Abstract
Individuals with germline mutations in the tumour suppressor gene CYLD are at high risk of developing disfiguring cutaneous appendageal tumours, the defining tumour being the highly organised cylindroma. Here, we analysed CYLD mutant tumour genomes by array comparative genomic hybridisation (aCGH) and gene expression microarray analysis. CYLD mutant tumours were characterised by an absence of copy number aberrations apart from loss-of-heterozygosity at chromosome 16q, the genomic location of the CYLD gene. Gene expression profiling of CYLD mutant tumours revealed dysregulated tropomyosin kinase (TRK) signalling with overexpression of TRKB and TRKC in tumours when compared to perilesional skin. Immunohistochemical analysis of a tumour microarray demonstrated strong membranous TRKB and TRKC staining in cylindromas, as well as elevated levels of ERK phosphorylation and BCL2 expression. Membranous TRKC overexpression was also observed in 70% of sporadic basal cell carcinomas. RNA interference mediated silencing of TRKB and TRKC, as well as treatment with the small molecule TRK inhibitor lestaurtinib, reduced colony formation and proliferation in three-dimensional primary cell cultures established from CYLD mutant tumours. These results suggest that TRK inhibition could be used as a strategy to treat tumours with loss of functional CYLD.
Keywords: Cylindroma, tropomyosin receptor kinase, TRK, microarray, comparative genomic hybridization, CYLD
Introduction
Germline truncating mutations in the tumour suppressor gene CYLD have been associated with three disfiguring hair follicle tumour syndromes; (i) familial cylindromatosis, (ii) Brooke-Spiegler syndrome and (iii) multiple familial trichoepitheliomas (Bowen et al., 2005). CYLD mutant patients develop three distinct types of cutaneous tumours, namely cylindromas, spiradenomas and trichoepitheliomas. The presence of painful spiradenomas and truncal and genital tumours can have a significant impact on quality of life, as can the surgery used to remove lesions as this may culminate in entire scalp removal. As such, the development of non-surgical approaches to control tumour burden are required (Rajan et al., 2009). Novel therapeutic approaches with inhibitors of the nuclear factor kappa beta (NFκB) pathway such as aspirin were proposed when CYLD was found to encode an ubiquitin hydrolase that negatively regulated this pathway (Brummelkamp et al., 2003; Kovalenko et al., 2003; Trompouki et al., 2003). However, initial clinical studies with topical salicylic acid demonstrated a significant response in only a small proportion of treated tumours (Oosterkamp et al., 2006). Therefore, a more detailed understanding of the molecular dysregulation caused by loss of functional CYLD is warranted and this may inform the development of novel therapeutic approaches.
CYLD encodes a ubiquitin hydrolase that cleaves lysine 63 (K63) linked ubiquitin chains (Brummelkamp et al., 2003; Kovalenko et al., 2003; Trompouki et al., 2003), the specificity of which depends on the presence of a B-box domain within the CYLD protein (Komander et al., 2008). In patients with germline CYLD mutations, the majority (94%) of mutations result in premature termination codons and predict translated truncated proteins that have reduced catalytic activity (Saggar et al., 2008) which in turn is thought to perturb regulation of the NFκB pathway. CYLD substrates including TRAF2, TRAF6 and NEMO which regulate canonical NFκB signalling are regulated by K63 ubiquitin tagging, and lack of functional CYLD results in constitutively active NFκB signalling (Brummelkamp et al., 2003; Kovalenko et al., 2003; Trompouki et al., 2003). CYLD interacts with and negatively regulates TAK1, reducing TAK1 mediated stimulation of IKK and hence activation of NFκB (Reiley et al., 2007). TAK1 also phosphorylates and activates MKK6 and MKK7, leading to the activation of p38 and JNK kinase pathways, which may also contribute to disease pathogenesis (Liu et al., 2006; Reiley et al., 2004). CYLD negatively regulates BCL3, preventing nuclear entry where it forms a dimer with p50/p52, resulting in transcription of genes involved in proliferation including cyclin D1 (Massoumi et al., 2006). CYLD was also shown to negatively regulate Wnt signalling, recognised to play a role in tumorigenesis, by deubiquitinating Dishevelled (Tauriello et al., 2010). Additional roles for CYLD have been suggested in the regulation of immunity, microtubule assembly, calcium ion channel regulation, facilitating entry into mitosis. (Gao et al., 2008; Reiley et al., 2006; Stegmeier et al., 2007; Stokes et al., 2006). How perturbation of these pathways by loss of functional CYLD contributes to tumour formation is not yet clear.
Apart from cylindromas, other cancers that lack functioning CYLD or have reduced expression of CYLD include hepatocellular and colonic carcinoma (Hellerbrand et al., 2007), multiple myeloma (Annunziata et al., 2007; Demchenko et al., 2010; Jenner et al., 2007; Keats et al., 2007), lung cancer (Zhong et al., 2007), prostate cancer (Kikuno et al., 2008) and malignant melanoma (Massoumi et al., 2009). In multiple myeloma, loss of heterozygosity (LOH) at 16q, the genomic location of the CYLD gene, is an indicator of poor prognosis and is associated with reduced overall survival (Jenner et al., 2007). Both deletions involving the locus of CYLD resulting in reduced copy number, as well as biallelic events, comprising a 16q deletion with mutations within the coding exons of CYLD in the remaining allele have been shown (Demchenko et al., 2010). Overexpression of NFκB target genes has been demonstrated in myeloma consistent with the proposed role of CYLD (Jenner et al., 2007).
CYLD deficient mouse models with complete loss of CYLD expression do not develop spontaneous tumours, but show an increased susceptibility to cancer. This includes an increased susceptibility to chemical carcinogen induced cutaneous tumours (Massoumi, 2006) as well as colorectal tumours following dextran sulphate induced colitis (Zhang et al., 2006). Mice expressing truncated CYLD mutations that mimic human mutations in contrast, are not viable beyond a few hours after birth (Trompouki et al., 2009). This difference in the two phenotypes is intriguing and suggests that further work is required to understand genotype-phenotype correlations.
CYLD has been shown to interact with TRKA and plays a role in receptor internalisation and signal transduction (Geetha et al., 2005). TRK receptors were first recognised to have a tumourigenic role when an oncogenic TRK fusion protein was found in a proportion of colonic carcinomas (Martin-Zanca et al., 1986) and subsequently thyroid carcinomas (Butti et al., 1995). Dysregulation of TRK signalling has also been demonstrated in a number of epithelial tumours (Lagadec et al., 2009; Ricci et al., 2001; Weeraratna et al., 2000).
Here, we have performed a detailed molecular analysis of tumours from patients with CYLD mutations. This revealed up-regulation of TRK signalling in CYLD defective tumours. We went on to develop a three dimensional cell culture from primary tumour cells to demonstrate the potential of therapeutically targeting TRK for the treatment of these tumours.
RESULTS
Cylindroma and spiradenomas tumours are similar and are genomically stable
Fresh frozen tumours from patients with germline CYLD mutations were microdissected to isolate tumour cells and perilesional control cells. Genomic analysis of cylindroma and spiradenoma tissue was performed on 12 tumours and 7 perilesional skin samples using a 32K bacterial artificial chromosome (BAC) tiling path array (Natrajan et al., 2009). This BAC array platform has been shown to be as robust as, and to have comparable resolution with high density oligonucleotide arrays (Coe et al., 2007; Gunnarsson et al., 2008; Tan et al., 2007). Comparative genomic hybridisation was performed using DNA extracted from peripheral blood leukocytes from the same patient as the reference. No obvious regional genomic amplification or deletion was noted in 12 tumour samples (Figure 1a). Moreover, no difference was seen between cylindromas (n= 7) and spiradenomas (n=5).
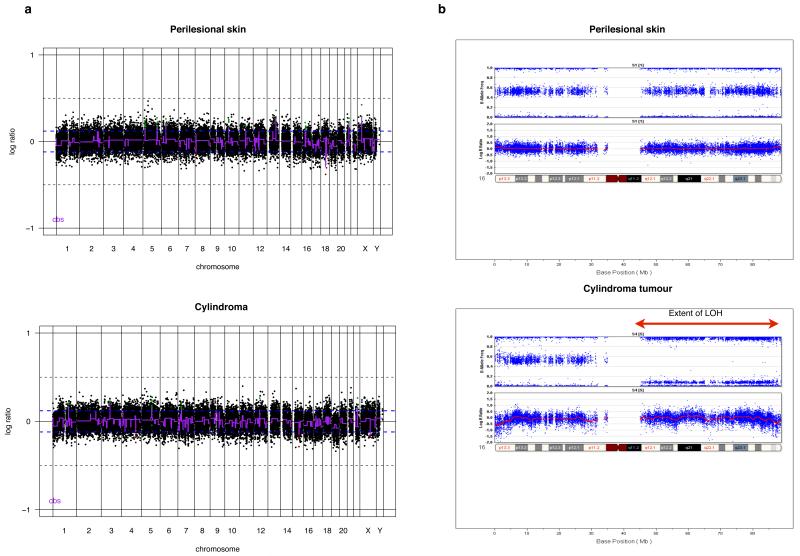
Figure 1. Cylindroma and spiradenomas tumours are genomically similar and show stability.
(a) Genomic DNA was extracted from 7 cylindroma and 5 spiradenoma microdissected tumours and competitively hybridised against patient matched peripheral lymphocyte DNA on a 32K BAC tiling path array. No significant copy number change was seen between tumours and perilesional skin; an indicative result is shown. (b) To examine for genomic changes at higher resolution, single nucleotide polymorphism (SNP) typing was performed using a 550K SNP array in a further 4 cylindromas. LOH was seen across the entire arm of chromosome 16q in all 4 samples, indicated by the red arrow.
In (a), circular binary segmentation (cbs)-smoothed Log2 ratios are plotted on the Y axis against each bacterial artificial chromosome (BAC) clone according to genomic location on the X axis. BACs categorised as displaying genomic gains or amplification are plotted in green and those categorised as genomic losses in red.
Given the apparent absence of genomic copy number variation observed using BAC arrays, we went on to perform high resolution SNP typing using a 550K SNP array platform (Illumina) on four cylindroma tumours and one control sample. This revealed copy number silent LOH for the entire arm of chromosome 16q in all four tumours but no other significant changes (Figure 1b).
LOH status at the site of the mutation was assessed in a further 27 tumours used in this study, which was possible as the genotype of each patient was known. LOH was assessed using genomic DNA from microdissected tumours by using restriction enzymes that specifically were unable to cut only the mutated allele in each tumour. Polymerase chain reaction (PCR) amplification was performed using primers that flanked exon 18, and the PCR product was subject to restriction enzyme digestion using enzymes chosen for pedigree specific mutations: HpyCH4V (c.2460delc) and Sml I (c.2469+1G>A). This revealed LOH at the site of the mutation in 75% of the 27 tumours, suggesting the mechanism resulting in LOH to be similar in the majority of tumours.
TRK signalling is dysregulated in cylindroma and spiradenoma tissue
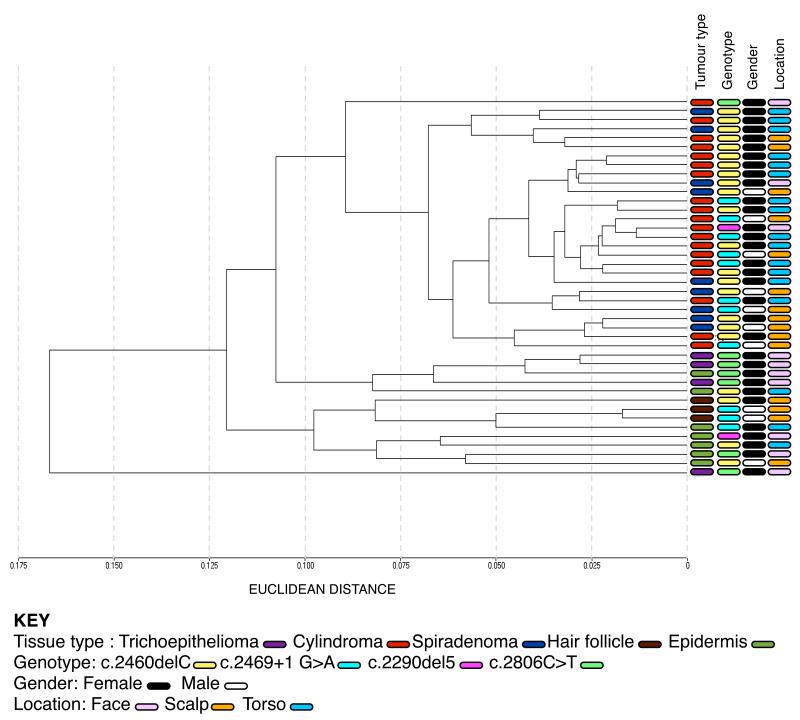
Gene expression profiling was performed on total RNA extracted from 42 microdissected samples, consisting of 19 cylindromas, 9 spiradenomas, 4 trichoepitheliomas and 10 perilesional controls. A bead array platform (Illumina) that is capable of assaying expression of 24,526 transcripts for each sample was used (April et al., 2009). Average signal values from replicate beads were summarised, log transformed, and normalised using quantile normalisation across all samples (Dunning et al., 2008; Workman et al., 2002). Unsupervised clustering analysis of the individual transcriptomes (24526 probes) by Euclidean distance revealed close clustering of the majority of cylindroma and spiradenoma tissue, and distinct clustering of the perilesional skin and trichoepithelioma tissue (Figure 2). This was corroborated with clustering by an independent technique that used multiple bootstrapping to determine the robustness of the clusters. Clustering of transcriptomes using individual probes (24526) was employed and dissimilarity was measured using the correlation method using a statistical package (pvclust) (Suzuki and Shimodaira, 2006). Clusters were maintained, however due to the small numbers of tumours used, statistical significance was seen between tumours and controls (p<0.01), but not between tumour subsets (Supplementary Figure 3). The unsupervised clustering analysis was further corroborated by a supervised clustering analysis, allowing class comparison between cylindroma and spiradenoma tumours. 19 cylindromas and 9 spiradenomas were subject to significance of microarrays analysis (SAM), which showed no significant difference between the tumour types when standard criteria were applied (Supplementary Figure 4). (Differentially expressed genes between the two groups had to pass a significance threshold of p<0.01 and 100 permutations of the data were analysed)
Figure 2. Cylindromas and spiradenomas share similar transcriptomes, and cluster distinctly from trichoepitheliomas and perilesional control skin.
Total RNA was extracted from 32 microdissected tumours and 10 perilesional controls and gene expression levels of 24526 transcripts for each sample were assayed. Unsupervised clustering of the 42 transcriptomes was performed, and cylindromas and spiradenomas clustered together, with trichoepitheliomas and perilesional skin clustering separately. No clustering by gender, genotype or tumour location was seen.
Quantitation of differential gene expression was determined using the bead array manufacturers’ custom algorithm (Illumina custom method- Illumina Inc, 2009). The statistical significance of differential gene expression between tumours and samples was calculated as a differential score. This calculation incorporated the difference in signal values between the two groups compared, corrected for appropriate negative control bead values, and was corrected for multiple hypothesis testing using a Bonferroni post test. We used stringent thresholds for filtering genes, with only transcripts that were differentially expressed with a p value of < 0.01 included for subsequent analysis (n=4492). Given the genomic and transcriptomic similarity seen in cylindromas and spiradenomas, these were pooled and analysed against pooled controls to provide enhanced statistical robustness.
Transcripts of proteins that were previously noted to be overexpressed in cylindromas were also present and supported the enrichment of cylindroma tissue following microdissection. These included numerous laminins (Table 1) (Tunggal et al., 2002), collagens 4 and 7 (Bruckner-Tuderman et al., 1991; Timpl et al., 1984) and cytokeratins (Meybehm and Fischer, 1997; Tellechea et al., 1995) which were preferentially expressed in the tumours compared to perilesional epidermis.
Table 1. NFκB target genes are overexpressed in cylindroma tumours.
Transcripts that were differentially expressed between tumours and perilesional controls were filtered with the criteria of a 4x fold change and p value of less than 0.01. Transcripts that fulfilled these criteria were compared against known NFkB target genes and tabulated. The purity of tumour microdissection was supported by overexpression of transcripts in cylindroma tissue described previously in the literature.
| NFKB target genes that are upregulated in tumour tissue (p<0.01) | ||
|---|---|---|
| Gene | Fold change | Gene name |
| SAA1 | 5.92 | Serum amyloid a2 |
| BCL2A1 | 5.74 | BCL2-related protein a1 |
| SAA2 | 4.58 | Serum amyloid a2 |
| BDNF | 2.72 | Brain-derived neurotrophic factor |
| OLR1 | 4.53 | Oxidized low density lipoprotein (lectin-like) receptor 1 |
| CD83 | 2.53 | Cd83 molecule |
| TIFA | 2.54 | Traf-interacting protein with forkhead-associated domain |
| TF | 2.77 | Transferrin |
| CXCL10 | 2.67 | Chemokine (c-x-c motif) ligand 10 |
| HLA-G | 3.44 | Major histocompatibility complex, class i, g |
| RELB | 2.36 | V-rel reticuloendotheliosis viral oncogene homolog b |
| MYB | 2.17 | V-myb myeloblastosis viral oncogene homolog (avian) |
|
Genes that are upregulated in tumour
tissue that have been previously described to be overexpressed in cylindroma tumours (p<0.01) | ||
| KRT18 | 1.74 | Keratin 18 |
| KRT13 | 4.6 | Keratin 13 |
| KRT8 | 3.71 | Keratin 8 pseudogene 9; similar to keratin 8 |
| KRT7 | 2.1 | Keratin 7 |
| LAMA1 | 4.14 | Laminin, alpha 1 |
| LAMA3 | 3.08 | Laminin, alpha 3 |
| LAMB3 | 2.79 | Laminin, beta 3 |
| LAMC2 | 1.86 | Laminin, gamma 2 |
| COL7A1 | 1.94 | Collagen, type vii, alpha 1 |
| COL4A1 | 1.5 | Collagen, type iv, alpha 1 |
| COL4A2 | 1.73 | Collagen, type iv, alpha 2 |
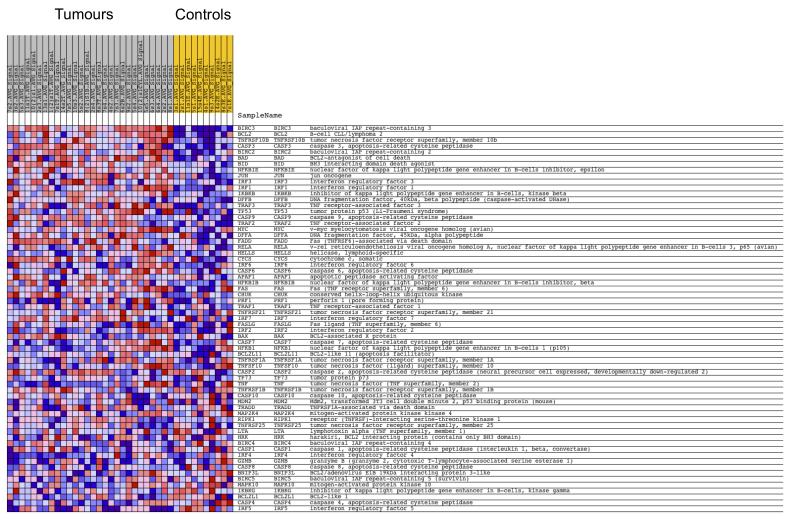
Upregulation of NFκB target genes (http://people.bu.edu/gilmore/nfkb/target/index.html) was present as expected and served as an internal positive control. All tumours were compared to all controls, and the NFκB target genes that were differentially expressed with a p-value of less than 0.01 were tabulated in Table 1. To detect low level changes in gene expression between all tumours and all control tissue, gene set enrichment analysis (GSEA) was performed on data filtered at a threshold of p< 0.05 (Subramanian et al., 2005). This highlighted multiple gene sets that were involved in apoptosis and genes that were common to these sets were found to be members of or target genes of NFκB and JNK signalling pathways (Figure 3, Supplementary Figure 1).
Figure 3. NFκB target genes are overexpressed in CYLD mutant tumours.
Expression levels of 24526 genes from 32 microdissected tumours were pooled and compared against 10 pooled perilesional controls and subject to gene set expression analysis. Sets of genes recognised to modulate apoptosis were found to be overexpressed in the tumour tissue (see also Supplementary figure 1). A heat map of the members of one set (HSA04210_APOPTOSIS) is shown here, with overexpressed genes shown in red and underexpressed genes in blue. Overexpressed genes included several NFκB target genes.
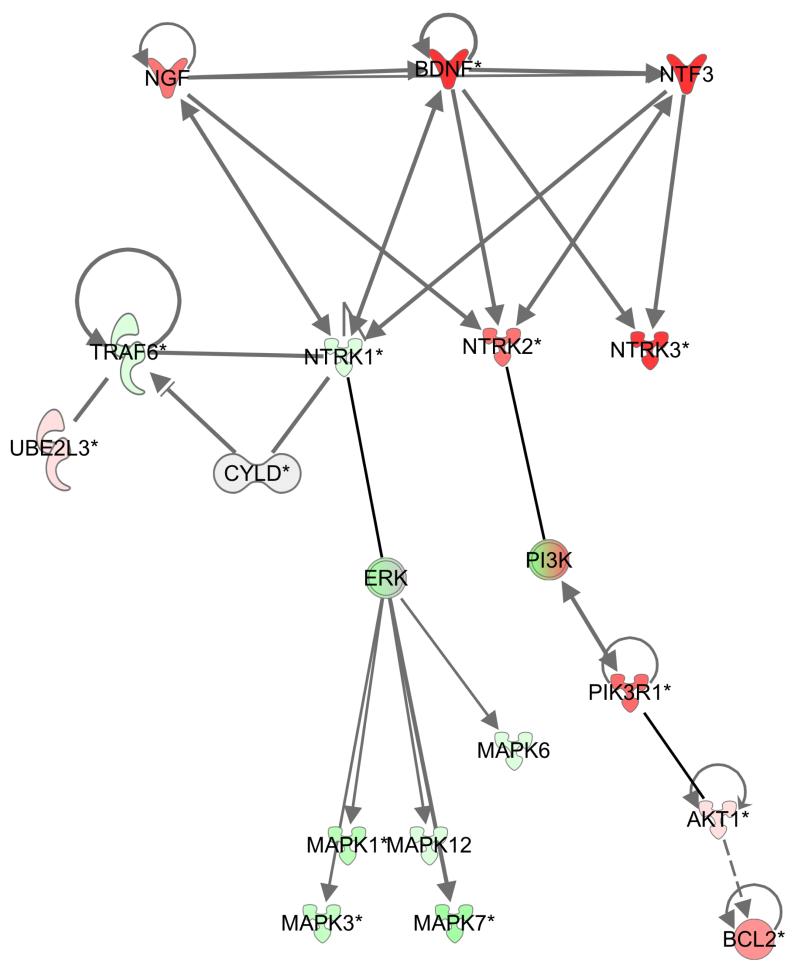
Fold change (FC) analysis (Table 2) revealed overexpression of several members of the TRK signalling pathway compared to perilesional tissue, namely, TRKC (FC = 4.75x), Neurotrophin 3 (FC=3.00x) and Neurotrophin 4/5 (FC=3.15x) in the tumour group when compared to perilesional skin. This finding was corroborated by connectivity mapping analysis (Lamb, 2006) using Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com). This highlighted the concerted overexpression of multiple members of the Neurotrophin/TRK signalling pathway in tumour tissue compared to control tissue (p = 4.02 × 10−3). In addition to the transcripts related to the TRK signalling pathway detected by FC analysis above, TRKB, brain derived neurotrophic factor and PIK3R1, which encodes a regulatory subunit of PI3 kinase, a downstream mediator of TRK signalling, were elevated in expression (Figure 4). Moreover, TRKA, a previously described CYLD interacting protein (Geetha et al., 2005), was found to be downregulated, whilst the cognate ligand nerve growth factor (NGF) was upregulated. No obvious changes in copy number were found at the chromosomal location of the different TRK receptors and their respective cognate ligands suggesting that genomic amplification or deletion was not directly responsible for these changes in transcript level.
Table 2. Altered gene expression in a human CYLDtrunc/trunc model.
Transcripts that were differentially expressed between tumours and perilesional controls were filtered with the criteria of a 4x fold change and p value of less than 0.01. Overexpressed transcripts are shown in red and underexpressed transcripts are shown in green.
| Gene | Fold change |
Gene name |
|---|---|---|
| FGF3 | 8.38 | Fibroblast growth factor 3 (murine mammary tumour virus integration site (v-int-2) oncogene homolog) (FGF3), mrna. |
| APIN | 6.74 | Apin protein (APIN), mrna. |
| ABTB2 | 5.65 | Ankyrin repeat and BTB (POZ) domain containing 2 (ABTB2), mrna. |
| IQCA | 5.53 | IQ motif containing with AAA domain (IQCA), mrna. |
| SAA1 | 5.13 | Serum amyloid A1 (SAA1), transcript variant 2, mrna. |
| RPRM | 5.07 | Reprimo, TP53 dependent G2 arrest mediator candidate (RPRM), mrna. |
| RARB | 5.06 | Retinoic acid receptor, beta (RARB), transcript variant 1, mrna. |
| COL9A2 | 4.97 | Collagen, type IX, alpha 2 (COL9A2), mrna. |
| USP6 | 4.84 | Ubiquitin specific peptidase 6 (Tre-2 oncogene) (USP6), mrna. |
| IL17RB | 4.83 | Interleukin 17 receptor B (IL17RB), transcript variant 2, mrna. |
| RARB | 4.82 | Retinoic acid receptor, beta (RARB), transcript variant 2, mrna. |
| C1ORF61 | 4.82 | Chromosome 1 open reading frame 61 (c1orf61), mrna. |
| BCL2A1 | 4.81 | BCL2-related protein A1 (BCL2A1), mrna. |
| REPS2 | 4.76 | RALBP1 associated Eps domain containing 2 (REPS2), mrna. |
| CPA6 | 4.70 | Carboxypeptidase A6 (CPA6), mrna. |
| NTRK3 | 4.66 | Neurotrophic tyrosine kinase, receptor, type 3 (NTRK3), transcript variant 3, mrna. |
| RAB6B | 4.64 | RAB6B, member RAS oncogene family (RAB6B), mrna. |
| LRRN5 | 4.59 | Leucine rich repeat neuronal 2 (LRRN2), transcript variant 1, mrna. |
| FLJ45803 | 4.58 | FLJ45803 protein (FLJ45803), mrna. |
| ARNT2 | 4.51 | Aryl-hydrocarbon receptor nuclear translocator 2 (ARNT2), mrna. |
| EDAR | 4.37 | Ectodysplasin A receptor (EDAR), mrna. |
| KIAA1622 | 4.34 | KIAA1622 (KIAA1622), transcript variant 1, mrna. |
| MIA | 4.34 | Melanoma inhibitory activity (MIA), mrna. |
| CLUL1 | 4.30 | Clusterin-like 1 (retinal) (CLUL1), transcript variant 2, mrna. |
| DACT2 | 4.30 | Dapper, antagonist of beta-catenin, homolog 2 (Xenopus laevis) (DACT2), mrna. |
| ADM2 | 4.25 | Adrenomedullin 2 (ADM2), mrna. |
| SNX22 | 4.15 | Sorting nexin 22 (SNX22), mrna. |
| ADAMTS18 | 4.12 | ADAM metallopeptidase with thrombospondin type 1 motif, 18 (ADAMTS18), transcript variant 1, mrna. |
| LOC130940 | 4.10 | Hypothetical protein BC015395 (LOC130940), mrna. |
| COL22A1 | 4.06 | Collagen, type XXII, alpha 1 (COL22A1), mrna. |
| MMP7 | 4.02 | Matrix metallopeptidase 7 (matrilysin, uterine) (MMP7), mrna. |
| ENPP6 | 4.00 | Ectonucleotide pyrophosphatase/phosphodiesterase 6 (ENPP6), mrna. |
| C6ORF85 | 0.25 | Chromosome 6 open reading frame 85 (c6orf85), mrna. |
| GRHL3 | 0.25 | Grainyhead-like 3 (Drosophila) (GRHL3), transcript variant 1, mrna. |
| KRT10 | 0.24 | Keratin 10 (epidermolytic hyperkeratosis; keratosis palmaris et plantaris) (KRT10), mrna. |
| ZD52F10 | 0.24 | Dermokine (DMKN), transcript variant 2, mrna. |
| ZD52F10 | 0.24 | Dermokine (DMKN), transcript variant 2, mrna. |
| TLCD1 | 0.24 | TLC domain containing 1 (TLCD1), mrna. |
| MUC15 | 0.23 | Mucin 15, cell surface associated (MUC15), mrna. |
| SLC19A3 | 0.23 | Solute carrier family 19, member 3 (SLC19A3), mrna. |
|
RP11- 19J3.3 |
0.23 | Centromere protein P (CENPP), mrna. |
| CPA4 | 0.23 | Carboxypeptidase A4 (CPA4), mrna. |
| LGALS7 | 0.23 | Lectin, galactoside-binding, soluble, 7 (galectin 7) (LGALS7), mrna. |
| ALDH3A1 | 0.22 | Aldehyde dehydrogenase 3 family, membera1 (ALDH3A1), mrna. |
| XKRX | 0.22 | XK, Kell blood group complex subunit-related, X-linked (XKRX), mrna. |
| BNIPL | 0.22 | BCL2/adenovirus E1B 19kd interacting protein like (BNIPL), transcript variant 1, mrna. |
| ALDH3B2 | 0.21 | Aldehyde dehydrogenase 3 family, member B2 (ALDH3B2), transcript variant 2, mrna. |
| ANXA8 | 0.21 | Annexin A8-like 2 (ANXA8L2), mrna. |
| LOC144501 | 0.20 | Hypothetical protein LOC144501 (LOC144501), mrna. |
| CST6 | 0.20 | Cystatin E/M (CST6), mrna. |
| MICALCL | 0.20 | MICAL C-terminal like (MICALCL), mrna. |
| HOP | 0.20 | Homeodomain-only protein (HOP), transcript variant 3, mrna. |
| MAP2 | 0.20 | Microtubule-associated protein 2 (MAP2), transcript variant 2, mrna. |
| GGT6 | 0.20 | Gamma-glutamyltransferase 6 homolog (rat) (GGT6), mrna. |
| PSORS1C2 | 0.20 | Psoriasis susceptibility 1 candidate 2 (PSORS1C2), mrna. |
| DCT | 0.20 | Dopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2) (DCT), mrna. |
| SERPINB13 | 0.20 | Serpin peptidase inhibitor, clade B (ovalbumin), member 13 (SERPINB13), mrna. |
| MAP2 | 0.20 | Microtubule-associated protein 2 (MAP2), transcript variant 1, mrna. |
| TMEM79 | 0.19 | Transmembrane protein 79 (TMEM79), mrna. |
| FGFR3 | 0.19 | Fibroblast growth factor receptor 3 (achondroplasia, thanatophoric dwarfism) (FGFR3), transcript variant 1, mrna. |
| C14ORF29 | 0.19 | Abhydrolase domain containing 12B (ABHD12B), transcript variant 2, mrna. |
| DSG1 | 0.19 | Desmoglein 1 (DSG1), mrna. |
| KRT1 | 0.19 | Keratin 1 (epidermolytic hyperkeratosis) (KRT1), mrna. |
| TRIM7 | 0.19 | Tripartite motif-containing 7 (TRIM7), transcript variant 6, mrna. |
| SLC39A2 | 0.18 | Solute carrier family 39 (zinc transporter), member 2 (SLC39A2), mrna. |
| FLJ10847 | 0.17 | Solute carrier family 47, member 1 (SLC47A1), mrna. |
| CST6 | 0.17 | Cystatin E/M (CST6), mrna. |
| LOC342897 | 0.16 | Similar to F-box only protein 2 (LOC342897), mrna. |
| ATP13A4 | 0.16 | Atpase type 13A4 (ATP13A4), mrna. |
| GPR115 | 0.15 | G protein-coupled receptor 115 (GPR115), mrna. |
| UNC93A | 0.15 | Unc-93 homolog A (C. Elegans) (UNC93A), mrna. |
| SBSN | 0.15 | Suprabasin (SBSN), mrna. |
| LOC144501 | 0.14 | Hypothetical protein LOC144501 (LOC144501), mrna. |
| TYRP1 | 0.14 | Tyrosinase-related protein 1 (TYRP1), mrna. |
| GSDM1 | 0.14 | Gasdermin 1 (GSDM1), mrna. |
| LOC130576 | 0.13 | Hypothetical protein LOC130576 (LOC130576), mrna. |
| FLJ21511 | 0.13 | Hypothetical protein FLJ21511 (FLJ21511), mrna. |
| C3ORF52 | 0.13 | Chromosome 3 open reading frame 52 (c3orf52), mrna. |
| WFDC5 | 0.12 | WAP four-disulfide core domain 5 (WFDC5), mrna. |
| SLC5A1 | 0.12 | Solute carrier family 5 (sodium/glucose cotransporter), member 1 (SLC5A1), mrna. |
| MLANA | 0.12 | Melan-A (MLANA), mrna. |
| TGM5 | 0.11 | Transglutaminase 5 (TGM5), transcript variant 1, mrna. |
| CYP3A5 | 0.10 | Cytochrome P450, family 3, subfamily A, polypeptide 5 (CYP3A5), mrna. |
| AADACL2 | 0.10 | Arylacetamide deacetylase-like 2 (AADACL2), mrna. |
| BPIL2 | 0.10 | Bactericidal/permeability-increasing protein-like 2 (BPIL2), mrna. |
| CDSN | 0.10 | Corneodesmosin (CDSN), mrna. |
| K5B | 0.10 | Keratin 5b (K5B), mrna. |
| RDH12 | 0.09 | Retinol dehydrogenase 12 (all-trans/9-cis/11-cis) (RDH12), mrna. |
| DSC1 | 0.08 | Desmocollin 1 (DSC1), transcript variant Dsc1b, mrna. |
| TGM3 | 0.07 | Transglutaminase 3 (E polypeptide, protein-glutamine-gamma-glutamyltransferase) (TGM3), mrna. |
| PPP2R2C | 0.07 | Protein phosphatase 2 (formerly 2A), regulatory subunit B, gamma isoform (PPP2R2C), transcript variant 2, mrna. |
| IL1F10 | 0.07 | Interleukin 1 family, member 10 (theta) (IL1F10), transcript variant 2, mrna. |
| CA12 | 0.07 | Carbonic anhydrase XII (CA12), transcript variant 1, mrna. |
| CCL27 | 0.07 | Chemokine (C-C motif) ligand 27 (CCL27), mrna. |
| PLA2G4F | 0.07 | Phospholipase A2, group IVF (PLA2G4F), mrna. |
| K6IRS3 | 0.06 | Keratin 73 (KRT73), mrna. |
Figure 4. TRK signalling is dysregulated in cylindroma and spiradenoma tissue.
Expression levels of 24526 genes from 32 microdissected tumours were pooled and compared against 10 pooled perilesional controls, and genes that were differentially expressed (p value <0.01) were analysed using Ingenuity pathway analysis. Overexpressed transcripts are shown in red and underexpressed transcripts are shown in green. TRKB (NTRK2) and TRKC (NTRK3), members of the TRK signalling pathway, were overexpressed in tumour tissue compared to control samples. Expression of TRKA (NTRK1) however was reduced. Downstream members such as PI3K and AKT and BCL2 were also over expressed.
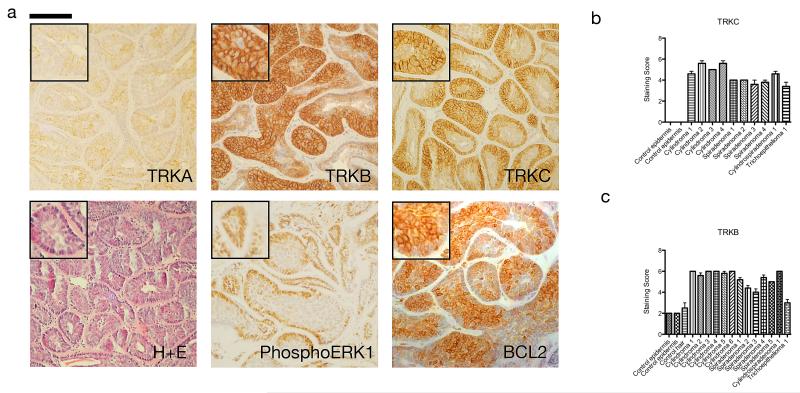
TRKB and TRKC proteins are overexpressed in human cylindromas and spiradenomas
We investigated whether the overexpression of TRKB and TRKC observed at the mRNA level was reflected in elevated protein expression in CYLD tumours. We constructed a tumour tissue microarray (TMA) of CYLD related tumours - cylindromas, spiradenomas, trichoepitheliomas and perilesional skin and analysed this using immunohistochemistry. Expression of TRKB and TRKC was elevated in cylindroma, spiradenoma and to a lesser extent in trichoepithelioma tissue compared to perilesional skin (Figure 5a). Strong membranous staining was apparent in cylindroma cells, whilst diffuse cytoplasmic staining was noted in overlying keratinocytes. TRKC expression was mainly noted over the basophilic peripheral cells in cylindroma islands, whilst TRKB overexpression was non-discriminatory within tumour islands (Figure 5a). Quantification of membranous TRK expression was performed on each core of the TMA. A scoring scale, which assessed the intensity of membranous TRK staining and the proportion of tumour cells stained was used, (Figure 5 b-c), and the difference in expression between tumours and perilesional keratinocytes was found to be statistically significant p<0.05 (Tables 3-4). Furthermore, cylindroma cells were strongly positive with antibodies against phosphorylated ERK (p44/42), a marker of active TRK signalling, mainly at the periphery of each tumour island. The antiapoptotic protein BCL2, a downstream target of TRK signalling, was also overexpressed in cylindroma tissue, showing strong perinuclear staining (Figure 5a)
Figure 5. TRKB and TRKC proteins are over expressed in human cylindromas and spiradenomas.
(a) Tissue microarrays of cylindromas and spiradenomas were probed with antibodies as indicated, and visualised using a horseradish peroxidase – diaminobenzene system, with haematoxylin used as a nuclear counterstain. Membranous TRKB and TRKC were overexpressed in the tumours, exemplified by the cylindroma tumours illustrated here. Patchy perinuclear TRKA staining was seen in some tumour cells. Expression of downstream targets phosphoERK and BCL2 was also seen. Scale bars indicate 100μm; insets are shown to demonstrate specific membranous staining of TRKB and TRKC (2x enlargement of related image). (b-c) Membranous immunostaining scores of TRKC and TRKB were made on all cores on the array, with p-values of the difference in score (t-test) between tumour and control indicated in Table 3. Error bars indicate standard error of the mean.
Table 3. Membranous TRKB is highly expressed in CYLD mutant tumours.
Membranous TRKB expression scores in cylindroma tumours compared to perilesional unaffected tissue. A custom tumour microarray was scored for membranous expression of TRKB. Mean scores were grouped by tumour type and compared to the scores of unaffected perilesional tissue.
| Membranous TRKB | Mean score and SEM | p value |
|---|---|---|
| Normal tissue (N=3) | 2.125 ± 0.1250 | |
| Cylindroma (N=6) | 5.900 ± 0.05571 | < 0.0001 |
| Spiradenoma (N=5) | 4.800 ± 0.1414 | < 0.0001 |
| Cylindrospiradenoma (N=1) | 5.800 ± 0.2000 | < 0.0001 |
| Trichoepithelioma (N=1) | 3.000 ± 0.3162 | 0.012 |
Table 4. Membranous TRKC is highly expressed in CYLD mutant tumours.
Membranous TRKC expression scores in cylindroma tumours compared to perilesional unaffected tissue. A custom tumour microarray was scored for membranous expression of TRKC. Mean scores were grouped by tumour type and compared to the scores of unaffected perilesional tissue.
| Membranous TRKC | Mean score and SEM | p value |
|---|---|---|
| Normal tissue (N=2) | 0.0005 ± 0.0005 | |
| Cylindroma (N=4) | 5.350 ± 0.1500 | < 0.0001 |
| Spiradenoma (N=4) | 3.850 ± 0.1094 | < 0.0001 |
| Cylindrospiradenoma (N=1) | 4.600 ± 0.2449 | < 0.0001 |
| Trichoepithelioma (N=1) | 3.400 ± 0.4000 | 0.0038 |
TRKB and TRKC silencing reduces cell viability and colony formation
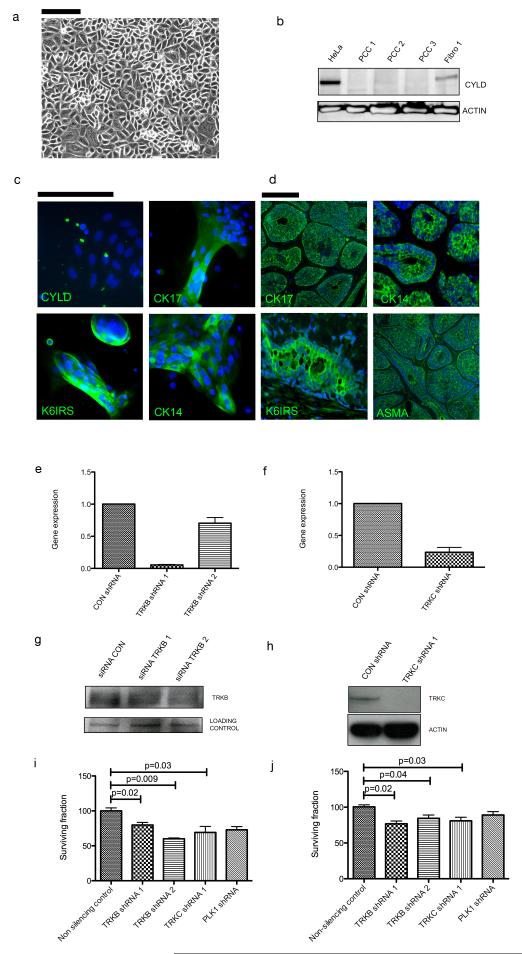
To assess the functional significance of TRKB and TRKC in cylindromas, primary cell cultures were established from fresh tumour samples from the patients that contributed samples used for the molecular profiling and used for functional studies (Figure 6a). Cylindroma tissue was microdissected from normal perilesional tissue under a stereomicroscope, subject to enzymatic digestion to obtain a single cell suspension, and cultured under standard conditions (5% CO2, 21% O2). These primary cell cultures were used within four weeks of surgery to protect against culture induced genomic alterations. To validate these cultures, CYLD expression was characterised and these cells did not express full length CYLD, which was observed in perilesional fibroblast culture (Figure 6b). Cylindroma cells in culture expressed cytokeratins 6, 14, 17 and smooth muscle actin, markers that were shown to be present in cylindroma tumours in vivo, confirming the purity of the cultures (Figure 6c-6d). These primary cells were transduced with lentiviruses expressing short hairpin (sh) RNAs targeting TRKB and TRKC. The shRNA vectors carried a GFP reporter allowing monitoring of delivery and a puromycin gene allowing selection - cells were grown in media containing puromycin for 48 hours post transfection resulting in cultures that were approximately 100% GFP positive when analysed. TRKB and TRKC knockdown was assayed using immunoblotting (Figure 6e-6f) and after 10-14 days in culture, cells were fixed, stained and colonies were counted. Primary cells with knockdown of TRKB and TRKC demonstrated a modest reduction (20-40%; p<0.05) in colony formation (Figure 6g).
Figure 6. TRKB and TRKC knockdown reduces primary cell culture colony formation.
(a) Cylindroma tumour tissue was enzymatically digested and converted to a single cell suspension, and grown on collagen coated tissue culture plastic. (b) Total cell lysates from control HeLa cells, three cylindroma primary cell cultures and a matched patient fibroblast culture were subject to SDS-PAGE, and probed with anti-CYLD antibodies. Lack of full length CYLD is seen in primary cell cultures. (c) Cylindroma primary cell cultures and tissue were fixed and probed with antibodies against the cytokeratins indicated and labelled with fluorescent secondary antibodies for visualization; DAPI was used as a nuclear counterstain. CYLD expression is not seen in primary cell culture. Cylindroma primary cell culture cells expressed cytokeratins that are seen (d) in cylindroma tumours. (e-f) Cylindroma cells were subject to lentiviral mediated short hairpin RNA knockdown of TRKB, TRKC and these were compared against a control hairpin. Knockdown was assessed at the protein level using immunoblotting. (g-h) Transcript expression of TRKB and TRKC is reduced following lentiviral knockdown. (i) Following puromycin selection, GFP positive cells were plated and grown over 12 days. TRKB and TRKC knockdown resulted in reduced colony formation. (j) Similar results were seen in a short-term cell viability assay. All experiments were repeated at least three times. Error bars indicate standard error of the mean and scale bars represent a distance of 100μm.
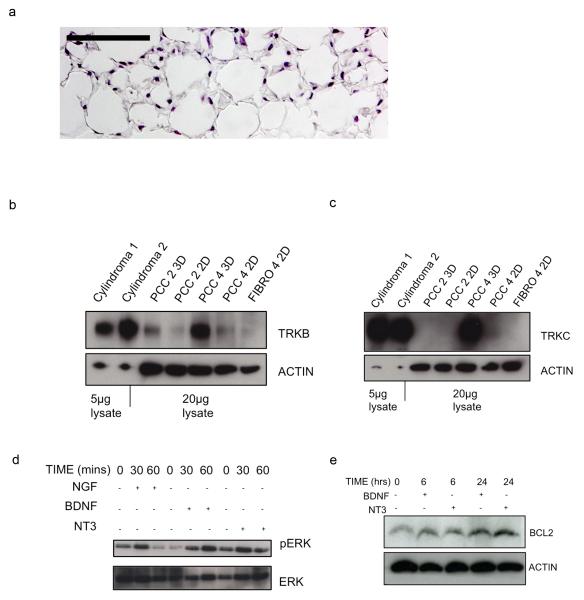
Three-dimensional primary cell cultures of cylindroma cells are sensitive to TRK inhibitors
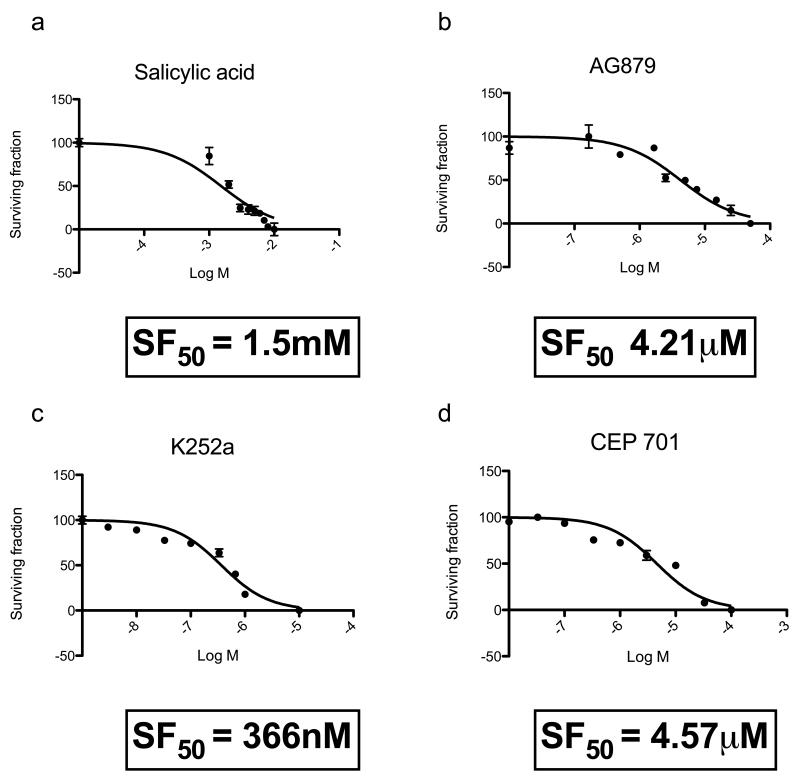
The primary cell cultures described above expressed only low levels of TRK, an observation made in previous studies (Lagadec et al., 2009). To establish a more biologically relevant cylindroma culture model we seeded primary cells on three-dimensional (3D) polystyrene tissue culture scaffolds to allow enhanced cell–cell contact (Figure 7a). This caused increased expression of TRKB and TRKC when compared to matched cells grown in standard two dimensional (2D) culture (Figures 7b and 7c). To determine if TRK signalling was active, we examined phosphorylated ERK and BCL2 after stimulating the cultured cells with cognate ligands of the TRK receptors (NGF, BDNF and NT3). Stimulation indeed resulted in increased phosphorylated ERK above basal levels (Figure 7d). Antiapoptotic factor BCL2 has been shown to be upregulated in response to NT3 in oligodendrocyte progenitor cells (Saini et al., 2004). This was recapitulated in our model, where BCL2 expression was increased following stimulation with TRK receptor ligands (Figure 7e). To determine if these cultures were dependent on TRK signalling for survival, they were treated with small molecule TRK kinase inhibitors. As a control they were also exposed to salicylic acid which has previously been shown to effect survival in CYLD deficient cells (Brummelkamp et al., 2003). Cells were only modestly sensitive to salicylic acid (surviving fraction (SF50) 1.5mM), but were much more so to the pan-TRK inhibitors, lestaurtinib (CEP-701) (SF50, 4.57μM), AG879 (SF50, 4.21 μM) and K252a (SF50, 366 nM) (Figure 8). It seems possible that loss of CYLD catalytic activity accounts for the TRK dependency. To address this, we introduced a cDNA construct expressing CYLD into primary tumour cells, with the aim of assessing whether CYLD expression could rescue the phenotypes observed. However, as is often the case when genes are artificially expressed from cDNA constructs, the reintroduction of CYLD cDNA resulted in reduced cell viability, precluding further mechanistic studies (Supplementary Figure 5).
Figure 7. Three-dimensional primary cell culture of cylindroma cells express TRK proteins and respond to stimulation with cognate TRK ligands.
(a) Cylindroma primary cell cultures were plated onto collagen coated tissue culture scaffolds (3D cell culture) and grown for 28 days in the same conditions as standard 2D cell culture. Tissue culture scaffolds were fixed and stained with haematoxylin and eosin (20x original magnification expression – scale bar 100μm) (b-c) Total cell lysates from fresh tumours, 3D and 2D cell cultures were separated using SDS-PAGE, and immunoblots were probed using (b) TRKB and (c) TRKC antibodies. Lanes 1 and 2 were loaded with 5μg of cell lysate from fresh tumours, labelled “Cylindroma 1 and 2”, rest 20μg of lysate from primary cell cultures in 3D (PCC 2,4 3D) and cultures in 2D (PCC 2,4 2D). TRKB and TRKC were expressed at higher levels in 3D cell cultures (Lanes 3 and 5) compared to matched 2D cultures (Lanes 4 and 6). (d-e) Cylindroma cells grown on 3D scaffolds and stimulated with NGF, BDNF and NT3 for the periods indicated, and then transferred to ice cold PBS, and cell lysates were extracted for immunoblotting. Membranes were probed with antibodies to phosphorylated ERK and BCL2. Increased levels of phosphoERK and BCL2 were seen. All experiments were repeated at least three times.
Figure 8. Cylindroma 3D primary cell cultures demonstrate an increased sensitivity to TRK inhibitors relative to aspirin.
(a-d) Cylindroma 3D cell cultures were grown for two weeks in the presence of salicylic acid, K252a, Tyrphostin AG879 and lestaurtinib (CEP-701) in triplicate in a 96 well format. This was repeated using at least three different tumours, from different body sites in three patients. Cell viability was assessed using a luminescent cell viability assay at the end of this period and dose response curves were generated. Cylindroma cells were sensitive to lestaurtinib at micromolar levels, whilst salicylic acid had a similar effect at milimolar levels.
Membrane-localised TRKC is overexpressed in sporadic basal cell carcinoma
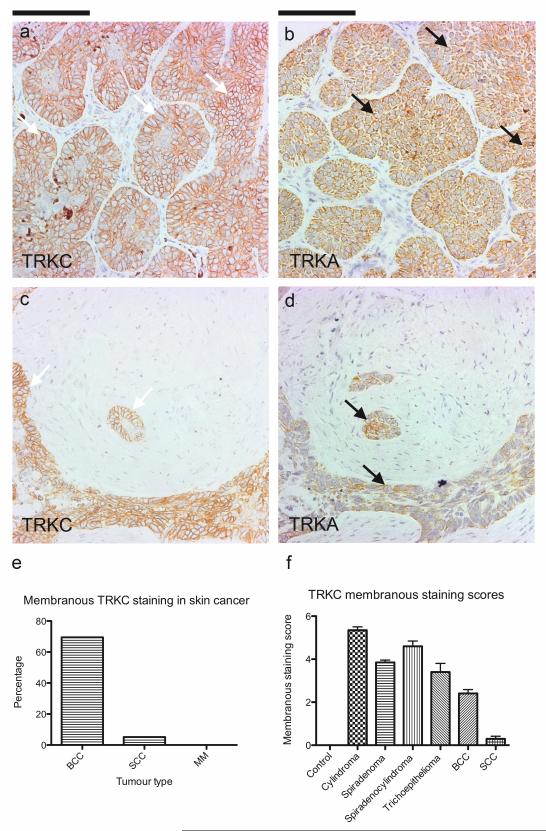
Cutaneous appendageal tumours seen in CYLD mutation carriers share similarities in cytokeratin profiles with sporadic basal cell carcinoma (BCC), that in humans are thought to be also derived from the hair follicle (Donovan, 2009). Dysregulation of several mutual oncogenic pathways are seen in cylindromas and BCCs. Wnt signalling is thought to play an oncogenic role in both, with expression of nuclear β-catenin demonstrated in both cylindromas (Tauriello et al., 2010) and human basal cell carcinoma (Kriegl et al., 2009). Also, the Sonic hedgehog pathway, recognised to depend on such canonical Wnt signalling for cutaneous tumour formation (Hoseong Yang et al., 2008), is an important final common pathway in both tumours. In a murine model that overexpresses GLI1 (Nilsson et al., 2000), tumours that had histological appearances that resembled both human basal cell carcinoma and cylindroma were seen. Diffuse light expression of TRK receptors has previously been noted in a small series of basal cell carcinoma using a pan TRK antibody (Chen-Tsai et al., 2004). Therefore, we went on to investigate if the membranous TRKB and TRKC overexpression we observed in CYLD mutant tumours was a feature that extended to sporadic skin cancers. We examined expression of TRK receptors in a human skin cancer TMA that included samples of squamous cell carcinoma (SCC; n=19), melanoma (MM; n=5) and basal cell carcinoma (BCC; n=23) using TRKA, TRKB and TRKC specific antibodies. Interestingly, membranous TRKC was overexpressed in 70% of BCC tumours (Figure 9), 5% of SCC but not melanoma. In some samples, TRKC staining was strongest at the invasive front where the tumour cells apposed stroma (Supplementary Figure 2). No membranous TRKA staining was seen, however some perinuclear staining, a feature associated with activated TRKA signalling (Wu et al., 2007) and was seen in a 39% of cases of BCC and 16% of cases of SCC. TRKB staining was seen in the cytoplasm in 13% of cases of BCC only, where it was seen in a few cells, but not at the cell membrane.
Figure 9. Membranous TRKC is overexpressed in basal cell carcinoma.
A skin cancer tumour microarray with 23 BCCs, 19 SCCs and 5 malignant melanomas with two cores per case was immunostained with antibodies against TRKA, TRKB and TRKC. (a) Strong circumferential membranous immunostaining of TRKC is seen in 16 out of 23 cases (70%) of basal cell carcinoma on a tissue microarray, whilst there is an absence of membranous TRKA and TRKB staining. (b) One squamous cell carcinoma demonstrated a similar pattern of staining. Malignant melanomas did not display membranous staining. (c-d) 39% of BCC and 16% of SCC also demonstrated perinuclear TRKA staining, and 13% of BCCs only demonstrated faint cytoplasmic TRKB staining. (e) Percentage of cases with membranous TRKC staining. (f) Mean membranous staining scores for TRKC were performed. Membranous TRKC staining is seen in 70% of BCC in this series, 5% of SCC, but not in malignant melanoma. The error bars indicate the standard error of the mean.
Discussion
CYLD mutation carriers face repeated, disfiguring surgery to control tumour burden, and in a study of two large pedigrees, up to one in four affected patients underwent total scalp removal (Rajan et al., 2009). Painful eccrine spiradenomas, the development of conductive deafness caused by occlusive tumours in the ear canal and the risk of malignant transformation (Gerretsen et al., 1993) justify the development of non-surgical alternatives for this patient group. Here, we performed an unbiased approach using genomic and transcriptomic analysis of CYLD defective tumours. We demonstrated that cylindroma and spiradenoma tumours were genomically similar and stable, with LOH at 16q found in the majority of tumours in this study. The genomic stability seen across 16 tumours could suggest homogeneity in the mechanisms of pathogenesis and potentially in the response to therapeutic agents. Moreover, it seems possible that genomically stable tumours may be less liable to the acquisition of resistance mechanisms that arise through genomic diversity. We detected upregulation of transcripts of TRKB and TRKC and their cognate ligands NT3/NT4 and BDNF in cylindroma and spiradenoma samples. This was of particular interest as CYLD, a deubiquitinase with K63 linked ubiquitin specificity, has previously been shown to deubiquitinate TRKA and inhibit its internalisation from the cell membrane and subsequent downstream signalling via the phosphatidylinositol-3-kinase (PI3K) pathway (Geetha et al., 2005). Furthermore, recent studies have shown that BDNF is a NFκB target gene that may afford a neuroprotective role in astrocytes, supporting the finding that this TRK ligand is overexpressed by the cylindroma cells resulting in an autocrine loop (Saha et al., 2006). Validation of protein expression using TRK specific antibodies demonstrated strong membranous expression of TRKB and TRKC in cylindromas and spiradenomas, when compared to perilesional epidermis and hair follicles. Finally, downstream targets of TRK signalling such as phosphorylated ERK and BCL2 were overexpressed in the tumours.
TRK receptors have received considerable attention as mediators of neuronal growth, survival and maintenance (Huang and Reichardt, 2001), and have been shown to be expressed in hair follicles (Adly et al., 2005; Blasing et al., 2005). Their role in cancer was initially shown by uncommon oncogenic fusions involving C-terminal sequences of the NTRK1/NGF receptor gene with 5′ terminal sequences of various activating genes, such as TPM3, TPR and TFG in colon and papillary thyroid carcinoma (Butti et al., 1995; Martin-Zanca et al., 1986). Exploitation of the constitutively active prosurvival signalling that is usually restricted to neurons was thought to confer an advantage for the cancer cells. Recent advances in the understanding of complex signalling pathways involved in cancer (Luo et al., 2009) has suggested that some neural derived cancer cells, such as neuroblastoma, may become dependent on TRK signalling for survival, independent of mutations in TRK (Matsumoto et al., 1995; Scala et al., 1996). Subsequent demonstration of TRK expression associated with a survival advantage in epithelial tumours including breast (Lagadec et al., 2009), prostate (Weeraratna et al., 2000), lung (Ricci et al., 2001), extended the role of TRK signalling to non-neural derived tumours. At a genomic level, we established that neither TRKB and TRKC amplification, nor that of BDNF and NT3/4 was demonstrated to account for the overexpression seen in cylindroma tumours, suggesting that CYLD dysfunction perturbs normal TRK homeostasis.
The differential expression of TRKA, TRKB and TRKC in these tumours is intriguing. Whilst TRKA is regulated by CYLD, the interaction of CYLD with TRKB and TRKC remain to be clarified. The low perinuclear expression of TRKA, but conversely the high membranous expression of TRKB and TRKC suggests that not all TRKs are regulated by CYLD in the same manner. This has been shown to be true with other forms of ubiquitin modification including monoubiquitunation (Arévalo et al., 2006) or Lys48 polyubiquitination (Geetha et al., 2005). Arévalo et al. demonstrated that an E3 ubiquitin ligase, Nedd4-2, associated with the TRKA receptor and was phosphorylated upon NGF binding whilst Nedd4-2 did not bind or ubiquitinate related TRKB receptors, due to the lack of a consensus PPXY motif. These results indicate that TRK receptors are differentially regulated by ubiquitination to modulate the survival of neurons (Arévalo et al., 2006). Interestingly, Geetha et al. have demonstrated that the scaffold protein p62 which facilitates the interaction between CYLD and TRKA, is able to bind to specific lysine residues on TRKB (Lys 811) and TRKC (Lys 602 and 815) and mutation of these sites disrupted downstream signalling (Geetha et al., 2008). This suggests the possibility that CYLD could interact with TRKB and TRKC, but the precise mechanism of TRK specific homeostasis remains to be clarified.
We established TRK signalling was effective in cylindroma primary cell culture using recombinant human BDNF and NT3 for TRKB and TRKC respectively, and demonstrated increased expression of phosphorylated ERK and BCL2. To delineate the advantage conferred by TRKB and TRKC overexpression, we used a lentiviral mediated knockdown system. This demonstrated reduced colony formation in TRKB and TRKC deficient cells supporting the pathogenic role of these receptors.
We attempted to investigate if the catalytic activity of CYLD was implicated in the perturbed TRK signalling seen in these cultures. However, reintroduction of wild type-CYLD resulted in reduced cell viability preventing further investigation in this model (Supplementary Figure 5). The dependence of perturbed TRK signalling seen on NFκB and JNK signalling was investigated by growing primary cultures in NFκB and JNK inhibitors, however we were unable to conclusively show a clear effect on TRK levels (data not shown).
To investigate whether dysregulated TRK signalling in patients with truncating CYLD mutations represented a druggable pathway, we employed a 3D primary cell culture model. As seen in breast cancer (Lagadec et al., 2009), TRK expression in vitro was low when 2D cylindroma cell culture was compared to tumours. 3D culture was associated with an increase in expression of TRKB and TRKC, allowing us to use this as an in vitro model to assay cell viability in the presence of TRK inhibitors. The reduction in viability at micromolar concentrations of TRK inhibitors is encouraging, particularly as these tumours are amenable to transcutaneous or intralesional drug delivery. We attempted to determine whether the mechanism of action of TRK inhibition in our primary cell culture model was mediated by ERK or BCL2. This analysis proved not to be possible because of the high sensitivity of the primary cells to TRK inhibitors, resulting in only very few live cells after TRK inhibitor exposure. The result seen with lestaurtinib is particularly interesting as this agent was recently found in a screen of 2800 compounds to be a potent inhibitor of NFκB, suggesting it may target two key pathways in CYLD mutant tumours (Miller et al., 2010). Lestaurtinib is currently in phase 3 clinical trials (Shabbir and Stuart, 2010), and serum levels in patients on a 80 mg oral dose twice a day was 17-25 μm after taking the drug for 28 days (Marshall et al., 2005), suggesting that the sensitivity of CYLD deficient tumours suggested by our model may be achievable in patients. As novel agents with increased pan TRK specificity such as AZ-23 become available (Thresset al., 2009), patients with truncating CYLD mutations may be another step closer to non-surgical alternatives for this disfiguring disease.
We chose to investigate the expression of TRK in other human skin cancer models. The over expression of membranous TRKC in a high proportion of basal cell carcinoma is interesting. In sporadic BCC, LOH at 9p, the locus for PTCH1, has been found in the majority (93% in this series) of basal cell carcinoma (Teh et al., 2005), whilst 16q, the locus for CYLD, appears rarely affected. TRKC signalling in non melanoma skin cancer has not been characterised, but there are data from other models to support that TRK signalling could confer a proliferative advantage to cells where constitutively active SHH signalling following loss of functional PTCH could occur. Synergy between NT3, the cognate ligand for TRKC, and SHH has already been demonstrated in murine motorneurons where both are important for growth, and use of NT3 antibodies in this system reduces growth of these neurons (Dutton et al., 1999). TRKC also directly interacts with bone morphogenetic signalling receptors (BMPRI) in murine epithelial cells, and inhibits the tumour suppressor effects of BMP signalling (Jin et al., 2007) in a mouse colon cancer model. BMP and SHH signalling are tightly balanced, and BMP2, the ligand of BMPRI has been shown to oppose SHH mediated proliferation in murine cerebellar granular precursor cells via regulation of N-myc (Alvarez-Rodríguez et al., 2007). Furthermore, BMP signalling is protective in cutaneous carcinoma, and a SMAD 4 conditional knockout expressed in murine keratinocytes has indeed shown to result in spontaneous squamous and basal cell carcinomas (Qiao et al., 2006). It is hence conceivable that TRKC mediated inhibition of BMP signalling, on a mutated PTCH background in basal cell carcinoma may play a pathogenic role. The localisation of TRKC at the invasive margin of basal cell carcinoma tumour islands suggests that this role may be in invasion (Supplementary Figure 2). The mechanism that results in TRKC overexpression in basal cell carcinoma however remains to be explored. However, TRK receptors may represent a novel therapeutic target both in tumours with loss of CYLD function but also for basal cell carcinoma, the commonest cancer in humans.
Materials and Methods
Tumour and blood samples
Tumour tissue was obtained from patients undergoing surgery indicated to control tumour burden or for symptomatic relief, under regional ethical committee approval (REC REF:06/1001/59). Patients provided blood samples from which genomic DNA was extracted. Patients had been genotyped from all three pedigrees, and were confirmed to have the same mutation when entered into the study. These mutations were c.2460delc, c.2469+1G>A and c.2290del5 (Accession No. NM_015247).
Sample microdissection
All samples for microarray studies were snap frozen immediately after surgery, with detailed location maps of the tumours made at the time. Tumour blocks were mounted in optimal cutting temperature compound and sectioned on a cryostat at 12 μm onto RNAse free Superfrost coated slides (VWR). All subsequent steps were performed in RNAse free solutions. Tissue sections were fixed in 100% ethanol at −20°C for 2 minutes and then washed in 70% DEPC treated ethanol and 50% DEPC treated ethanol for 2 further minutes. Sections were stained with crystal violet solution (Ambion) for 40 seconds and then dehydrated in increasing concentrations of ethanol. Slides were then stored on dry ice until needle microdissection was performed. Microdissection was performed at 4°C using a Leica dissecting microscope. The dissected samples were placed directly into Trizol ® (Invitrogen UK). Needle homogenisation was performed and the sample was frozen at −80°C until RNA and DNA extraction was performed.
RNA and DNA extraction from tissue
Trizol® (Invitrogen UK) was used to extract RNA and DNA from the samples. The standard supplied protocol was used, with glycogen added to aid precipitation of RNA into the aqueous phase. RNA was quantified using a Nanodrop spectrophotometer (Nanodrop). RNA integrity analysis was performed using a chip based electrophoresis kit to confirm preservation of the 18S and 28S ribosomal bands (RNA 6000 Nano and Pico Chip kits; Agilent 2100 Bioanalyzer).
DNA extraction from peripheral blood
Genomic DNA from peripheral blood was extracted using the QIAGEN whole blood kit according to manufacturer’s protocols.
Array CGH
The 32K BAC re-array collection (CHORI) tiling path aCGH platform was constructed at the Breakthrough Breast Cancer Research Centre, as described earlier (Marchio et al., 2008). This type of BAC array platform has been shown to be as robust as and to have comparable resolution with high- density oligonucleotide arrays. DNA labelling, array hybridisations and image acquisition were performed as described earlier (Turner et al., 2010). aCGH data were pre-processed and analysed using an in-house R script (BACE.R) in R version 2.9.0 as previously described(Natrajan et al., 2009). After filtering polymorphic BACs, a final data set of 31544 clones with unambiguous mapping information according to the August 2009 build (hg19) of the human genome (http://www.ensembl.org) was smoothed using the circular binary segmentation (cbs) algorithm (Natrajan et al., 2009). A categorical analysis was applied to the BACs after classifying them as representing amplification (>0.45), gain (>0.12 and <0.44), loss (< −0.12), or no-change according to their cbs-smoothed Log2 ratio values. ArrayCGH data were deposited at ROCK (http://brcabase.icr.ac.uk/index.jsp), at URL: http://rock.icr.ac.uk/collaborations/Rajan/Cylindroma.
SNP typing
Genomic DNA was extracted from microdissected tissue and assayed on an Illumina 550K SNP array according to manufacturers protocols. SNP array data were deposited at ROCK (http://brcabase.icr.ac.uk/index.jsp), at URL: http://rock.icr.ac.uk/collaborations/Rajan/Cylindroma.
Restriction enzyme allelotyping
Polymerase chain reaction (PCR) was performed using primers (5′-3)′ CYLD18F –GAGAGCTTAAGCAGATGGAA and CYLD 18R –TAAACAGAAAAGGCAAAAGC that flanked exon 18, and the PCR product was subject to restriction enzyme digestion using HpyCH4V (c.2460delc) and Sml I (c.2469+1G>A). Digested products were separated on an agarose gel and visualised using ethidium bromide on an ultraviolet transilluminator. LOH was determined by the absence of a band that corresponded with loss of the wild-type allele in both cases.
Gene expression profiling
The Illumina WG-DASL platform was used for each sample that allowed for monitoring of gene expression of 24,526 transcripts. 50ng of total RNA from each sample was converted to cDNA as per the supplied protocol and then hybridised to Illumina Human 8 v3 chips. A confocal Illumina Infinuim bead reader then scanned the chips and the results were extracted using Illumina Beadstudio software. Microarray gene expression profiles were deposited at Arrayexpress (www.ebi.ac.uk) at accession number E-MTAB-352.
Data normalisation and analysis
Raw gene expression values were normalised using quantile normalisation. Differential expression analysis was performed using Beadstudio software to detect significantly different expressed genes. Differences in signal values for each gene was scored with a differential score (DS). Bonferroni testing was performed to correct for multiple hypothesis testing, and the results were ranked in order of differential score with a differential score ranging from +374 to −374, with +/−65 corresponding to a p value cut off of <0.01. Signal values from genes that fulfilled this level of significance were analysed for fold change, and results were tabulated in Table 1. Pathway discovery was further aided by Ingenuity pathway analysis software. Finally, a parallel analysis was carried out using gene set expression analysis (GSEA). For this analysis, probe values (24526) were collapsed to gene symbols (18630), with the max probe value used for replicate probes. After filtering, 867 gene sets were used in the analysis. Gene sets were permutated 1000 times, before being ranked in order using a “difference of classes” metric, with a weighted enrichment statistic applied. 56 gene sets were significantly enriched at a nominal p-value of <0.01.
Quantitative PCR
Total RNA was extracted from cylindroma primary cell cultures as described above, subject to DNAse digestion and then reverse transcribed using a Superscript III kit (Invitrogen) according to manufacturers instructions. cDNA was then used for quantitative PCR with the following primers (5′-3)′: (GAPDH F – ATGGGGAAGGTGAAGGTCG, GAPDH R –GGGGTCATTGATGGCAACAATA, NTRK2 F – TGTTCAGCACATCAAGCGACA, NTRK2 R – GCTCAGGACAGAGGTTATAGCAT, NTRK3 F TCCGTCAGGGACACAACTG, NTRK3 R –GCACACTCCATAGAACTTGACA) with a SYBR green containg Taq polymerase master mix (Sigma), in an ABI 7900HT thermal cycler. Gene expression was normalised to GAPDH and comparisons in expression were made using the 2−ΔΔCT formula as the PCR reactions had similar efficiencies of amplification.
Immunoblotting
Homogenised tumour samples, and lysates were sonicated in lysis buffer (65mM Tris, 50mM NaCl, 5mM EDTA, 1mM PMSF, 1mM Na orthovanadate, 0.1%SDS, 2% TritonX and 1% NP-40) with added phosphatase and protease inhibitors (Roche). 5-20 ug of total protein was loaded onto SDS-PAGE gels, and then transferred to PVDF membranes. Immunoblots were probed with the following antibodies: TRKB, TRKC, BCL2, p44/42, β-actin (Cell signalling Technology) and visualised with appropriate HRP conjugated secondary antibodies using ECL-Plus (GE healthcare). Immunoblots were exposed to Biomax XR film and processed routinely.
Tissue microarray (TMA)
A custom made tissue microarray was made from single 2mm cores from representative tumour areas and was used to validate protein expression in tissue sections. The TMA contained 10 cylindroma samples, 4 spiradenoma samples and 2 trichoepithelioma samples. Cylindromas and spiradenomas were chosen from both scalp and torso, and trichoepitheliomas were from the face. Human skin cancer TMA slides (A216) were purchased from Cepheid UK.
Immunohistochemistry (IHC) on paraffin embedded sections and TMAs
Tissue sections from the TMA were dewaxed with xylene and rehydrated in graded ethanols, before undergoing microwave antigen retrieval in sodium citrate buffer (pH 6.0). Tissue sections were then blocked with peroxidase blocking agent (DAKO) and then incubated with the following primary antibodies: TRKA, TRKB, TRKC, p44/42 (phospho ERK) and BCL2. Tissue sections were then washed in phosphate buffered saline and probed with secondary HRP-conjugated antibodies, and staining was visualised with 3,3′-Diaminobenzidine (DAB). Haematoxylin was used as a nuclear counterstain. Perilesional epidermis was used as a negative control, and anagen hair follicles in perilesional tissue served as a positive control for TRK antibodies.
IHC scoring of TRK expression
Tissue sections were assessed for staining in 5 randomly selected high power fields per core on the TMA. A modified version of an established score to assess membranous staining of Her-2 was developed (Kavanagh et al., 2009).To quantify TRK staining, tumour cells were scored as having circumferential strong membranous staining (3), strong non-circumferential staining (2), weak staining (1) or no staining (0). Proportion of such cells stained were counted per HPF on a 63x objective on a Leica DMR microscope and scored as follows: 75-100% of cells (3), 50-75% (2), and less than 50% (1). A total score of up to 6 was possible per field per core and scores were compared to perilesional keratinocytes for statistical significance using a t-test.
Cylindroma primary cell culture and drug inhibition experiments
Cylindroma samples were micro dissected using a Zeiss microscope following surgery and tumour pieces were enzymatically digested in 2.5% Trypsin. A cell suspension was then counted for viable cells using Trypan blue and the cells were plated in collagen coated tissue culture plasticware and grown in keratinocyte serum free medium. Lentiviral transfection was performed with shRNA against TRKB, TRKC and compared to a non-silencing control (Openbiosystems). Cells were grown in puromycin selection media till all cells were green fluorescent protein positive and then used for experiments. For colony forming assays, cells were plated at a low density of 4000 cells per well of a 6 well plate. These cells were maintained for 10-14 days, before cells were fixed in 3.3% trichloroacetic acid, and stained with 0.57% sulphorhodamine in 1% acetic acid. Plates were then scanned and colonies with more than 10 cells were counted. For short term viability assays, cells were grown in a 96 well format for 96 hours, before cell viability was assayed using a luminescent assay that lysed cells and used liberated ATP as readout for viability (CellTiter Glo - Promega). This was carried out according to standard manufacturers protocols and luminescence was read on a luminometer (Luminoskan – Ascent). For 3D cultures, cells were seeded on a polystyrene collagen coated scaffold (Reinnervate, Durham, UK), grown for 28 days and then treated with drugs. Cells were grown in the following inhibitors were used, diluted in DMSO, with a final concentration of DMSO at 0.1%: K252a and Tyrphostin AG879, Salicylic acid (SIGMA), Lestaurtinib (Toronto Research Chemicals). After 14 days, CellTiter Glo (Promega) was added to the media according to standard manufacturers protocols and luminescence was read on a luminometer (Luminoskan – Ascent). All experiments were done in triplicate.
FACS analysis
Cells were trypsinised, resuspended in PBS and fixed by adding ice cold ethanol whilst gently vortexing the tube, until a final concentration of 70% ethanol was reached. Cells were fixed for 1 hour at 4° C. Cells were then pelleted and resuspended in Cystain Solution (Partech, UK) which stained the nuclear DNA with DAPI. Cells were stained for 1 hour in the dark at 4° C, before analysis on a FACS Canto II flow cytometer (Becton Dickson, UK) using a UV laser. Cells were analysed for total DNA content and a minimum of 10000 events were used for cell cycle data. All cell cycle data was analysed using Modfit (Verity Software House, USA)
Supplementary Material
Expression levels of 24K genes from 32 microdissected tumours were pooled and compared against 10 pooled perilesional controls and subject to gene set expression analysis. Sets of genes recognised to modulate apoptosis were found to be enriched in the tumour sample. Leading edge analysis of these gene sets revealed NFκB target genes to be frequently present in these samples.
A skin cancer tissue microarray containing 23 BCCs, 19 SCCs and 5 malignant melanomas with two cores per case was immunostained with antibodies against TRKA, TRKB and TRKC. (a-j) TRKA and TRKC staining varied across basal cell carcinoma tumour samples, with membranous TRKC seen at the invasive edge adjacent to stroma, indicated with white arrows Figures S2 (a,c,f and h) are enlarged in (d,e,i and j). TRKA staining was cytoplasmic, but predominantly perinuclear at the invasive front, indicated with black arrows. Figures S2 (a-c, f-h) were taken at 5x original magnification and Figures S2 (d-e, i-j) were taken at 20x original magnification. Scale bars represent a distance of 100μm.
Clustering of transcriptomes using individual probes (24526) was employed and dissimilarity was measured using the correlation method using a statistical package (pvclust) (Suzuki and Shimodaira, 2006). Clusters seen in Figure 2 were maintained, however due to the small numbers of tumours used, statistical significance was seen between tumours and controls (p<0.01), but not between tumour subsets. pvclust provides two types of p-values: AU (Approximately Unbiased – numbers in red) p-value and BP (Bootstrap Probability – numbers in green) value. AU p-value, which is computed by multiscale bootstrap resampling, is a better approximation to the true unbiased p-value than the BP value computed by normal bootstrap resampling. Numbers in grey are edge values.
Significance of microarrays analysis, a form of supervised clustering analysis was performed between 19 cylindroma and 9 spiradenoma samples, and showed no significant difference between samples types.
Cells were plated in equal numbers and transfected with empty vector, wild type CYLD and red fluorescent protein, and selected with puromycin. At Day 5 post selection, cells were trypsinised and viable cells counted. (a) Expression of CYLD was assessed using immunoblotting. (b) Small numbers of viable cells were seen with CYLD reintroduction. (c) FACS analysis was performed to determine changes in cell cycle and highlighted a sub G0/G1 peak suggestive of apoptosis. The experiments were performed in three different primary cell cultures derived from separate tumours.
Table 5. Membranous TRKA, TRKB and TRKC expression in sporadic skin cancers.
A skin cancer tissue microarray was scored for the presence of membranous TRK staining.
| BCC | SCC | MM | |
|---|---|---|---|
| TRKA | 0/23 | 0/19 | 0/5 |
| TRKB | 0/23 | 0/19 | 0/5 |
| TRKC | 16/23 | 1/19 | 0/5 |
Acknowledgements
This work was funded in part by grants from the Newcastle Healthcare Charities Trust, the North East Skin Research Fund, Breakthrough Breast Cancer, Cancer Research UK and the Medical Research Council. Neil Rajan is a MRC Clinical Training Fellow.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest
References
- Adly MA, Assaf HA, Nada EA, Soliman M, Hussein M. Human scalp skin and hair follicles express neurotrophin-3 and its high-affinity receptor tyrosine kinase C, and show hair cycle-dependent alterations in expression. Br J Dermatol. 2005;153:514–20. doi: 10.1111/j.1365-2133.2005.06763.x. [DOI] [PubMed] [Google Scholar]
- Alvarez-Rodríguez R, Barzi M, Berenguer J, Pons S. Bone morphogenetic protein 2 opposes Shh-mediated proliferation in cerebellar granule cells through a TIEG-1-based regulation of Nmyc. J Biol Chem. 2007;282:37170–80. doi: 10.1074/jbc.M705414200. [DOI] [PubMed] [Google Scholar]
- Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–30. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- April C, Klotzle B, Royce T, Wickham-Garcia E, Boyaniwsky T, Izzo J, et al. Whole-genome gene expression profiling of formalin-fixed, paraffin-embedded tissue samples. PLoS ONE. 2009;4:e8162. doi: 10.1371/journal.pone.0008162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arévalo JC, Waite J, Rajagopal R, Beyna M, Chen Z-Y, Lee FS, et al. Cell survival through Trk neurotrophin receptors is differentially regulated by ubiquitination. Neuron. 2006;50:549–59. doi: 10.1016/j.neuron.2006.03.044. [DOI] [PubMed] [Google Scholar]
- Blasing H, Hendrix S, Paus R. Pro-inflammatory cytokines upregulate the skin immunoreactivity for NGF, NT-3, NT-4 and their receptor, p75NTR in vivo: a preliminary report. Arch Dermatol Res. 2005;296:580–4. doi: 10.1007/s00403-005-0563-y. [DOI] [PubMed] [Google Scholar]
- Bowen S, Gill M, Lee DA, Fisher G, Geronemus RG, Vazquez ME, et al. Mutations in the CYLD gene in Brooke-Spiegler syndrome, familial cylindromatosis, and multiple familial trichoepithelioma: lack of genotype-phenotype correlation. J Invest Dermatol. 2005;124:919–20. doi: 10.1111/j.0022-202X.2005.23688.x. [DOI] [PubMed] [Google Scholar]
- Bruckner-Tuderman L, Pfaltz M, Schnyder UW. Cylindroma overexpresses collagen VII, the major anchoring fibril protein. J Invest Dermatol. 1991;96:729–34. doi: 10.1111/1523-1747.ep12470965. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- Butti MG, Bongarzone I, Ferraresi G, Mondellini P, Borrello MG, Pierotti MA. A sequence analysis of the genomic regions involved in the rearrangements between TPM3 and NTRK1 genes producing TRK oncogenes in papillary thyroid carcinomas. Genomics. 1995;28:15–24. doi: 10.1006/geno.1995.1100. [DOI] [PubMed] [Google Scholar]
- Chen-Tsai CP, Colome-Grimmer M, Wagner RF. Correlations among neural cell adhesion molecule, nerve growth factor, and its receptors, TrkA, TrkB, TrkC, and p75, in perineural invasion by basal cell and cutaneous squamous cell carcinomas. Dermatologic surgery : official publication for American Society for Dermatologic Surgery [et al] 2004;30:1009–16. doi: 10.1111/j.1524-4725.2004.30306.x. [DOI] [PubMed] [Google Scholar]
- Coe BP, Ylstra B, Carvalho B, Meijer GA, Macaulay C, Lam WL. Resolving the resolution of array CGH. Genomics. 2007;89:647–53. doi: 10.1016/j.ygeno.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Demchenko YN, Glebov OK, Zingone A, Keats JJ, Bergsagel PL, Kuehl WM. Classical and/or alternative NF B pathway activation in multiple myeloma. Blood. 2010;115:3541–52. doi: 10.1182/blood-2009-09-243535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan J. Review of the hair follicle origin hypothesis for basal cell carcinoma. Dermatologic surgery : official publication for American Society for Dermatologic Surgery [et al] 2009;35:1311–23. doi: 10.1111/j.1524-4725.2009.01236.x. [DOI] [PubMed] [Google Scholar]
- Dunning MJ, Barbosa-Morais NL, Lynch AG, Tavaré S, Ritchie ME. Statistical issues in the analysis of Illumina data. BMC Bioinformatics. 2008;9:85. doi: 10.1186/1471-2105-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton R, Yamada T, Turnley A, Bartlett PF, Murphy M. Regulation of spinal motoneuron differentiation by the combined action of Sonic hedgehog and neurotrophin 3. Clin Exp Pharmacol Physiol. 1999;26:746–8. doi: 10.1046/j.1440-1681.1999.03108.x. [DOI] [PubMed] [Google Scholar]
- Gao J, Huo L, Sun X, Liu M, Li D, Dong JT, et al. The tumor suppressor CYLD regulates microtubule dynamics and plays a role in cell migration. J Biol Chem. 2008;283:8802–9. doi: 10.1074/jbc.M708470200. [DOI] [PubMed] [Google Scholar]
- Geetha T, Jiang J, Wooten MW. Lysine 63 polyubiquitination of the nerve growth factor receptor TrkA directs internalization and signaling. Mol Cell. 2005;20:301–12. doi: 10.1016/j.molcel.2005.09.014. [DOI] [PubMed] [Google Scholar]
- Geetha T, Seibenhener ML, Chen L, Madura K, Wooten MW. p62 serves as a shuttling factor for TrkA interaction with the proteasome. Biochem Biophys Res Commun. 2008;374:33–7. doi: 10.1016/j.bbrc.2008.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerretsen AL, van der Putte SC, Deenstra W, van Vloten WA. Cutaneous cylindroma with malignant transformation. Cancer. 1993;72:1618–23. doi: 10.1002/1097-0142(19930901)72:5<1618::aid-cncr2820720521>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Gunnarsson R, Staaf J, Jansson M, Ottesen AM, Goransson H, Liljedahl U, et al. Screening for copy-number alterations and loss of heterozygosity in chronic lymphocytic leukemia--a comparative study of four differently designed, high resolution microarray platforms. Genes Chromosomes Cancer. 2008;47:697–711. doi: 10.1002/gcc.20575. [DOI] [PubMed] [Google Scholar]
- Hellerbrand C, Bumes E, Bataille F, Dietmaier W, Massoumi R, Bosserhoff AK. Reduced expression of CYLD in human colon and hepatocellular carcinomas. Carcinogenesis. 2007;28:21–7. doi: 10.1093/carcin/bgl081. [DOI] [PubMed] [Google Scholar]
- Hoseong Yang S, Andl T, Grachtchouk V, Wang A, Liu J, Syu L, et al. Pathological responses to oncogenic Hedgehog signaling in skin are dependent on canonical Wnt/β-catenin signaling. Nat Genet. 2008;40:1130–1135. doi: 10.1038/ng.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illumina Inc. Beadstudio Gene Expression Module v3.4 User Guide (113172615 Rev A) San Diego, CA: 2009. [Google Scholar]
- Jenner, Leone, Walker, Ross, Johnson, Gonzalez, et al. Gene mapping and expression analysis of 16q loss of heterozygosity identifies WWOX and CYLD as being important in determining clinical outcome in multiple myeloma. Blood. 2007;110:3291–300. doi: 10.1182/blood-2007-02-075069. [DOI] [PubMed] [Google Scholar]
- Jin W, Yun C, Kim H-S, Kim S-J. TrkC binds to the bone morphogenetic protein type II receptor to suppress bone morphogenetic protein signaling. Cancer Res. 2007;67:9869–77. doi: 10.1158/0008-5472.CAN-07-0436. [DOI] [PubMed] [Google Scholar]
- Kavanagh DO, Chambers G, O’Grady L, Barry KM, Waldron RP, Bennani F, et al. Is overexpression of HER-2 a predictor of prognosis in colorectal cancer? BMC Cancer. 2009;9:1. doi: 10.1186/1471-2407-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keats J, Fonseca R, Chesi M, Schop R, Baker A, Chng W, et al. Promiscuous Mutations Activate the Noncanonical NF-κB Pathway in Multiple Myeloma. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuno N, Shiina H, Urakami S, Kawamoto K, Hirata H, Tanaka Y, et al. Genistein mediated histone acetylation and demethylation activates tumor suppressor genes in prostate cancer cells. Int J Cancer. 2008;123:552–60. doi: 10.1002/ijc.23590. [DOI] [PubMed] [Google Scholar]
- Komander D, Lord CJ, Scheel H, Swift S, Hofmann K, Ashworth A, et al. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol Cell. 2008;29:451–64. doi: 10.1016/j.molcel.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–5. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- Kriegl L, Horst D, Kirchner T, Jung A. LEF-1 expression in basal cell carcinomas. Br J Dermatol. 2009;160:1353–6. doi: 10.1111/j.1365-2133.2009.09144.x. [DOI] [PubMed] [Google Scholar]
- Lagadec C, Meignan S, Adriaenssens E, Foveau B, Vanhecke E, Romon R, et al. TrkA overexpression enhances growth and metastasis of breast cancer cells. 2009;28:1960–1970. doi: 10.1038/onc.2009.61. [DOI] [PubMed] [Google Scholar]
- Lamb J. The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Liu H-H, Xie M, Schneider MD, Chen ZJ. Essential role of TAK1 in thymocyte development and activation. Proc Natl Acad Sci USA. 2006;103:11677–82. doi: 10.1073/pnas.0603089103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–37. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchio C, Natrajan R, Shiu KK, Lambros MB, Rodriguez-Pinilla SM, Tan DS, et al. The genomic profile of HER2-amplified breast cancers: the influence of ER status. J Pathol. 2008;216:399–407. doi: 10.1002/path.2423. [DOI] [PubMed] [Google Scholar]
- Marshall JL, Kindler H, Deeken J, Bhargava P, Vogelzang NJ, Rizvi N, et al. Phase I trial of orally administered CEP-701, a novel neurotrophin receptor-linked tyrosine kinase inhibitor. Invest New Drugs. 2005;23:31–7. doi: 10.1023/B:DRUG.0000047103.64335.b0. [DOI] [PubMed] [Google Scholar]
- Martin-Zanca D, Hughes SH, Barbacid M. A human oncogene formed by the fusion of truncated tropomyosin and protein tyrosine kinase sequences. Nature. 1986;319:743–8. doi: 10.1038/319743a0. [DOI] [PubMed] [Google Scholar]
- Massoumi R. Cyld Inhibits Tumor Cell Proliferation by Blocking Bcl-3-Dependent NF-κB Signaling. Cell. 2006;125:665–677. doi: 10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell. 2006;125:665–77. doi: 10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- Massoumi R, Kuphal S, Hellerbrand C, Haas B, Wild P, Spruss T, et al. Down-regulation of CYLD expression by Snail promotes tumor progression in malignant melanoma. Journal of Experimental Medicine. 2009;206:221–232. doi: 10.1084/jem.20082044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Wada RK, Yamashiro JM, Kaplan DR, Thiele CJ. Expression of brain-derived neurotrophic factor and p145TrkB affects survival, differentiation, and invasiveness of human neuroblastoma cells. Cancer Res. 1995;55:1798–806. [PubMed] [Google Scholar]
- Meybehm M, Fischer HP. Spiradenoma and dermal cylindroma: comparative immunohistochemical analysis and histogenetic considerations. Am J Dermatopathol. 1997;19:154–61. doi: 10.1097/00000372-199704000-00009. [DOI] [PubMed] [Google Scholar]
- Miller SC, Huang R, Sakamuru S, Shukla SJ, Attene-Ramos MS, Shinn P, et al. Identification of known drugs that act as inhibitors of NF-kappaB signaling and their mechanism of action. Biochem Pharmacol. 2010;79:1272–80. doi: 10.1016/j.bcp.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natrajan R, Weigelt B, Mackay A, Geyer FC, Grigoriadis A, Tan DSP, et al. An integrative genomic and transcriptomic analysis reveals molecular pathways and networks regulated by copy number aberrations in basal-like, HER2 and luminal cancers. Breast Cancer Res Treat. 2009;121:575–589. doi: 10.1007/s10549-009-0501-3. [DOI] [PubMed] [Google Scholar]
- Nilsson M, Undèn AB, Krause D, Malmqwist U, Raza K, Zaphiropoulos PG, et al. Induction of basal cell carcinomas and trichoepitheliomas in mice overexpressing GLI-1. Proc Natl Acad Sci USA. 2000;97:3438–43. doi: 10.1073/pnas.050467397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterkamp HM, Neering H, Nijman SM, Dirac AM, Mooi WJ, Bernards R, et al. An evaluation of the efficacy of topical application of salicylic acid for the treatment of familial cylindromatosis. Br J Dermatol. 2006;155:182–5. doi: 10.1111/j.1365-2133.2006.07224.x. [DOI] [PubMed] [Google Scholar]
- Qiao W, Li AG, Owens P, Xu X, Wang X-J, Deng C-X. Hair follicle defects and squamous cell carcinoma formation in Smad4 conditional knockout mouse skin. Oncogene. 2006;25:207–17. doi: 10.1038/sj.onc.1209029. [DOI] [PubMed] [Google Scholar]
- Rajan N, Langtry JAA, Ashworth A, Roberts C, Chapman P, Burn J, et al. Tumor mapping in 2 large multigenerational families with CYLD mutations: implications for disease management and tumor induction. Archives of dermatology. 2009;145:1277–84. doi: 10.1001/archdermatol.2009.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiley W, Zhang M, Sun S-C. Negative regulation of JNK signaling by the tumor suppressor CYLD. J Biol Chem. 2004;279:55161–7. doi: 10.1074/jbc.M411049200. [DOI] [PubMed] [Google Scholar]
- Reiley WW, Jin W, Lee AJ, Wright A, Wu X, Tewalt EF, et al. Deubiquitinating enzyme CYLD negatively regulates the ubiquitin-dependent kinase Tak1 and prevents abnormal T cell responses. J Exp Med. 2007;204:1475–85. doi: 10.1084/jem.20062694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiley WW, Zhang M, Jin W, Losiewicz M, Donohue KB, Norbury CC, et al. Regulation of T cell development by the deubiquitinating enzyme CYLD. Nat Immunol. 2006;7:411–7. doi: 10.1038/ni1315. [DOI] [PubMed] [Google Scholar]
- Ricci A, Greco S, Mariotta S, Felici L, Bronzetti E, Cavazzana A, et al. Neurotrophins and neurotrophin receptors in human lung cancer. Am J Respir Cell Mol Biol. 2001;25:439–46. doi: 10.1165/ajrcmb.25.4.4470. [DOI] [PubMed] [Google Scholar]
- Saggar S, Chernoff KA, Lodha S, Horev L, Kohl S, Honjo RS, et al. CYLD mutations in familial skin appendage tumours. J Med Genet. 2008;45:298–302. doi: 10.1136/jmg.2007.056127. [DOI] [PubMed] [Google Scholar]
- Saha RN, Liu X, Pahan K. Up-regulation of BDNF in astrocytes by TNF-alpha: a case for the neuroprotective role of cytokine. J Neuroimmune Pharmacol. 2006;1:212–22. doi: 10.1007/s11481-006-9020-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini HS, Gorse KM, Boxer LM, Sato-Bigbee C. Neurotrophin-3 and a CREB-mediated signaling pathway regulate Bcl-2 expression in oligodendrocyte progenitor cells. J Neurochem. 2004;89:951–61. doi: 10.1111/j.1471-4159.2004.02365.x. [DOI] [PubMed] [Google Scholar]
- Scala S, Wosikowski K, Giannakakou P, Valle P, Biedler JL, Spengler BA, et al. Brain-derived neurotrophic factor protects neuroblastoma cells from vinblastine toxicity. Cancer Res. 1996;56:3737–42. [PubMed] [Google Scholar]
- Shabbir M, Stuart R. Lestaurtinib, a multitargeted tyrosine kinase inhibitor: from bench to bedside. Expert Opin Investig Drugs. 2010;19:427–36. doi: 10.1517/13543781003598862. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Sowa ME, Nalepa G, Gygi SP, Harper JW, Elledge SJ. The tumor suppressor CYLD regulates entry into mitosis. Proc Natl Acad Sci U S A. 2007;104:8869–74. doi: 10.1073/pnas.0703268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes A, Wakano C, Koblan-Huberson M, Adra CN, Fleig A, Turner H. TRPA1 is a substrate for de-ubiquitination by the tumor suppressor CYLD. Cell Signal. 2006;18:1584–94. doi: 10.1016/j.cellsig.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–2. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- Tan DS, Lambros MB, Natrajan R, Reis-Filho JS. Getting it right: designing microarray (and not ‘microawry’) comparative genomic hybridization studies for cancer research. Lab Invest. 2007;87:737–54. doi: 10.1038/labinvest.3700593. [DOI] [PubMed] [Google Scholar]
- Tauriello DVF, Haegebarth A, Kuper I, Edelmann MJ, Henraat M, Canninga-van Dijk MR, et al. Loss of the tumor suppressor CYLD enhances Wnt/beta-catenin signaling through K63-linked ubiquitination of Dvl. Mol Cell. 2010;37:607–19. doi: 10.1016/j.molcel.2010.01.035. [DOI] [PubMed] [Google Scholar]
- Teh M-T, Blaydon D, Chaplin T, Foot NJ, Skoulakis S, Raghavan M, et al. Genomewide single nucleotide polymorphism microarray mapping in basal cell carcinomas unveils uniparental disomy as a key somatic event. Cancer Res. 2005;65:8597–603. doi: 10.1158/0008-5472.CAN-05-0842. [DOI] [PubMed] [Google Scholar]
- Tellechea O, Reis JP, Ilheu O, Baptista AP. Dermal cylindroma. An immunohistochemical study of thirteen cases. Am J Dermatopathol. 1995;17:260–5. [PubMed] [Google Scholar]
- Thress K, Macintyre T, Wang H, Whitston D, Liu ZY, Hoffmann E, et al. Identification and preclinical characterization of AZ-23, a novel, selective, and orally bioavailable inhibitor of the Trk kinase pathway. Mol Cancer Ther. 2009;8:1818–27. doi: 10.1158/1535-7163.MCT-09-0036. [DOI] [PubMed] [Google Scholar]
- Timpl R, Fujiwara S, Dziadek M, Aumailley M, Weber S, Engel J. Laminin, proteoglycan, nidogen and collagen IV: structural models and molecular interactions. Ciba Found Symp. 1984;108:25–43. doi: 10.1002/9780470720899.ch3. [DOI] [PubMed] [Google Scholar]
- Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424:793–6. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- Trompouki E, Tsagaratou A, Kosmidis SK, Dollé P, Qian J, Kontoyiannis DL, et al. Truncation of the catalytic domain of the cylindromatosis tumor suppressor impairs lung maturation. Neoplasia. 2009;11:469–76. doi: 10.1593/neo.81424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunggal L, Ravaux J, Pesch M, Smola H, Krieg T, Gaill F, et al. Defective laminin 5 processing in cylindroma cells. Am J Pathol. 2002;160:459–68. doi: 10.1016/S0002-9440(10)64865-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N, Lambros MB, Horlings HM, Pearson A, Sharpe R, Natrajan R, et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010;29:2013–2023. doi: 10.1038/onc.2009.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeraratna AT, Arnold JT, George DJ, DeMarzo A, Isaacs JT. Rational basis for Trk inhibition therapy for prostate cancer. Prostate. 2000;45:140–8. doi: 10.1002/1097-0045(20001001)45:2<140::aid-pros8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Workman C, Jensen LJ, Jarmer H, Berka R, Gautier L, Nielser HB, et al. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-9-research0048. research0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Ramirez A, Cui B, Ding J, Delcroix J-DM, Valletta JS, et al. A functional dynein-microtubule network is required for NGF signaling through the Rap1/MAPK pathway. Traffic. 2007;8:1503–20. doi: 10.1111/j.1600-0854.2007.00636.x. [DOI] [PubMed] [Google Scholar]
- Zhang J, Stirling B, Temmerman ST, Ma CA, Fuss IJ, Derry JMJ, et al. Impaired regulation of NF-κB and increased susceptibility to colitis-associated tumorigenesis in CYLD-deficient mice. J Clin Invest. 2006;116:3042–3049. doi: 10.1172/JCI28746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Fields CR, Su N, Pan Y-X, Robertson KD. Pharmacologic inhibition of epigenetic modifications, coupled with gene expression profiling, reveals novel targets of aberrant DNA methylation and histone deacetylation in lung cancer. Oncogene. 2007;26:2621–34. doi: 10.1038/sj.onc.1210041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression levels of 24K genes from 32 microdissected tumours were pooled and compared against 10 pooled perilesional controls and subject to gene set expression analysis. Sets of genes recognised to modulate apoptosis were found to be enriched in the tumour sample. Leading edge analysis of these gene sets revealed NFκB target genes to be frequently present in these samples.
A skin cancer tissue microarray containing 23 BCCs, 19 SCCs and 5 malignant melanomas with two cores per case was immunostained with antibodies against TRKA, TRKB and TRKC. (a-j) TRKA and TRKC staining varied across basal cell carcinoma tumour samples, with membranous TRKC seen at the invasive edge adjacent to stroma, indicated with white arrows Figures S2 (a,c,f and h) are enlarged in (d,e,i and j). TRKA staining was cytoplasmic, but predominantly perinuclear at the invasive front, indicated with black arrows. Figures S2 (a-c, f-h) were taken at 5x original magnification and Figures S2 (d-e, i-j) were taken at 20x original magnification. Scale bars represent a distance of 100μm.
Clustering of transcriptomes using individual probes (24526) was employed and dissimilarity was measured using the correlation method using a statistical package (pvclust) (Suzuki and Shimodaira, 2006). Clusters seen in Figure 2 were maintained, however due to the small numbers of tumours used, statistical significance was seen between tumours and controls (p<0.01), but not between tumour subsets. pvclust provides two types of p-values: AU (Approximately Unbiased – numbers in red) p-value and BP (Bootstrap Probability – numbers in green) value. AU p-value, which is computed by multiscale bootstrap resampling, is a better approximation to the true unbiased p-value than the BP value computed by normal bootstrap resampling. Numbers in grey are edge values.
Significance of microarrays analysis, a form of supervised clustering analysis was performed between 19 cylindroma and 9 spiradenoma samples, and showed no significant difference between samples types.
Cells were plated in equal numbers and transfected with empty vector, wild type CYLD and red fluorescent protein, and selected with puromycin. At Day 5 post selection, cells were trypsinised and viable cells counted. (a) Expression of CYLD was assessed using immunoblotting. (b) Small numbers of viable cells were seen with CYLD reintroduction. (c) FACS analysis was performed to determine changes in cell cycle and highlighted a sub G0/G1 peak suggestive of apoptosis. The experiments were performed in three different primary cell cultures derived from separate tumours.