Abstract
An increasing number of anti-cancer therapeutic agents target specific mutant proteins that are expressed by many different tumor types. Recent evidence suggests that the selection of patients whose tumors harbor specific genetic alterations identifies the subset of patients that are most likely to benefit from the use of such agents. As the number of genetic alterations that provide diagnostic and/or therapeutic information increases, the comprehensive characterization of cancer genomes will be necessary to understand the spectrum of distinct genomic alterations in cancer, to identify patients who are likely to respond to particular therapies and to facilitate the selection of treatment modalities. Rapid developments in new technologies for genomic analysis now provide the means to perform comprehensive analyses of cancer genomes. Here we review the current state of cancer genome analysis and discuss the challenges and opportunities necessary to implement these technologies in a clinical setting.
Introduction
The Case for Individualized Cancer Medicine
Work from many laboratories has identified genetic alterations that occur at an appreciable frequency in specific types of cancers. For example, a reciprocal translocation between chromosome 9 and 22 known as the Philadelphia chromosome, resulting in the BCR-ABL fusion gene, occurs in ~95% of chronic myelogenous leukemias (CMLs) (1, 2); oncogenic KIT mutations are present in ~85% of gastrointestinal stromal tumors (GISTs) (3, 4); mutations in the serine-threonine kinase BRAF are present in >50% of cutaneous melanoma (5); activating mutations in the epidermal growth factor receptor (EGFR) have been identified in ~15% of non-small cell lung cancers (NSCLC) (6–8), and ERBB2 is amplified in 15–20% of breast cancers (9–12). Biochemical studies confirmed that these genetic alterations result in constitutively active molecules, and tumors that harbor such mutations depend on the activity of these proteins for survival.
Based on these observations, efforts to target these molecules with either small molecule inhibitors or antibodies have led to several agents that induce significant clinical responses. For example, the tyrosine kinase inhibitor (TKI) imatinib induces clinical responses in Philadelphia chromosome-positive CML (13) and GISTs that harbor KIT mutations (14, 15); PLX4032 has been shown to induce responses in cutaneous melanomas with BRAF mutations (16, 17); the EGFR TKIs erlotinib and gefitinib show activity in non-small cell lung cancers that harbor activating mutations and small insertions/deletions in EGFR (6–8); and the ERBB2 inhibitors trastuzumab and lapatinib show clinical responses in breast cancers with amplification/overexpression of ERBB2 (18). Moreover, the presence of mutations in proteins other than the intended therapeutic target can affect the response to a particular therapeutic regimen. As an example, lung and colorectal cancers that harbor mutations in EGFR as well as KRAS or BRAF fail to respond to treatment with anti-EGFR directed agents (19–22).
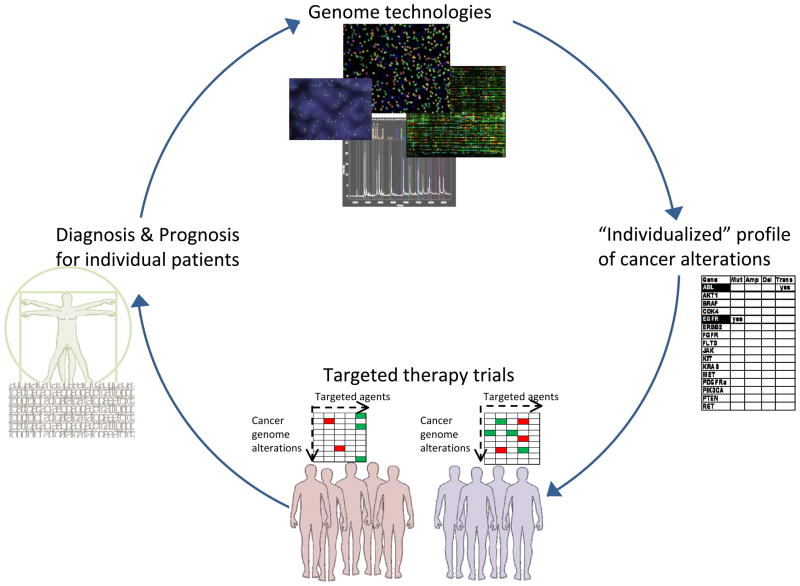
The field of molecularly-based individualized cancer care will thus be enabled and reinforced by a cyclical process (depicted in Figure 1) of selecting treatment for an individual patient based on the genetic expression, proteomic profiles, deregulated cellular pathways and/or somatic mutations in cancer cells of each individual patient, using this profile to accurately define the prognosis in these patients, and suggesting treatment options or clinical trials that are most likely to succeed (23, 24). However, further research and systematic screening of the cancer genome is needed to uncover the full spectrum of mutations that occur in both primary and recurrent tumors. Moreover, since few of the targeted agents developed to-date induce durable remissions, it is likely that combination therapies based on rational combinations of targeted agents will be necessary. In this review article, we focus on both the opportunities and the challenges presented by the implementation of new genomic technologies that provide the potential to interrogate cancer genomes in the clinical setting and transform the current cancer treatment paradigm.
Figure 1. Cycle of personalized cancer medicine.
Genomic technologies will increasingly be used in the generation of a profile of cancer alterations for an individual. This profile can be used (for example) to stratify patients for clinical trials with agents targeted to these specific alterations, thus leading to more effective treatment strategies. The paradigm of personalized cancer medicine cycles through to improve diagnosis and prognosis for each individual patient.
Current Molecular Diagnostic Strategies
Although the number of molecular tests required for clinical selection of patients for targeted therapy is increasing, the available technologies are limiting and demonstration of the benefit of selecting patients based on their molecular subtype in clinical trials is a lengthy process. Currently most cancers are categorized in terms of their tissues of origin, size of the primary lesion and the presence of metastatic lesions. For solid tumors, anatomic origin of a tumor is generally the decisive factor in assessing treatment options, and treatment choices are matched to patients based on factors such as primary tumor diagnosis or site, nodal status, histologic subtype, or hormone receptor status. In hematological malignancies, blood count and microscopy are standard diagnostic tools used to categorize lymphomas and leukemias by cell lineage; a number of these diseases can also be classified by cytogenetics [acute myeloid leukemia, CML) or immunophenotyping of malignant cells [lymphoma, myeloma, chronic lymphocytic leukemia (CLL)].
Currently technologies to profile samples for the clinical selection of patients for targeted therapies assess the mutational status of one or a few genes (capillary sequencing, pyrosequencing), or interrogate a specific histological or pathological phenotype (immunohistochemistry, fluorescence in-situ hybridization (FISH). Figure 2 highlights current and emerging clinical technologies to detect various cancer DNA alterations. For example, FISH is used to detect the translocation and resultant fusion of BCR and ABL in CML in clinical settings (25, 26). The BCR-ABL translocation is also found in acute lymphoblastic leukemia (ALL) at a lower frequency (25–30%); thus molecular characterization of BCR-ABL is of critical diagnostic importance in ALL where the presence or absence of this alteration mutation will dictate therapy. In a similar manner, amplification of ERBB2 in breast cancers (27) and fusions involving the anaplastic lymphoma kinase (ALK) gene (28) are detected by FISH and identify patients who are likely to respond to anti-ERBB2 agents or small molecule inhibitors of ALK, respectively.
Figure 2. Genome alterations, current tests and future technologies.
The major classes of genomic alterations that give rise to cancer, exemplary cancer genes for each category, and the current and emerging clinical technologies to detect these various types of alterations. TS- tumor suppressor; CML- chronic myelogenous leukemia; PCR- polymerase chain reaction; FISH- fluorescence in situ hybridization; IHC- immunohistochemistry.
Several additional tests have been developed for other oncogenes. Detection of nucleotide substitution mutations, insertion or deletions within the kinase domain of EGFR for gefitinib treatment is currently determined by capillary gel sequencing. KIT mutations in GISTs can be detected in the clinical setting using sequencing or quantitative PCR (4, 29). Recently, sequencing assays have been developed to detect mutations in BRAF (specifically V600 alterations), and agents targeting this alteration are currently being evaluated in clinical trials (30, 31). Table 1 lists the targeted therapeutic agents (available or in clinical trials) for exemplary cancer genes in each category of genomic alteration.
Beyond interrogating specific genes, several groups have explored the use of gene expression profiling to identify signatures that identify specific subtypes of cancers or predict the response to therapy. For example, Staudt and colleagues showed that expression profiling can distinguish between germinal center B-like (GCB) and activated B-cell-like (ABC) lymphoma as well as to identify poor prognosis in each group of patients (32, 33). Guidelines established by the American Society of Clinical Oncology (ASCO) and the National Comprehensive Cancer Network (NCCN) include the Oncotype DX assay, which interrogates a 21-gene signature, to predict benefit from chemotherapy as well as risk of recurrence in breast cancer patients (34). MammaPrint is a microarray test (35, 36) approved by the US Food and Drug Administration (FDA) to classify breast tumors for risk of recurrence. Further work is necessary to determine whether these and other signatures will become commonplace in clinical practice, as such tests require substantially different protocols for the preparation of samples than is usually employed by pathology laboratories and the full predictive value of these tests require further evaluation.
The technology revolution in genomics
The technological revolution in the fields of genomics began over 30 years ago with the development of the Sanger method for DNA sequencing (37), followed twenty years later by the development of microarrays (38). Mass-spectrometric genotyping emerged as a useful technology shortly afterwards (39), and initial next-generation sequencing methodologies were reported in 2005 (40, 41) (see Figure 3 for a timeline of technological advances and concomitant cancer genome landmark discoveries). Concomitant advances in the field of sequencing resulted in the emergence of several massively parallel sequencing technologies, which yield vastly greater amounts of data. These sequencing technologies have advanced DNA sequencing capacity at an unprecedented rate (40–43) (for a comprehensive review on second-generation sequencing, see Shendure and Ji, (44)), and lowered the cost per base of sequencing data by 10,000 fold in the last decade (45).
Figure 3. Technological advances and concomitant developments in cancer biology.
A timeline of the technological advances impacting cancer research in the past 100 years. The landmark discoveries enabled by these technologies are indicated above the line, and the number of these landmarks has increased dramatically in the past 10 years, due in large part to next-generation sequencing capabilities.
The completion of a highly-refined human genome sequence (46, 47) was a landmark achievement in establishing a baseline reference genome to which other sequences could be compared. The availability of such reference genomes has enabled the concomitant characterization of genomic alterations from many different diseases, including cancers (48). Technological advances in experimental and informatics methodologies over the past 10 years have allowed the characterization of cancer genomes. In initial studies, a gene-focused approach was employed using first-generation (Sanger) sequencing, which resulted in the identification of many driver events in cancers such as mutations of BRAF in melanoma (5), EGFR in NSCLC (6–8), JAK2 in myeloproliferative disorders (49, 50), FGFR2 in endometrial carcinoma (51), ALK in neuroblastoma (52, 53), and PIK3CA mutations in several cancers (54). However, with the development of the “second-generation” sequencing technologies, it is now feasible to sequence exomes (known exons in the genome), transcriptomes (expressed genes the genome) or whole genomes of cancer samples. Massively parallel DNA sequencing platforms have become widely available, and the number of both normal and cancer genomes that have been completed now numbers in the thousands (42, 55–59). International efforts to sequence normal [the 1000 Genomes Project (http://www.1000genomes.org)] as well as cancer genomes (ICGC) promises to provide a growing number of reference whole genome sequences. In the past 4 years for example, whole cancer genomes derived from AML (60, 61), lung (62–64), breast (65–67), melanoma (68) and multiple myeloma (69) have been reported in detail. Large scale cancer genome studies, such as The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC), are applying next-generation sequencing technologies to tumors from 50 different cancer types to generate more than 25,000 genomes at the genomic, transcriptomic, and epigenomic level, and will provide the foundation for a complete catalog of oncogenic mutations (70).
Translating Individualized Medicine to the Clinic
The result of this explosion in molecular characterization of cancer is an emerging vision for personalized cancer medicine. In this paradigm, patients will have the specific genomic alterations driving their tumor analyzed, and a targeted therapy or therapies will be recommended based on the genomic characterization of each tumor. This will maximize efficacy of treatment while minimizing undesirable side-effects. Significant challenges currently exist that will need to be overcome to realize the goal of tailored treatments based on tumor genomics. The challenges and opportunities facing scientists, researchers, clinicians and oncologists in the near future are the subject of this review. Three pivotal components will be required to enable a paradigm shift to personalized cancer medicine:
Every patient is profiled to identify genetic alterations present in a specific cancer.
Caregivers have access to this information in a form that provides relevant contextual information.
This information can be obtained in a timescale that permits the incorporation of this information into the decision-making process.
An increased understanding of the biological driver events for some cancers, coupled with advances in technologies used to detect somatic cancer alterations, have led to the establishment of some initial personalized cancer medicine programs at several cancer centers including the Dana-Farber/Brigham and Women’s Cancer Center (71, 72), Massachusetts General Hospital (73, 74), Memorial Sloan-Kettering Cancer Center (75–77), MD Anderson Cancer Center (78), Oregon Health & Science University (79), and Vanderbilt Cancer Center (73). Each of these programs uses a genotyping platform to profile patient samples for alterations in a panel of potentially “actionable” or druggable gene mutations that may inform a therapeutic paradigm for patients. Currently, these initiatives are limited by the number of genes interrogated, the type of somatic alteration investigated (mostly mutations and small insertions/deletions), throughput and cost of technology, and types of cancers being screened. Additional efforts are underway to translate the recent advances in genome technologies (such as massively parallel, whole-genome sequencing) into the clinical setting to guide patient treatment options.
Although advances in technology and the biological understanding of cancer pathogenesis has brought the goal of personalized cancer medicine closer, significant challenges remain, pertaining to each step described above, before this vision of individualized cancer care is actualized. These challenges are discussed in detail below.
Technological challenges in sample preparation
Diagnostic interventions that successfully introduce tumor mutation profiling to clinical practice must circumvent several technical and logistical hurdles. Important limitations in the deployment of genomic technologies in the clinic are the minute amounts of tumor material available for genomic profiling as the availability of tissue in forms amendable to available technologies.
A challenging barrier to large-scale genomic interrogation is the specimen itself. Many of the research-oriented, large-scale cancer sequencing efforts thus far focus on surgically resected samples that yield a large amount of tissue for analysis. In the absence of a surgical resection however, biopsies taken for diagnostic purposes are often small and are usual composed of both normal and malignant cells in a variable ratio. Moreover, for some common cancers such as breast and prostate cancers, primary tumors are often identified at early stage when tumors are small. Incorporation of genomic profiling into clinical decision-making will therefore require reliable genomic profiling of small specimens, diffuse biopsy samples, and fine-needle aspirates.
In addition to the availability of limiting quantities of material for genomic profiling, the quality of nucleic acids can also vary greatly from sample to sample. The standard practice of banking formalin-fixed, paraffin-embedded (FFPE) clinical specimens (whether surgical resections or biopsies) is advantageous for a clinical laboratory that performs histological diagnoses. The majority of specimens are fixed for purposes other than genetic characterization, and this process often results in degraded and/or chemically modified nucleic acids (80–82) that are unlikely to yield the quality or quantity of DNA necessary for many high-throughput genomic technologies (83). Thus, any clinical test must be able to accept as input nucleic acid material derived from FFPE and/or archival tumor specimens.
In addition, samples derived from tumors are rarely homogeneous. For example, many tumors contain large areas of necrotic tissue that reduces the overall amount of usable DNA. In addition, specimens contain a mixture of normal cells and cancer cells, and thus the identification of mutations must have sufficient sensitivity to detect mutations in samples where the tumor often represent less than half of the material. In addition, many cancers are populations of heterogeneous cells (84), with events that occur throughout the timeline of evolution of a tumor (85). Finally, the emergence of resistance mutations is a clinically relevant genotype that may impact a therapeutic regimen, and is in some cases driven by a mutation that may be present initially (whether in the target of therapy or an entirely different gene) in a small subclone of cancer cells (86–89). As these mutations may be present in only a fraction of the tumor cells examined (90), even more sensitivity is required to detect low level events (91).
Technological challenges: the selection of genomic approach and platform
Until recently, one of the main limiting factors inherent in the detection of cancer genomic alterations has been the availability of technologies to assay samples for all types of alterations; however, recent major technological innovations have all but eliminated this as a bottleneck in the translation of individualized cancer profiling to the clinical setting.
Capillary sequencing, used to detect point mutations, insertions, deletions, and substitutions at the DNA level, is still considered the “gold standard” in clinical laboratories, and is used to detect EGFR mutations in lung cancer and KRAS in colorectal cancer, among others. More sensitive sequencing technologies, such as pyrosequencing, are also routinely used in the clinical laboratory for detection of small base-pair changes in genes. Comparative genomic hybridization (CGH) (92), FISH and array CGH (aCGH) (93) allow the detection of genomic imbalances, including copy number alterations and deletions at a low-resolution. Mass-spectrometric analysis of DNA allows for the detection of base pair changes, and small (<50 bp) insertions and deletions in genes (71, 72, 94, 95).
However, the implementation of these technologies in a clinical setting will be challenging for several reasons. Some limitations can be attributed to the inherent technical aspects of PCR and DNA manipulations in general. Moreover, the data output from hybridization-based tests is subject to error, and both comparators (such as wild-type or normal) and replicates are necessary to inform a conclusive test result. In the case of mass-spectrometric tests and sequencing, the signal generated is plotted as a composite intensity peak and (similar to hybridization-based methodologies) is subject to saturation limits and necessitates a subjective interpretation.
Second-generation sequencing technologies (such as Illumina and SOLiD) are better candidates to incorporate in the clinical setting. They allow for the detection of the full range of genomic alterations, including mutations, structural rearrangements, and copy number changes, using a single approach (60–64, 66, 68, 96) (for a comprehensive review of genomic sequencing strategies and technologies, see (96) and references therein), to detect epigenetic modifications using chromatin immunoprecipitation (ChIP-seq) (97) and alterations in RNA (RNA seq) (98) such as transcript expression, allele-specific expression, and alternative splicing. The theoretical sensitivity of second generation technology platforms can be increased by sequencing to a higher coverage. In addition, protocols are available to analyze samples derived from FFPE (83).
Current challenges of these sequencing platforms include the lack of validation efforts, high running costs and slow turn-around-time. The reliability and reproducibility of second generation sequencing platforms have yet to be established. International comparative studies may therefore need to be organized, similar to the MicroArray Quality Control project (MAQC) for the assessment of quality and reproducibility between microarray platforms and microarray data generating laboratories (99). The MAQC effort was instrumental for the acceptance and confidence in microarray data by the scientific community.
Costs and throughput in sequencing the full genome will need to decrease by an order of magnitude before these technologies can be used routinely in clinical laboratories. Many research and diagnostic goals, however, may be achieved much sooner by sequencing a specific subset of the genome in large numbers of individuals or at a greater sequencing depth. Several advances including barcoding DNA for multiplex sequencing (100, 101) and hybridization-based sequence capture methods reduce the complexity of the total genome (102–105) and decrease the cost, increase the number of applications of next generation sequencing, and retain sensitivity sufficient to detect low abundant somatic mutations (106). Transcriptome sequencing is a sensitive application to detect intragenic fusions (including in-frame fusion events that lead to oncogene activation (107, 108)) and to generate gene expression profiles (109).
Second-generation sequencing technologies currently have a slow turnaround time (1–2 weeks) and are still quite costly. To be amenable for a clinical setting, technological advances are necessary to achieve performance necessary for clinical implementation, including short turn-around-time (in the order of days), high accuracy and sensitivity, inter-and intra-laboratory reproducibility and cost-effectiveness. Additional modifications in chemistries, detection methodologies and decreasing the experimental surface areas may enable such advances. Recently, smaller, faster versions of available technologies have been released. These “personal sequencers” (produced by Intelligent Biosystems and Illumina, among others) have outputs sufficient for analysis of one exome that can be generated within a day. Going forward, faster (third-generation) sequencing technologies may have the ability to interrogate cancer genome alterations on a single molecule or per-cell basis, and possibly enable non-invasive methods for cancer diagnosis and monitoring.
Analytical and Informatics Issues in Cancer Genome Diagnostics
A challenge deriving from the interrogation of low quality tumor samples subject to the biological confounders detailed above is the analysis of data from cancer genome characterization efforts. Due to issues such as tumor heterogeneity, ploidy, and stromal contamination, a heterozygous somatic mutation in a tumor specimen with 60% admixed normal cells will be detected at an overall allele frequency of 20%. Thus mutation calling algorithms can be confounded by the presence of additional (normal) signal, and disambiguating a somatic mutation signal from background can be difficult. Similar issues apply to the detection of other types of alterations such as copy number changes and chromosomal rearrangements. Looking to current technologies, massively parallel sequencing can overcome or mitigate some of these issues. The data output from these technologies is essentially digital – i.e. one can count the number of reads, alleles or nucleotides at any position on the aligned sequence. The advantage of this digital read-out of data is that counting the number of reads that identify mutant alleles can help to overcome challenges of low tumor quantity, variable ploidy, tumor heterogeneity and normal cell admixture. Challenges in the detection of somatic alterations in tumor specimens can thus be addressed using statistically rigorous algorithms and bioinformatics approaches (for a comprehensive review of considerations in the analysis of cancer genomes, see (96)). Complete cancer genome sequencing requires that both the tumor and the matched normal sample be significantly oversampled. Specifically, the nucleotide sequence of the same fragment of DNA is sequenced repeatedly in order to gain a confident high-quality readout sequence (42), which allows the identification of somatic variants (as opposed to germ line alterations which may be private or common) that can be selected as candidates for further interrogation. From a diagnostic perspective, it may be prudent to have a high quality cutoff for clinically relevant somatic mutations, or candidate events can be validated using an orthogonal technology such as genotyping, pyrosequencing or capillary sequencing, preferably in CLIA-certified (Clinical Laboratory Improvement Amendments) laboratories.
The vast amount of data produced from whole-genome sequencing studies presents a challenge of its own. Indeed, new algorithms are currently being developed to translate raw sequence data into meaningful reads that can be used to analyze the variety of genomic endpoints. A number of groups and approaches are currently in development including mrFAST for CNV by Alkan et al. (110) the genome analysis toolkit (GATK) developed by DePristo and co-workers for SNP (111) and genotyping discovery and several ChipSeq analysis tools reviewed by Kim et al. (112).
Besides new algorithm development, the computational cost for storage and processing of data is another challenge and may approach the cost of sequencing itself. These factors suggest that it may be more parsimonious to keep a record of only the deviations of a patient’s tumor (and normal) from a reference genome. Another important consideration for clinical sequencing is a robust sample tracking, management and LIMS system, as maintaining the identity of a sample is of critical importance. The myriad of sequence information that will be collected using a sequencing technology in the clinical setting is advantageous, in that it enables identification of somatic tumor alterations, and matching of tumor with the corresponding normal (or germline) events that will be seen in both matched samples, thus ensuring that sample identity can be maintained.
Interpretation of Biological Data
Whole genome studies have shown that the number and type of alterations in cancer genomes are often diverse and complex. Signals of driver alterations (those that dysregulate growth or promote tumorigenesis) are often difficult to distinguish from passenger events (alterations that are inconsequential and do not contribute to tumorigenesis). The significance of a somatic event must therefore be assessed in the context of the background of additional somatic events that may not have physiological consequence. Integrating data sets and genomic endpoints may help in identifying driver mutations or pathways. The initial findings of TCGA, for example, demonstrated the utility of this approach by using integrative analyses of DNA sequence, copy number, gene expression and DNA methylation in glioblastomas to provide a network view of altered pathways (113). However, this approach requires sufficient biological knowledge of the relevant pathways, relationships and interactions to be able to interpret and understand signals. Nevertheless, for diagnostic purposes distinguishing driver from passenger mutations may not be that relevant, but for prognosis it is important to understand the master regulating genes or pathways to predict disease progression and response to targeted personalized therapy. Determining driver events or molecular targets is an emerging area recognized by the National Cancer Institute (NCI) in efforts such as the Cancer Target Discovery and Development (CTD2) Network to prospectively pursue this path and to test strategies for selecting therapeutic targets (114).
A related question concerning the use of genomics to guide cancer therapeutic decisions is if mutations predictive of pharmacologic sensitivity in one tumor type can be extended to predict sensitivity in other tumor types. Many oncogenic KIT mutations have been identified that are predictive of imatinib response in GISTs. The subsequent discovery of identical KIT mutations in some acral and mucosal melanomas (115) allowed assessment of this hypothesis. Several reports indicate that inhibition of KIT using imatinib or nilotinib can elicit clinical responses in KIT-mutant melanoma (116, 117). These observations suggest that known driver genomic events may have the potential to denote therapeutic vulnerability regardless of the tissue type in which they occur, though clinical studies will be necessary to confirm this in many cases. Other reports, however, have contradictory indications, and indicate that cellular context does play a role: ovarian cancer patients with PIK3CA and either KRAS or BRAF mutations respond to treatment with a PI3-kinase pathway inhibitor, while colorectal cancer patients with PIK3CA and KRAS mutations do not respond (118).
A critical step toward defining a correct personalized anti-cancer therapy is the identification of the additional genes and pathways altered in the tumor, and the elucidation of their particular oncogenic role. Genetic interactions and compensatory mechanisms may therefore be challenging for determining cancer treatment ((119) for review) and the treatment paradigm for a patient thus may not be decided on the mutational status of single gene, but instead in the context in which that mutation is found. As detailed above, mutations in genes other than the predicted therapeutic target can dramatically affect the response of a patient to a specific therapy, as in the case of RAS and RAF mutations indicating non-response to EFGR inhibitor therapy in colorectal cancer and NSCLC (19–22). Treating metastatic melanoma patients with a BRAF-selective inhibitor, in the presence of RAS mutations, may unintentionally activate the MEK-ERK signaling pathway, instead of inhibiting tumor formation (120). The presence of additional mutations in intracellular signaling pathways activated by RTKs can exhibit patterns of “cross-talk”. To this point, it has been reported that inhibition of certain pathways, such as the mTOR route may lead to increased MAPK activity (121). Therefore, ablation of signals through both routes may be required for efficient antitumoral activity. Moreover, combined inhibition of PI3K and MAPK routes has shown superior anti-tumoral effect compared to individual targeting of either pathway (122). Another example is sensitivity of BRCA1/2 deficient tumor cells to PARP inhibitors. PARP has been shown to be critical to cell survival in a compensatory mechanism for BRCA1 or BRCA2 loss-of-function mutations (123).
Accurate representation of the significance of a variant is of critical importance-translating this information to physicians, clinicians and oncologists requires a thoughtful analysis of the utility of available data. As the current approach to clinical characteristics that drive analyses today transitions to a more nuanced and realistic representation of the complexity of individual human physiology, robust decision support systems will be critical as an adjunct to a clinician’s evaluation. A system based on levels-of-evidence, for example, might indicate to a provider how much information has been collated for a specific mutation, tissue type, and therapeutic target. This would aid in treatment decisions; as more exome-based and genome-based sequencing approaches translate to the clinical arena, a database linking to approved indications and guidelines may assist in collating the mass of information into a more interpretable framework. Regardless of the sophistication of informatics support, thoughtfully designed and carefully monitored clinical trials will be necessary to safely and effectively disambiguate the complex network of molecular alterations, signaling pathways, cellular context, and response to a particular therapeutic regimen.
Disease Monitoring
The molecular signature of a tumor during progression can be dynamic, one consequence of which is the emergence of resistance during therapy. Clinical responses to targeted anticancer therapeutics are frequently confounded by de novo or acquired resistance (15, 124, 125). Logistically then, it would be ideal to sample a tumor at diagnosis, and then track progression of disease by recurrent sampling. This would aid in the management of disease, and might indicate the emergence of resistance mutations. The clinical promise of selective RAF inhibitors for example, has widespread ramifications for patient treatment, yet single-agent targeted therapy is almost invariably followed by relapse due to acquired drug resistance. Identification of resistance mechanisms in a manner that elucidates alternative ‘druggable’ targets may inform effective long-term treatment strategies (126).
Recent studies have demonstrated that mutant BRAF-specific inhibitors can activate CRAF through the formation of dimeric RAF complexes, and this process is enhanced by the presence of an oncogenic RAS mutation. Thus these inhibitors should be used for treating cancers driven by BRAF, but should be avoided in cancers caused by RAS mutations (127, 128). Similarly, resistance to RAF inhibition can be achieved by multiple MAP3K-dependent mechanisms of MEK/ERK reactivation (129) but might be intercepted through combined therapeutic modalities for MAPK pathway inhibition. Thus, identification the correct combination of therapeutic agents targeting several key steps in signaling pathways will be a new paradigm for cancer management.
However, whether acquired resistance is due to compensatory mechanisms/mutations or due to the selection of existing mutations within a heterogenic tumor is not yet clear. Several recent studies suggest that cancer genomes evolve as the cancer progresses. Deep sequencing of two primary breast cancers and subsequent metastases showed either novel mutations, or enrichment for low-frequency mutations in the metastases, indicating that analysis of such tissues in some cancers might identify additional mutations that may be important drug targets (65, 66).
In many cases, it may be convenient to diagnose or detect the molecular signature of a tumor using non-invasive procedures. To this end, much effort is focused on the development of technologies to detect genomic alterations in circulating tumor cells (CTCs) (130–132) or plasma DNA (133–135). The efficacy of using tumor-derived CTCs in the monitoring of disease and the early detection of resistance mutations during treatment has been reported for EGFR in lung cancer (130) as well as other solid tumor metastases (132); prospective clinical trials are underway in metastatic breast cancer. An FDA-approved technology for detection has also shown promise in characterizing the molecular profile (specifically HER2 expression) of CTCs in breast cancer patients (136). Next Generation Sequencing could be the ideal technology to screen CTCs now that enough DNA can be extracted from flow sorted single nuclei (137).
Clinical trial design
As our knowledge of the myriad alterations that drive cancer biology increases, our ability to accurately interpret these somatic alterations (and transfer patients to the appropriate targeted therapy) will become more refined. Thus, personalized diagnostic technologies may aid in the stratification and enrollment of patients with specific molecular alterations for a clinical trial with a targeted therapeutic regimen. One might therefore design clinical trials based on a targeted genotype rather than the cancer type, and to select patients based on that genotype. It is clear that selecting patients in different cancers by mutation will be challenging due to the heterogeneity of the natural history of the cancers, as well as the question of whether different responses will be found in tumors derived from different tissues even if they harbor the same genetic alteration.
To date, the success of molecularly-characterized targeted therapeutic trials has occurred in cancer types where the hallmark mutation is present in a large subset of specimens as defined by tissue type or histological classification. For example, KIT mutations occur in ~85% of GIST (14, 138) for example, and targeted therapies such as imatinib mesylate are effective only in patients with alterations in the intended targets (mutated KIT or PDGFRα) (139, 140). There is increasing evidence however, that these potentially actionable events occur at lower frequencies in other cancer types, and thus patients harboring these alterations are potentially suitable candidates for a targeted therapy; this is the basis for the paradigm of personalized or individualized cancer medicine. NSCLC patients for example, can harbor activating mutations in EGFR, but also KRAS, BRAF, PIK3CA, HER2, or translocations involving ALK (28, 141, 142). Similarly, SLC45A3-BRAF and ESRP1-RAF1 fusions were observed in frequencies below 2% in patients with advanced prostate cancer, gastric cancer and melanoma, and may be therefore be sensitive to RAF or MEK inhibitors (143).
Improvements in our understanding of chemical biology and drug targets should also aid in the interpretation of clinical trial data and inform our understanding and interpretation of biological response to specific treatment paradigms. The first BRAF-targeting drug to enter the clinic, for example, was sorafenib, a type II multi-kinase inhibitor (144–146). In clinical trials, sorafenib was ineffective in BRAF-mutated melanoma (147), but effective in renal cell (148) and hepatocellular carcinoma (149). Thus the interpretation of clinical trial data can be confounded by the relative specificity and potency of inhibitors in specific cancer types as well as off-target effects. The efficacy of a specific BRAF V600E inhibitor in clinical trials demonstrates the power of using targeted, mechanistic inhibitors (30, 150); the necessity to add combination therapies to offset the cellular resistance to such drugs (151) indicates that our ability to interpret this data is still limited; carefully designed and controlled clinical trials in combination with robust genomic profiling technologies will be needed to define the effective mutation-drug combinations.
It is imperative that new tests, drugs and procedures in patients be subjected to rigorous appraisal, and that new signatures of relapse risk, such as that described for colon cancer, should also be tested in prospective clinical trials (152). The efficacy of such clinical trials has been demonstrated in stratification of NSCLC patients based on the mutational status of EGFR (153, 154). Additional large-scale, prospective randomized clinical trials are underway for breast cancer gene expression tests.
With the cost of genome sequencing decreasing dramatically, it may be feasible in the not-too-distant future to augment clinical trials of cancer drugs with complete cancer genome or transcriptome sequencing in order to identify these determinants. It has yet to be determined, however, if clinical trials will be required to show the value of comprehensive cancer genome profiling, and what form those trials should take. We refer the reader to recent reviews that discuss new approaches for clinical trial design and development of new therapeutics (155–158)
Ethical, Legal, Social Implications and IP Challenges
Patients and subjects must be consented properly, as well as educated as to the import and impact of genomic testing. Privacy issues may also be a concern for the patient. The United States FDA recently updated its requirements for informed consent documents used in clinical trials of drugs, biologics, and medical devices to include a statement to inform participants that information from trials will be entered into a databank. For somatic mutations and current clinical tests assaying a small number of DNA changes, this information can be linked to the patient’s medical record when contained within in a secure database with restricted access. Looking ahead to sequencing-based approaches, there may be a necessity to protect this patient data and confidentiality. In the US, the Genetic Information Nondiscrimination Act (GINA) was passed in 2008 to protect against discrimination (health insurance and employment) based on genetic information, and was seen as a landmark achievement in enabling patients to take advantage of personalized medicine without fear of discrimination.
As more genome-wide association studies are completed across populations, individual germline differences between patients will also be factored into the equation-lower penetrance alleles in many coding and non-coding regions can help inform progression of disease, response to therapy, and metabolism of specific compounds. Testing for disease-causing mutations in the BRCA1 and BRCA2 genes (implicated in familial breast and ovarian cancer syndromes) is an early example of the contribution of germline features to disease prognosis. Discovery of a disease-causing mutation in a family can inform at-risk individuals as to whether they are at higher risk for cancer and may prompt individualized prophylactic therapy. Another consequence of using sequencing-based technologies is the generation of ancillary or unintentional data. This might come in the form of sequence alterations that were not being screened or tested for, but may impact the health or treatment of the patient (and possibly related individuals) in other ways.
Public education on the potential benefits of personalized cancer medicine and individualized treatments will be an important facet of its widespread acceptance. In addition, the implementation of these technologies will require several levels of additional oversight, such as obtaining Institutional Review Board (IRB) approved consent for appropriate testing, with a view to ensuring ethical research conduct and appropriate subject protection (for a review on electronic health records and healthcare information technology considerations in personalized medicine, see (159)). Considerations such as privacy concerns and the unknown clinical significance of genome sequence data must be weighed against the ethical principles of respect for autonomy, and the right of every patient to receive relevant personal medical information. As next-generation sequencing technologies and genomic science filter into clinical medicine and standard-of-care, there will be a need for guidance from programs such as the DOE- and NIH-funded ethical, legal and social issues (ELSI) program, in order to adequately protect subjects and patients.
A number of additional hurdles currently confound the process of implementing personalized cancer profiling in the clinical setting including pre-existing intellectual property (IP) environment that gives a company the sole right to genetic tests of specific genes and mutations. The arena of gene-based IP is currently in flux with regard to clinical diagnostics and patented genes. Addressing these changes in the architecture of cancer care may require a substantial shift in the cultural understanding of individualized cancer medicine and how it affects each party in the arena of cancer care.
Oversight and Regulatory Challenges
In the US, several large stakeholders currently use the output from clinical research to make decisions and set policy. Three of the largest are the US FDA, Agency for Healthcare Research and Quality (AHRQ) and CMS. While the FDA regulates diagnostic devices sold as kits (Medical Device Amendments of 1976), the CMS regulates diagnostic tests that are developed and performed in clinical laboratories under CLIA certification. Generally, FDA clearance of in vitro diagnostic devices (IVD) includes evaluation of the performance claims of the assay, while CLIA laboratory inspections focus on reference laboratory quality standards. In addition, many state health departments require their own certifications for tests performed on patients from their state, and these inspections review assay validation reports for in-house developed tests in detail. The Clinical and Laboratory Standards Institute (CLSI) publishes standards on assay validation and performance (160). Furthermore, large reference laboratories generally have adopted assay validation and acceptance policies to comply with pharmaceutical industry guidelines and expectations, including ICH, GCP and GLP, and ISO accreditation.
The proliferation of a consensus interpretative framework that will provide recommendations to clinicians and oncologists would be a helpful framework for personalized cancer medicine. One possible structure would be a form similar to the NCCN or ASCO, where a centralized body (or bodies) collates information on clinical trials and issue guidelines on clinical policy in oncology based on a system akin to “levels of evidence”. Guidelines on personalized cancer medicine could be issued in the form of an annual guide, for example, and available in an affirmed database. These guidelines would be based on the strength of evidence available from preclinical and clinical trials. The result of this interface between cancer centers would be a gradual shift from empirical cancer treatment options to more individualized (and effective) therapies.
An unintentional but nonetheless important consequence of creating a cross-disciplinary, integrated framework is the empowerment of any participating cancer center with clinical trials of sufficient volume. A linked database could then act a venue to interpret the findings of clinical trials, and to determine the therapeutic significance of an individual mutation or a spectrum of alterations. This framework can enable the emergence of professional and patient-centric networks and support groups, and function as an interface between individual cancer centers, and a more global network of participants.
Overall, realizing the goal of personalized cancer medicine will require the systematic engagement of all interested parties: patients, oncologists, cancer centers, large cooperative clinical trials groups (such as the National Surgical Adjuvant Breast and Bowel Project, NSABP), pharmaceutical companies, insurance providers and government and regulatory agencies. There has been some tentative movement in this direction with the emergence of the drug-diagnostic co-development paradigm (161), where drug development groups team with diagnostic entities to develop a companion test for their therapeutic. The proliferation of targeted agents in development and clinical practice necessitates concomitant implementation of companion diagnostic approaches that enrich for subpopulations most likely to respond to a drug.
Concluding Remarks
Ongoing global genome characterization efforts are revolutionizing both tumor biology and the optimal paradigm for cancer treatment at an unprecedented scope. The pace of advance is empowered in large part through technological innovations that render complete cancer genome characterization feasible on a large scale. The technologies available in the research setting today will become the clinical tests of tomorrow. In the near future, traditional diagnostic and prognostic factors will be supplemented by tumor genomic signatures, well-characterized molecular therapeutic targets, chemotherapy response assays, and the identification of deranged molecular pathways. Despite the inherent complexity of cancer genomics, incorporating this knowledge of the molecular basis of cancer into clinical decision-making will speed the advent of more effective anticancer therapies. Realizing the goal of individualized cancer medicine necessitates the concomitant engagement of academic thought leaders, clinicians, oncologists, cancer centers, government and regulatory agencies, and patient advocacy groups. Molecularly based individualized cancer care will have a tremendous impact of the future of oncology. Personalized cancer genomics, once widely available and translated into the clinical setting in an appropriate and thoughtful manner, will greatly improve the lives of many cancer patients.
Key Concepts and Relevance.
-
Genomic Strategies in Cancer Medicine
Cancer is a genomic disease; newly developed targeted therapies in cancer medicine are based on the mutational status of one or a set of particular genes in the tumor sample.
-
Personalized Cancer Medicine
With the advent of new technological breakthroughs, analyzing a complete cancer genome in high detail at a reasonable cost is now feasible and can be used for personalized treatment and disease monitoring approaches.
-
Disruptive Genomic Technologies
The current and emerging technologies enable comprehensive genome-wide analysis of the alterations that are a hallmark of cancer.
-
Clinical Implementation Challenges
Despite the inherent complexity of cancer genomics, incorporating the knowledge of the molecular basis of cancer into clinical decision-making will speed the advent of more effective anticancer therapies. The challenges (in many different fields) related to the implementation of comprehensive screening of a patient’s cancer genome for diagnostic and/or therapeutic use. These challenges lie in the areas of technologies, clinical trial design, legal and social implications, healthcare information technology, and insurance and reimbursement.
Acknowledgments
We apologize to our many colleagues whose work could not be cited due to space limitations.
Footnotes
Conflict of interests: Laura E MacConaill: No Conflicts of Interests
Paul Van Hummelen: No Conflicts of Interests
Matthew Meyerson: Consultant for and received research support from Novartis. Founding advisor of, consultant for, and equity holder in Foundation Medicine. Co-inventor of patent on use of EGFR mutations for lung cancer diagnosis, licensed to Genzyme Genetics/Labcorp.
William C. Hahn: Consultant for Novartis and Thermo-Fisher. Received research support from Novartis
References
- 1.de Klein A, van Kessel AG, Grosveld G, Bartram CR, Hagemeijer A, Bootsma D, et al. A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1982;300(5894):765–7. doi: 10.1038/300765a0. [DOI] [PubMed] [Google Scholar]
- 2.Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243(5405):290–3. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 3.Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279(5350):577–80. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 4.Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, Joensuu H, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21(23):4342–9. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 5.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 6.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 7.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 8.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101(36):13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang XR, Chang-Claude J, Goode EL, Couch FJ, Nevanlinna H, Milne RL, et al. Associations of breast cancer risk factors with tumor subtypes: a pooled analysis from the Breast Cancer Association Consortium studies. J Natl Cancer Inst. 2011;103(3):250–63. doi: 10.1093/jnci/djq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamberg P, Bos MM, Braun HJ, Stouthard JM, van Deijk GA, Erdkamp FL, et al. Randomized Phase II Study Comparing Efficacy and Safety of Combination-Therapy Trastuzumab and Docetaxel vs. Sequential Therapy of Trastuzumab Followed by Docetaxel Alone at Progression As First-Line Chemotherapy in Patients with HER2(+) Metastatic Breast Cancer: HERTAX Trial. Clin Breast Cancer. 2011;11(2):103–13. doi: 10.1016/j.clbc.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 11.Owens MA, Horten BC, Da Silva MM. HER2 amplification ratios by fluorescence in situ hybridization and correlation with immunohistochemistry in a cohort of 6556 breast cancer tissues. Clin Breast Cancer. 2004;5(1):63–9. doi: 10.3816/cbc.2004.n.011. [DOI] [PubMed] [Google Scholar]
- 12.Lee AH, Key HP, Bell JA, Hodi Z, Ellis IO. Breast carcinomas with borderline (2+) HER2 immunohistochemistry: percentage of cells with complete membrane staining for HER2 and the frequency of HER2 amplification. J Clin Pathol. 2011;64(6):490–2. doi: 10.1136/jcp.2011.089177. [DOI] [PubMed] [Google Scholar]
- 13.Buchdunger E, Cioffi CL, Law N, Stover D, Ohno-Jones S, Druker BJ, et al. Abl protein-tyrosine kinase inhibitor STI571 inhibits in vitro signal transduction mediated by c-kit and platelet-derived growth factor receptors. J Pharmacol Exp Ther. 2000;295(1):139–45. [PubMed] [Google Scholar]
- 14.Demetri GD, von Mehren M, Blanke CD, Van den Abbeele AD, Eisenberg B, Roberts PJ, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347(7):472–80. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 15.Heinrich MC, Corless CL, Blanke CD, Demetri GD, Joensuu H, Roberts PJ, et al. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J Clin Oncol. 2006;24(29):4764–74. doi: 10.1200/JCO.2006.06.2265. [DOI] [PubMed] [Google Scholar]
- 16.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burris HA, 3rd, Hurwitz HI, Dees EC, Dowlati A, Blackwell KL, O’Neil B, et al. Phase I safety, pharmacokinetics, and clinical activity study of lapatinib (GW572016), a reversible dual inhibitor of epidermal growth factor receptor tyrosine kinases, in heavily pretreated patients with metastatic carcinomas. J Clin Oncol. 2005;23(23):5305–13. doi: 10.1200/JCO.2005.16.584. [DOI] [PubMed] [Google Scholar]
- 19.Khambata-Ford S, Garrett CR, Meropol NJ, Basik M, Harbison CT, Wu S, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25(22):3230–7. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 20.Jackman DM, Miller VA, Cioffredi LA, Yeap BY, Janne PA, Riely GJ, et al. Impact of epidermal growth factor receptor and KRAS mutations on clinical outcomes in previously untreated non-small cell lung cancer patients: results of an online tumor registry of clinical trials. Clin Cancer Res. 2009;15(16):5267–73. doi: 10.1158/1078-0432.CCR-09-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loupakis F, Ruzzo A, Cremolini C, Vincenzi B, Salvatore L, Santini D, et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101(4):715–21. doi: 10.1038/sj.bjc.6605177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Cutsem E, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360(14):1408–17. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 23.Mansour JC, Schwarz RE. Molecular mechanisms for individualized cancer care. J Am Coll Surg. 2008;207(2):250–8. doi: 10.1016/j.jamcollsurg.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 24.van’t Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452(7187):564–70. doi: 10.1038/nature06915. [DOI] [PubMed] [Google Scholar]
- 25.Yanagi M, Shinjo K, Takeshita A, Tobita T, Yano K, Kobayashi M, et al. Simple and reliably sensitive diagnosis and monitoring of Philadelphia chromosome-positive cells in chronic myeloid leukemia by interphase fluorescence in situ hybridization of peripheral blood cells. Leukemia. 1999;13(4):542–52. doi: 10.1038/sj.leu.2401383. [DOI] [PubMed] [Google Scholar]
- 26.Reinhold U, Hennig E, Leiblein S, Niederwieser D, Deininger MW. FISH for BCR-ABL on interphases of peripheral blood neutrophils but not of unselected white cells correlates with bone marrow cytogenetics in CML patients treated with imatinib. Leukemia. 2003;17(10):1925–9. doi: 10.1038/sj.leu.2403077. [DOI] [PubMed] [Google Scholar]
- 27.Kallioniemi OP, Kallioniemi A, Kurisu W, Thor A, Chen LC, Smith HS, et al. ERBB2 amplification in breast cancer analyzed by fluorescence in situ hybridization. Proc Natl Acad Sci U S A. 1992;89(12):5321–5. doi: 10.1073/pnas.89.12.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363(18):1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Debiec-Rychter M, Sciot R, Le Cesne A, Schlemmer M, Hohenberger P, van Oosterom AT, et al. KIT mutations and dose selection for imatinib in patients with advanced gastrointestinal stromal tumours. Eur J Cancer. 2006;42(8):1093–103. doi: 10.1016/j.ejca.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 30.Flaherty K, Puzanov I, Sosman J, Kim K, Ribas A, McArthur G, et al. Phase I study of PLX4032: Proof of concept for V600E BRAF mutation as a therapeutic target in human cancer. J Clin Oncol. 2009;27(15S):9000. [Google Scholar]
- 31.Wilhelm SM, Adnane L, Newell P, Villanueva A, Llovet JM, Lynch M. Preclinical overview of sorafenib, a multikinase inhibitor that targets both Raf and VEGF and PDGF receptor tyrosine kinase signaling. Mol Cancer Ther. 2008;7(10):3129–40. doi: 10.1158/1535-7163.MCT-08-0013. [DOI] [PubMed] [Google Scholar]
- 32.Rosenwald A, Staudt LM. Clinical translation of gene expression profiling in lymphomas and leukemias. Semin Oncol. 2002;29(3):258–63. doi: 10.1053/sonc.2002.32901. [DOI] [PubMed] [Google Scholar]
- 33.Rosenwald A, Staudt LM. Gene expression profiling of diffuse large B-cell lymphoma. Leuk Lymphoma. 2003;44 (Suppl 3):S41–7. doi: 10.1080/10428190310001623775. [DOI] [PubMed] [Google Scholar]
- 34.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, et al. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25(33):5287–312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 35.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 36.van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 37.Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975;94(3):441–8. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- 38.Pease AC, Solas D, Sullivan EJ, Cronin MT, Holmes CP, Fodor SP. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc Natl Acad Sci U S A. 1994;91(11):5022–6. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ross P, Hall L, Smirnov I, Haff L. High level multiplex genotyping by MALDI-TOF mass spectrometry. Nat Biotechnol. 1998;16(13):1347–51. doi: 10.1038/4328. [DOI] [PubMed] [Google Scholar]
- 40.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437(7057):376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309(5741):1728–32. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 42.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456(7218):53–9. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith DR, Quinlan AR, Peckham HE, Makowsky K, Tao W, Woolf B, et al. Rapid whole-genome mutational profiling using next-generation sequencing technologies. Genome Res. 2008;18(10):1638–42. doi: 10.1101/gr.077776.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26(10):1135–45. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 45.Pettersson E, Lundeberg J, Ahmadian A. Generations of sequencing technologies. Genomics. 2009;93(2):105–11. doi: 10.1016/j.ygeno.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 46.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 47.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291(5507):1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 48.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470(7333):187–97. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 49.Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365(9464):1054–61. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 50.Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7(4):387–97. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 51.Dutt A, Salvesen HB, Chen TH, Ramos AH, Onofrio RC, Hatton C, et al. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc Natl Acad Sci U S A. 2008;105(25):8713–7. doi: 10.1073/pnas.0803379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455(7215):971–4. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 53.George RE, Sanda T, Hanna M, Frohling S, Luther W, 2nd, Zhang J, et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455(7215):975–8. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 55.Ahn SM, Kim TH, Lee S, Kim D, Ghang H, Kim DS, et al. The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res. 2009;19(9):1622–9. doi: 10.1101/gr.092197.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456(7218):60–5. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452(7189):872–6. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 58.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5(10):e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim JI, Ju YS, Park H, Kim S, Lee S, Yi JH, et al. A highly annotated whole-genome sequence of a Korean individual. Nature. 2009;460(7258):1011–5. doi: 10.1038/nature08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456(7218):66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–66. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campbell PJ, Stephens PJ, Pleasance ED, O’Meara S, Li H, Santarius T, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40(6):722–9. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 463(7278):184–90. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee W, Jiang Z, Liu J, Haverty PM, Guan Y, Stinson J, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465(7297):473–7. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 65.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464(7291):999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461(7265):809–13. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 67.Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462(7276):1005–10. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 463(7278):191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–72. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, Bernabe RR, et al. International network of cancer genome projects. Nature. 2010;464(7291):993–8. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.MacConaill LE, Campbell CD, Kehoe SM, Bass AJ, Hatton C, Niu L, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS One. 2009;4(11):e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39(3):347–51. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 73.Su Z, Dias-Santagata D, Duke M, Hutchinson K, Lin YL, Borger DR, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn. 2011;13(1):74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dias-Santagata D, Akhavanfard S, David SS, Vernovsky K, Kuhlmann G, Boisvert SL, et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2(5):146–58. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arcila M, Lau C, Nafa K, Ladanyi M. Detection of KRAS and BRAF mutations in colorectal carcinoma roles for high-sensitivity locked nucleic acid-PCR sequencing and broad-spectrum mass spectrometry genotyping. J Mol Diagn. 2011;13(1):64–73. doi: 10.1016/j.jmoldx.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lau C, Ang D, Brzostowski EB, Riely GJ, Rusch VR, Zakowski MF, Kris MG, Ladanyi M. LC-MAP: a pilot study of prospective profiling of clinical tumor specimens for the presence of key mutations in targetable pathways in patients with lung adenocarcinoma and metastatic colorectal cancer using Sequenom genotyping. J Mol Diagn. 2010;12:910. [Google Scholar]
- 77.Arcila M, Lau CY, Jhanwar SC, Zakowski MF, Kris MG, Ladanyi M. Comprehensive analysis for clinically relevant oncogenic driver mutations in 1131 consecutive lung adenocarcinomas. Mod Pathol. 2011;24(Suppl 1):404a. [Google Scholar]
- 78.Gonzalez-Angulo AM, Hennessy BT, Mills GB. Future of personalized medicine in oncology: a systems biology approach. J Clin Oncol. 2010;28(16):2777–83. doi: 10.1200/JCO.2009.27.0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beadling CHM, Warrick A, Forbes EM, Nelson D, Justusson E, Levine J, Neff TL, Patterson J, Presnell A, McKinley A, Winter LJ, Dewey C, Harlow A, Barney O, Druker BJ, Schuff KG, Corless CL. Multiplex Mutation Screening by Mass Spectrometry: Evaluation of 820 Cases from a Personalized Cancer Medicine Registry. J Mol Diagn. 2011 doi: 10.1016/j.jmoldx.2011.04.003. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gilbert MT, Haselkorn T, Bunce M, Sanchez JJ, Lucas SB, Jewell LD, et al. The isolation of nucleic acids from fixed, paraffin-embedded tissues-which methods are useful when? PLoS One. 2007;2(6):e537. doi: 10.1371/journal.pone.0000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gallegos Ruiz MI, Floor K, Rijmen F, Grunberg K, Rodriguez JA, Giaccone G. EGFR and K-ras mutation analysis in non-small cell lung cancer: comparison of paraffin embedded versus frozen specimens. Cell Oncol. 2007;29(3):257–64. doi: 10.1155/2007/568205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161(6):1961–71. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wood HM, Belvedere O, Conway C, Daly C, Chalkley R, Bickerdike M, et al. Using next-generation sequencing for high resolution multiplex analysis of copy number variation from nanogram quantities of DNA from formalin-fixed paraffin-embedded specimens. Nucleic Acids Res. 2010;38(14):e151. doi: 10.1093/nar/gkq510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Navin N, Krasnitz A, Rodgers L, Cook K, Meth J, Kendall J, et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010;20(1):68–80. doi: 10.1101/gr.099622.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jones S, Chen WD, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105(11):4283–8. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.O’Hare T, Eide CA, Deininger MW. Bcr-Abl kinase domain mutations, drug resistance, and the road to a cure for chronic myeloid leukemia. Blood. 2007;110(7):2242–9. doi: 10.1182/blood-2007-03-066936. [DOI] [PubMed] [Google Scholar]
- 87.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Antonescu CR, Besmer P, Guo T, Arkun K, Hom G, Koryotowski B, et al. Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutation. Clin Cancer Res. 2005;11(11):4182–90. doi: 10.1158/1078-0432.CCR-04-2245. [DOI] [PubMed] [Google Scholar]
- 89.Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci U S A. 2009;106(48):20411–6. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kwak EL, Sordella R, Bell DW, Godin-Heymann N, Okimoto RA, Brannigan BW, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci U S A. 2005;102(21):7665–70. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thomas RK, Nickerson E, Simons JF, Janne PA, Tengs T, Yuza Y, et al. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat Med. 2006;12(7):852–5. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- 92.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258(5083):818–21. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 93.Van Buggenhout G, Melotte C, Dutta B, Froyen G, Van Hummelen P, Marynen P, et al. Mild Wolf-Hirschhorn syndrome: micro-array CGH analysis of atypical 4p16.3 deletions enables refinement of the genotype-phenotype map. J Med Genet. 2004;41(9):691–8. doi: 10.1136/jmg.2003.016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Oeth P, del Mistro G, Marnellos G, Shi T, van den Boom D. Qualitative and quantitative genotyping using single base primer extension coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MassARRAY) Methods Mol Biol. 2009;578:307–43. doi: 10.1007/978-1-60327-411-1_20. [DOI] [PubMed] [Google Scholar]
- 95.Ehrlich KC, Montalbano BG, Cotty PJ. Analysis of single nucleotide polymorphisms in three genes shows evidence for genetic isolation of certain Aspergillus flavus vegetative compatibility groups. FEMS Microbiol Lett. 2007;268(2):231–6. doi: 10.1111/j.1574-6968.2006.00588.x. [DOI] [PubMed] [Google Scholar]
- 96.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11(10):685–96. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 97.Mardis ER. ChIP-seq: welcome to the new frontier. Nat Methods. 2007;4(8):613–4. doi: 10.1038/nmeth0807-613. [DOI] [PubMed] [Google Scholar]
- 98.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24(9):1151–61. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Quail MA, Swerdlow H, Turner DJ. Curr Protoc Hum Genet. Unit 18. Chapter 18. 2009. Improved protocols for the illumina genome analyzer sequencing system; p. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lennon NJ, Lintner RE, Anderson S, Alvarez P, Barry A, Brockman W, et al. A scalable, fully automated process for construction of sequence-ready barcoded libraries for 454. Genome Biol. 2010;11(2):R15. doi: 10.1186/gb-2010-11-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4(11):903–5. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 103.Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, et al. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39(12):1522–7. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 104.Porreca GJ, Zhang K, Li JB, Xie B, Austin D, Vassallo SL, et al. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4(11):931–6. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 105.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27(2):182–9. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. Dissecting Therapeutic Resistance to RAF Inhibition in Melanoma by Tumor Genomic Profiling. J Clin Oncol. 2011 doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Maher CA, Palanisamy N, Brenner JC, Cao X, Kalyana-Sundaram S, Luo S, et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci U S A. 2009;106(30):12353–8. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458(7234):97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Morrissy AS, Morin RD, Delaney A, Zeng T, McDonald H, Jones S, et al. Next-generation tag sequencing for cancer gene expression profiling. Genome Res. 2009;19(10):1825–35. doi: 10.1101/gr.094482.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Alkan C, Kidd JM, Marques-Bonet T, Aksay G, Antonacci F, Hormozdiari F, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009;41(10):1061–7. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kim H, Kim J, Selby H, Gao D, Tong T, Phang TL, et al. A short survey of computational analysis methods in analysing ChIP-seq data. Hum Genomics. 2011;5(2):117–23. doi: 10.1186/1479-7364-5-2-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schreiber SL, Shamji AF, Clemons PA, Hon C, Koehler AN, Munoz B, et al. Towards patient-based cancer therapeutics. Nat Biotechnol. 2010;28(9):904–6. doi: 10.1038/nbt0910-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24(26):4340–6. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 116.Hodi FS, Friedlander P, Corless CL, Heinrich MC, Mac Rae S, Kruse A, et al. Major response to imatinib mesylate in KIT-mutated melanoma. J Clin Oncol. 2008;26(12):2046–51. doi: 10.1200/JCO.2007.14.0707. [DOI] [PubMed] [Google Scholar]
- 117.Lutzky J, Bauer J, Bastian BC. Dose-dependent, complete response to imatinib of a metastatic mucosal melanoma with a K642E KIT mutation. Pigment Cell Melanoma Res. 2008;21(4):492–3. doi: 10.1111/j.1755-148X.2008.00475.x. [DOI] [PubMed] [Google Scholar]
- 118.Janku F, Tsimberidou AM, Garrido-Laguna I, Wang X, Luthra R, Hong DS, et al. PIK3CA Mutations in Patients with Advanced Cancers Treated with PI3K/AKT/mTOR Axis Inhibitors. Mol Cancer Ther. 2011;10(3):558–65. doi: 10.1158/1535-7163.MCT-10-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ashworth A, Lord CJ, Reis-Filho JS. Genetic interactions in cancer progression and treatment. Cell. 2011;145(1):30–8. doi: 10.1016/j.cell.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 120.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140(2):209–21. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118(9):3065–74. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hoeflich KP, O’Brien C, Boyd Z, Cavet G, Guerrero S, Jung K, et al. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin Cancer Res. 2009;15(14):4649–64. doi: 10.1158/1078-0432.CCR-09-0317. [DOI] [PubMed] [Google Scholar]
- 123.Ashworth A. Drug resistance caused by reversion mutation. Cancer Res. 2008;68(24):10021–3. doi: 10.1158/0008-5472.CAN-08-2287. [DOI] [PubMed] [Google Scholar]
- 124.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 125.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293(5531):876–80. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 126.Daub H, Specht K, Ullrich A. Strategies to overcome resistance to targeted protein kinase inhibitors. Nat Rev Drug Discov. 2004;3(12):1001–10. doi: 10.1038/nrd1579. [DOI] [PubMed] [Google Scholar]
- 127.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464(7287):427–30. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464(7287):431–5. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 129.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468(7326):968–72. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359(4):366–77. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Paterlini-Brechot P, Benali NL. Circulating tumor cells (CTC) detection: clinical impact and future directions. Cancer Lett. 2007;253(2):180–204. doi: 10.1016/j.canlet.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 132.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450(7173):1235–9. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2(20):20ra14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Brevet M, Johnson ML, Azzoli CG, Ladanyi M. Detection of EGFR mutations in plasma DNA from lung cancer patients by mass spectrometry genotyping is predictive of tumor EGFR status and response to EGFR inhibitors. Lung Cancer. 2010 doi: 10.1016/j.lungcan.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.McBride DJ, Orpana AK, Sotiriou C, Joensuu H, Stephens PJ, Mudie LJ, et al. Use of cancer-specific genomic rearrangements to quantify disease burden in plasma from patients with solid tumors. Genes Chromosomes Cancer. 2010;49(11):1062–9. doi: 10.1002/gcc.20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Riethdorf S, Muller V, Zhang L, Rau T, Loibl S, Komor M, et al. Detection and HER2 expression of circulating tumor cells: prospective monitoring in breast cancer patients treated in the neoadjuvant GeparQuattro trial. Clin Cancer Res. 2010;16(9):2634–45. doi: 10.1158/1078-0432.CCR-09-2042. [DOI] [PubMed] [Google Scholar]
- 137.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–4. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Corless CL, McGreevey L, Haley A, Town A, Heinrich MC. KIT mutations are common in incidental gastrointestinal stromal tumors one centimeter or less in size. Am J Pathol. 2002;160(5):1567–72. doi: 10.1016/S0002-9440(10)61103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.de Jong FA, Verweij J. Role of imatinib mesylate (Gleevec/Glivec) in gastrointestinal stromal tumors. Expert Rev Anticancer Ther. 2003;3(6):757–66. doi: 10.1586/14737140.3.6.757. [DOI] [PubMed] [Google Scholar]
- 140.Verweij J, van Oosterom A, Blay JY, Judson I, Rodenhuis S, van der Graaf W, et al. Imatinib mesylate (STI-571 Glivec, Gleevec) is an active agent for gastrointestinal stromal tumours, but does not yield responses in other soft-tissue sarcomas that are unselected for a molecular target. Results from an EORTC Soft Tissue and Bone Sarcoma Group phase II study. Eur J Cancer. 2003;39(14):2006–11. [PubMed] [Google Scholar]
- 141.Gandhi J, Zhang J, Xie Y, Soh J, Shigematsu H, Zhang W, et al. Alterations in genes of the EGFR signaling pathway and their relationship to EGFR tyrosine kinase inhibitor sensitivity in lung cancer cell lines. PLoS One. 2009;4(2):e4576. doi: 10.1371/journal.pone.0004576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 143.Palanisamy N, Ateeq B, Kalyana-Sundaram S, Pflueger D, Ramnarayanan K, Shankar S, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med. 2010;16(7):793–8. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Karasarides M, Chiloeches A, Hayward R, Niculescu-Duvaz D, Scanlon I, Friedlos F, et al. B-RAF is a therapeutic target in melanoma. Oncogene. 2004;23(37):6292–8. doi: 10.1038/sj.onc.1207785. [DOI] [PubMed] [Google Scholar]
- 145.Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H, et al. BAY 43–9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004;64(19):7099–109. doi: 10.1158/0008-5472.CAN-04-1443. [DOI] [PubMed] [Google Scholar]
- 146.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116(6):855–67. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 147.Eisen T, Ahmad T, Flaherty KT, Gore M, Kaye S, Marais R, et al. Sorafenib in advanced melanoma: a Phase II randomised discontinuation trial analysis. Br J Cancer. 2006;95(5):581–6. doi: 10.1038/sj.bjc.6603291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kane RC, Farrell AT, Saber H, Tang S, Williams G, Jee JM, et al. Sorafenib for the treatment of advanced renal cell carcinoma. Clin Cancer Res. 2006;12(24):7271–8. doi: 10.1158/1078-0432.CCR-06-1249. [DOI] [PubMed] [Google Scholar]
- 149.Lang L. FDA approves sorafenib for patients with inoperable liver cancer. Gastroenterology. 2008;134(2):379. doi: 10.1053/j.gastro.2007.12.037. [DOI] [PubMed] [Google Scholar]
- 150.Algazi AP, Soon CW, Daud AI. Treatment of cutaneous melanoma: current approaches and future prospects. Cancer Manag Res. 2010;2:197–211. doi: 10.2147/CMR.S6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Whittaker S, Kirk R, Hayward R, Zambon A, Viros A, Cantarino N, et al. Gatekeeper mutations mediate resistance to BRAF-targeted therapies. Sci Transl Med. 2010;2(35):35ra41. doi: 10.1126/scitranslmed.3000758. [DOI] [PubMed] [Google Scholar]
- 152.O’Connell MJ, Lavery I, Yothers G, Paik S, Clark-Langone KM, Lopatin M, et al. Relationship between tumor gene expression and recurrence in four independent studies of patients with stage II/III colon cancer treated with surgery alone or surgery plus adjuvant fluorouracil plus leucovorin. J Clin Oncol. 2010;28(25):3937–44. doi: 10.1200/JCO.2010.28.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 154.Douillard JY, Shepherd FA, Hirsh V, Mok T, Socinski MA, Gervais R, et al. Molecular predictors of outcome with gefitinib and docetaxel in previously treated non-small-cell lung cancer: data from the randomized phase III INTEREST trial. J Clin Oncol. 2010;28(5):744–52. doi: 10.1200/JCO.2009.24.3030. [DOI] [PubMed] [Google Scholar]
- 155.Haber DA, Gray NS, Baselga J. The evolving war on cancer. Cell. 2011;145(1):19–24. doi: 10.1016/j.cell.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 156.Yap TA, Sandhu SK, Workman P, de Bono JS. Envisioning the future of early anticancer drug development. Nat Rev Cancer. 2010;10(7):514–23. doi: 10.1038/nrc2870. [DOI] [PubMed] [Google Scholar]
- 157.McDermott U, Downing JR, Stratton MR. Genomics and thecontinuum of cancer care. N Engl J Med. 2011;364(4):340–50. doi: 10.1056/NEJMra0907178. [DOI] [PubMed] [Google Scholar]
- 158.de Bono JS, Ashworth A. Translating cancer research into targeted therapeutics. Nature. 2010;467(7315):543–9. doi: 10.1038/nature09339. [DOI] [PubMed] [Google Scholar]
- 159.Ullman-Cullere MH, Mathew JP. Emerging landscape of genomics in the electronic health record for personalized medicine. Hum Mutat. 2011 doi: 10.1002/humu.21456. [DOI] [PubMed] [Google Scholar]
- 160.Clark L, Garrett PE, Martin R, Meier KL. User Protocol. For evaluation of qualitative test performances; Approval Guidelines. NCCLS document EP12-A. 2002 [Google Scholar]
- 161.Hinman LM, Carl KM, Spear BB, Salerno RA, Becker RL, Abbott BM, et al. Development and regulatory strategies for drug and diagnostic co-development. Pharmacogenomics. 2010;11(12):1669–75. doi: 10.2217/pgs.10.141. [DOI] [PubMed] [Google Scholar]