Abstract
No methods proposed thus far have the sensitivity to measure the transport of single molecules through single nuclear pore complexes (NPCs) in intact cells. Here we demonstrate that fluorescence correlation spectroscopy (FCS) combined with real-time tracking of the center of mass of single NPCs in live, unperturbed cells allows us to detect the transport of single molecules in a reference system of a pore with high temporal (millisecond) and spatial (limited by diffraction) resolution. We find that the transport of the classical receptor karyopherin-β1 (Kapβ1) is regulated so as to produce a peculiar distribution of characteristic times at the NPC. This regulation, which is spatially restricted to the pore, depends on the properties and metabolic energy of Kapβ1. As such, this method provides a powerful tool for studying nucleocytoplasmic shuttling at the nanometer scale under physiological conditions.
Measurements of molecular transport through nuclear pore complexes (NPCs) provide valuable information that can be used to unravel the mechanisms of communication between the nucleus and the cytoplasm (1,2). Unfortunately, because single molecules undergo very rapid transport, it is challenging to follow their motion in a physiological state in live cells. Because of the highly dynamic nature of this process, investigators have developed a variety of camera-based single particle tracking (SPT) approaches to study it (3–7). Such methods have been used to analyze transit times, trajectories, and interactions between transport receptors and model cargo molecules at the NPC. However, SPT measurements rely on the tracking of bright and isolated particles, and measurements must be repeated many times to provide enough statistics. Furthermore, most SPT experiments require complex experimental procedures because the molecule of interest must be purified, labeled, and introduced into the cytoplasm by microinjection or cell permeabilization. Consequently, it is unclear whether the correct pore function is preserved under classical SPT experimental conditions. By contrast, fluorescence correlation spectroscopy (FCS) provides single-molecule sensitivity in the presence of many molecules in unperturbed cells (8,9). Along with other investigators (10,11), we recently used FCS to study nucleocytoplasmic transport (12). In that study, we measured the transit times of GFP-tagged molecules over several microns across the nuclear envelope (NE), but without single-pore resolution.
Here we overcome those limitations by measuring nucleocytoplasmic transport at the single-molecule and single-pore level in intact cells. Because the entire NPC can perform local nanometer diffusion within the NE or follow global rearrangements of the cell (13), it is crucial to subtract that motion if we want to distinguish between the diffusion of the molecules and the overall thermal motion of the NPC. We need to track the average position of the pore so that only the motion of the molecules relative to the entire NPC is measured. This method compensates for temperature-driven local diffusion of the large unit represented by the NPC. In this reference frame, the NPC would be stationary and mainly within the same orientation. We propose a real-time, feedback-based fluorescence tracking method that we previously developed to track the motion of a particle in a chromosome (14). The method is based on rapid orbiting of a laser beam around the object to be tracked. This method is capable of tracking an object with a response time of ∼8–16 ms and a location precision in the nanometer range (14). Thus, we are able to jump onto the reference system of the pore and follow the translocation of molecules with great precision and a time resolution that is faster than the motion of the overall NPC. Because we are only measuring the changes in fluorescence intensity along the orbit with very high time resolution, we can determine the correlations in space and time of single molecules binding and leaving the NPC. For example, if molecules are leaving one location of the NPC and reappearing in another area of the same NPC, the intensity along the orbit will change in a time- and space-correlated way. Because we are measuring single-molecule events, we can measure the exact location of the event and the time course of the process.
To test our method, we use the classical karyopherin-β1 (Kapβ1) nuclear receptor tagged to GFP and transiently transfected into CHO-K1 cells. Kapβ1-GFP serves as a dynamic marker of the pore as it shuttles across the NE and locally accumulates on single NPCs (Fig. 1 A), where it performs bidirectional transport. According to the accepted molecular model (2), Kapβ1 recognizes the cargo molecule on the cytoplasmic side of the NPC and translocates to the nucleoplasmic basket, where it releases the cargo and binds Ran guanosine triphosphate (RanGTP). The Kapβ1-RanGTP complex is readily exported back to the cytoplasm. This bidirectional process is sustained by the chemical gradient of RanGTP across the NE. Because it is ∼500 times more concentrated in the nucleus, RanGTP acts as a molecular ratchet imparting directionality to cargo-binding and cargo-release reactions across the NE.
Figure 1.
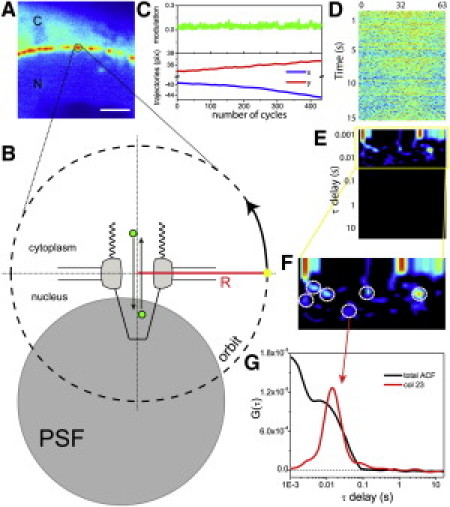
(A) Cell expressing Kapβ1-GFP (scale bar: 2 μm). (B) The PSF is scanned along a 64-point orbit of 180 nm in radius (R) around the pore (the dimensions of the pore in the drawing are in scale with the orbit radius). The first half of the orbit (points 0–32) passes over the cytoplasmic side of the pore, and the second half (points 32–63) passes over the nucleoplasmic side. (C) Modulation and xy trajectories over time (one cycle = 16 orbits). (D) Fluorescence intensity along the orbit versus time. (E and F) Total ACF carpet and zoom on the region containing humps (dashed circles). (G) Average ACF plot (black) and ACF of column 23 (red).
A schematic representation of the experiment is reported in Fig. 1 B. A circular light envelope is formed around the pore by the scanning point spread function (PSF), whereas the tracking routine keeps the center of mass of the pore always at the center of the orbit (Fig. 1 C; see Supporting Material).
Given the size of the NPC (15), the orbit radius is set to 180 nm to allow proper imaging of both the cytoplasmic and nucleoplasmic sides of the NPC. Along the orbit, 64 points are sampled. The orientation of the NE is selected in such a way that the scan start point (yellow dot in Fig. 1 B) is located on the NE. Thus, the first half of the orbit (points 0–32) corresponds to one compartment (e.g., the nucleus) and the second half (points 32–63) corresponds to the other compartment. The fluorescence intensity is recorded along the orbit and then constructed in a two-dimensional carpet (Fig. 1 D), where the x axis represents the total number of points along the orbit (points 0–63) and the y axis represents the time of acquisition. Typically, 6.4 × 105 orbits are scanned, corresponding to ∼32 s of total acquisition (orbit time = 0.5 ms). FCS analysis is performed to extract information about the Kapβ1 dynamics at the single-molecule level. First we calculate the autocorrelation function (ACF) at each point along the orbit to quantify the corresponding local diffusivity of Kapβ1-GFP. The ACF is constructed in a two-dimensional carpet, where the x axis represents the number of pixels (0–63) and the y axis represents the correlation time in log scale (Fig. 1 E). Careful inspection of the ACF carpet reveals the dynamics of Kapβ1-GFP at the pore to be characterized by both a fast-decaying component (<3 ms, typical of diffusion) and a substantial hump component (dashed circles in Fig. 1 F) at later times. The hump identifies a characteristic correlation time of the fluorescence fluctuations detected at the pore (see also the average and local ACFs in Fig. 1 G). This effect can be readily explained in terms of bidirectional Kapβ1-GFP movement through the pore if it is assumed that the same molecule is detected at one position at time t and again at the same location with a certain delay τ, after a complete transport cycle. Consequently, the humps can be at different locations (and times) because molecules can reasonably start and stop at different points in the pore structure. For 25 analyzed cells, the average peak position is 9.6 ± 3.5 ms (mean ± standard deviation (SD)), with a full width at half-maximum of 13 ± 6.5 ms (mean ± SD). This average time of transport is approximately two times higher than the typical translocation times measured by SPT (3–7), in keeping with the idea that we are measuring a complete bidirectional cycle. The two components of Kapβ1 shuttling can be separated by spatial pair correlation between points 16 and 48 (and the reverse) of the orbit. This analysis gives very similar results for the two directions (3.3 ± 1.8 ms from cytoplasm to nucleus, 3.2 ± 1.5 ms from nucleus to cytoplasm; mean ± SD). As expected, these transport times are close to half of the bidirectional one measured by ACF. If our interpretation of the ACF profile is correct, the hump effect should prove to be 1), dependent on the presence of Kapβ1 (i.e., reflecting Kapβ1 activity); 2), characteristic of the pore proximity (i.e., reflecting pore activity); and 3), dependent on metabolic energy. To address these issues, we set a two-channel experiment in which we simultaneously assessed Kapβ1-GFP active transport and mCherry unbiased diffusion through the pore (Fig. 2 A). Here the GFP signal is exploited to track the pore position while the shuttling of mCherry is concomitantly recorded within the same reference system. First, the ACF analysis confirms the expected hump effect for Kapβ1-GFP and reveals the absence of any specific time of correlation at the pore for a molecule (untagged mCherry) that is not expected to specifically interact with the pore structure (Fig. 2, B and C). Thus, the hump effect measured through the ACF reflects specific Kapβ1 activity at the pore. Second, if an orbit much larger than the NPC size (e.g., with a radius of ∼1 μm; red circle in Fig. 2 A) is scanned around the pore, the hump effect vanishes from the first channel, and only Kapβ1-GFP isotropic diffusion can be measured (Fig. 2, D and E). If a small-radius orbit is scanned far from the pore (e.g., in the nucleoplasm), only isotropic intracompartment diffusivity can be measured (see Supporting Material). This demonstrates that the hump is visible only in proximity to the pore. The distribution of the hump is also dependent on the orientation of the pore with respect to the scan start point (see Supporting Material).
Figure 2.
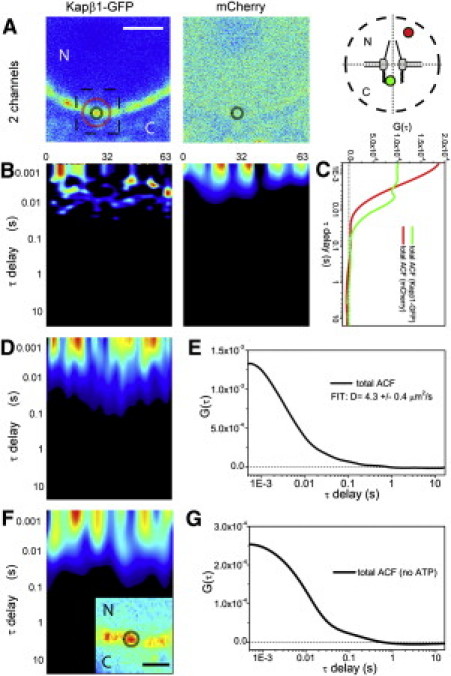
(A) Two-channel acquisition of a cell coexpressing Kapβ1-GFP and mCherry (scale bar: 2 μm). (B) ACF carpets obtained in the two channels. (C) The average ACF curves show the different behaviors of Kapβ1-GFP and mCherry at the pore. (D and E) If the tracking routine is applied far from the pore (orbit radius ∼1 μm, as depicted in A, red circle), no detectable humps are observed in the ACF carpet (left) or the corresponding average ACF curve (right). (F and G) The same NPC in energy-depleting conditions (dashed square in A is shown in the inset; scale bar: 1 μm) yields no detectable humps in the ACF carpet (left) or the corresponding average ACF curve (right).
Finally, we depleted the energy content of the cell as described previously (16). This treatment equilibrates Kapβ1-GFP across the NE but maintains its local accumulation on single NPCs (inset in Fig. 2 F). Its shuttling yields no detectable humps in the ACF (Fig. 2, F and G). This is consistent with receptor-mediated transport (described above), in which the metabolic energy acts as a ratchet in coupling Kapβ1-dependent cargo binding and release (17) but is not essential for its shuttling per se (18).
In summary, the combination of FCS with orbital tracking allows us to extract the characteristic correlations of transport events in the reference system of the pore, and in the presence of many molecules under physiological conditions. This information is somewhat hidden in the trajectories of isolated molecules, but it becomes evident when the fluctuation analysis is applied to many events. We expect that this approach, in combination with other spectroscopic techniques such as spectral information, lifetime, and polarization, will become a valuable new tool for dissecting the kinetic and molecular details of nucleocytoplasmic transport in live cells.
Acknowledgments
This work was supported in part by the National Institutes of Health (grants P41-RRO3155, P50-GM076516, and RO1 DK066029).
Supporting Material
References and Footnotes
- 1.Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 2.Fahrenkrog B., Aebi U. The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat. Rev. Mol. Cell Biol. 2003;4:757–766. doi: 10.1038/nrm1230. [DOI] [PubMed] [Google Scholar]
- 3.Yang W., Gelles J., Musser S.M. Imaging of single-molecule translocation through nuclear pore complexes. Proc. Natl. Acad. Sci. USA. 2004;101:12887–12892. doi: 10.1073/pnas.0403675101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang W., Musser S.M. Visualizing single molecules interacting with nuclear pore complexes by narrow-field epifluorescence microscopy. Methods. 2006;39:316–328. doi: 10.1016/j.ymeth.2006.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kubitscheck U., Grünwald D., Peters R. Nuclear transport of single molecules: dwell times at the nuclear pore complex. J. Cell Biol. 2005;168:233–243. doi: 10.1083/jcb.200411005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dange T., Grünwald D., Kubitscheck U. Autonomy and robustness of translocation through the nuclear pore complex: a single-molecule study. J. Cell Biol. 2008;183:77–86. doi: 10.1083/jcb.200806173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma J., Yang W. Three-dimensional distribution of transient interactions in the nuclear pore complex obtained from single-molecule snapshots. Proc. Natl. Acad. Sci. USA. 2010;107:7305–7310. doi: 10.1073/pnas.0908269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim S.A., Heinze K.G., Schwille P. Fluorescence correlation spectroscopy in living cells. Nat. Methods. 2007;4:963–973. doi: 10.1038/nmeth1104. [DOI] [PubMed] [Google Scholar]
- 9.Schwille P., Korlach J., Webb W.W. Fluorescence correlation spectroscopy with single-molecule sensitivity on cell and model membranes. Cytometry. 1999;36:176–182. doi: 10.1002/(sici)1097-0320(19990701)36:3<176::aid-cyto5>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 10.Abu-Arish A., Kalab P., Fradin C. Spatial distribution and mobility of the Ran GTPase in live interphase cells. Biophys. J. 2009;97:2164–2178. doi: 10.1016/j.bpj.2009.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrmann M., Neuberth N., Naber A. Near-field optical study of protein transport kinetics at a single nuclear pore. Nano Lett. 2009;9:3330–3336. doi: 10.1021/nl901598z. [DOI] [PubMed] [Google Scholar]
- 12.Cardarelli F., Gratton E. In vivo imaging of single-molecule translocation through nuclear pore complexes by pair correlation functions. PLoS ONE. 2010;5:e10475. doi: 10.1371/journal.pone.0010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daigle N., Beaudouin J., Ellenberg J. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 2001;154:71–84. doi: 10.1083/jcb.200101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levi V., Ruan Q., Gratton E. 3-D particle tracking in a two-photon microscope: application to the study of molecular dynamics in cells. Biophys. J. 2005;88:2919–2928. doi: 10.1529/biophysj.104.044230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rout M.P., Aitchison J.D. The nuclear pore complex as a transport machine. J. Biol. Chem. 2001;276:16593–16596. doi: 10.1074/jbc.R100015200. [DOI] [PubMed] [Google Scholar]
- 16.Cardarelli F., Serresi M., Beltram F. In vivo study of HIV-1 Tat arginine-rich motif unveils its transport properties. Mol. Ther. 2007;15:1313–1322. doi: 10.1038/sj.mt.6300172. [DOI] [PubMed] [Google Scholar]
- 17.Izaurralde E., Kutay U., Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kose S., Imamoto N., Yoneda Y. Ran-unassisted nuclear migration of a 97-kD component of nuclear pore-targeting complex. J. Cell Biol. 1997;139:841–849. doi: 10.1083/jcb.139.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


