4, 8). It is similar to the so-called finite-element approach, which instead of minibeads uses platelets or other elements to describe the surface (9–12).
In HYDROPRO and our other shell-modeling programs, the number of beads Ns in the shell is varied between a minimum of ∼200–400 to a maximum (currently) of ∼2000. This maximum is limited by the computational cost, which is proportional to Ns3 (we emphasize that this is an internal, working parameter that does not limit the number of elements in the PHM). Nevertheless, we think that this is sufficient for describing the details that may be significant with regard to hydrodynamic coefficients (and we recall that the extrapolation to , i.e., to , further improves the accuracy of the final, extrapolated results. Thus, the shell model has been shown to predict very accurately the numerical values of arrays of spheres obtained from either rigorous fluid-dynamics calculation (4) or experiments with real, very well-defined multibead nanoparticles (13).
However, other authors have preferred to perform hydrodynamic calculations with bead models that somehow fill the interior of the protein. In such models (14–17), each Nr amino acid residue is represented by two or more beads (one at the skeleton and the others for the side chains). A sophisticated procedure is applied to avoid bead overlaps by changing some bead sizes, fusing overlapping beads, removing very internal beads, etc. (details can be found in the original references). With such delicate transformations performed before the hydrodynamic calculations, results of a quality comparable to that obtained with HYDROPRO calculations have been achieved in some instances. In such approaches, the number of beads finally used in the bead-model, HYDRO-like calculation is slightly larger but on the order of Nr, typically several hundreds for proteins of a few hundred residues. This is well below the largest number of minibeads that, regardless of the protein size, are employed by HYDROPRO (Ns ≈ 2000). Because the computing time is proportional to the third power of the number of beads (N3 = Nr3) in these methods, they are computationally faster than our previous version of HYDROPRO for a protein such as bovine pancreatic trypsin inhibitor (BPTI) with Nr = 58. However, what is an advantage for such small proteins (in comparison with the shell/bead HYDROPRO internal method) turns out to be a disadvantage for proteins with >1000 residues (>100–200 kDa), and the calculation with such methods becomes very time-consuming for large proteins (e.g., GroEL, which has Nr ≈ 7300 residues), whereas the computational cost of HYDROPRO is the same for GroEL and BPTI.
Materials and Methods
Apart from other computational improvements mentioned further below, the main purpose of this work is to report advances in modeling strategies that refine or further develop the applicability of HYDROPRO. We conducted our previous analysis of this methodology (1) on a set of proteins for which mainly translational data (sedimentation coefficient, s, and diffusion coefficient, Dt) and intrinsic viscosity [η] were available (1), and later complemented the data with rotational relaxation times, τc (2, 18). The optimization of the element radius in the atomic PHM, a, carried out in those successive stages yielded values in the range of 3.0–3.3 Å (slightly lower for Dt and s, and higher for τc). We recently developed a scheme for a systematic procedure to simultaneously analyze various properties of a set of many samples (19, 20). Instead of the measured properties Rg, Dt or s, [η], and τc, the procedure employs, in more coherent manner, equivalent radii, aG, aT, aI, and aR, which are the values of the radius of a sphere that would have that value of the property (19). Then, for a set of values of different properties of a series of samples, in this global-fit approach, we seek the minimization of a target function:
| (1) |
where the outermost summation is over the diverse samples (proteins) and the innermost runs over the properties, X, available for each sample. Note that is the root mean-square relative difference between the experimental and calculated equivalent radii, aX. Thus, 100Δ can be regarded as a typical percent error that characterizes the accurateness of the prediction procedure. It is possible to specifically evaluate the deviation for a specific property over a set of samples. Then Eq. 1 would be reduced to
| (2) |
and 100ΔX would be a typical percent error associated with property X. In our previous work (1, 2, 18), we compiled properties for ∼20 proteins. Even though this could be a sufficiently representative set, we now intend to extend the collection of experimental and structural data used in our new (to our knowledge) analysis methodology based on Eq. 1.
Also, although the modeling procedure and the computer program were initially intended for atomic-level protein structures, it is desirable to have a scheme for situations where resolution is limited to the residue level. This may be the case because of a lack of more-detailed, atomic-level information or just because in coarse-grained modeling a reduction in detail is adopted for simplicity or convenience. This was pioneered by our group for the case of nucleic acids (21). Some authors have tried to make hydrodynamic calculations based on residue-level (Cα-only) models, in which each residue is represented in the PHM by a single bead, usually placed at the Cα atom (22–28). In this work, we evaluate this alternative procedure for all of the cases in which the atomic-level description is also applied, so that we can compare the outcome of the two approaches.
As indicated above, most previous studies involving models with one or a few beads per residue employed a simple hydrodynamic treatment that treats each bead as a frictional element in our HYDRO program, in spite of the appreciable bead overlapping and its possible effects. In more-recent versions (e.g., HYDRO++ (4, 29)), we introduced physical and computational modifications that allow an adequate handling of bead overlapping, particularly regarding the so-called volume corrections in the calculation of [η] and τc. We considered that it was also worthwhile to include in our work the bead-model computational scheme to evaluate its performance. Finally, in previous works, most of the detailed (atomic- or residue-level) hydrodynamic modeling was applied to small proteins and nucleic acids (25, 30). Thus, another new aspect considered here is the extension of the applicability to large proteins and macromolecular complexes.
Results
Small and medium-sized proteins
As in our previous work, we used the new version of HYDROPRO to evaluate the hydrodynamic properties of a number of small and medium-sized proteins, with varying values of the hydrodynamic radius of the elements, a, in the PHM. We looked for an optimal value that would fit best different properties, this time using the global fitting method based on Eq. 1. In principle, we employed the set of data obtained in our previous studies on HYDROPRO (1) and HYDRONMR (2), and compiled in a previous review (18), which we term the GT set. This set includes values for the radii of gyration, translational coefficients, intrinsic viscosities, and rotational correlation times of 13, 19, 13, and 16 proteins, respectively. A list of the atomic structures and experimental values of the properties is given in Table S1 of the Supporting Material.
We first carried out the calculation with atomic-level PHMs employing the usual shell-model procedure of HYDROPRO, and compared the results with experimental data to obtain a best-fitting value a ≈ 2.9 Å, with Δ ≈ 0.04, meaning that with that choice of a, the typical error in the prediction of the hydrodynamic radii is ∼4%. The present value of a for the atomic PHM model is very slightly lower than that previously proposed (3.1 Å in the previous version of HYDROPRO), but this difference does not cause an essential difference in the quality of the fit, as judged by the Δ values (vide infra). Assuming a typical van der Waals radius of the nonhydrogen atoms of 1.8 Å, this amounts to an increase due to hydration of ∼1.1 Å, which coincides with the thickness increase employed by other authors (9, 12). It is also noteworthy that a ≈ 2.9 Å is also the value that fits the atomic PHMs of nucleic acids, as determined in a study of quasirigid oligonucleotides (30). Therefore, the methodology can be applied to protein-nucleic acid complexes with the same parameter for the atoms in both components.
We note the success of the atomic-level shell-model (HYDROPRO) calculation of the radius of gyration, Rg, which for such moderately sized macromolecules is mostly determined by small-angle x-ray scattering (SAXS). For some time, there was some doubt about how the hydration layer could contribute to SAXS intensities. A detailed study by Perkins (31) indicated that the electron density of water in the hydration layer is rather close to that of the protein itself, and, as in hydrodynamic modeling, the layer can be considered as an expansion of the protein. Thus, the same PHM can be used to evaluate Rg (and considered as an acceptable model for predicting the scattering intensities) as is considered in HYDROPRO.
Next, we took as the PHM the one constructed with one bead per residue, placed at the Cα atom, and performed the calculation again with the HYDROPRO/shell-model procedure. We then found the radius of the elements (residues) to be ∼5.0 Å. Of note, the typical errors were practically the same as with the atomic PHM. Furthermore, we ran the calculations of the residue-level PHM using the classical bead (not shell) model procedure implemented in HYDRO++, which, thanks to the Rotne-Prager interaction tensor, is known to account adequately for bead overlapping in the case of translational properties, switching off all the corrections for intrinsic viscosity and rotational diffusion that are not compatible with overlapping. We found that even with this supposedly less accurate hydrodynamic treatment, the results were quite similar to those of the other approaches. We obtained a ≈ 6.1 Å, and a just slightly higher overall error, mainly due to the worse prediction of the radius of gyration in this case, as the quality of fit for the other properties was similar.
As stated in the Introduction, subsequent to our earlier work (1, 2), other authors have approached the problem of predicting hydrodynamic properties of proteins from atomic- or residue-level models, compiling for that purpose other sets of experimental properties. A particularly large set of data for translational coefficients and intrinsic viscosities was collected by Hahn and Aragon (12) for 31 proteins. Brookes et al. (17) recently compiled the translational coefficients of 14 proteins (eight viscosity and six rotational values). We unified the various sets into a WHOLE superset containing 57 values of translational coefficients, 48 values of the intrinsic viscosity, and 28 values of the rotational correlation time. A list of the structures and experimental data for the WHOLE set is given in Table S1. We repeated the optimization procedure with the various PHMs and calculation modes, and found that the results of the WHOLE superset were nearly identical to those of the GT set. These results are displayed in Table 1.
Table 1.
Results of global analysis
| PHM, Calc. mode | Set | a (Å) | < % dif. > | % dif. aG | % dif. aT | % dif. aI | % dif. aR |
|---|---|---|---|---|---|---|---|
| Atomic, shell model | GT | 2.9 | 4.7 | 4.0 | 4.3 | 3.2 | 5.8 |
| Atomic, shell model | WHOLE | 2.9 | 4.5 | 3.9 | 4.7 | 3.8 | 5.3 |
| Residue/Cα, shell model | GT | 5.2 | 4.5 | 4.8 | 4.3 | 4.0 | 4.7 |
| Residue/Cα, shell model | WHOLE | 4.8 | 4.4 | 4.0 | 4.7 | 3.5 | 5.3 |
| Residue/Cα, bead model | GT | 6.1 | 5.1 | 6.7 | 4.4 | 3.6 | 5.2 |
| Residue/Cα, bead model | WHOLE | 6.1 | 4.8 | 6.7 | 5.0 | 3.9 | 5.0 |
Shown are the optimal radii in the PHM for the various calculation procedures, and typical errors for each property, for the GT and WHOLE experimental data sets.
Apart from their use as a target minimization function for optimization of the a parameter, the values of Δ deserve some comment. Their dependence on a is displayed in Fig. 1. It is noteworthy that the ΔX-values for each individual properties show a similar trend. For the different properties, the minimum is located at very near values of a, e.g., 2.6–3.3 Å for the atomic-level shell-model calculation, 4.5–5.5 Å for the residue-level shell-model calculation, and 5.5–6.5 Å for the residue-level bead-model calculation. Also, the typical errors for the various properties vary scarcely, from ∼4% for the translational (Stokes) radius to ∼5% for the rotational radius, and this trend is nearly the same for the three modeling and computational approaches.
Figure 1.
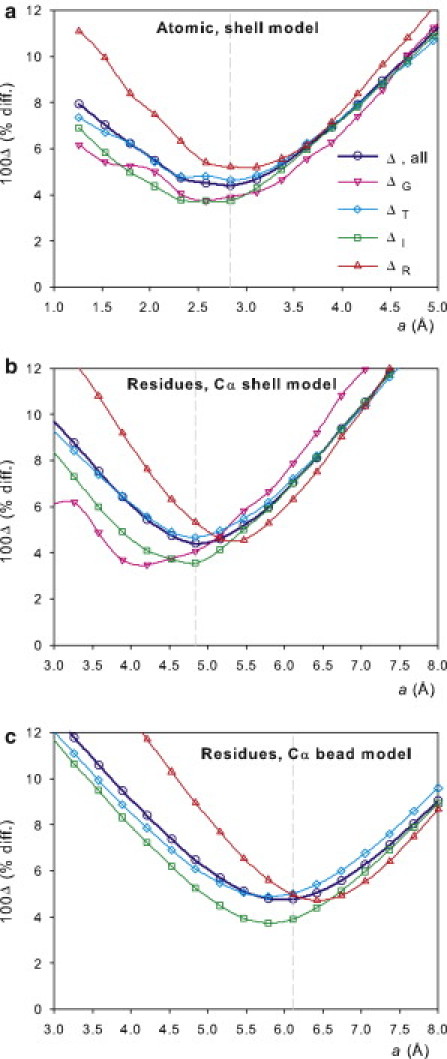
Values of 100ΔX and 100Δ as a function of the radius of elements in the PHM, a, calculated for small and medium-sized proteins, with the WHOLE set of experimental data. (a) Atomic-level PHM, HYDROPRO shell calculation. (b) Residue-level PHM, HYDROPRO shell calculation. (c) Residue-level PHM, HYDRO bead-model calculation.
Large proteins and macromolecular complexes
The application of detailed hydrodynamic modeling to proteins is usually done for moderately sized molecules (i.e., <200 kDa). However, there is no restriction about the number or elements in the PHM (either Na or Nr). We note that if one uses the shell-model approach, regardless of the number of elements, the number of beads to be used in the hydrodynamic calculation is that of the shell models, Ns, which is up to 2000 in the present implementation. Alternatively, if one uses the bead-model calculation, N = Nr, again, and there is no limit to the number of beads allowed by the program (the present versions of HYDRO employ dynamic memory allocation), the limitation may be dictated by the long computing time that will be required for macromolecules with a large number of residues.
We performed calculations for the following protein structures:
GroEL
The largest component of the GroEL-GroES proteic complex is the chaperone GroEL, with a molecular mass of 800 kDa. The structure is hydrodynamically peculiar because it has noticeable holes between the subunits.
Urease
This hexameric protein, of 480 kDa, is composed by two identical and noncovalently bound trimers. Experimental data are available for the whole hexameric protein and the trimer half-unit.
IgM antibody
IgM is a large (905 kDa) and peculiar antibody molecule constituted by an arrangement of five subunits. Unlike other protein structures, IgM is remarkably open and extended.
Ribosomal subunits
Of the various substructures that comprise the assembly of the whole ribosome, we selected the 30S and 50S subunits and the whole 70S ribosome from Escherichia coli. These are very large protein-nucleic acid complexes, having molecular masses of 1000, 1700, and 2500 kDa. Experimental data are available for the sedimentation coefficient (we recall that the classical notation xxS for proteins, particularly employed to name the ribosomal subunits, is based on the value of the sedimentation coefficient). Furthermore, this example provides an illustration on how to handle protein-nucleic acid complexes with our hydrodynamic modeling. In the case of the atomic-level model, the fact that the element radius in the atomic PHM found for proteins and oligonucleotides is the same (∼2.9 Å) avoids any distinction. If the PHM is a residue-level, Cα-only model, then the value assigned to the nucleotide residues must be different from the value adopted for amino acids. From early studies of the hydrodynamics of small, quasirigid oligonucleotides (21, 32), we know that nucleotide unit can be represented by a bead with radius 3.5 Å, centered at the P atom. Calculations from the atomic coordinates of the ribosome and its subunits were carried out at the level of atoms and residues.
Fig. S1 and Table S2 provide full experimental data regarding the structures and properties, along with literature references for these large proteins and macromolecular complexes. The results from the calculations, from each of the three computational schemes, are presented in Table 2. In spite of the complexity and large size of these macromolecules, the agreement with experiments is quite satisfactory. The only remarkable discrepancy is the intrinsic viscosity of the ribosomal subunits (see Supporting Material). Other results are in acceptable and in some cases very good accordance with experimental data. Apart from the ability of the three procedures to predict experimental properties, a remarkable finding is that the quality of the results from each of the three procedures is very similar. This confirms the trend noted in the first part of this work for small proteins.
Table 2.
Summary of calculated values, compared with experimental data, for large proteins and macromolecular complexes
| Protein, Nr | Experimental∗ | Atomic shell model N < 2000 | Residue-Cα shell model N < 2000 | Residue-Cα bead model N = Nr |
|---|---|---|---|---|
| GroEL, 7273 | s = 22.13 | 21.5 | 22.3 | 21.5 |
| Dt = 2.59 | 2.58 | 2.68 | 2.58 | |
| Rg = 67 | 66.1 | 64.4 | 65.0 | |
| Urease, 4996 | Dt = 3.5 | 3.3 | 3.3 | 3.3 |
| s = 18.3 | 18.0 | 18.1 | 17.4 | |
| Half urease, 2504 | s = 11.5 | 11.3 | 11.3 | 10.6 |
| Ribosome 30S, 3883 | Rg = 69 | 67.5 | 67.4 | 65.9 |
| s = 31.8 | 34.0 | 33.4 | 38.4 | |
| Dt = 2.18 | 2.55 | 2.50 | 2.62 | |
| [η] = 8.1 | 4.2 | 4.4 | 3.8 | |
| Ribosome 50S, 6336 | Rg = 77 | 71.7 | 74.3 | 77.6 |
| s = 50.2 | 52.0 | 50.4 | 50.8 | |
| Dt = 1.90 | 2.22 | 2.15 | 2.21 | |
| [η] = 5.6 | 3.6 | 3.9 | 3.6 | |
| Ribosome 70S, 10219 | Rg= 91.5 | 87.0 | 89.3 | 91.0 |
| s = 70.5 | 67.6 | 67.2 | 74.9 | |
| Dt = 1.72 | 1.82 | 1.81 | 1.66 | |
| [η] = 5.8 | 4.1 | 4.1 | 4.6 | |
| IgM, 7514 | Rg = 121 | † | 127 | 126 |
| Dt = 1.82 | † | 1.73 | 1.70 | |
| s = 17.5 | † | 18.4 | 18.4 | |
| [η] = 13.4 | † | 13.7 | 13.8 |
s expressed in Svedberg units (S); Dt in units of 10−7 cm2/s; Rg in Å, [η] in cm3/g.
Atomic structure not available.
In all cases, the typical deviation of the calculated values from experimental data is 4%, which is close to what could be considered a typical error. Plots of calculated versus experimental data illustrate the quality of the agreement. Such plots are displayed in Fig. 2 for the shell-model calculation of residue-level structures. It is clear that the quality of the agreement is similar for the four properties, for both small and large proteins. Fig. S4 and Fig. S5 display similar plots for shell-model calculations with atomic-level structures and bead-model calculations with residue-level structures.
Figure 2.
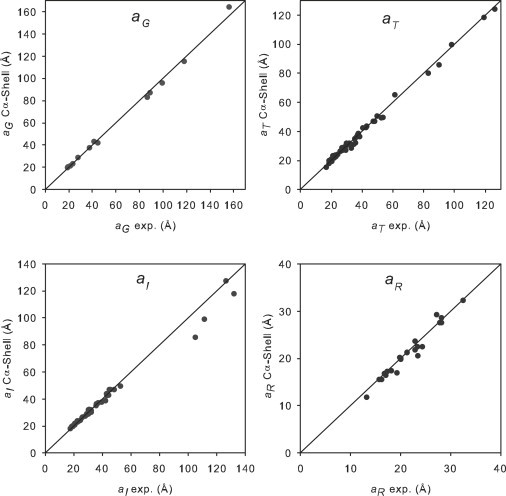
Representation of the values calculated by HYDROPRO shell-model from the Cα coordinates (y axis) versus experimental values (x axis) of the four equivalent radii employed in this work, for the WHOLE set of proteins as well as the large proteins and macromolecular complexes.
Conclusions
We have shown that the shell-model calculation implemented in HYDROPRO enables one to predict the solution properties of rigid biomacromolecules, with practically the same accuracy from residue-level as for atomic-level models. This indicates that a residue-level description suffices for evaluating overall properties such as the hydrodynamic coefficient and tensors, and the radius of gyration. Furthermore, we have shown that this methodology is equally able to predict the properties of very large proteins and macromolecular complexes.
We also explored the possibility of making predictions from a bead-model calculation directly from a residue-level PHM (in spite of bead overlapping) with a number of beads proportional to the number of residues, Nr. With adequate choices for the hydrodynamic interactions and a properly parameterized element radius, the quality of the results is close to that achieved with the shell-model calculation. Whereas for the former the computing time is proportional to Nr3, for the latter it is proportional to Ns3, where Ns is always up to ∼2000 independently of protein size. Thus, the bead-model calculation is a suitable choice (when computing time matters and a slight sacrifice in accuracy is acceptable) in the case of proteins with <2000 residues (i.e., <200 kDa). For larger proteins, the shell-model calculation will be far more efficient. Thus, the shell-model calculation, from either the atomic- or residue-level model, seems to be the generally advisable choice.
Computer programs
A new version of HYDROPRO is available that includes the three working methods considered in this work: 1), shell-model calculation from an atomic-level PHM (this was the only mode in the previous version); 2), shell-model calculation from a residue-level PHM; and 3), bead-model calculation from a residue-level PHM. Also, important improvements have been introduced in the numerical calculations that greatly improve its computational efficiency as a result of careful optimization and parallelization (for currently available, conventional multicore computers), and consequently the new version is much faster than the previous one. Furthermore, we have also constructed a graphical user interface that may assist some casual users in handling the program. Information on HYDROPRO CPU benchmarking and the graphical user interface is provided in the Supporting Material. This new version and related computer programs can be freely downloaded from our web site (http://leonardo.inf.um.es/macromol/).
Acknowledgments
This work was supported by the Grupo de Excelencia de la Región de Murcia (grant 04531/GERM/06) and the Ministerio de Educación y Ciencia (grant CTQ-2009-06831), including FEDER funds. A.O. received a postdoctoral fellowship from Universidad de Murcia, and D.A. received a predoctoral fellowship from the Ministerio de Educación y Ciencia. Computing time was provided by the Fundación Parque Científico de Murcia.
Supporting Material
References
- 1.García De La Torre J., Huertas M.L., Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys. J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.García de la Torre J., Huertas M.L., Carrasco B. HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J. Magn. Reson. 2000;147:138–146. doi: 10.1006/jmre.2000.2170. [DOI] [PubMed] [Google Scholar]
- 3.Ortega A., García de la Torre J. Efficient, accurate calculation of rotational diffusion and NMR relaxation of globular proteins from atomic-level structures and approximate hydrodynamic calculations. J. Am. Chem. Soc. 2005;127:12764–12765. doi: 10.1021/ja053080l. [DOI] [PubMed] [Google Scholar]
- 4.García de la Torre J., del Río Echenique G., Ortega A. Improved calculation of rotational diffusion and intrinsic viscosity of bead models for macromolecules and nanoparticles. J. Phys. Chem. B. 2007;111:955–961. doi: 10.1021/jp0647941. [DOI] [PubMed] [Google Scholar]
- 5.Bloomfield V.A., Dalton W.O., Van Holde K.E. Frictional coefficients of multisubunit structures. I. Theory. Biopolymers. 1967;5:135–148. doi: 10.1002/bip.1967.360050202. [DOI] [PubMed] [Google Scholar]
- 6.Bloomfield V.A., Filson D.P. Shell model calculations of translational and rotational frictional coefficients. J. Polym. Sci. C Polym. Symp. 1968;25:73–83. [Google Scholar]
- 7.Teller D.C., Swanson E., de Haën C. The translational friction coefficient of proteins. Methods Enzymol. 1979;61:103–124. doi: 10.1016/0076-6879(79)61010-8. [DOI] [PubMed] [Google Scholar]
- 8.Carrasco B., García de la Torre J. Hydrodynamic properties of rigid particles: comparison of different modeling and computational procedures. Biophys. J. 1999;76:3044–3057. doi: 10.1016/S0006-3495(99)77457-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou X.Z. Calculation of translational friction and intrinsic viscosity. II. Application to globular proteins. Biophys. J. 1995;69:2298–2303. doi: 10.1016/S0006-3495(95)80100-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allison S.A., Tran V.T. Modeling the electrophoresis of rigid polyions: application to lysozyme. Biophys. J. 1995;68:2261–2270. doi: 10.1016/S0006-3495(95)80408-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brune D., Kim S. Predicting protein diffusion coefficients. Proc. Natl. Acad. Sci. USA. 1993;90:3835–3839. doi: 10.1073/pnas.90.9.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hahn D., Aragon S. Intrinsic viscosity of proteins and platonic solids by boundary element methods. J. Chem. Theory Comput. 2006;2:1416–1428. doi: 10.1021/ct600062y. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann M., Wagner C.S., Wittemann A. 3D Brownian diffusion of submicron-sized particle clusters. ACS Nano. 2009;3:3326–3334. doi: 10.1021/nn900902b. [DOI] [PubMed] [Google Scholar]
- 14.Byron O. Hydrodynamic modeling: the solution conformation of macromolecules and their complexes. Methods Cell Biol. 2008;84:327–373. doi: 10.1016/S0091-679X(07)84012-X. [DOI] [PubMed] [Google Scholar]
- 15.Byron O. Hydrodynamic bead modeling of biological macromolecules. Methods Enzymol. 2000;321:278–304. doi: 10.1016/s0076-6879(00)21199-3. [DOI] [PubMed] [Google Scholar]
- 16.Rai N., Nollman M., Rocco M. SOMO (Solution Modeller) differences between X-ray and NMR-derived bead models suggest a role for side chain flexibility in protein dynamics. Structure. 2005;13:722–734. doi: 10.1016/j.str.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 17.Brookes E., Demeler B., Rocco M. Developments in the US-SOMO bead modeling suite: new features in the direct residue-to-bead method, improved grid routines, and influence of accessible surface area screening. Macromol. Biosci. 2010;10:746–753. doi: 10.1002/mabi.200900474. [DOI] [PubMed] [Google Scholar]
- 18.García de la Torre J. Hydration from hydrodynamics. general considerations and aplications of bead modelling to globular proteins. Biophys. Chem. 2001;93:150–159. doi: 10.1016/s0301-4622(01)00218-6. [DOI] [PubMed] [Google Scholar]
- 19.Ortega A., García de la Torre J. Equivalent radii and ratios of radii from solution properties as indicators of macromolecular conformation, shape, and flexibility. Biomacromolecules. 2007;8:2464–2475. doi: 10.1021/bm700473f. [DOI] [PubMed] [Google Scholar]
- 20.Ortega A., Amorós D., García de la Torre J. Global fit and structure optimization of flexible and rigid macromolecules and nanoparticles from analytical ultracentrifugation and other dilute solution properties. Methods. 2011;54:115–123. doi: 10.1016/j.ymeth.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 21.García de la Torre J., Navarro S., López Martinez M.C. Hydrodynamic properties of a double-helical model for DNA. Biophys. J. 1994;66:1573–1579. doi: 10.1016/S0006-3495(94)80949-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antonsiewicz J., Porschke D. Volume correction for bead model simulations of rotational frictions. J. Phys. Chem. 1989;93:5301–5305. [Google Scholar]
- 23.Antosiewicz J., Porschke D. Electrostatics of hemoglobins from measurements of the electric dichroism and computer simulations. Biophys. J. 1995;68:655–664. doi: 10.1016/S0006-3495(95)80226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hellweg T., Eimer W., Müller A. Hydrodynamic properties of nitrogenase—the MoFe protein from Azotobacter vinelandii studied by dynamic light scattering and hydrodynamic modelling. Biochim. Biophys. Acta. 1997;1337:311–318. doi: 10.1016/s0167-4838(96)00179-3. [DOI] [PubMed] [Google Scholar]
- 25.Banachowicz E., Gapiński J., Patkowski A. Solution structure of biopolymers: a new method of constructing a bead model. Biophys. J. 2000;78:70–78. doi: 10.1016/S0006-3495(00)76573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osborne M.J., Wright P.E. Anisotropic rotational diffusion in model-free analysis for a ternary DHFR complex. J. Biomol. NMR. 2001;19:209–230. doi: 10.1023/a:1011283809984. [DOI] [PubMed] [Google Scholar]
- 27.Frembgen-Kesner T., Elcock A.H. Striking effects of hydrodynamic interactions on the simuulated diffusion and folding of proteins. J. Chem. Theory Comput. 2009;5:242–256. doi: 10.1021/ct800499p. [DOI] [PubMed] [Google Scholar]
- 28.Frembgen-Kesner T., Elcock A.H. Absolute protein-protein association rate constants from flexible, coarse-grained Brownian dynamics simulations: the role of intermolecular hydrodynamic interactions in barnase-barstar association. Biophys. J. 2010;99:L75–L77. doi: 10.1016/j.bpj.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.García de la Torre J., Amorós D., Ortega A. Intrinsic viscosity of bead models for macromolecules and nanoparticles. Eur. Biophys. J. 2010;39:381–388. doi: 10.1007/s00249-009-0405-5. [DOI] [PubMed] [Google Scholar]
- 30.Fernandes M.X., Ortega A., García de la Torre J. Calculation of hydrodynamic properties of small nucleic acids from their atomic structure. Nucleic Acids Res. 2002;30:1782–1788. doi: 10.1093/nar/30.8.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perkins S.J. X-ray and neutron scattering analyses of hydration shells: a molecular interpretation based on sequence predictions and modelling fits. Biophys. Chem. 2001;93:129–139. doi: 10.1016/s0301-4622(01)00216-2. [DOI] [PubMed] [Google Scholar]
- 32.Huertas M.L., Navarro S., García de la Torre J. Simulation of the conformation and dynamics of a double-helical model for DNA. Biophys. J. 1997;73:3142–3153. doi: 10.1016/S0006-3495(97)78340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


