Abstract
The physical attributes of the extracellular matrix play a key role in endothelium function by modulating the morphology and phenotype of endothelial cells. Despite the recognized importance of matrix-cell interactions, it is currently not known how the arrangement of adhesive ligands affects the morphology, signal transduction processes, and migration of endothelial cells. We aimed to study how endothelial cells respond to the average spatial arrangement of integrin ligands. We designed functionalized silicon surfaces with average spacing ranging from nanometers to micrometers of the peptide arginine-glycine-aspartic acid (RGD). We found that endothelial cells adhered to and spread on surfaces independently of RGD-to-RGD spacing. In contrast, organization within focal adhesions (FAs) was extremely sensitive to ligand spacing, requiring a nanoscaled average RGD spacing of 44 nm to form lipid raft domains at FAs. The localized membrane organization strongly correlated with the signaling efficiencies of integrin activation and regulated vascular endothelial growth factor (VEGF)-induced signaling events. Importantly, this modulation in signal transduction directly affected the migratory ability of endothelial cells. We conclude that endothelial cells sense nanoscaled variations in the spacing of integrin ligands, which in turn influences signal transduction processes. Average RGD spacing similar to that found in fibronectin leads to lipid raft accumulation at FAs, enhances sensitivity to VEGF stimulation, and controls migration in endothelial cells.
Introduction
Cells sense and respond to the physical attributes of their local environment, a concept embodied by the terms mechanosensing and mechanotransduction. Advances in surface chemistry and nanotechnology have provided unique insights into the ability of cells to adjust their shape and motility to minute changes in the chemical and physical features of their immediate surroundings (1). A remarkable discovery is that cells can sense nanometer-scale variations in the average spacing of randomly organized integrin ligands (2,3). Fibroblasts adhere, migrate, and proliferate on surfaces with average spacings of the tripeptide arginine-glycine-aspartic acid (RGD) of <70 nm, whereas they adhere poorly and migrate erratically when integrin ligands are spaced farther apart (2). Importantly, the 10–200 nm scale of average ligand spacing is physiologically relevant because the nanoscaled and periodic spacing is similar to that found in fibronectin and collagen fibers (4–6).
The concept of mechanotransduction appears to be particularly relevant for endothelial cells. Interactions between endothelial cells and the extracellular matrix (ECM) control many vascular processes (7), including permeability (8), sensitivity to growth factors (e.g., responsiveness to vascular endothelial growth factor (VEGF) stimulation (9)), and transformation into a proliferative and invasive phenotype that is characteristic of angiogenesis (10). However, although the importance of cell-matrix interactions for the functioning of the endothelium is recognized, little is known about how fundamental physical features of the matrix, such as the average spacing of integrin ligands, affects the behavior of endothelial cells.
Endothelial adhesion to the ECM is facilitated by integrins (11). Engaged integrins cluster together with cytoskeletal and signaling proteins to form focal adhesions (FAs) and complexes (11). These complexes control a range of cell activation responses, including cell polarization and migration, membrane trafficking, cell cycle progression, gene expression, and oncogenic transformation (7,12–14). Signaling at FAs also includes VEGF-induced intracellular calcium fluxes, activation of phosphatydylinositol-3 (PI3) kinase and mitogen-activated protein (MAP) kinases, and, further downstream, activation of endothelial nitric oxide synthase (eNOS) (15). Curiously, although integrins have no intrinsic enzymatic activity (14), in many cases they enable growth factor signals, that is, growth factor signaling does not occur unless integrins are occupied (9,10). Hence, VEGF and integrin form a functional partnership in endothelial cells; however, how integrin spacing and FA organization influence VEGF signaling is currently not known.
The exposure of endothelial cells to RGD peptides, which are found in fibronectin and recognized by the integrins αvβ3 and α5β1, sensitizes endothelial cells to angiogenic transformation (10,16). In this study, we sought to determine how endothelial cells respond to the average spacing of randomly distributed RGD ligands by assessing FAs, integrin activation, and VEGF-induced signaling and migration. To that end, we functionalized silicon surfaces with average RGD spacing from nano- to micrometers. We found that nanoscaled variations in integrin ligand spacing govern membrane order within FAs, which in turn determines signaling efficiency and cell migration. Taken together, our results suggest that the spatial arrangement of the local cellular environment may significantly contribute to the proangiogenic behavior of endothelial cells.
Materials and Methods
Preparation of silicon surfaces with monolayer modification
Silicon wafers (p-type (100), 0.07–0.09 Ω.cm resistivity) were cleaned in piranha solution (concentrated sulphuric acid/30% hydrogen peroxide = 3:1, v/v) and then etched in 2.5% hydrofluoric (HF) acid for 90 s to remove the native oxide layer. The freshly etched samples were then immersed in undecenoic acid that had previously been deoxygenated. The surfaces were left to react at 120°C for 12 h. The wafers were rinsed with ethanol, ethyl acetate, and dichloromethane, and blown dry under argon. (Caution: Piranha solution reacts violently with organic materials. HF acid is extremely corrosive, and dilute HF solutions can cause delayed serious tissue damage. Both should be handled with extreme care.)
Coupling of the 1-amino hexa(ethylene oxide) moieties and peptide immobilization
The surfaces were immersed for 1 h in a 0.1 M/0.05 M aqueous solution of 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS), and rinsed in ultrapure water and ethanol. The samples were then incubated 12 h in 20 mM solutions in dimethylformamide containing various ratios of 1-amino hexa(ethylene oxide) to 1-amino hexa(ethylene oxide) monomethyl ether followed by rinsing with dichloromethane and ethyl acetate, and dried under a stream of argon. The hydroxyl terminated hexa(ethylene oxide) molecules were activated in a 0.1 M/0.1 M solution of dry dimethylformamide of N,N′-disuccinimidyl carbonate (DSC) and 4-dimethyl aminopyridine (DMAP) for 12 h. After rinsing with dichloromethane and ethyl acetate, and drying, the samples were immersed in 1 mM aqueous Gly-Arg-Gly-Asp-Ser peptide solution for 12 h. Finally, the samples were rinsed with ultrapure water and ethanol, and dried.
Cell culture
Bovine aortic endothelial cells (BAECs) were cultured in endothelial basal medium supplemented with 0.1% human epidermal growth factor, 0.1% gentamycin, 0.4% bovine brain extract, and 5% fetal bovine serum (FBS) at 37°C in 5% CO2. The BAECs were transfected with enhanced green fluorescent protein tagged paxillin (eGFP-paxillin) using the Lipofectamine LTX reagent (Invitrogen, Carlsbad, CA) and selected in medium containing 1 mg/ml of G418 antibiotics. Where indicated, adherent cells were incubated with 10 mM methyl-β-cyclodextrin (mβCD; Sigma-Alrich, St. Louis, MO) in serum-free media for 25 min at 37°C.
Fluorescence microscopy
BAECs were serum-starved for 18 h and replated onto silicon surfaces for 30 min or 3 h in endothelial basal medium with 10% FBS. Adherent cells were stained with the membrane dye FM 1-43FX or fixed with 4% paraformaldehyde. Fixed cells were stained with phalloidin-Alexa555, anti-vinculin antibodies (Sigma-Aldrich), anti-paxillin antibodies (BD Biosciences, Franklin Lakes, NJ), anti-phospho-caveolin-1 antibodies (BD Biosciences), and appropriate fluorophore-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Philadelphia, PA) (17,18). Cells were imaged on an epifluorescence microscope (Nikon Eclipse TE 2000-S, Yokohama, Japan).
Membrane order
Membrane order was determined as described previously (17,19). Briefly, adherent endothelial cells were labeled with 10 μM Laurdan (6-dodecanoyl-2-dimethylaminonaphthalene, Invitrogen) for 30 min at 37°C, fixed, and immunostained as outlined above (17). Images were obtained with a 2-photon microscope (TCS SP5; Leica Microsystems, Mannheim, Germany). Laurdan was excited at 800 nm with a Ti:Sapphire laser (Mai-Tai; Spectra Physics, Santa Clara, CA) and emissions were recorded simultaneously in the range of 400–460 nm and 470–530 nm. The two intensity images were converted into generalized polarization (GP) images (ImageJ) according to
The final GP images were pseudo-colored in Adobe Photoshop. Confocal images were taken at the same focal depth as the Laurdan intensity images. To determine GP values at FA, the confocal images defined the regions of interest within the GP image, and the mean GP value of these regions of interest was determined for each image. Neither fixation nor permeabilization affects the measurement of GP values (17).
Signaling assays
Serum-starved cells were replated onto functionalized silicon surfaces for 3 h, mβCD-treated where indicated, stimulated with VEGF (25 ng/ml), and lysed in radio-immunoprecipitation assay buffer. Equal proteins were loaded and separated in 10% SDS-PAGE gels, transferred onto nitrocellulose (Invitrogen), and probed for anti-phospho-Akt (Ser473), anti-Akt, anti-phospho-Src (Tyr416), anti-Src, anti-phospho-ERK (Thr202/Tyr204), and anti-ERK, anti-phospho-PLCγ1 (Tyr783) (all from Cell Signaling, Beverly, MA); anti-PLCδ, anti-phospho-FAK (pY397), anti-FAK, and anti-Paxillin (all from BD Biosciences); anti-phospho-eNOS (Ser1177) and anti-eNOS (both from Upstate Biotechnology/Millipore, Billerica, MA); anti-vinculin (Sigma); and appropriate horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories). Band intensities were quantified by densitometry with ImageJ.
Integrin activation assay
Fusion protein GST-FN9-11 containing glutathione S-transferase (GST) and fibronectin type III repeats 9, 10, and 11 were isolated from bacteria (17). Serum-starved cells were replated onto silicon surfaces for 3 h in 5% FBS. Cells were incubated with 20 μg/ml GST-FN9-11 in the absence or presence of 1 mM Mn2+ and 1 mM Ca2+ for 30 min (17,20). The cells were washed and then lysed in radio-immunoprecipitation assay buffer. The amount of bound fibronectin 9-11 GST was then determined by immunoblotting with anti-GST antibody (Santa Cruz, Santa Cruz, CA).
Migration
BAECs were replated on the silicon surfaces in a confluent monolayer. A wound was created by scraping cells off the surface with a plastic pipette tip (diameter: 300 μm). After 0–5 h, the cells were fixed, stained, and imaged as outlined above. The width of the wound was measured with ImageJ.
Statistics
Statistical analysis was performed by analysis of variance with Tukey's multiple-comparison tests.
Results
We developed a strategy for controlling the average spatial arrangement of integrin ligands on silicon surfaces using self-assembled monolayers. Traditionally, investigators have prepared surfaces by either adsorbing proteins onto substrates or covalently attaching peptides to polymers. These approaches fail to give an unambiguous presentation of adhesive ligands, which is vital for controlling FA composition (2). Prime and Whitesides (21) achieved an unequivocal presentation of ligands by attaching ligands to gold surfaces modified with oligo(ethylene oxide)-terminated alkanethiol self-assembled monolayers. Subsequently, Singhvi et al. (22) showed that these surfaces are effective mimics of the ECM in cell adhesion studies by preventing nonspecific protein absorption (21), hence ensuring specific interaction with the immobilized ligand. Other studies have demonstrated the impact of the average density (23–25), affinity (26), and spatial organization (24,27,28) of RGD ligands on cell adhesion and spreading has been. However, because gold can quench fluorescence, the use of gold surfaces has limited studies of adherent cells by fluorescence microscopy (29).
We exploited the precise and stable monolayer chemistry on silicon to synthesize surfaces with various densities of randomly distributed RGD peptides (30). To attach the pentapeptide GRGDS (referred to as RGD) to the silicon surfaces (see Fig. S1 in the Supporting Material), we first modified silicon, without an oxide layer, with a base undecenoic acid layer to which 1-amino-hexa(ethylene oxide) molecules (green in Fig. 1 A) were attached. We achieved various RGD densities by altering the ratio of two different 1-amino-hexa(ethylene oxide) molecules: one hydroxyl terminated to which the RGD could be attached (blue in Fig. 1 A) and one terminated with a methoxy group (referred to as EO6) to which peptide coupling cannot occur (31). The purpose of the ethylene oxide components was to provide a layer that resists the nonspecific adherence of cells and proteins, such that cell adhesion is controlled by the RGD peptides. Hence, the surface layer was designed in such a way that only the adhesive peptides protruded from an otherwise cell-resistant layer. We controlled the average spacing of RGD peptides by altering the ratio of the two hexa(ethylene oxide) components in solution during attachment to the surface.
Figure 1.
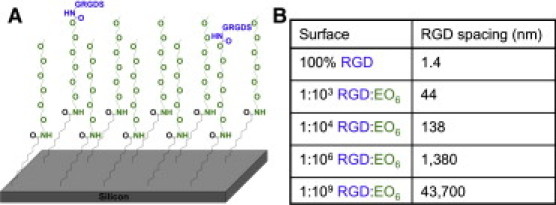
Schematic representation of functionalized silicon surfaces. (A) Silicon surfaces were modified with undecenoic acid and various ratios of 1-amino hexa(ethylene oxide) monomethyl ether (EO6) to 1-amino hexa(ethylene oxide). The alcohol terminus of the hexa(ethylene oxide) was activated to a succinimide ester using DSC and DMAP such that the pentapeptide RGD (blue) could be coupled to the surface. (B) The table summarizes the average RGD-to-RGD spacings for the various ratios of surface-coupled RGD/EO6-terminated molecules.
We used high-resolution x-ray photoelectron spectroscopy (Fig. S2) to determine the chemical composition of the surface throughout the multistep fabrication (32,33,34,35). We estimated the coupling yields from the area ratios of peaks in the carbon 1s spectrum assigned to the different chemical moieties on the derivatized Si surface, taking the attenuation of photoelectrons into account (34). We then determined the average density of RGD peptides by multiplying the overall coupling yield for RGD with the known density of alkyl chains in Si-C linked monolayers (36). Using this approach, we estimated that the 100% RGD surface had an average density of ∼6 × 1011 RGD per mm2. To obtain lower average RGD densities, we diluted the hydroxyl-terminated hexa(ethylene glycol) species with a methoxy-terminated species. Because the chemical structures of the two molecules are almost identical, it is assumed that the rates of the coupling reaction are identical. Hence, the ratio of these two molecules in solution is expected to be faithfully represented on the surface. This assumption is supported by the observation that the coupling yields of the two hexa(ethylene glycol) species to the base monolayer were determined to be the same by x-ray photoelectron spectroscopy (Fig. S2). Importantly, there is no control over the distribution of these two molecules, and hence any attached RGD ligands will be randomly distributed across the surface. Clustering of the hydroxyl terminated species is not expected because of the almost identical chemistry of the two molecules. The calculated average RGD densities and the corresponding RGD spacings for the surfaces used in this study are shown in Fig. 1 B.
We adhered serum-starved endothelial cells onto the surfaces and imaged the shape and morphology of the cells by staining for membranes, F-actin (Fig. 2 A), and the FA marker vinculin (Fig. 2, B and C). The images in Fig. 2 show that endothelial cells form FAs with a distinct dash-like morphology at the cell edge. We found FAs on surfaces with 100% RGD, and 1:103 and 1:106 RGD/EO6. When the average RGD peptide spacing was greater (1:109 RGD/EO6), FAs/complexes were still visible, whereas no FAs were found on the antifouling surfaces (100% EO6). Similarly, observations were made with endothelial cells expressing the adhesion protein paxillin-GFP (Fig. S3).
Figure 2.
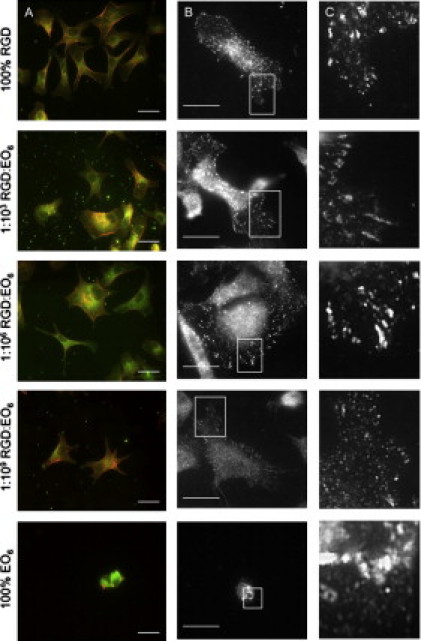
Endothelial cells on functionalized silicon surfaces. Serum-starved endothelial cells were replated onto silicon surfaces with various ratios of RGD/EO6 (as indicated) for 3 h in serum-containing media, fixed, and stained with (A) the membrane dye FM1-43X (green) and phalloidin-Alexa555 (red) or (B and C) anti-vinculin antibodies. Fluorescence images were taken with epifluorescence (A) and 2-photon (B and C) microscopes. Panel C shows zoomed sections of FAs at the cell edges. Scale bars = 20 μm.
Importantly, we observed that on the antifouling EO6-modified silicon surfaces, a small number of endothelial cells adhered (Fig. 3 A) but did not spread (Fig. 3 B) when incubated for 3 h. Cell viability is limited on EO6-modified silicon surfaces, which is in agreement with the lack of FAs in these cells. This observation provides evidence that EO6 modification is a sufficiently inert background for our study.
Figure 3.
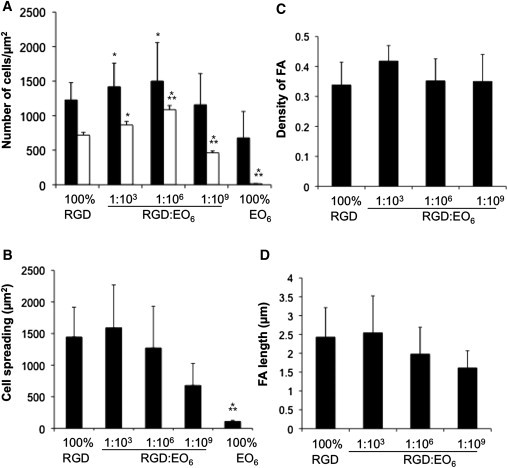
Serum-starved endothelial cells were re-plated onto functionalized silicon surfaces for 30 min (white bars) or 3 h (black bars), fixed, stained and imaged as described for Fig. 2. The fluorescence images were analyzed to determine (A) the average number of adherent cells per area (μm2) after serum-starved cells were adhered for 30 min (white bars) and 3 h (black bars) in serum-containing media to the indicated functionalized silicon surfaces, (B) cell spreading, (C) the number of vinculin-containing FAs per area (μm2), and (D) length of vinculin-containing FAs after cells adhered for 3 h on functionalized surfaces. Error bars represent standard deviations (SDs) of (A) >100 cells and (B–D) >15 cells. ∗p < 0.05 between the indicated surface and 100% RGD; ∗∗p < 0.05 relative to 1:103 RGD/EO6 surfaces.
On the RGD-modified surfaces, we observed a biphasic trend, with most cells adhering onto 1:106 RGD/EO6 surfaces (Fig. 3 A) and cell spreading occurring over a wide range of average RGD spacings (Fig. 3 B). Higher densities of RGD peptides did not enhance cell adhesion or cell spreading. As expected, an average RGD spacing of >10 μm limited cell adhesion and spreading. We examined FA characteristics using vinculin as a marker (Fig. 3, C and D) but found no differences in FA density (Fig. 3 C) or length (Fig. 3 D) on RGD-containing surfaces. In summary, endothelial cells on functionalized silicon surfaces adhere, spread, and form FAs with a wide range of average RGD-to-RGD spacings. This allows us to examine how RGD spacing influences FA-dependent parameters such as membrane order and growth factor signaling.
We previously reported that FAs reside in highly ordered membrane domains that biophysically resemble lipid rafts (17). Gaus et al. (19) examined membrane order at FAs by using the spectral properties of the fluorescent membrane dye Laurdan. The peak emission of Laurdan shifts from ∼500 nm in fluid membranes to ∼440 nm in ordered membranes (37). A normalized ratio of the two emission regions, termed the GP, is therefore an index of membrane fluidity that is independent of membrane ruffles and dye concentration. GP values range from −1 (most fluid) to +1 (most ordered), with the cell membrane of endothelial cells having a mean GP value of 0.2 ± 0.15 (colored green on the color scale in Fig. 4) (17). To image and quantify membrane order at FAs, we first recorded a confocal image of the FA protein paxillin, followed by the Laurdan GP image at the identical focal plane (Fig. 4). The GP images indicate that FAs in cells on 1:103 RGD/EO6 surfaces (colored yellow/red) are more ordered than FAs in cells on 100% RGD and 1:106 RGD/EO6 surfaces (colored predominantly green). Indeed, when membrane order is quantified over many cells (Fig. 4 E), we observe a biphasic behavior similar to that described above for cell adhesion: FAs in cells on 1:103 RGD/EO6 surfaces are significantly more ordered than FAs in cells on 1:104–1:109 RGD/EO6 or 100% RGD surfaces. Hence, membrane order at FAs is highly sensitive to nanoscale variations in average RGD spacing. Even an increase in average RGD-to-RGD spacing from an average of 44 nm (1:103 RGD/EO6) to 138 nm (1:104 RGD/EO6) resulted in a significant decrease in membrane order at FAs from GP = 0.482 ± 0.059 to 0.385 ± 0.058 (p < 0.05), whereas FAs in cells on 1:109 RGD/EO6 surfaces were as fluid as the plasma membrane in endothelial cells. Global membrane order was similar in all cells adherent to RGD-modified surfaces, suggesting that average RGD spacing specifically influences membrane order at the site of FA only. When endothelial cells were plated onto a fibronectin matrix, the degree of membrane order did not significantly differ from that found in cells on 1:103 RGD/EO6 surfaces (Fig. S4). Taken together, these data suggest that average RGD spacing is a critical determinant of membrane order at FAs, with an average RGD spacing of 44 nm (the same as that found in fibronectin) achieving the highest FA membrane order.
Figure 4.
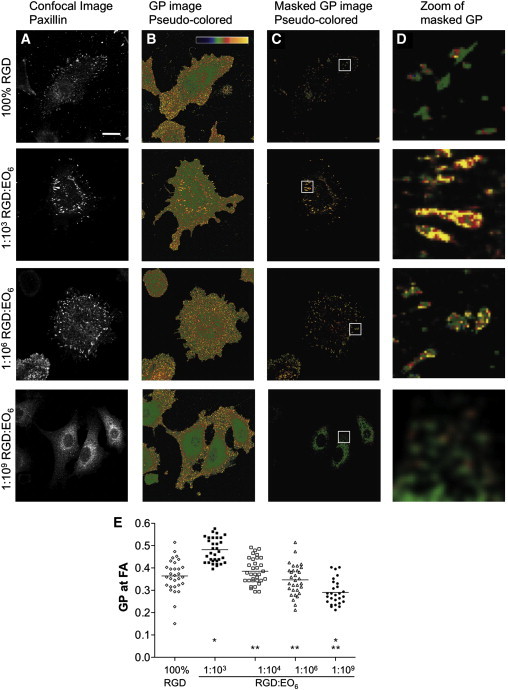
Membrane order at FAs. Endothelial cells were plated on functionalized surfaces, labeled with the fluorescent membrane dye Laurdan, fixed, and immunostained with anti-paxillin antibodies. Laurdan's spectral properties were exploited to determine membrane order defined as the fluidity index GP. GP values range from −1 (fluid, blue in B–D) to +1 (ordered, yellow). (A) Confocal images of paxillin staining. (B) Pseudo-colored GP images as indicated by the color scale. (C) Masked GP images showing only GP values (same coloring as in B) for the pixels that were positive for paxillin (A) was above background. (D) Magnified regions of the masked GP images in C. Bars = 20 μm. (E) GP values of paxillin-positive pixels. Each symbol represents the mean GP value of one cell; horizontal bars indicate the means of GP values; ∗p < 0.05 between a given surface and 100% RGD; ∗∗p < 0.05 relative to 1:103 RGD/EO6 surfaces.
Given the differences in FA organization, we examined integrin α5β1 activation (Fig. 5) and VEGF-induced signaling (Fig. 6). To probe for integrin configuration as a measure of integrin activation, we made use of the fibronectin fragment GST-FNIII9-11,which binds only to the open or high-affinity conformation of α5β1 integrins. When we probed for the extent of GST-FNIII9-11 binding to cells adhering to the silicon surfaces (Fig. 5 A), we found that cells on 1:103 RGD/EO6 surfaces had ∼2.4 times more activated α5β1 than cells adhering to 100% RGD (p < 0.05), whereas cells on 1:106 RGD/EO6 surfaces displayed a similar degree of integrin activation compared with 100% RGD (p > 0.05). This difference cannot be attributed to the number of α5β1 integrins expressed. This is shown by incubating cells with Mn2+, which causes all integrins to be converted to the open configuration so that GST-FNIII9-11 binding provides an estimate of the total α5β1 integrin expression (20). Under this condition, no detectable difference in α5β1 integrin expression was found. By normalizing activated integrins to the equivalence of total α5β1 integrins, we confirmed that integrin activation was indeed significantly higher in cells on 1:103 RGD/EO6 surfaces compared with cells on any of the other surfaces (Fig. 5 B). To summarize, we found that the optimum average RGD spacing for integrin α5β1 activation was 44 nm, although the cells expressed similar levels of total α5β1 integrins.
Figure 5.
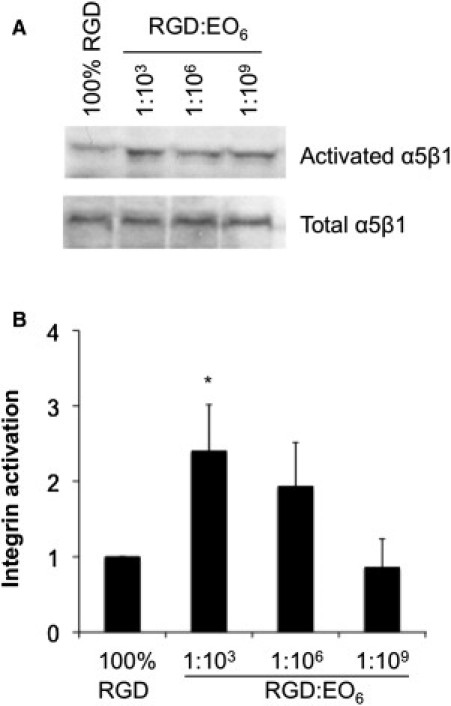
Integrin activation. Integrin activation was determined with GST-FN9-11, which specifically binds to activated α5β1. (A) Endothelial cells that adhered to functionalized silicon surfaces for 3 h were incubated with GST-FN9-11 for 30 min in the absence or presence of 1 mM Mn2+, which activates integrins (total α5β1). Cell lysates were probed for GST. (B) Quantification of integrin activation was expressed as the ratio of activated/total α5β1, and the ratio was normalized to cells on 100% RGD surfaces. Data and error bars represent the mean ± SD of four independent experiments; ∗p < 0.05 between 100% RGD and 1:103 RGD/EO6 surfaces, with none of the other surfaces being significantly different from the 100% RGD surface.
Figure 6.
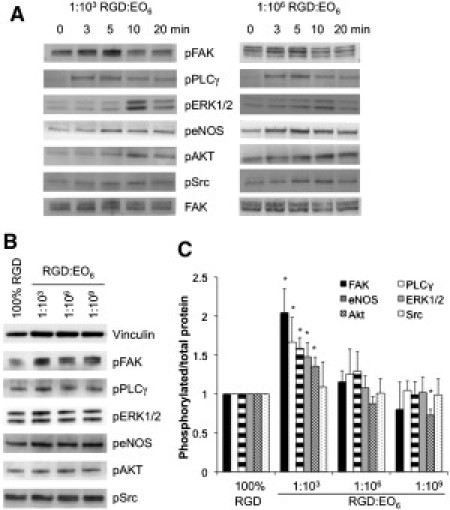
VEGF-induced signaling. (A) Endothelial cells that adhered to 1:103 RGD/EO6 and 1:106 RGD/EO6 surfaces for 3 h were stimulated with 25 ng/ml VEGF for 0–20 min, lysed, and analyzed by immunoblotting for the phosphorylation of FAK, PLCγ, ERK1/2, eNOS, Akt, and Src on the same blot. Probing for nonphosphorylated proteins (total), as shown for FAK, acted as a loading control. (B and C) Adherent cells on various surfaces were stimulated with VEGF stimulation for 10 min. Immunoblots were quantified by expressing the ratio of phosphorylated/total protein and normalizing the ratio to cell lysates from 100% RGD surfaces of the same blot. Data and error bars represent the mean ± SD of four independent experiments; ∗p < 0.05 relative to 100% RGD surfaces.
Next, we examined VEGF stimulation of endothelial cells on these surfaces. We probed for the phosphorylation of adhesion proteins (FAK at Y397), receptor-activated kinases (Src at Y416), signaling proteins that regulate intracellular calcium fluxes (PLCγ at Y783), and proteins that are involved in the PI3 kinase (Akt at S473) and MAP kinase (ERK1/2 at T202/Y204) pathway, as well as the activation of eNOS (at S1177). A time course showed that all of the selected proteins reached their peak of phosphorylation within the first 5–10 min of stimulation, with only minor differences between cells plated on 1:103 RGD/EO6 and 1:106 RGD/EO6 surfaces (Fig. 6 A). To assess the signaling activities in quantitative terms, we compared the extent of phosphorylated signaling proteins after 10 min of VEGF stimulation across different functionalized surfaces (Fig. 6 B) and normalized each signaling protein to its nonphosphorylated form (total) of the same blot. The ratio of phosphorylated/total signaling protein was normalized to the ratio found in cells on 100% RGD-modified surfaces (Fig. 6 C). We conclude that phosphorylation of FAK, PLCγ, eNOS, ERK1/2, and AKT was significantly higher in cells on 1:103 RGD/EO6 surfaces than on 100% RGD (p < 0.05), whereas Src phosphorylation was independent of average RGD spacing. The latter is consistent with the notion that Src activation is triggered by VEGF receptor ligation, which was constant in all of the samples. The higher signaling efficiency in cells on 1:103 RGD/EO6 surfaces correlates extremely well with FA membrane order (Fig. 4 E) and integrin activation (Fig. 5 B).
We next assessed whether the enhanced signaling efficiency on 1:103 RGD/EO6 surfaces was caused by the high membrane order of FAs in these cells. After cells adhered to functional surfaces, we depleted cellular cholesterol using methyl-β-cyclodextrin (mβCD). Cholesterol depletion significantly decreased FA membrane order in cells on 1:103 RGD/EO6 and 1:106 RGD/EO6 surfaces, such that the FA organization was now similar in both conditions (p < 0.05 in Fig. 7 A). Similarly, VEGF-induced signaling activity also decreased in cholesterol-depleted cells (Fig. 7 B). The same quantification described above demonstrated that FA membrane order regulated VEGF-induced signal activity, because cholesterol depletion caused a greater decrease in phosphorylation levels in cells on 1:103 RGD/EO6 surfaces, where membrane order was higher in nontreated control cells (Fig. 4 E) compared with cells on 1:106 RGD/EO6 surfaces. The similar FA membrane order in cholesterol-depleted cells on 1:103 RGD/EO6 and 1:106 RGD/EO6 surfaces also resulted in a similar VEGF signaling activity. Hence, we conclude that average RGD-RGD spacing regulates FA membrane order, which in turn controls VEGF signaling efficiency.
Figure 7.
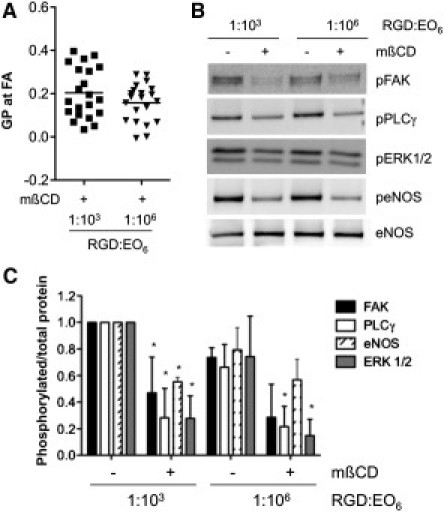
Membrane order regulates VEGF signaling. (A) Endothelial cells that adhered to 1:103 RGD/EO6 and 1:106 RGD/EO6 surfaces for 3 h were cholesterol-depleted with 10 mM mβCD for 25 min. Cholesterol depletion significantly decreased membrane order at FAs in cells plated on either surface, such that the mean GP values of FAs of cholesterol-depleted cells (indicated by the horizontal bar) are not significantly different. (B) Immunoblots for the phosphorylation of FAK, PLCγ, ERK1/2, and eNOS for control and cholesterol-depleted cells treated with 25 ng/ml VEGF for 10 min. Probing for nonphosphorylated proteins, as shown for eNOS, was used as loading control. (C) Quantification of VEGF-induced signaling activity expressed as the ratio of phosphorylated/total protein, and normalizing the ratio to cell lysates from 1:103 RGD/EO6 surfaces. Data and error bars represent the mean ± SD of three independent experiments; ∗p < 0.05 relative to non-mβCD-treated cells on equivalent surfaces. Signaling activity in cholesterol-depleted cells on 1:103 RGD/EO6 versus 1:106 RGD/EO6 is not significantly different.
Given the differences in signaling efficiencies in cells on surfaces with different average RGD spacings, we sought to determine whether these differences would influence the ability of endothelial cells to migrate, as assessed in a wound-healing assay. A monolayer of endothelial cells was plated onto the functionalized silicon surfaces and scraped to induce a wound with a width of 310 ± 25 μm that removed cells but left the surface chemistry intact (Fig. 8, A and B). The cells were incubated for a further 3 h and the width of the wound was determined (Fig. 8 C). In the absence of VEGF (Fig. 8 A and white bars in Fig. 8 C), cells at the cell front extended membrane protrusions and migrated into the cell-free gap on all of the RGD-modified surfaces. Under these conditions, cells migrated fastest on 1:106 RGD/EO6 surfaces. In the presence of VEGF (Fig. 8 B and black bars in Fig. 8 C), endothelial cells again displayed the classical migration phenotype, with elongated cell shape and extension of membrane protrusions on RGD-modified surfaces. Under VEGF stimulation, cells migrated fastest on 1:103 RGD/EO6 surfaces. In fact, VEGF-enhanced cell migration on these surfaces was 1.6-fold faster than without VEGF stimulation, whereas VEGF-stimulated cell migration on 1:106 RGD/EO6 surfaces was reduced compared with migration in the absence of VEGF. The shift of fastest cell migration from 1:106 to 1:103 RGD/EO6 surfaces when endothelial cells were stimulated with VEGF correlates well with the enhanced signaling efficiency observed in cells on these surfaces. Irrespectively of VEGF, cells on 100% EO6 surfaces did not form membrane protrusions and barely migrated into the wound. Together, these results show that average RGD spacing influences membrane order at FAs, integrin activation, VEGF-induced signaling efficiency, and migration speed, and suggest that the spatial arrangement of integrin ligand may be an important determinant of endothelial cell phenotype.
Figure 8.
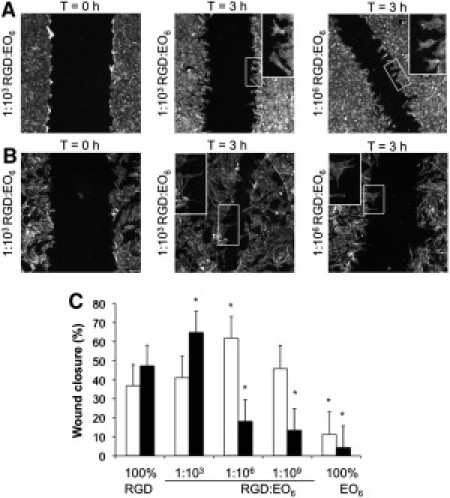
Migration of endothelial cells. A monolayer of endothelial cells cultured on silicon surfaces was scratched to introduce a wound with an average width of 310 ± 25 μm. After a further 3 h in the absence (A) or presence (B) of 25 ng/ml VEGF, individual cells started to migrate into the wound. Zoomed images show the elongated shape of migrating cells and the extension of filopodia (C). The distance between the cell fronts was measured for >50 different sites. The data show the mean ± SD of wound closure in the absence (white bars) and presence (black bars) of VEGF; ∗p < 0.05 relative to 100% RGD surfaces.
Discussion
Recent advances in nanopatterning and surface chemistry, including microcontact printing (38), self-assembled monolayer chemistries (39), and block copolymer nanolithography (2,3), have provided unique insights into how cells sense and respond to their local environment (1). Cell adhesion onto two-dimensional surfaces revealed that fibroblasts and osteoblasts adhere and spread differently if the density (2), patterning (3), or degree of ordering (28) of the integrin ligands is altered on the nanometer to micrometer scale (40). Despite the recognition that matrix interactions determine the function of endothelial cells (8–10), it remains to be determined how endothelial cells respond to the spatial arrangement of ligand presentation.
By plating endothelial cells on silicon surfaces with nanometer to micrometer spacings of randomly distributed RGD peptides, we found that the average RGD spacing on the nanometer scale regulates the membrane organization within FA, which in turn determines the signaling efficiency and migratory ability of endothelial cells. An average RGD spacing of ∼44 nm appears to mimic fibronectin, resulting in highly ordered FA, and this shifts the balance of signaling pathways toward FAK, eNOS, Akt, and ERK activation in the MAP kinase pathway. The latter are critical proliferation signals that have been widely implicated in angiogenesis. Therefore, the average spacing of integrin ligands and the concomitant nanoscaled organization of FA significantly influence the phenotype of endothelial cells. This is physiologically important because it demonstrates that there is an optimal spatial arrangement of ECM motifs on the nanometer scale at which the synergy between integrin and growth factor signaling can drive angiogenic transformation.
Our work has three important implications for endothelial cell biology: First, although endothelial cells adhere to and spread on surfaces with a wide range of average RGD-to-RGD spacings, the organization within FA is highly sensitive to nanoscaled variations in the average RGD spacing. Confirming previous observations in other cell types, we found that ∼1000 RGD ligands per cell or average RGD spacings well in excess of 44 nm are optimal for endothelial cell spreading (23,24) and migration, possible due to FA instability (2). In contrast, membrane order within FAs was highly sensitive to average RGD spacing and was highest on surfaces with an average RGD spacing of 44 nm, matching FA order in cells on fibronectin-coated surfaces. Because membrane order is the biophysical characteristic of lipid rafts, our experiments infer that lipid raft accumulation at FA is dependent on RGD spacing. Taken together, our data strongly suggest that endothelial cells sense nanoscaled variations in their local environment and restructure their FAs accordingly.
Second, membrane order at FAs correlated extremely well with integrin activation and VEGF-induced signaling. Our data suggest a model in which sensing, engagement, and activation of integrin confer a specific membrane order at that site (17), which subsequently aids the recruitment and retention of proteins and lipids that have a greater affinity for that membrane milieu (41). This hypothesis is supported by single-cell force measurements that showed that the distance to the nearest integrin-binding sites determines binding strength (42). The notion that lipid raft accumulation at FA reflects the mechanical properties of endothelial cells (e.g., responds to shear stress) would be an interesting area for further research.
Although the function of lipid rafts in signal transduction is still debated (43,44), our data show that lipid raft accumulation at FAs critically determines VEGF-induced signaling efficiencies. This study proposes that the coordination of integrin and growth factor signaling occurs at the level of membrane domains. This could be physiologically relevant because even a minor shift in the balance of different signaling pathways, mediated by the degree of lipid raft accumulation, may alter the sensitivity of endothelial cells to growth factors and shift the signaling hierarchies when cells are stimulated with multiple stimuli, thereby influencing the overall response of endothelial cells.
Third, we find that the efficiency in signal transduction facilitated by the FA organization directly affects the migratory ability of endothelial cells. Our data therefore strongly suggest that the average spacing of integrin ligands significantly influences the phenotype of endothelial cells. This is of physiological importance because it implies that a specific spatial arrangement of integrin ligands induces a synergy between integrin and growth factor signaling that may critically determine the behavior of endothelial cells. Thus, it may be possible to control endothelial cell phenotypes by engineering an appropriate cellular environment in medical implants (40), biosensors (45), and tissue engineering materials.
Acknowledgments
This work was supported by a grant from the National Health and Medical Research Council of Australia (K.G. and J.J.G.), the Australian Research Council (K.G. and J.J.G.), and the Human Frontier Science Program Organization (T.B.).
Contributor Information
J. Justin Gooding, Email: justin.gooding@unsw.edu.au.
Katharina Gaus, Email: k.gaus@unsw.edu.au.
Supporting Material
References
- 1.Geiger B., Spatz J.P., Bershadsky A.D. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 2.Cavalcanti-Adam E.A., Volberg T., Spatz J.P. Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands. Biophys. J. 2007;92:2964–2974. doi: 10.1529/biophysj.106.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold M., Hirschfeld-Warneken V.C., Spatz J.P. Induction of cell polarization and migration by a gradient of nanoscale variations in adhesive ligand spacing. Nano Lett. 2008;8:2063–2069. doi: 10.1021/nl801483w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carreiras F., Thiébot B., Darbeida H. Involvement of αvβ 3 integrin and disruption of endothelial fibronectin network during the adhesion of the human ovarian adenocarcinoma cell line IGROV1 on the human umbilical vein cell extracellular matrix. Int. J. Cancer. 2002;99:800–808. doi: 10.1002/ijc.10433. [DOI] [PubMed] [Google Scholar]
- 5.Vogel V. Mechanotransduction involving multimodular proteins: converting force into biochemical signals. Annu. Rev. Biophys. Biomol. Struct. 2006;35:459–488. doi: 10.1146/annurev.biophys.35.040405.102013. [DOI] [PubMed] [Google Scholar]
- 6.Davidson W.S., Gillotte K.L., Phillips M.C. The effect of high density lipoprotein phospholipid acyl chain composition on the efflux of cellular free cholesterol. J. Biol. Chem. 1995;270:5882–5890. doi: 10.1074/jbc.270.11.5882. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz M.A., Ginsberg M.H. Networks and crosstalk: integrin signalling spreads. Nat. Cell Biol. 2002;4:E65–E68. doi: 10.1038/ncb0402-e65. [DOI] [PubMed] [Google Scholar]
- 8.Mehta D., Malik A.B. Signaling mechanisms regulating endothelial permeability. Physiol. Rev. 2006;86:279–367. doi: 10.1152/physrev.00012.2005. [DOI] [PubMed] [Google Scholar]
- 9.Eliceiri B.P. Integrin and growth factor receptor crosstalk. Circ. Res. 2001;89:1104–1110. doi: 10.1161/hh2401.101084. [DOI] [PubMed] [Google Scholar]
- 10.Stupack D.G., Cheresh D.A. Integrins and angiogenesis. Curr. Top. Dev. Biol. 2004;64:207–238. doi: 10.1016/S0070-2153(04)64009-9. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz M.A. Integrins, oncogenes, and anchorage independence. J. Cell Biol. 1997;139:575–578. doi: 10.1083/jcb.139.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahai E., Marshall C.J. RHO-GTPases and cancer. Nat. Rev. Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz M.A., Assoian R.K. Integrins and cell proliferation: regulation of cyclin-dependent kinases via cytoplasmic signaling pathways. J. Cell Sci. 2001;114:2553–2560. doi: 10.1242/jcs.114.14.2553. [DOI] [PubMed] [Google Scholar]
- 14.Hynes R.O. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 15.Zachary I. VEGF signalling: integration and multi-tasking in endothelial cell biology. Biochem. Soc. Trans. 2003;31:1171–1177. doi: 10.1042/bst0311171. [DOI] [PubMed] [Google Scholar]
- 16.Brooks P.C., Clark R.A., Cheresh D.A. Requirement of vascular integrin α v β 3 for angiogenesis. Science. 1994;264:569–571. doi: 10.1126/science.7512751. [DOI] [PubMed] [Google Scholar]
- 17.Gaus K., Le Lay S., Schwartz M.A. Integrin-mediated adhesion regulates membrane order. J. Cell Biol. 2006;174:725–734. doi: 10.1083/jcb.200603034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rentero C., Zech T., Gaus K. Functional implications of plasma membrane condensation for T cell activation. PLoS ONE. 2008;3:e2262. doi: 10.1371/journal.pone.0002262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaus K., Gratton E., Jessup W. Visualizing lipid structure and raft domains in living cells with two-photon microscopy. Proc. Natl. Acad. Sci. USA. 2003;100:15554–15559. doi: 10.1073/pnas.2534386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orr A.W., Ginsberg M.H., Schwartz M.A. Matrix-specific suppression of integrin activation in shear stress signaling. Mol. Biol. Cell. 2006;17:4686–4697. doi: 10.1091/mbc.E06-04-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prime K.L., Whitesides G.M. Self-assembled organic monolayers: model systems for studying adsorption of proteins at surfaces. Science. 1991;252:1164–1167. doi: 10.1126/science.252.5009.1164. [DOI] [PubMed] [Google Scholar]
- 22.Singhvi R., Kumar A., Ingber D.E. Engineering cell shape and function. Science. 1994;264:696–698. doi: 10.1126/science.8171320. [DOI] [PubMed] [Google Scholar]
- 23.Massia S.P., Hubbell J.A. An RGD spacing of 440 nm is sufficient for integrin α v β 3-mediated fibroblast spreading and 140 nm for focal contact and stress fiber formation. J. Cell Biol. 1991;114:1089–1100. doi: 10.1083/jcb.114.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maheshwari G., Brown G., Griffith L.G. Cell adhesion and motility depend on nanoscale RGD clustering. J. Cell Sci. 2000;113:1677–1686. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 25.Koo L.Y., Irvine D.J., Griffith L.G. Co-regulation of cell adhesion by nanoscale RGD organization and mechanical stimulus. J. Cell Sci. 2002;115:1423–1433. doi: 10.1242/jcs.115.7.1423. [DOI] [PubMed] [Google Scholar]
- 26.Kato M., Mrksich M. Using model substrates to study the dependence of focal adhesion formation on the affinity of integrin-ligand complexes. Biochemistry. 2004;43:2699–2707. doi: 10.1021/bi0352670. [DOI] [PubMed] [Google Scholar]
- 27.Houseman B.T., Mrksich M. The microenvironment of immobilized Arg-Gly-Asp peptides is an important determinant of cell adhesion. Biomaterials. 2001;22:943–955. doi: 10.1016/s0142-9612(00)00259-3. [DOI] [PubMed] [Google Scholar]
- 28.Huang J., Grater S.V., Spatz J.P. Impact of order and disorder in RGD nanopatterns on cell adhesion. Nano Lett. 2009;9:1111–1116. doi: 10.1021/nl803548b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kandere-Grzybowska K., Campbell C., Borisy G.G. Molecular dynamics imaging in micropatterned living cells. Nat. Methods. 2005;2:739–741. doi: 10.1038/nmeth796. [DOI] [PubMed] [Google Scholar]
- 30.Kilian K.A., Böcking T., Gooding J.J. Si-C linked oligo(ethylene glycol) layers in silicon-based photonic crystals: optimization for implantable optical materials. Biomaterials. 2007;28:3055–3062. doi: 10.1016/j.biomaterials.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 31.Böcking T., Kilian K.A., Gooding J.J. Formation of tetra(ethylene oxide) terminated Si-C linked monolayers and their derivatization with glycine: an example of a generic strategy for the immobilization of biomolecules on silicon. Langmuir. 2005;21:10522–10529. doi: 10.1021/la051191s. [DOI] [PubMed] [Google Scholar]
- 32.Kilian K.A., Böcking T., Gooding J.J. Peptide-modified optical filters for detecting protease activity. ACS Nano. 2007;1:355–361. doi: 10.1021/nn700141n. [DOI] [PubMed] [Google Scholar]
- 33.Böcking T., James M., Barrow K.D. Structural characterization of organic multilayers on silicon(111) formed by immobilization of molecular films on functionalized Si-C linked monolayers. Langmuir. 2004;20:9227–9235. doi: 10.1021/la048474p. [DOI] [PubMed] [Google Scholar]
- 34.Böcking T., Wong E.L.S., Coster H.G.L. Immobilization of dendrimers on Si-C linked carboxylic acid-terminated monolayers on silicon(111) Thin Solid Films. 2006;515:1857–1863. [Google Scholar]
- 35.Kilian K.A., Böcking T., Gooding J.J. Introducing distinctly different chemical functionalities onto the internal and external surfaces of mesoporous materials. Angew. Chem. Int. Ed. Engl. 2008;47:2697–2699. doi: 10.1002/anie.200704784. [DOI] [PubMed] [Google Scholar]
- 36.Sieval A.B., Linke R., Sudhölter E.J.R. High-quality alkyl monolayers on silicon surfaces. Adv. Mater. 2000;12:1457–1460. [Google Scholar]
- 37.Gaus K., Zech T., Harder T. Visualizing membrane microdomains by Laurdan 2-photon microscopy. Mol. Membr. Biol. 2006;23:41–48. doi: 10.1080/09687860500466857. [DOI] [PubMed] [Google Scholar]
- 38.Chen C.S., Mrksich M., Ingber D.E. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 39.Mrksich M., Chen C.S., Whitesides G.M. Controlling cell attachment on contoured surfaces with self-assembled monolayers of alkanethiolates on gold. Proc. Natl. Acad. Sci. USA. 1996;93:10775–10778. doi: 10.1073/pnas.93.20.10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevens M.M., George J.H. Exploring and engineering the cell surface interface. Science. 2005;310:1135–1138. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- 41.Brown D.A. Lipid rafts, detergent-resistant membranes, and raft targeting signals. Physiology (Bethesda) 2006;21:430–439. doi: 10.1152/physiol.00032.2006. [DOI] [PubMed] [Google Scholar]
- 42.Selhuber-Unkel C., López-García M., Spatz J.P. Cooperativity in adhesion cluster formation during initial cell adhesion. Biophys. J. 2008;95:5424–5431. doi: 10.1529/biophysj.108.139584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simons K., Toomre D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 44.Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 45.Kilian K.A., Lai L.M., Gooding J.J. Smart tissue culture: in situ monitoring of the activity of protease enzymes secreted from live cells using nanostructured photonic crystals. Nano Lett. 2009;9:2021–2025. doi: 10.1021/nl900283j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


