Abstract
ΔEGFR, an in-frame deletion mutant of the extracellular ligand-binding domain, which occurs in about 30% of glioblastoma, is a potent oncogene that promotes tumor growth and progression. The signaling of ΔEGFR is ligand-independent and low intensity, allowing it to evade the normal mechanisms of internalization and degradation by the endocytic machinery, and hence is persistent. The basis of the oncogenic potential of ΔEGFR remains incompletely understood, including whether dimerization plays an important role in its signal and whether its oncogenic potential is dependent on its relatively low intensity, when compared to the acutely activated wild-type receptor. To examine these two important questions we have generated a chimeric ΔEGFR that allows forced dimerization via domains derived from variants of the FKBP12 protein that are brought together by FK506-derivatives. Forced dimerization of chimeric ΔEGFR significantly increased the intensity of its signal, as measured by receptor phosphorylation levels, suggesting that the naturally occurring ΔEGFR does not form strong or stable dimers as part of its low level signal. Interestingly the increased activity of dimerized, chimeric ΔEGFR did not promote receptor internalization, implying that ΔEGFR’s reduced rate of endocytic downregulation is an inherent characteristic. Significantly, forced dimerization enhanced the oncogenic signal of the receptor, implying that the ΔEGFR is a potent oncogene despite, not because of its low intensity.
Keywords: ΔEGFR, EGFR, Forced dimerization, Oncogenic Signaling, Glioma
Introduction
Aberrant Receptor Tyrosine Kinase signaling is a major contributor to cancer including glioma, where EGFR gene amplification is frequently accompanied by gene rearrangements, with the most common mutation, ΔEGFR, leading to deletion of exon 2–7 (1, 2). This deletion results in the loss of 267 amino acids from the extracellular domain and renders the ΔEGFR unable to bind ligand (3). ΔEGFR is tumor-specific and associated with advanced disease and resistance to therapy (4, 5). ΔEGFR occurs in glioma in the context of an overexpressed EGFR (6), and so presumably high levels of EGFR signaling, but nevertheless makes an important contribution to glioblastoma growth. Patients with ΔEGFR-expressing tumors have a shorter interval to clinical relapse and poorer survival than patients with ΔEGFR-negative tumors. For GBM patients who survive one year or longer after diagnosis, the expression of ΔEGFR is also an independent negative prognostic indicator of survival (7, 8). In xenograft models, ΔEGFR is also capable of enhancing the tumorigenicity of glioma cells, lending greater take and growth rates (9, 10), which it dose by reducing apoptosis and increasing proliferation (11). Astrocytes or neural stem cells from INK4A/Arf deficient mice can be transformed by ΔEGFR and induce high-grade glioma when implanted (12). Therefore, ΔEGFR is a potent glioma oncogene, and attenuating its signal is important.
ΔEGFR differs from EGFR in a number of ways. The signaling of ΔEGFR is ligand-independent and as a result is low intensity and also evades the normal mechanisms of internalization and degradation by the endocytic machinery, and hence its signaling is persistent (13, 14). The markedly different contributors of ΔEGFR and EGFR to glioma formation suggested that their signals are different (15, 16). Analysis of downstream targets has led to the finding that certain elements in the EGFR pathway are activated to a greater degree or in a more sustained fashion by ΔEGFR. Early studies showed that ΔEGFR associates strongly with adaptor proteins Shc1 and Grb2 (17, 18). The pronounced and preferential activation of PI3K in ΔEGFR-expressing cells (19) was recently substantiated by a broad proteomics screen (20). In addition, c-Met and STAT pathways have been identified as an important characteristic of ΔEGFR signaling. The activating phosphorylation site on the c-Met receptor was found to be highly responsive to ΔEGFR levels, indicating cross-activation of c-Met by ΔEGFR (21). Our recent phosphoproteomic screen also found this strong connection to c-Met signaling, and identified STAT5 as a novel downstream target of ΔEGFR (22).
The lack of interaction with ligand, as well as the lack of internalization, suggests that ΔEGFR may not dimerize efficiently, a hypothesis that has however not been tested experimentally before. Observations of ΔEGFR in cell lines have not found strong evidence for dimerized ΔEGFR by crosslinking and western blot analysis in transiently transfected fibroblasts or in glioma cells (13, 23). In order to examine this question experimentally, we have generated a chimeric ΔEGFR that allows forced dimerization via domains derived from variants of the FKBP12 that are brought together by FK506-derivatives (24, 25). This approach has been used to induce dimerization of EGFR/ErbB1 and ErbB2/Her2/Neu where it resulted in signaling indistinguishable from that caused by physiological ligand, and also for the study of other signaling interactions, such as those of JNK2, Raf-1, and receptors for insulin, PDGF and G-CSF (26–29). While chimeric wild-type EGFR showed similar levels of activity when dimerized by EGF or the dimerization agent, forcing the dimerization of chimeric ΔEGFR significantly increased the intensity of its signal, as measured by autophosphorylation. Forced dimerization of ΔEGFR did not noticeably alter the signal generated by the receptor, either at the level of downstream targets or at the level of cellular responses. Interestingly increasing the activity of ΔEGFR did not enhance its internalization and downregulation, but it did increase the oncogenic impact of the receptor.
Materials and Methods
Cell Lines and Constructs
Glioma cell lines, U87, LN428, and LNZ308, were a kind gift from Dr. W.K. Alfred Yung (UT MD Anderson Cancer Center, USA) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS. Cells were proven to be mycoplasma free by routine testing, authenticated by STR fingerprinting.
Chimeric EGFR constructs were made by modification of pCLEGFRv2E (kind gift from Dr. Victor Rivera, Ariad Pharmaceuticals), which encodes a C-terminal fusion of two FKBPF36V domains, termed Fv2 (25). Then fusion cDNA was recloned into pLRNL containing either wild-type EGFR or ΔEGFR to generate two plasmids pL(EFv2)RNL encoding the chimeric wild-type EGFR, and pL(ΔEFv2)RNL encoding chimeric ΔEGFR.
Immunoprecipitation and Western Blot
Cells were washed with ice-cold PBS and were lysed inTriton-X-100 lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1% Triton-X100, 1 mM EDTA, 1 mM EGTA, 20 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 2 μg/ml of aprotinin and leupeptin, and a phosphatase inhibitor cocktail). For the immunoprecipitation, lysates were incubated with anti-EGFR at 4°C overnight. Ab–protein complexes were precipitated with Protein A/G-Agarose, washed, and analyzed by Western blotting. For Western blotting, samples were separated by SDS/PAGE using NuPAGE Bis-Tris gels (Invitrogen), blotted to PVDF and incubated with primary Ab for overnight. Ab complex was visualized by chemiluminescence. All antibodies for signaling study were purchased from Cell Signaling.
BrdU incorporation
Five thousand cells were seeded in 96 well culture plates. After 24h serum starvation, cells were incubated with 20 ng/ml EGF (Sigma) or 50 nM AP20187 in the presence of 100 μM BrdU for 24h. After labeling, BrdU incorporation was measured by colorimetric immunoassay using a commercially available cell proliferation ELISA kit (Roche).
Cell surface biotinylation
All the biotinylation procedures were conducted on ice. The cells were serum-starved, were incubated with 0.25 mg/ml EZ-Link NHS-SS-biotin (Pierce) for 1hr, and unreacted biotin was quenched with 20 mM glycine for 10min. After washing, cells were either subjected to 15 min post-stimulation with 20 ng/ml EGF or 50 nM AP20187 at 37°C (+ internalization) or proceeded to the next step immediately ( internalization). One dish of cells was treated with cleavage buffer (50 mM glutathione, 90 mM NaCl, 1 mM MgCl2, 0.1 mM CaCl2, 60 mM NaOH and 0.2% BSA, pH 8.6) for 20min twice. Another dish of cells was left untreated with cleavage buffer and directly processed for extraction to define total biotinylated proteins. Cells were lysed and the cleared lysates were incubated with streptavidin–conjugated beads (Amersham Biosciences) for overnight to isolate biotinylated proteins. And precipitated proteins were analyzed by western blot with FKBP antibody (Abcam).
Wound-Healing Assay
The cells were plated onto 6-well dishes and were allowed to grow to complete confluence. After serum starvation, wounds were then created using pipette tip, and EGF or AP20187 were added to each well. The pictures of same area were taken for each wound every day until EGF/AP20187 wells had healed using an inverted microscope at 5x magnification; photos were taken immediately after a wound was inflicted to the cell monolayer, the distances between the edges of the wound were measured. The degree of motility is expressed as percent of wound closure as compared with the zero time point.
Animal Experiments
All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of MD Anderson Cancer Center. Four groups of 10 animals each were implanted intracranially with 2×105 chEGFR/chΔEGFR expressing U87 cells. At the same time, animals were implanted with Alzet osmotic minipumps (DURECT Corp) that maintain a constant flow for 2 weeks. The reservoir of the each pump was filled with AP20187 or with vehicle. Animals were sacrificed when moribund and tumors were harvested, and snap frozen and stored in liquid nitrogen. Lysates were made by homogenizing the tumor tissue in the lysis buffer. The brains were fixed in 4% paraformaldehyde, were then performed H&E staining. Survival data were analyzed with Gehan-Breslow-Wilcoxon Test using Prism software (GraphPad Software, Inc.) and p-values of less than 0.05 were considered statistically significant.
Results
ΔEGFR is activated by forced dimerization without interfering the normal function of the receptor
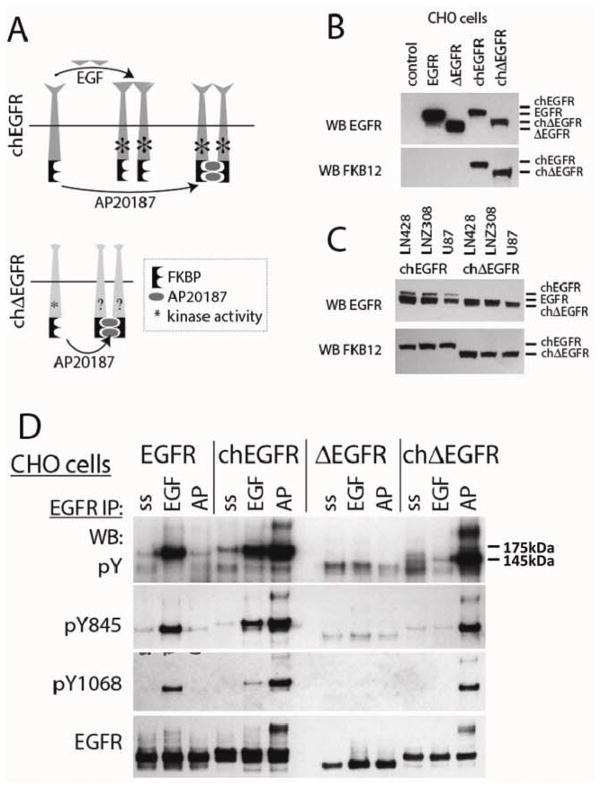
Chimeric receptors were created which had either a wild-type EGFR sequence, or the in-frame deletion mutant ΔEGFR, and two FKBP-F36V domains (24, 25) added at the C-terminus (Fig. 1A). Based on previous reports we anticipated that the wild-type chimeric receptor, chEGFR, would be activated both by EGF and by the chemical inducer of dimerization, AP20187 (30), while the activity of the mutant chΔEGFR was unknown. The chimeric receptors are slightly larger than their non-chimeric counterparts, and can be selectively detected using an antibody to the FKBP domains (Fig. 1B). When expressed in glioma cells, which have endogeneous EGFR, the chEGFR is readily distinguishable as it migrates at a higher position in the gel. However, the chΔEGFR migrates very close to the endogeneous EGFR, requiring detection with anti-FKBP antibody (Fig. 1C). Expression of the chimeric receptors was lower than the endogenous EGFR in glioma cells, as demonstrated by the relative intensity of the endogenous and chEGFR bands in the EGFR western blot, and the comparable level of expression of chEGFR and chΔEGFR in the FKBP12 blot (Fig. 1C). The simplest model in which to examine the relative activity of these receptors are cells that lack endogenous EGFR, and so we first tested their activation in CHO cells. EGFR and chEGFR (Fig. 1D), but not ΔEGFR or chΔEGF responded to EGF stimulation with increased levels of phosphorylation, as measured by both pan-phosphotyrosine and site-specific antibodies. Furthermore, as expected chEGFR was activated by AP20187, but neither of the non-chimeric receptors was. Interestingly, chΔEGFR was strongly stimulated by AP20187 treatment, to levels comparable to AP20187-stimulated chEGFR. The dimerization induced by AP20187 in the chimeric receptors could be detected on western blots as a band that migrated well above EGFR monomers with anti-EGFR antibodies. Similar, but fainter bands can be seen in EGF stimulated EGFR and chEGFR, but not in any of the other samples where ΔEGFR was expressed. These data demonstrate that forced dimerization of chΔEGFR significantly enhances the phosphorylation level of this receptor and does so in association with the apparent formation of dimers.
Figure 1.
Forced Dimerization Leads to Activation of chΔEGFR. (A) Schematic representation of the chimeric EGFR constructs that allow chemically induced dimerization. Chimeric receptors were constructed by fusing EGFR or ΔEGFR to twin, modified FKBP (FKBPv) domains. Chemical compound AP20187 was used for the dimerization of FKBPv. (B and C) Chimeric EGFR and ΔEGFR constructs were transiently transfected into CHO and glioma cells. Lysates were immunoblotted with anti-EGFR and anti-FKBP as indicated. In CHO cells, only the transfected EGFRs are detected. In glioma cells, endogenous EGFR is also detected with the anti-EGFR antibody. Endogenous EGFR and chΔEGFR migrate at approximately same level, and the bands often overlap. (D) CHO cells stably expressing chimeric receptors were stimulated with 50 ng/ml EGF or 50 nM AP20187 after serum starvation. Lysates were immunoprecipitated with anti- EGFR antibody, and blotted with anti-pTyr, anti-pEGFR (Y845, Y1068), and anti-EGFR antibodies as indicated.
Forced dimerization increases but does not fundamentally alter downstream signaling of EGF receptors
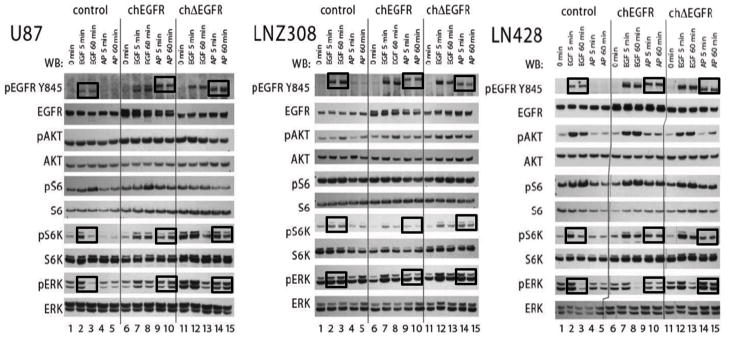
To examine the signaling of the chimeric receptors in glioma cells, we measured phosphorylation of downstream targets in the PI3K and MAPK pathways that are commonly associated with EGFR (Fig. 2). It is important to stress that in these cells EGF stimulation activates the endogenous EGFR, as well as chEGFR when present, but not chΔEGFR; AP20187 activates the two chimeric receptors selectively. EGF stimulation of cells expressing ch EGFR can therefore be used to compare signaling of the endogenous EGFR with elevated ch EGFR via stimulation with AP20187. In the PTEN negative cell line U87, we observed elevated levels of pAkt under all conditions, likely due to lack of regulation in the PI3K pathway. In LNZ308 cells, which also lack PTEN an increase in pAkt was observed in cells where endogenous EGFR, chEGFR or chΔEGFR was stimulated when compared to serum-starved cells. In LN428 cells, which express wild-type PTEN, pAkt levels were elevated by stimulation of endogenous EGFR by EGF, but not when both chimeric receptors were stimulated with AP20187. We also observed elevated pS6 under all conditions in all three of cell lines. A slightly different pattern emerged with pS6K, which showed lower basal levels in serum-starved cells, including the lines that lack PTEN. For this node, a positive response was registered whenever endogenous EGFR, chEGFR or chΔEGFR was stimulated. Upon PI3K activation, S6K is phosphorylated at T389 site by one of key effectors PI3K pathway, PDK1. Here we observed prolonged phosphorylation of S6K at T398 site, suggesting that forced dimerization is able to increase the signal received by pS6K. The pERK signals, in general, responded rapidly to EGF stimulation, with elevations at 5 min and a return to lower levels at 60 min. Elevations of pERK after AP20187 treatment were less pronounced than those caused by EGF (compare lanes 7 and 9 and lanes 12 and 14 in all three panels in Fig. 2), but also more sustained (compare lanes 8 and 10 and lanes 13 and 15 in all three panels in Fig. 2). In summary, these data show that chimeric receptors activate the same downstream targets in the PI3K and MAPK pathways as non-chimeric receptors, with somewhat slower and more sustained kinetics. Importantly they show that chemically induced dimerization of chΔEGFR causes an increase in downstream signaling to levels resembling those seen with acutely stimulated EGFR and significantly higher than those observed in cells expressing chΔEGFR alone.
Figure 2.
Signaling by chEGFR and chΔEGFR Receptors. Glioma cells U87, LNZ308, and LN428 stably expressing control vector, chEGFR or chΔEGFR were serum starved and then stimulated with 20 ng/ml EGF or 50 nM AP20187 and analyzed at two time points, 5 and 60 min. EGFR downstream signaling was analyzed by immunoblotting with indicated pan- and phospho antibodies; pEGFR(Tyr 845), pAKT(Ser473), pS6K(Thr389), pS6(Ser235/236), and pERK1/2(Thr202/Tyr204).
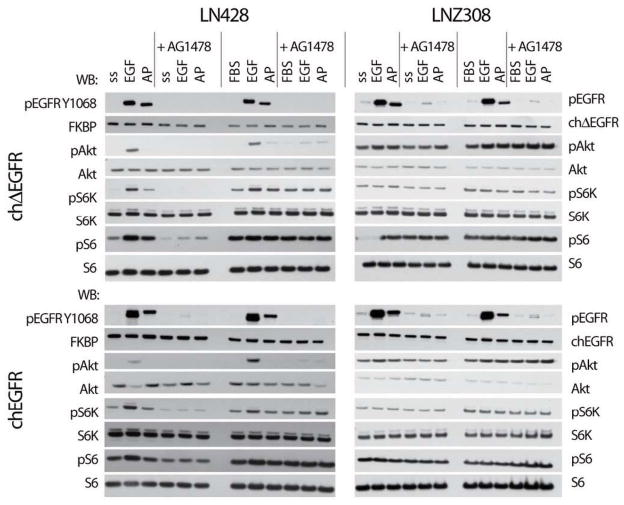
EGFR and ΔEGFR are both sensitive to inhibition by active site inhibitors, such as AG1478/tyrphostin. To confirm that the chimeric receptors were also susceptible to inhibition by this drug, we treated cells with AG1478 before stimulating them with EGF or AP20187, and assessed activity in EGFRs and downstream targets. At the level of the receptor, endogenous EGFR and chEGFR stimulated by EGF, or chEGFR and chΔEGFR stimulated by AP20187 were inhibited by AG1478, regardless of whether they were cultured in the presence of serum or not and in both LN428 and LNZ308 cells (Fig. 3). In terms of downstream pathways, cells in serum showed activation of Akt, S6 and S6K even when AG1478 was present, suggesting EGFR-independent pathways were active, as expected. Interestingly, one exception to this was levels of pAkt in the PTEN wild-type LN428 cells stimulated by EGF, which were lowered by AG1478 even in the presence of serum, though not to baseline, suggesting that Akt is more closely linked to EGFR in these cells, but that other serum ligands can stimulate pS6 and pS6K by other pathways. As before, stimulation of chEGFR or chΔEGFR with AP20187 did not lead to an increase in pAkt levels. In the absence of serum, AG1478 was able to suppress the activation of Akt, S6 and S6K in LN428 cells, but had no impact in LNZ308 cells. Importantly, the effect of AG1478 on EGF-stimulated and AP20187-stimulated cells was similar, suggesting that chimeric receptors responded to the inhibitor in a manner similar to the endogenous EGFR, and so that chemically induced dimerization did not fundamentally alter the mechanism of action of receptor.
Figure 3.
Chimeric Receptors Are Inhibited By AG1478. LN428 and LNZ308 glioma cells expressing either chEGFR or chΔEGFR were incubated in media containing no serum (serum starved, ss) or with regular growth medium containing 10% FBS, as indicated. They were left untreated or treated with 10 μM AG1478 for 2h before stimulation with 20 ng/ml EGF or 50 nM AP20187 for another 15 min. Signaling was analyzed by immunoblotting of lysates with the indicated antibodies.
STAT5 was identified as an important target of ΔEGFR and so we measured STAT5 phosphorylation status along with that of the signaling proteins GAB1, SHP2, SRC and STAT3 (Supplementary Fig. S1). Even in the absence of stimulation, the cells displayed a high level of phosphorylation of these signaling proteins, making it difficult to detect increases, although the level of pSTAT5 was higher in ch EGFR cells than cells expressing chEGFR, consistent with our previous work (22). We did see an increase in phosphorylated GAB1 when ch EGFR expressing cells were stimulated with AP20187 or when chEGFR cells were stimulated with EGF, with the chemically induced dimers generating a more sustained signal as before. These data further support the conclusion that increasing the activity of EGFR does not lead to the engagement of fundamentally different pathways.
A more comprehensive and open approach to comparing signaling patterns emanating from ch EGFR is to use phosphotyrosine directed shotgun proteomics. We employed an immunoaffinity based method to enrich phosphotyrosine containing peptides and analyzed them on an ion trap mass spectrometer. We identified 89 tyrosine-phosphorylated peptides and quantified 75 of those peptides (Supplementary Fig. S4 and Table S1). Similar to the observations made with specific signaling nodes by western blot, we did not detect many peptides with difference in the phosphorylation intensity when we compared AP20187 stimulated ch EGFR with AP20187 stimulated chEGFR, suggesting that when both receptors are maximally active that they resemble each other more in the overall downstream signal. Nevertheless, some specific differences remained, including SHC1, CDC2 and Paxillin which were phosphorylated at a higher level in AP20187-treated cells expressing ch EGFR than chEGFR, and are known downstream targets of EGFR (31, 32). We did observe a larger group of peptides, including one derived from EGFR, which show increased phosphorylation upon stimulation with AP20187 in ch EGFR cells, as was expected from the finding presented above.
The increased activity of ΔEGFR has no effect on its internalization
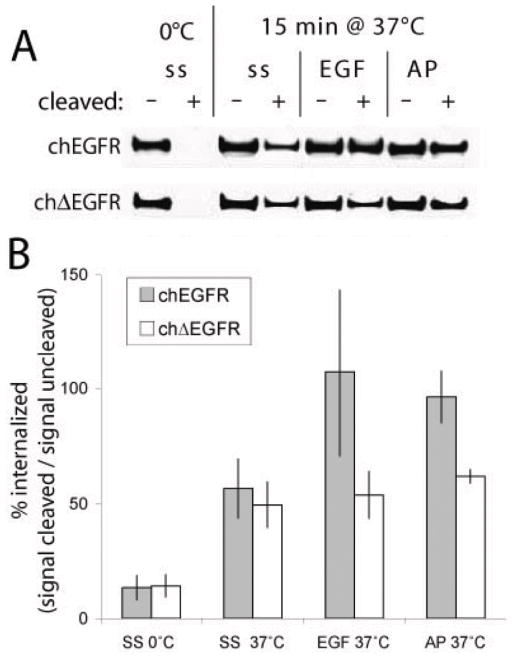
One characteristics of ΔEGFR is a reduced rate of internalization when compared to acutely stimulated EGFR, which we attributed to its relatively low level of activity (14). To determine whether increasing the activity level of chΔEGFR increased its rate of internalization, we labeled extracellular proteins with biotin, stimulated the cells with EGF or AP20187 at 37°C for 15 min, and then cleaved the remaining surface exposed biotin. Recovery of proteins with streptavidin and western blotting for FKBP selective revealed chimeric EGFRs that had been internalized (Fig. 4A). When cells were kept at 0°C throughout the experiment very little receptor was detected after cleavage, as expected because little membrane trafficking occurs at this temperature, effectively preventing internalization. The simple elevation of cells to 37°C for 15 min leads to a basal level of signal, as membranes resume normal turnover. In the case of chEGFR stimulation with either EGF or AP20187 further increased the level of internalized receptor, in accordance with the activity dependent internalization that this receptor undergoes. The chΔEGFR did not respond to EGF as expected, but it also showed no increase in internalization in response to AP20187 (all these data are quantified across three experiments in Fig. 4B). This suggests that ΔEGFR’s reduced internalization rate is not solely due to its inherently lower level of activity as previously proposed, but may reflect an inherent difference in how this receptor interacts with the endocytosis machinery. These data do suggest that the increase in signal we obtain with chemically induced dimerization of chΔEGFR is not able to overcome this aspect of the mutant receptor’s behavior.
Figure 4.
Chemically Induced Dimerization Does Not Promote chΔEGFR Internalization. (A) Cells were cooled to 4°C on ice, and surface proteins were biotinylated, before being subjected to a variety of conditions and then exposed to a biotin-cleavage agent incapable of crossing the membrane, or left uncleaved. Biotinylated proteins were recovered with streptavidin-conjugated beads, and the resulting precipitates immunoblotted with anti-FKBP. Conditions prior to cleavage included leaving the cells on ice, incubation with pre-warmed media containing no serum (ss), 20 ng/ml EGF or 50 nM AP20187 for 15 min at 37°C. As internalization protects proteins from cleavage, the degree of internalization can be determined by the signal after cleavage divided by the signal in the absence of cleavage. A typical experiment is shown. (B) Graphical representation of quantification of western blots as in (A), and showing the average percent of internalized receptors from three independent experiments.
Cell motility is not dependent on ΔEGFR activity
At the cellular level we examined two behaviors, DNA synthesis in S-phase and motility. Cells were serum starved, grown in FBS or stimulated with EGF or AP20187, and BrdU incorporation determined (Fig. 5). As expected, cells expressing chEGFR or chΔEGFR responded with increased BrdU incorporation when stimulated with any of these agents (Fig. 5, left hand panels). In the case of chΔEGFR-expressing cells this demonstrates that increasing the signal from the mutant receptor has impact at the cellular level. In LN428 cells we also observed higher basal BrdU incorporation in the absence of stimulation, suggesting that the ligand-independent activity of chΔEGFR was also manifest at this level. In contrast to DNA synthesis, motility as measured by a scratch assay was not enhanced by chΔEGFR even when it was stimulated by AP20187 (Fig. 5, right hand panel). LN428 cells expressing chΔEGFR showed migration rates similar to the parental cells, while chEGFR expressing cells showed elevated motility in the presence of either EGF or AP20187. This implies that elevated activity chEGFR, but not chΔEGFR could accelerate cell movement, consistent with a demonstrated role of EGFR in cell motility, but none for ΔEGFR. These data therefore also support that chemically induced dimerization enhanced the chΔEGFR signal but did not alter it from that of ΔEGFR at the level of cell behavior.
Figure 5.
Dimerization of chΔEGFR Induces Cell Division but Not Migration. (A) LN428 or LNZ308 glioma cells were serum starved and stimulated with 20 ng/ml EGF or 50 nM AP20187 in the presence of 100 μM BrdU for 24h. BrdU incorporation was measured by a colorimetric immunoassay. Data are from three independent experiments (*p < 0.05; **p < 0.01; t-test). (B) A scratch was made in a confluent cell culture of LN428 cells, that were grown in the absence of serum supplemented with 20 ng/ml EGF or 50 nM AP20187 as indicated, and the degree of scratch closure was measured at 24h using a digital photograph and XCAP-Lite software. Graph shows the average ± standard deviation from 4 independent experiments.
Increased activity of ΔEGFR shortens survival time in a xenograft model
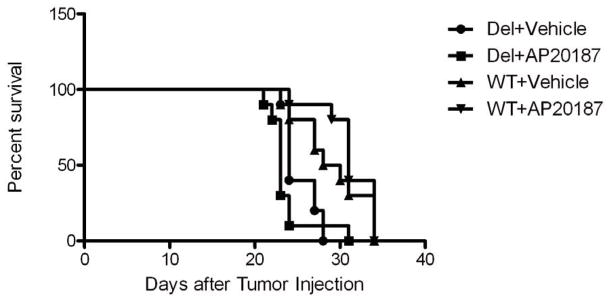
To evaluate whether increasing the signal from chΔEGFR strengthens its tumorigenic signal, chimeric receptor expressing U87 cells were intracranially implanted into nude mice and the tumors treated with AP20187 or vehicle for 2 weeks using Osmotic pumps. We confirmed expression of chimeric receptors in tumors and observed that the chimeric receptors were signaling in vivo as they had in cultured cells by immunoblotting (Supplementary Fig. S2). AP20187-stimulation of cells expressing chΔEGFR shortened median survival by one day, from 24 to 23 days (p = 0.0145 Wilcoxon Test), when compared to vehicle treated mice (Fig. 6). Furthermore histological examination of H&E stained tumor sections showed the AP20187 and vehicle tumors had a similar appearance, typical of U87 intracranial tumors, suggesting that the shortening of survival was not related to changes in tumor morphology such as invasion caused by AP20187 (Supplementary Fig. S3). Therefore, increasing the intensity of the chΔEGFR signal also enhances its already potent oncogenic signal in vivo.
Figure 6.
Comparison of the survival curves. AP20187 treatment cause early death in chΔEGFR-implanted mice. Data were analyzed using Gehan-Breslow-Wilcoxon tests of the statistical package Prism version 5.0 (GraphPad Software Inc.). P values of < 0.05 were taken as statistically significant.
Discussion
The absence of an intact ligand-binding domain in ΔEGFR, as well as the lack of response to EGF stimulation, suggested that it is defective in dimerization either as a homodimer or with family members capable of being activated by EGF (33, 34). Nevertheless, under some experimental circumstances, dimers have been reported, and it has been suggested that dimerization is required for ΔEGFR function (23). Here we report for the first time the experimentally induced dimerization of a chimeric version of ΔEGFR, and observed a very significant enhancement of its phosphorylation levels. It is worth noting that the strategy that we used to force dimerization of EGFRs, first established by Dr. David Spencer and colleagues, did not result in abnormally high levels of phosphorylation of chEGFR when compared to EGF stimulation, suggesting that at least the initial impact of dimerization was similar regardless of whether it was mediated by the natural, extracellular domain and ligand or an artificial, intracellular domain and ligand. We conclude that if chΔEGFR was dimerized to any significant degree in terms of duration and stability in the absence of AP20187, the addition of this compound would not have been able to induce such a significant increased in phosphorylation and hence propose that naturally occurring ΔEGFR does not dimerize to the extent of the ligand-activated wild-type receptor. Interestingly, a recent report showed that a small subpopulation of ΔEGFR is dimerized, via a disulfide bond, and that the dimerized receptors are the most active as judged by tyrosine phosphorylation (35). Taking these data together allows us to suggest that forced dimerization increases the activity of the ΔEGFR pool overall by bringing more receptors into this state, and stabilizing them there.
That ΔEGFR generates a signal that is different in some aspect from its wild-type counterpart is likely, as it is a much more potent oncogene in many model systems and is associated with worse outcomes for patients with glioblastoma. Our previous analysis comparing the signal of ΔEGFR and EGFR using phosphotyrosine directed shotgun phosphoproteomics suggested that there are no absolute differences, in that we were not able to identify any downstream signaling components that were exclusive to mutant or wild-type receptor. However, we did identify several phosphorylated tyrosines in target proteins that showed a stronger association with ΔEGFR, with signals reaching levels similar to what was observed when wild-type EGFR was acutely stimulated in serum starved cells, a likely non-physiological maximization of its signal. One of these targets was Y699 on pSTAT5b (22), and we saw a similar preference for phosphorylation of this residue here, in that the signal in chΔEGFR cells was higher than in serum starved chEGFR cells (Supplementary Fig. S1). However, the level of pSTAT5 was not increased further by addition of AP20187 to cells expressing chΔEGFR, suggesting that it may be maximal, and not limited by the low level signal of unstimulated ch EGFR. In other words, ΔEGFR may signal at an overall lower level, but reach near-maximal levels with a few select downstream targets, including STAT5. Other signaling components, such as GAB1 may still be preferentially connected (18, 22) but not maximally so (Supplementary Fig. S1). Comparison of overall patterns of phosphorylation of a spectrum of tyrosine residues by an open approach (Supplementary Fig. S4 and Table. S1) did not reveal any profound differences, further suggesting that changing signal strength does not noticeably change signal content, and so that the two are not tightly coupled. Indeed several of phosphorylations that we did observe to increase with forced dimerization of ch EGFR, such as those on paxillin, might be related to signaling through established downstream targets, in this case c-Met. This conclusion is supported by the analysis of cell behaviors. Increasing the signal of ch EGFR by forced dimerization enhances the existing stimulation of mitosis, but does not convert a non-migration/invasion-inducing ΔEGFR signal to a migration/invasion-inducing EGFR signal. While the close connection of c-Met and EGFR observed by others and us might suggest that EGFR should promote cell motility, a behavior associated mesenchymal transition of epithelial tumors, we do not observe it in glioma cells.
ΔEGFR signaling is constitutive with little receptor internalization. We previously hypothesized that the lack of ΔEGFR downregulation is due to the inability of the weakly active ΔEGFR to recruit the CIN85-Cbl complex that initiates endocytosis in EGFR (36). This hypothesis would predict that enhancing the signal by forced dimerization would reestablish the active downregulation of the receptor, but we did not observe this for chΔEGFR, while the controls for chEGFR behaved as expected (Fig. 4). Our results strongly suggest that increased ΔEGFR activity is not sufficient to reestablish ΔEGFR downregulation, and so we conclude that the impaired downregulation of ΔEGFR is not due to its lower level of activity or lack of dimerization.
In summary our data provide the first direct, experimental test of ΔEGFR dimerization, and imply that it does not form strong, durable dimers under normal circumstances, which may be related to its low level of activity. Increasing the ΔEGFR activity by forced dimerization of chΔEGFR also provided the first opportunity to compare signals of ΔEGFR and EGFR at similar amplitudes by an open phosphoproteomic approach, which revealed no major qualitative differences. This in turn suggests that the differences that were observed by others and us previously (21, 22) related to the signals that the naturally occurring, overall low-activity ΔEGFR maintains at an elevated, near-maximal level; they stood out because they were high by comparison to the bulk of EGFR targets. Increasing the chΔEGFR signal did increase its ability to stimulate entry into S-phase and tumor growth rate as measured by survival of tumor bearing mice. Interestingly, however, the lack of downregulation of chΔEGFR was not altered by enhancing its activity, suggesting that this may be a fundamental characteristic of this mutant. The picture of ΔEGFR that emerges is of a receptor whose oncogenicity relies on a sustained signal, which although overall of low intensity, is capable of focusing on several select downstream nodes which are significantly activated. Raising its activity enhances the signal overall profoundly, but only to a modest degree on some of the already very active partners. The key to ΔEGFR’s oncogenicity may therefore very well be its inability to be downregulated, and this in turn suggests that altering this behavior may be key to targeting it therapeutically.
Supplementary Material
Acknowledgments
We thank Dr. Victor Rivera and ARIAD Pharmaceuticals, for providing EGFR chimera plasmids, Homodimerization Regulation Kits and AP20187 for our experiments. We are grateful to Dr. David Spencer, Baylor College of Medicine for advice on the use of the chemically induced dimerization technology and Dr. Gregory Fuller, Dept. of Neuropathology, UT MD Anderson Cancer Center for advice on histopathology. We thank Verlene Henry and Lindsay Holmes of Dept. of Neurosurgery, UT MD Anderson Cancer Center for their help in carrying out animal experiments.
Grant Support
These studies were supported in part by grants from the National Cancer Institute of the National Institutes of Health, RO1CA108500 (O.B.), P50CA127001 (O.B.) and through The University of Texas MD Anderson Cancer Centre Support Grant CA016672.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
References
- 1.Libermann TA, Nusbaum HR, Razon N, Kris R, Lax I, Soreq H, et al. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature. 1985;313:144–7. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 2.Wong AJ, Bigner SH, Bigner DD, Kinzler KW, Hamilton SR, Vogelstein B. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc Natl Acad Sci U S A. 1987;84:6899–903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ekstrand AJ, Sugawa N, James CD, Collins VP. Amplified and rearranged epidermal growth factor receptor genes in human glioblastomas reveal deletions of sequences encoding portions of the N- and/or C-terminal tails. Proc Natl Acad Sci U S A. 1992;89:4309–13. doi: 10.1073/pnas.89.10.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moscatello DK, Holgado-Madruga M, Godwin AK, Ramirez G, Gunn G, Zoltick PW, et al. Frequent expression of a mutant epidermal growth factor receptor in multiple human tumors. Cancer Res. 1995;55:5536–9. [PubMed] [Google Scholar]
- 5.Tang CK, Gong XQ, Moscatello DK, Wong AJ, Lippman ME. Epidermal growth factor receptor vIII enhances tumorigenicity in human breast cancer. Cancer Res. 2000;60:3081–7. [PubMed] [Google Scholar]
- 6.Shinojima N, Tada K, Shiraishi S, Kamiryo T, Kochi M, Nakamura H, et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003;63:6962–70. [PubMed] [Google Scholar]
- 7.Heimberger AB, Hlatky R, Suki D, Yang D, Weinberg J, Gilbert M, et al. Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clin Cancer Res. 2005;11:1462–6. doi: 10.1158/1078-0432.CCR-04-1737. [DOI] [PubMed] [Google Scholar]
- 8.Pelloski CE, Ballman KV, Furth AF, Zhang L, Lin E, Sulman EP, et al. Epidermal growth factor receptor variant III status defines clinically distinct subtypes of glioblastoma. J Clin Oncol. 2007;25:2288–94. doi: 10.1200/JCO.2006.08.0705. [DOI] [PubMed] [Google Scholar]
- 9.Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, et al. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci U S A. 1994;91:7727–31. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lal A, Glazer CA, Martinson HM, Friedman HS, Archer GE, Sampson JH, et al. Mutant epidermal growth factor receptor up-regulates molecular effectors of tumor invasion. Cancer Res. 2002;62:3335–9. [PubMed] [Google Scholar]
- 11.Nagane M, Coufal F, Lin H, Bogler O, Cavenee WK, Huang HJ. A common mutant epidermal growth factor receptor confers enhanced tumorigenicity on human glioblastoma cells by increasing proliferation and reducing apoptosis. Cancer Res. 1996;56:5079–86. [PubMed] [Google Scholar]
- 12.Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–77. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 13.Huang HS, Nagane M, Klingbeil CK, Lin H, Nishikawa R, Ji XD, et al. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem. 1997;272:2927–35. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt MH, Furnari FB, Cavenee WK, Bogler O. Epidermal growth factor receptor signaling intensity determines intracellular protein interactions, ubiquitination, and internalization. Proc Natl Acad Sci U S A. 2003;100:6505–10. doi: 10.1073/pnas.1031790100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pawson T, Hunter T. Signal transduction and growth control in normal and cancer cells. Curr Opin Genet Dev. 1994;4:1–4. doi: 10.1016/0959-437x(94)90084-1. [DOI] [PubMed] [Google Scholar]
- 16.Moscatello DK, Montgomery RB, Sundareshan P, McDanel H, Wong MY, Wong AJ. Transformational and altered signal transduction by a naturally occurring mutant EGF receptor. Oncogene. 1996;13:85–96. [PubMed] [Google Scholar]
- 17.Prigent SA, Nagane M, Lin H, Huvar I, Boss GR, Feramisco JR, et al. Enhanced tumorigenic behavior of glioblastoma cells expressing a truncated epidermal growth factor receptor is mediated through the Ras-Shc-Grb2 pathway. J Biol Chem. 1996;271:25639–45. doi: 10.1074/jbc.271.41.25639. [DOI] [PubMed] [Google Scholar]
- 18.Holgado-Madruga M, Wong AJ. Role of the Grb2-associated binder 1/SHP-2 interaction in cell growth and transformation. Cancer Res. 2004;64:2007–15. doi: 10.1158/0008-5472.can-03-2886. [DOI] [PubMed] [Google Scholar]
- 19.Moscatello DK, Holgado-Madruga M, Emlet DR, Montgomery RB, Wong AJ. Constitutive activation of phosphatidylinositol 3-kinase by a naturally occurring mutant epidermal growth factor receptor. J Biol Chem. 1998;273:200–6. doi: 10.1074/jbc.273.1.200. [DOI] [PubMed] [Google Scholar]
- 20.Huang PH, Cavenee WK, Furnari FB, White FM. Uncovering therapeutic targets for glioblastoma: a systems biology approach. Cell Cycle. 2007;6:2750–4. doi: 10.4161/cc.6.22.4922. [DOI] [PubMed] [Google Scholar]
- 21.Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, et al. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci U S A. 2007;104:12867–72. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chumbalkar V, Latha K, Hwang Y, Maywald R, Hawley L, Sawaya R, et al. Analysis of phosphotyrosine signaling in glioblastoma identifies STAT5 as a novel downstream target of DeltaEGFR. J Proteome Res. 2011;10:1343–52. doi: 10.1021/pr101075e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chu CT, Everiss KD, Wikstrand CJ, Batra SK, Kung HJ, Bigner DD. Receptor dimerization is not a factor in the signalling activity of a transforming variant epidermal growth factor receptor (EGFRvIII) Biochem J. 1997;324 ( Pt 3):855–61. doi: 10.1042/bj3240855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–24. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 25.Clackson T, Yang W, Rozamus LW, Hatada M, Amara JF, Rollins CT, et al. Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc Natl Acad Sci U S A. 1998;95:10437–42. doi: 10.1073/pnas.95.18.10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farrar MA, Alberol-Ila J, Perlmutter RM. Activation of the Raf-1 kinase cascade by coumermycin-induced dimerization. Nature. 1996;383:178–81. doi: 10.1038/383178a0. [DOI] [PubMed] [Google Scholar]
- 27.Yang J, Symes K, Mercola M, Schreiber SL. Small-molecule control of insulin and PDGF receptor signaling and the role of membrane attachment. Curr Biol. 1998;8:11–8. doi: 10.1016/s0960-9822(98)70015-6. [DOI] [PubMed] [Google Scholar]
- 28.Mohi MG, Arai K, Watanabe S. Activation and functional analysis of Janus kinase 2 in BA/F3 cells using the coumermycin/gyrase B system. Mol Biol Cell. 1998;9:3299–308. doi: 10.1091/mbc.9.12.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kume A, Ito K, Ueda Y, Hasegawa M, Urabe M, Mano H, et al. A G-CSF receptor-gyrase B fusion gene: A new type of molecular switch for expansion of genetically modified hematopoietic cells. Biochem Biophys Res Commun. 1999;260:9–12. doi: 10.1006/bbrc.1999.0859. [DOI] [PubMed] [Google Scholar]
- 30.Muthuswamy SK, Gilman M, Brugge JS. Controlled dimerization of ErbB receptors provides evidence for differential signaling by homo- and heterodimers. Mol Cell Biol. 1999;19:6845–57. doi: 10.1128/mcb.19.10.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sturla LM, Amorino G, Alexander MS, Mikkelsen RB, Valerie K, Schmidt-Ullrichr RK. Requirement of Tyr-992 and Tyr-1173 in phosphorylation of the epidermal growth factor receptor by ionizing radiation and modulation by SHP2. J Biol Chem. 2005;280:14597–604. doi: 10.1074/jbc.M413287200. [DOI] [PubMed] [Google Scholar]
- 32.Liang X, Fonnum G, Hajivandi M, Stene T, Kjus NH, Ragnhildstveit E, et al. Quantitative comparison of IMAC and TiO2 surfaces used in the study of regulated, dynamic protein phosphorylation. J Am Soc Mass Spectrom. 2007;18:1932–44. doi: 10.1016/j.jasms.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Chen WS, Lazar CS, Poenie M, Tsien RY, Gill GN, Rosenfeld MG. Requirement for intrinsic protein tyrosine kinase in the immediate and late actions of the EGF receptor. Nature. 1987;328:820–3. doi: 10.1038/328820a0. [DOI] [PubMed] [Google Scholar]
- 34.Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110:669–72. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- 35.Ymer SI, Greenall SA, Cvrljevic A, Cao DX, Donoghue JF, Epa VC, et al. Glioma Specific Extracellular Missense Mutations in the First Cysteine Rich Region of Epidermal Growth Factor Receptor (EGFR) Initiate Ligand Independent Activation. Cancers. 2011;3:2032–49. doi: 10.3390/cancers3022032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt MH, Hoeller D, Yu J, Furnari FB, Cavenee WK, Dikic I, et al. Alix/AIP1 antagonizes epidermal growth factor receptor downregulation by the Cbl-SETA/CIN85 complex. Mol Cell Biol. 2004;24:8981–93. doi: 10.1128/MCB.24.20.8981-8993.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.