Abstract
Autophagy is a lysosomal bulk degradation process for intracellular protein and organelles. FIP200 (200 kDa FAK-family interacting protein) is an essential component of mammalian autophagy that is implicated in breast cancer in recent studies. Here we show that inactivation of FIP200 resulted in deficient repair of DNA damage induced by ionizing radiation and anticancer agents in mouse embryonic fibroblasts (MEFs). The persistent DNA damage correlated to increased apoptosis and reduced survival of FIP200 knockout (KO) MEFs after treatments with camptothecin (CPT), a topoisomerase I inhibitor and chemotherapeutic agent. Re-expression of FIP200 in FIP200 KO MEFs restored both efficient DNA damage repair and cell survival. Furthermore, knock-down of the increased p62 expression in FIP200 KO MEFs rescued the impaired DNA damage repair and CPT-induced cell death. In contrast, treatment of cells with N-acetyl-cysteine did not affect these defects in FIP200 KO MEFs. Lastly, FIP200 KO MEFs also showed deficient DNA damage repair and increased cell death compared to control MEFs, when treated with etoposide, a topoisomerase II inhibitor and another anticancer agent. Together, these results identify a new function for FIP200 in the regulation of DNA damage response and cell survival through its activity in autophagy, and suggest the possibility of FIP200 or other autophagy proteins as a potential target for treatment to enhance the efficiency of cancer therapy using DNA damage-inducing agents.
Introduction
Autophagy is a conserved intracellular process for bulk degradation of proteins and organelles through the formation of the double-membrane-bound vesicles called autophagosomes and their fusions with lysosomes (1, 2). It is induced in response to nutrient starvation and other stress conditions and functions to maintain cellular homeostasis by removing large protein aggregates and damaged organelles and recycling the degraded cellular components for macromolecular synthesis in these cells. Both basal and starvation-induced autophagy has been shown to play critical roles in a variety of physiological and pathological processes, including adaptive response to starvation, quality control of intracellular proteins and organelles, anti-aging, suppression of tumor formation, antigen presentation, and elimination of intracellular microbes (3-7).
Previous studies have shown that autophagy can act both positively and negatively in cancer cells (8, 9). In response to various cellular stresses, activation of autophagy provides cellular protection by eliminating harmful cytosolic components/invading pathogens and maintaining energy balance. This pro-survival function of autophagy could promote tumor cell survival and growth in the tumor microenvironment of hypoxia and nutrient starvation. Indeed, pharmacological or genetic inhibition of autophagy has been shown to sensitize tumor cells to the cytotoxic effects of chemotherapy and ionizing radiation to enhance cancer treatments (10-15). On the other hand, defective autophagy has also been linked to increased tumorigenesis because mono-allelical deletion of the mammalian autophagy gene beclin1 is frequently found in sporadic human breast cancers and ovarian cancers (16) and heterozygous deletion of beclin1 promoted spontaneous malignancies including lung and liver cancers and lymphomas in mouse models (17-19). It was also demonstrated that in apoptosis-defective cells, inhibition of autophagy caused by heterozygous loss of beclin1 or homozygous deletion of Atg5 induced accumulation of p62, damaged mitochondria and reactive oxygen species (ROS), leading to genomic instability and tumorigenesis (14, 20, 21).
FIP200 (FAK-family Interacting Protein of 200 kDa) encodes a conserved protein characterized by a large coiled-coil region containing a leucine zipper motif, which was initially found through its interaction with focal adhesion kinase (FAK) and its related kinase Pyk2 (22, 23). Several recent studies identified FIP200 as a component of the ULK1-Atg13-FIP200 complex essential for induction of autophagy in mammalian cells (24-28). Earlier studies implicated a role of FIP200 in breast cancer as deletion of FIP200 gene was found in a fraction of primary mammary tumor samples (29) and overexpression of FIP200 inhibited cell cycle progression in several breast cancer cell lines (30). However, we found recently that heterozygous deletion of FIP200 did not lead to development of mammary or any other tumors, whereas homozygous deletion resulted in embryonic lethality (31). Moreover, conditional KO of FIP200 in mammary epithelial cells (MaECs) did not lead to spontaneous development of breast cancer (32), suggesting that, in contrast to the earlier suggestion (29, 30) and unlike the better characterized autophagy protein Beclin1 (17-19), FIP200 may not function as a suppressor for breast or other cancers. Thus it also remains to be determined whether inactivation of FIP200 could lead to increased DNA damage and genomic instability which often associate with tumorigenesis, as observed in the deletion of several other autophagy proteins including Beclin1 (14, 20, 21).
In this study, we investigated the potential role of FIP200 in DNA damage repair and cell death upon various genotoxic treatments. We found that FIP200 deletion led to a significant decrease in DNA damage repair in response to ionizing radiation as well as cancer chemotherapeutic agents camptothecin (CPT) and etoposide. FIP200-null cells also showed an increased sensitivity to cell death induced by CPT and etoposide, which correlated to the increased DNA damage of the cells. These studies also identified p62 as a critical mediator of FIP200 regulation of DNA damage repair and cell survival. These results implicate FIP200 in maintaining normal cellular response to DNA damage and also provide support for the idea of targeting FIP200 or other autophagy proteins in combination with chemotherapy for cancer.
Materials and Methods
Cell cultures and reagents
MEFs were cultured in DMEM from Gibco (Invitrogen, CA) with 10% fetal bovine serum (FBS) (Atlanta Biologicals, GA) and 100 U/ml penicillin-Streptomycin, and incubated in 5% CO2 incubator with 95% humidity at 37°C. All chemicals were purchased from Sigma (Sigma, St. Louis, MO) unless otherwise specified. Camptothecin and Etoposide were dissolved in 100% dimethyl sulfoxide (DMSO) and stored at -20°C until use, and those treatments were not exceeded over 0.1 % (v/v) media. N-acetyl-cysteine (NAC) and hydrogen peroxide (30% v/v) were freshly prepared daily to experimental concentrations in sterile water and filtrated and stored at 4°C.
Preparation of recombinant lentiviruses encoding p62 shRNA
pGIPZ lentiviral vectors (Openbiosystem, AL) encoding p62 shRNA or a scrambled control shRNA were obtained from the shRNA core facility at the University of Michigan. For lentivirus packaging, we followed the calcium phosphate transfection method as described in manufacturer's protocol (Openbiosystem). Efficient infection of the recipient cells by the recombinant lentiviruses was determined by GFP expression under an Olympus IX70 fluorescence microscope (Olympus, Center Valley, PA).
Immunofluorescence
Immunofluorescent staining was performed in accordance with the manufacturer's protocol (Cell Signaling Tech, Danvers, MA). Briefly, cells were grown on coverslips and fixed in 4% formaldehyde for 15 min at room temperature after a rinse with PBS. Each coverslip in cell-side up position was rinsed three times in PBS for 5 minutes each, and immersed in blocking buffer (5% normal goat serum and 0.3% Triton X-100 in PBS) for 60 minutes at room temperature. After blocking, cells were incubated overnight at 4°C or 2 h at room temperature with anti-p62 (Enzo Life Sciences, Plymouth Meeting, PA) or anti-γ H2A.X (Cell Signaling Technology, Danvers, MA) rabbit primary antibody properly diluted in antibody dilution buffer (10% bovine serum albumin and 0.3% Triton X-100 in PBS). Rinsed three times in PBS for 5 minutes each, then coverslips were incubated with FITC- and Texas red-conjugated goat anti-rabbit IgG secondary antibodies diluted in antibody dilution buffer for 1 h at room temperature in dark. For nuclei counterstaining, 4',6-diamidino-2-phenylindole(DAPI) (Invitrogen, Camarillo, CA) was used. Washed with three times with PBS for 5 minutes each, slides were mounted with Vectrashield® mounting medium (Vector, Burlingame, CA) and examined under an Olympus BX41 microscope (Olympus, Center Valley, PA).
Western blotting analysis
Cells were lysed with hot SDS lysis buffer (1% SDS, 1 mM sodium ortho-vanadate, 10 mM Tris pH [7.4]) with a brief sonication. They were centrifuged at 13000 rpm for 10 minutes at 4 °C and the supernatant of each sample was collected. Equal amount of proteins was subjected to electrophoresis on 6 to 12% SDS-PAGE and transferred to nitrocellulose membranes for overnight at 25 V or 1 h at 100 V. After transfer, membranes were blocked with 5% fat-free milk in TBST (0.1M Tris-HCl, pH8.0, 0.9% (wt/vol) NaCl, 0.1% (vol/vol) Tween-20) for 1 hr at room temperature and incubated with primary antibodies in 5% fat-free milk TBST at 4°C overnight or for 1 h at room temperature. And then, properly diluted HRP-conjugated secondary antibody was replaced and incubated for 1 h at room temperature. After 3X10 min washing in TBST, chemiluminescence generated by ECL solution (Thermo Scientific, IL) was captured by Foto/Analyst Luminary/FX systems (Fotodyne Inc, WI).
Neutral comet assay
DNA strand break was analyzed by single-cell agarose gel electrophoresis under neutral conditions, as described previously (33). N1 neutral lysis buffer (2% sarkosyl, 0.5M Na2EDTA, 0.5mg/ml proteinase K (pH 8.0) and N2 electrophoresis solution (90mM Tris buffer, 90mM boric acid, 2mM Na2EDTA (pH 8.5) were used. Lysis was performed in the 37°C incubator overnight. The images were analyzed for comet tail moment using CometScore software. At least 40 images were processed for each sample.
Clonogenic survival and cell viability and measurements
To determine clonogenic survival of cells after IR, cells (at about 70% confluence) were harvested immediately after irradiation and 500 cells were re-seeded onto 100 mm culture dishes in triplicates. After 2 weeks, cell colonies were fixed by 6.0% glutaraldehyde, and then stained with 0.5% crystal violet. The surviving fraction was calculated by counting the number of colonies compared with non-irradiated control.
Cell viability was determined by MTT (thiazolyl blue) assay, as described previously (34). Propidium iodide (PI) staining was also used to detect cell death. Briefly, cells were seeded into a 6 well plate and cultured at 37°C, 5% CO2 until over 60% confluency and then subjected to various treatments as described in the text. Two ug/ml PI solution was directly added into medium and incubated for 10 min. The number of PI-positive cells was determined using an Olympus IX70 fluorescence microscope (Olympus).
Flow cytometric analysis of ROS
Reactive oxygen species generation was determined by staining with DCFDA (Invitrogen) at 10 μm for 15 min at 37°C, according to manufacturer's instructions, followed by flow cytometry, as described previously (35).
Statistical analysis
Data are presented as means±SEM and analyzed with SigmaStat v 3.1 (Jandel Scientific, San Rafael, CA); p < 0.05 was considered significant.
Results
FIP200 deletion leads to defective repair of irradiation-induced DNA damage
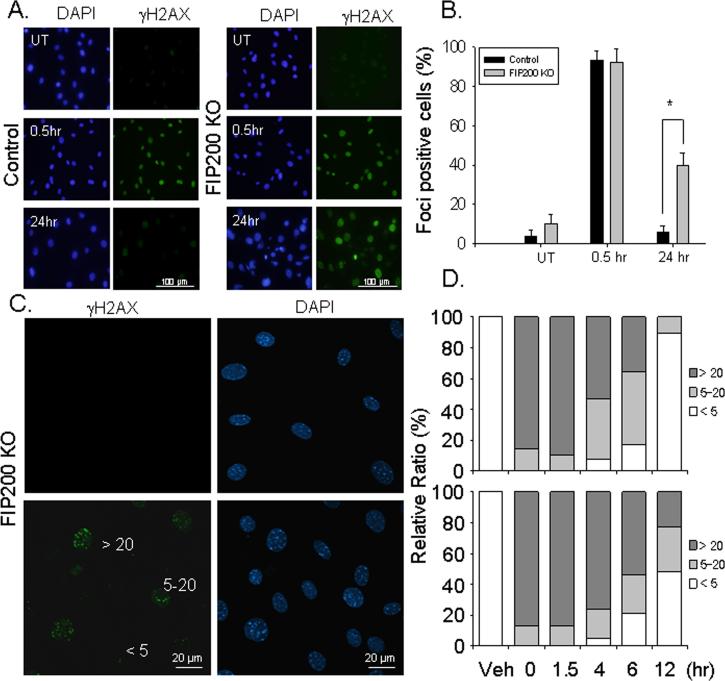
To investigate a potential role of FIP200 in the DNA damage response, FIP200 KO and wild type control mouse embryonic fibroblasts (MEFs) were subjected to ionizing radiation (IR) to induce DNA damage. At various times after exposure to 10 Gy of IR, the cells were immunostained by antibodies against S139 phosphorylated H2AX (γ-H2AX), a marker for DNA double-strand break formation (36). As shown in Figs. 1A and 1B, rapid induction of DNA damage was detected within 30 min in both control and FIP200 KO MEFs. In control MEFs, γ-H2AX signal was abolished at 24 hr after ionizing radiation, indicating repair of the DNA damage as expected (37). In contrast, γ-H2AX signal remained in a substantial fraction of FIP200 KO MEFs. Furthermore, examination of the cells at 24 hr after IR under higher magnification showed sustained γ-H2AX foci in the nucleus of FIP200 KO MEFs but not control MEFs (Data not shown). We next subjected the cells to a lower dose IR at1.5 Gy and examined the individual foci per cell at various times after IR exposure. As shown in Figs. 1C and 1D, similar level of DNA damage was induced by 1.5 Gy IR in both FIP200 KO and control MEFs. At 4 to 6 hr, approximately equal fractions of control MEFs contained 5-20 foci per cell and >20 foci per cell with a small fraction having <5 foci per cell, whereas the majority of FIP200 KO MEFs contained >20 foci per cell. By 12 hr, most of the control MEFs had <5 foci per cell, while significant fractions of FIP200 KO MEFs still contain 5-20 or >20 foci per cell. Double immunofluorescent staining showed co-localization of γ-H2AX and 53BP1 in the foci of the cells (Supplemental Fig. S1A), supporting that genuine DSBs were determined by γ-H2AX foci measurements. Furthermore, treatment of cells with an autophagy inhibitor 3MA also reduced the efficiency of DNA damage repair after IR (Supplemental Fig. S1), which is consistent with the idea that defective autophagy in FIP200 KO MEFs is responsible for the defective DNA damage repair.
Fig. 1. Analysis of ionizing radiation-induced DNA damage and repair in FIP200 KO MEFs.
(A-B) Control and FIP200 KO MEFs were exposed to 0 (UT) or 10 Gy of X-ray followed by 0.5 or 24 hr incubation at 37°C, as indicated. They were then fixed and immunostained with anti-γH2AX antibodies and DAPI (A). The percentage of cells with nuclear foci marked by γH2AX was determined under higher magnification. Mean ± SEM from three experiments are shown in panel B. *P<0.05. (C-D) Control and FIP200 KO MEFs were exposed to 0 (UT) or 1.5 Gy of X-ray followed by incubation at 37°C for various times, as indicated. The cells were then fixed, and immunostained with anti-γH2AX antibodies and DAPI, and viewed under higher magnification to visualize individual foci. Panel C shows representative images of cells with < 5, 5-20, or >20 foci. Panel D shows percentages of cells with <5, 5–20, or >20 foci per cell.
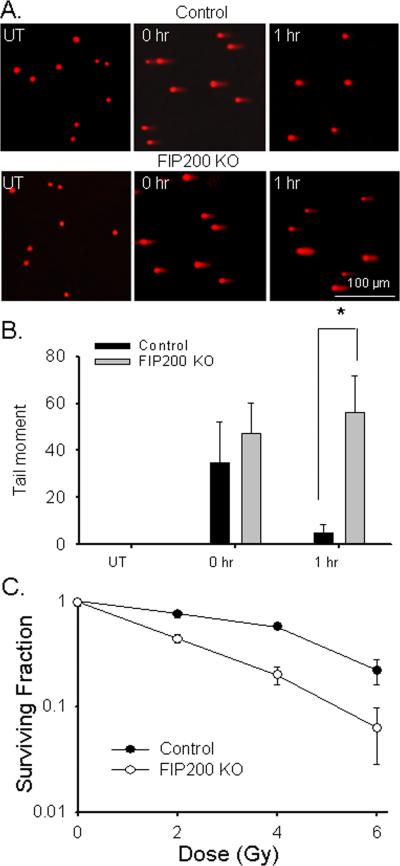
To further investigate whether the increased DNA damage is caused by reduced DNA repair capacity, the extent of DNA damage by IR was measured by neutral comet assay with quantification by tail moment as described in materials and methods. Consistent with previous reports (38, 39), IR induced short-lived DNA double-stranded breaks that were repaired within 1 hr post-incubation time after treatment with 10 Gy X-ray in control MEFs (Fig. 2A upper panels and Fig. 2B). In contrast, these DNA breaks persisted beyond 1 hr after post-incubation time (Fig. 2A lower panels and Fig. 2B) and only reduced to almost un-detectable level by 5 hr after resting (data not shown) in FIP200 KO MEFs after the same dose of IR. Consistent with the reduced DNA damage repair, FIP200 KO MEFs also showed a dose-dependent decrease in viability after IR compared to control MEFs (Fig. 2C). Taken together, these results demonstrated that that inhibition of autophagy by FIP200 deletion resulted in defective DNA damage repair and reduced cell survival in response to irradiation.
Fig. 2. Analysis of ionizing radiation-induced DNA double strand break and cell survival in FIP200 KO MEFs.
(A-B) Control and FIP200 KO MEFs were exposed to 0 (UT) or 10 Gy of X-ray followed by 0 or 1 hr incubation at 37°C, as indicated. They were then analyzed for DNA breaks by comet assay under neutral conditions. Representative images of cells with comet tails are shown in panel A. Tail moment was calculated by computerized image analysis with Comet Score™ software. Mean ± SEM from three experiments are shown in panel B. *P<0.05. (C) Control and FIP200 KO MEFs were exposed to various dose of IR, as indicated. Cells were then subjected to clonogenic survival assay as described in the Materials and Methods. The ratio between IR-treated and non-irradiated cells is shown. Results were from three independent experiments.
Reduced DNA damage repair leads to increased cell death in FIP200 KO cells
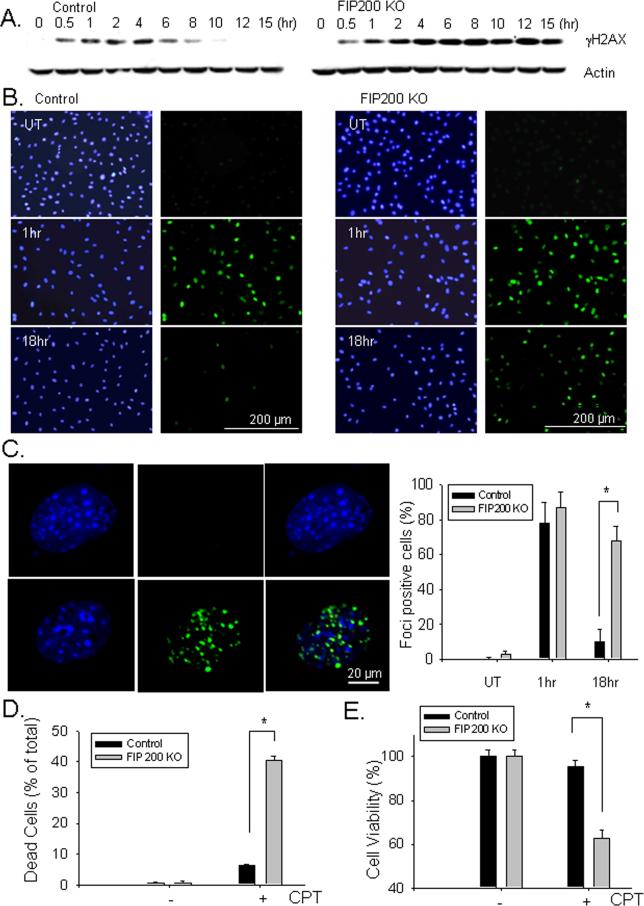
Deficiency in DNA damage repair could lead to genomic instability resulting in tumorigenesis (40), but excessively increased DNA damage may also lead to increased cell death, which is exploited for chemotherapy by DNA damaging inducing drugs (41). To further study the role of FIP200 in DNA damage repair and cell survival, FIP200 KO and control MEFs were treated with the chemotherapeutic agent camptothecin (CPT), a topoisomerase I inhibitor that can induce DNA damage and cell death (42). At various times after treatment with 5 μM CPT, lysates were prepared from control and FIP200 KO MEFs and analyzed by Western blotting using antibodies against γ-H2AX. As shown in Fig. 3A, induction of DNA damage was detected within 30 min and reached to maximal levels by about 4 hrs after CPT stimulation in both control and FIP200 KO MEFs. In control MEFs, the γ-H2AX level then gradually decreased to un-detectable level by 10 hrs after treatment, indicating repair of the DNA damage as expected (37). In contrast, the γ-H2AX level remained high and only started to decrease at 15 hrs after CPT treatment in FIP200 KO MEFs, suggesting a defect in the repair of CPT-induced DNA damage in these cells. Analysis of the samples from 0.5 to 4 hrs after CPT treatment indicated similarly increased levels of DNA-PK or ATR phosphorylation between FIP200 KO and control MEFs (Supplemental Fig. S2), suggesting that FIP200 deletion did not affect the cellular sensing of DNA damage or the phosphorylation of H2AX by ATR or DNA-PK (although ATM was not studied here), but rather only reduced DNA damage repair. The possible defective DNA damage repair in FIP200 KO MEFs was also assessed by immunofluorescent staining of anti-γ-H2AX antibodies. As shown in Fig. 3B, DNA damage was rapidly induced in the nuclei of majority of control and FIP200 KO MEFs within 1 hr of CPT treatment (middle panels). Consistent with results from Western blotting analysis, γ-H2AX signal was abolished in control MEFs, but was still present in a significant fraction of FIP200 KO MEFs, at 18 hrs after CPT treatments (lower panels). Furthermore, microscopic examination of the cells under higher magnification showed the presence of individual γ-H2AX foci in the nuclei of both cells in the earlier time point, but only FIP200 KO MEFs at the later time point (Fig. 3C). Together, these results indicated that deletion of FIP200 also resulted in defective DNA damage repair after CPT treatment.
Fig. 3. Analysis of DNA damage repair and cell survival in FIP200 KO MEFs after CPT treatment.
(A). Control and FIP200 KO MEFs were treated with 5 μM CPT for various times, as indicated. The cells were then lyzed and analyzed by Western blotting using anti-γH2AX or anti-β-actin antibodies. (B and C) Control and FIP200 KO MEFs were treated with mock (UT) or 5 μM CPT for 1 or 18 hr as indicated. They were then fixed and immunostained with anti-γH2AX antibodies and DAPI (B). In Panel C, the cells were viewed under higher magnification to visualize individual foci. The percentage of cells with > 5 foci per cell was determined. Representative images are shown on the left and Mean ± SEM from three experiments are shown on the right. *P<0.05. (D and E) The percentage of dead cells at 48 hr after treatment with 5 μM CPT are determined by PI staining (D) and the cell viability was measured by MTT assay (E). Mean ± SEM from three experiments are shown. *P<0.05.
We next examined the effect of reduced DNA damage repair upon deletion of FIP200 on cell survival in response to CPT. As reported previously (43), treatment of control MEFs by CPT induced cell death by apoptosis of these cells, as measured by propidium iodide staining (Fig. 3D) as well as apoptosis assay kit for DNA fragmentation (Supplemental Fig. S3). Interestingly, significantly more cell death and apoptosis were found for FIP200 KO MEFs after treatment with the same amount of CPT, suggesting an elevated sensitivity to CPT-induced apoptosis compared to control MEFs. The reduced survival of FIP200 KO MEFs after CPT treatment in comparison to control MEFs was also verified by MTT assays (Fig. 3E).
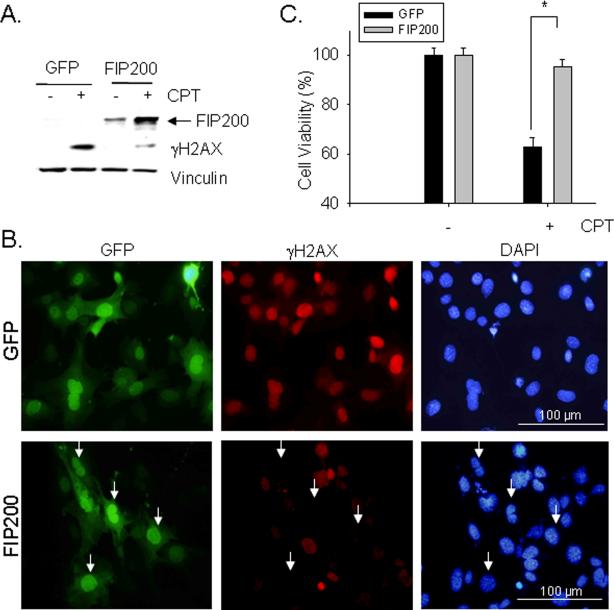
To further demonstrate that FIP200 deletion is responsible for the impaired DNA damage repair and consequent increase in cell death induced by CPT, we examined the effects of restoration of FIP200 expression in FIP200 KO MEFs by infection of recombinant adenovirus encoding FIP200. As shown in Fig. 4A, re-expression of FIP200 in FIP200 KO MEFs significantly reduced the level of γ-H2AX level induced by CPT-treatment of these cells, suggesting rescue of deficient DNA damage repair in these cells. FIP200 KO MEFs with re-expression of FIP200 or GFP as a control were also examined by immunofluorescent staining using anti-γ-H2AX antibodies (Fig. 4B). Expression of GFP control did not affect the impaired DNA damage repair in these cells after CPT-treatment, as cells with or without green fluorescent signals showed similar level of red γ-H2AX signal (top panels). In contrast, FIP200 KO MEFs with high expression of FIP200 virtually abolished γ-H2AX signal in individual cells (arrows, lower panels), suggesting restoration of efficient DNA damage repair in these cells. Re-expression of FIP200 in FIP200 KO MEFs also rescued the increased sensitivity of CPT-induced apoptosis of these cells (Fig 4C). Taken together, these results provide strong support that the impairment of DNA damage repair and consequent increased sensitivity to CPT-induced cell death was caused by FIP200 inactivation.
Fig. 4. FIP200 re-expression rescues impaired DNA damage repair and cell survival in FIP200 KO MEFs.
FIP200 KO MEFs were infected by recombinant adenoviruses encoding GFP or GFP-FIP200 fusion protein (FIP200) for two days, as indicated. (A) The cells were then treated with or without 5 μM CPT for 18 hr. The lysates were prepared and analyzed by Western blotting using various antibodies as indicated. (B) The infected cells were treated with 5 μM CPT for 18 hr and then examined by immunofluorescence using various antibodies as indicated. (C) The infected cells were incubated with 5 μM CPT for 48 hr and then cell viability was measured by MTT assay, as described in Fig. 3E. *P<0.05.
Increased p62 expression upon FIP200 deletion may be responsible for the reduced DNA damage repair and cell survival
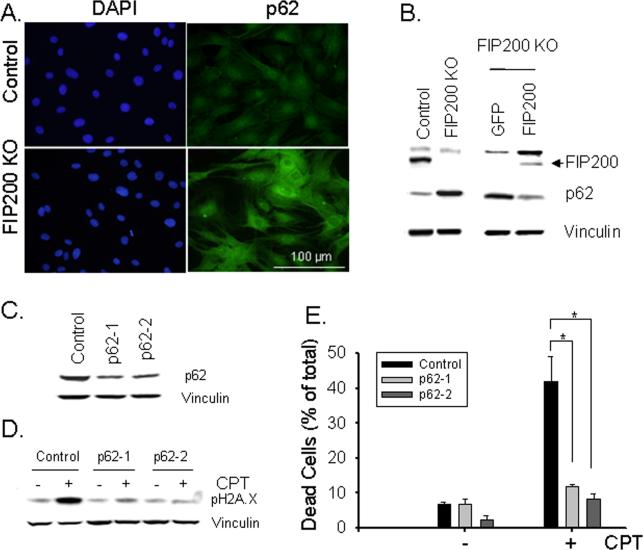
FIP200 has been identified as a key component of autophagy and its deletion has been shown to block autophagosome formation and induce p62 accumulation in mammalian cells (25). Consistent with these previous results, immunofluorescent analysis of FIP200 KO and control MEFs detected significant amount of p62-positive aggregates in FIP200 KO MEFs while little p62 staining was found in control MEFs (Fig. 5A). The increased p62 expression in FIP200 KO MEFs was also shown by Western blotting analysis of the lysates (Fig. 5B). Moreover, re-expression of FIP200, but not GFP as a control, in FIP200 KO MEFs significantly reversed the increase in p62 expression, indicating that defective autophagy upon FIP200 deletion caused accumulation of p62 in these cells.
Fig. 5. Increased expression of p62 and its role in the defective DNA damage repair and cell survival in FIP200 KO MEFs.
(A) Control and FIP200 KO MEFs were fixed and immunostained with anti-p62 antibodies and DAPI. (B) Lysates were prepared from Control and FIP200 KO MEFs (left) or FIP200 KO MEFs that had been infected with recombinant adenoviruses encoding GFP or GFP-FIP200 fusion protein (FIP200) for two days (right). They were then analyzed by Western blotting using various antibodies as indicated. (C-E) FIP200 KO MEFs were infected by recombinant lentiviruses encoding shRNA targeting two different part of p62, or a scramble sequence as a control, as indicated. The cells were then incubated with 5 μM CPT for 18 hr (D, + lanes), 48 hr (E, + bars), or left untreated (C, - lanes and bars in D and E). Lysates were prepared and analyzed by Western blotting using various antibodies as indicated (C and D). The percentage of dead cells are determined by PI staining (E), as described in Fig. 3D. *P<0.05.
We next examined the potential role of the increased p62 expression in the reduced DNA damage repair and cell survival by employing lentiviral shRNA to deplete p62 in FIP200 KO MEFs. As shown in Fig. 5C, transient transfection of FIP200 KO MEFs by lentiviral vectors encoding shRNA targeting two different p62 sequences both reduced the expression of p62 in these cells. Interestingly, knockdown of p62 expression restored efficient repair of DNA damage induced by CPT in FIP200 KO MEFs, as measured by the level of γ-H2AX at 18 hr after CPT treatment of these cells (Fig. 5D). Furthermore, the increased sensitivity of FIP200 KO MEFs to CPT-induced cell death was also rescued (Fig. 5E). Lastly, knockdown of p62 also partially rescued the defective DNA damage repair and reduced cell viability in FIP200 KO MEFs after IR (Supplemental Figs. S4 and S5). Together, these results suggested that increased p62 expression caused by deficient autophagy due to FIP200 deletion may be responsible for the defective DNA damage repair and consequent increase in cell death following CPT treatment.
Oxidative stress response has no effect on impaired DNA damage repair in FIP200 deficient cells
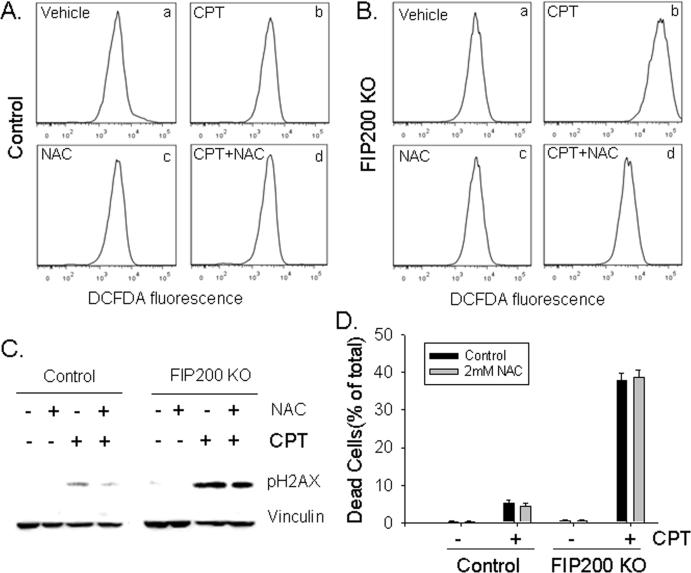
Defective autophagy upon deletion of several autophagy genes including FIP200 has been shown to result in increased ROS production in various cells (44, 45). More interestingly, a recent study showed that suppression of ROS accumulation reduced DNA damage in metabolically stressed autophagy-defective tumor cells (20). We therefore examined whether FIP200 KO MEFs exhibit increased ROS that could also contribute to the defective DNA damage repair and increased cell death induced by CPT. As shown in Figs. 6A and 6B, markedly increased ROS was detected in FIP200 KO MEFs compared to control MEFs after CPT treatment as measured by 2’-7’-dichlorofluorscein diacetate (DCFDA) staining, although no significant difference was found between these two cell types without treatment. As expected, the addition of the ROS scavenger, N-acetyl-cysteine (NAC), reversed the increased ROS level in FIP200 KO MEFs. Surprisingly, however, the presence of NAC during CPT treatment did not affect the increased γ-H2AX signal at 18 hr after stimulation by CPT (Fig. 6C). Moreover, NAC did not reduce CPT-induced cell death in FIP200 KO MEFs (Fig. 6D). The scavenger effectiveness of NAC was verified by the rescue of cell death after H2O2 treatment (Supplemental Fig. S6). These results suggested that oxidative stress induced by autophagy deficiency does not significantly affect CPT-induced DNA damage and cell death in FIP200 KO MEFs.
Fig. 6. ROS scavenging does not affect the impaired DNA damage repair and reduced survival of FIP200 KO MEFs after CPT treatment.
(A and B) Control and FIP200 KO MEFs were treated with 5 μM CPT, 2 mM NAC, or both, as indicated. They were then stained by DCFDA and analyzed by FACS, as described in the Materials and Methods. Representative FACS profiles of DCFDA staining are shown. (C and D) Control and FIP200 KO MEFs were pretreated with 2 mM NAC for 45 min, and then incubated with 5 μM CPT for 18 hr (C) or 48 hr (D), as indicated. Lysates were prepared and analyzed by Western blotting using various antibodies as indicated (C). The percentage of dead cells are determined by PI staining (D), as described in Fig. 3D. *P<0.05.
Reduced DNA damage repair and cell survival of FIP200 KO MEFs in response to etoposide
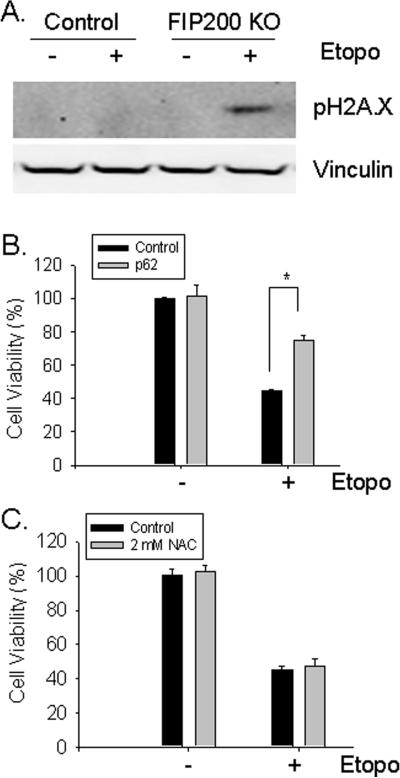
To further evaluate the role of FIP200 in DNA damage repair and cell survival, we examined the responses of FIP200 KO and control MEFs to etoposide, which is another important chemotherapeutic agent that induces double-stranded DNA break through inhibition of topoisomerase II (46, 47). At 24 hr after treatment with etoposide, lysates were prepared and analyzed for DNA damage by Western blotting. Similar to our observations of cells treated by CPT, a significant level of γ-H2AX was found in FIP200 KO MEFs but not in control MEFs (Fig. 7A), suggesting a reduced DNA damage repair upon FIP200 deletion. In consistent with these results, we also observed an increased cell death induced by etoposide in FIP200 KO MEFs compared to control MEFs (Supplemental Fig. S7). Knockdown of p62 by shRNA partially reversed etoposide-induced cell death of FIP200 KO MEFs (Fig. 7B). In contrast, suppression of ROS by NAC had little effect on the survival of these cells in response to etoposide treatment (Fig. 7C). These results provide support that deletion of FIP200 reduces DNA damage repair and cell survival in response to both double-stranded (by etoposide) and single-stranded (by CPT) DNA breaks in a p62-dependent but ROS-independent manner.
Fig. 7. Analysis of DNA damage repair and cell survival in response to etoposide.
(A) Control and FIP200 KO MEFs were stimulated with 25 μM etoposide for 24 hr. Cell lysates were prepared and analyzed by Western blotting using various antibodies as indicated. (B) FIP200 KO MEFs were infected by recombinant lentiviruses encoding shRNA targeting p62, or a scramble sequence as a control along with a puromycin-resistant marker, as indicated. The infected cells were selected in 5 μg/ml puromycin for two days, and then treated with or without 25 μM etoposide for two days. Cell viability was measured by MTT assay, as described in Fig. 3E. *P<0.05. (C) FIP200 KO MEFs were pretreated with 2 mM NAC for 45 min, and then incubated with 25 μM etoposide for two days, as indicated. Cell viability was measured by MTT assay, as described in Fig. 3E.
Discussion
Increasing evidence indicates intimate connections between autophagy and its abnormalities and cancer development and progression since the seminal discovery of Beclin 1, a haploinsufficient tumor suppressor, as a key component of autophagy (17-19). Remarkable advance has been made in recent years, which revealed the molecular mechanisms of Beclin 1 haploinsufficiency or inactivation of other autophagy proteins to promote tumorigenesis through increased accumulation of p62 and genomic instability (14, 20, 21). Interestingly, initial identification and earlier studies of FIP200 also suggested it as a putative tumor suppressor (29, 30), and more recent data showed FIP200 in a complex with ULK1/2 and Atg13 that are essential for autophagy induction (24, 26-28). Unlike Beclin 1, however, FIP200 inactivation in mouse models did not result in spontaneous development of breast cancer or augment lymphomagenesis induced by p53 deletion (32). These results question the earlier suggestion of FIP200 as a putative tumor suppressor and also highlight the potential complex role of autophagy in tumor development and progression (8, 9). Data presented here indicate that, contrary to a potential tumor suppression function, inactivation of FIP200 and subsequent deficiency in autophagy led to increased cell death upon stimulation with chemotherapeutic agents CPT and etoposide, thus may be exploited to enhance cancer treatments instead.
Our results are consistent with a number of previous reports that autophagy provides cell survival mechanism under different conditions and inhibition of autophagy sensitizes cells to various apoptosis-inducing agents (10-15). Moreover, our analysis suggested that defective DNA damage repair in FIP200-null cells may be responsible for the increased cell death. In contrast to the transient increase of γ-H2AX in control MEFs, FIP200 KO MEFs exhibited a more sustained γ-H2AX signal as measured by both Western blotting and nuclei foci staining upon treatment with CPT or etoposide as well as ionizing radiation. Furthermore, re-expression of FIP200 rescued defective DNA damage repair and the increased cell death. These results provide a plausible mechanism for the increased sensitivity to chemotherapy in autophagy defective cells observed here and in previous studies (10-15).
The mechanisms by which FIP200 deletion lead to the defective DNA damage repair are not well understood at present. Previous studies suggested a role of p62 accumulation in the increased genomic instability upon inhibition of autophagy in Beclin 1+/- or Atg5-/- cells that are defective in apoptosis (20). Interestingly, we also observed up-regulated p62 expression and p62-containing aggregates in FIP200 KO MEFs. Furthermore, re-expression of FIP200 in these cells suppressed p62 expression as well as CPT-induced cell death. Lastly, the direct inhibition of p62 expression using RNA interference also markedly suppressed sustained γ-H2AX signal and cell death induced by CPT or etoposide. These results suggested that, consistent with previous studies (20), up-regulated p62 expression caused by FIP200 deletion and autophagy deficiency could mediate the reduced DNA damage repair in FIP200 KO MEFs. In contrast to the increased tumorigenesis in the apoptosis deficient cells in previous studies (14, 20, 21), however, the sustained DNA damage likely contributed to the increased cell death in FIP200 KO MEFs due to their intact apoptosis mechanisms.
How does then up-regulated p62 expression impair DNA repair system? p62 is known to localize primarily in the cytoplasm as a cargo protein of ubiquitinated proteins for the autophagic degradation (48). However, a recent report showed that p62 has nuclear localization signals and a nuclear export signal, which allow it to shuttle between the cytoplasm and the nucleus, and that p62 is required for polyubiquitinated protein interaction with promyelocytic leukemia (PML) nuclear bodies (49). PML nuclear bodies are involved in DNA damage repair as they contain several DNA damage response proteins such as BLM/WRN DNA helicases, the Mre11 complex, or TopBP1 (50). Although these considerations suggest a potential direct connection between p62 and DNA damage repair processes in the nuclei, we did not detect any significant re-localization of p62 after CPT treatment in FIP200 KO or control MEFs. Given its function as a scaffolding protein, it is also likely that increased expression of p62 in the cytoplasm may influence DNA damage repair through its interactions with multiple other proteins in an indirect manner (48).
In addition to the increased p62 expression, the increased ROS production in autophagy deficient cells is also suggested to mediate increased DNA damage in previous studies (21). These studies also suggested a possible amplification loop between ROS and p62 in promoting genomic instability and tumorigenesis. Indeed, we also observed increased ROS production in hematopoietic cells after FIP200 deletion in recent studies (44). Surprisingly, however, NAC treatment did not significantly reverse the deficient DNA damage repair or increased cell death in FIP200 KO MEFs compared to control MEFs. The discrepancy between these studies and the previous studies employing apoptosis defective cells with deficiency in other autophagy proteins remains to be further investigated. It is also interesting to note that whereas previous studies showed that loss of autophagy survival promoted tumor growth (20, 21), deletion of FIP200 in mammary epithelial cells did not lead to spontaneous development of breast cancer (32). Thus, future studies will also be directed at the potential contribution of the defective DNA damage repair after FIP200 inactivation in tumor development and progression in vivo.
Supplementary Material
Acknowledgements
We are grateful to Dr. Benjamin Chen of UT Southwestern Medical School and Dr. Xiaochun Yu of University of Michigan for discussions, reagents, help throughout this project, and critical reading of the manuscript and helpful comments. We also thank members of Guan laboratory for critical reading of the manuscript and their suggestions. This research was supported by NIH grants GM052890 and CA150926 to J.-L. Guan.
References
- 1.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. 2009. pp. 773–82. [DOI] [PMC free article] [PubMed]
- 2.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annual review of genetics. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rabinowitz JD, White E. Autophagy and metabolism. Science. 330(6009):1344–8. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 40(2):280–93. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 12(9):823–30. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10(5):461–70. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15(17):5308–16. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen N, Debnath J. Autophagy and tumorigenesis. FEBS Lett. 584(7):1427–35. doi: 10.1016/j.febslet.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 2007;14(3):500–10. doi: 10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- 11.Amaravadi RK, Yu D, Lum JJ, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117(2):326–36. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang S, Sinicrope FA. Celecoxib-induced apoptosis is enhanced by ABT-737 and by inhibition of autophagy in human colorectal cancer cells. Autophagy. 2010;6(2):256–69. doi: 10.4161/auto.6.2.11124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez-Polo RA, Niso-Santano M, Ortiz-Ortiz MA, et al. Inhibition of paraquat-induced autophagy accelerates the apoptotic cell death in neuroblastoma SH-SY5Y cells. Toxicol Sci. 2007;97(2):448–58. doi: 10.1093/toxsci/kfm040. [DOI] [PubMed] [Google Scholar]
- 14.Degenhardt K, Mathew R, Beaudoin B, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer cell. 2006;10(1):51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishikawa T, Tsuno NH, Okaji Y, et al. The inhibition of autophagy potentiates anti-angiogenic effects of sulforaphane by inducing apoptosis. Angiogenesis. 2010 doi: 10.1007/s10456-010-9180-2. [DOI] [PubMed] [Google Scholar]
- 16.Aita VM, Liang XH, Murty VV, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59(1):59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 17.Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402(6762):672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 18.Qu X, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112(12):1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100(25):15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathew R, Karp CM, Beaudoin B, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137(6):1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathew R, Kongara S, Beaudoin B, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21(11):1367–81. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbi S, Ueda H, Zheng C, et al. Regulation of focal adhesion kinase by a novel protein inhibitor FIP200. Mol Biol Cell. 2002;13(9):3178–91. doi: 10.1091/mbc.E02-05-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ueda H, Abbi S, Zheng C, Guan JL. Suppression of Pyk2 Kinase and Cellular Activities by FIP200. J Cell Biol. 2000;149(2):423–30. doi: 10.1083/jcb.149.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hara T, Mizushima N. Role of ULK-FIP200 complex in mammalian autophagy: FIP200, a counterpart of yeast Atg17? Autophagy. 2009;5(1):85–7. doi: 10.4161/auto.5.1.7180. [DOI] [PubMed] [Google Scholar]
- 25.Hara T, Takamura A, Kishi C, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181(3):497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284(18):12297–305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung CH, Jun CB, Ro SH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20(7):1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981–91. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chano T, Kontani K, Teramoto K, Okabe H, Ikegawa S. Truncating mutations of RB1CC1 in human breast cancer. Nat Genet. 2002;31(3):285–8. doi: 10.1038/ng911. [DOI] [PubMed] [Google Scholar]
- 30.Melkoumian ZK, Peng X, Gan B, Wu X, Guan JL. Mechanism of cell cycle regulation by FIP200 in human breast cancer cells. Cancer Res. 2005;65(15):6676–84. doi: 10.1158/0008-5472.CAN-04-4142. [DOI] [PubMed] [Google Scholar]
- 31.Gan B, Peng X, Nagy T, Alcaraz A, Gu H, Guan JL. Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J Cell Biol. 2006;175(1):121–33. doi: 10.1083/jcb.200604129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei H, Gan B, Wu X, Guan JL. Inactivation of FIP200 Leads to Inflammatory Skin Disorder, but Not Tumorigenesis, in Conditional Knock-out Mouse Models. J Biol Chem. 2009;284(9):6004–13. doi: 10.1074/jbc.M806375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olive PL, Banath JP. The comet assay: a method to measure DNA damage in individual cells. Nat Protocols. 2006;1(1):23–9. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 34.Shi Y, Porter K, Parameswaran N, Bae HK, Pestka JJ. Role of GRP78/BiP degradation and ER stress in deoxynivalenol-induced interleukin-6 upregulation in the macrophage. Toxicol Sci. 2009;109(2):247–55. doi: 10.1093/toxsci/kfp060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, Azad MB, Gibson SB. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009;16(7):1040–52. doi: 10.1038/cdd.2009.49. [DOI] [PubMed] [Google Scholar]
- 36.Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36(17):5678–94. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lobrich M, Shibata A, Beucher A, et al. gammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle. 2010;9(4):662–9. doi: 10.4161/cc.9.4.10764. [DOI] [PubMed] [Google Scholar]
- 38.Djuzenova CS, Rothfuss A, Oppitz U, et al. Response to X-irradiation of Fanconi anemia homozygous and heterozygous cells assessed by the single-cell gel electrophoresis (comet) assay. Lab Invest. 2001;81(2):185–92. doi: 10.1038/labinvest.3780226. [DOI] [PubMed] [Google Scholar]
- 39.Gabai VL, Sherman MY, Yaglom JA. HSP72 depletion suppresses gammaH2AX activation by genotoxic stresses via p53/p21 signaling. Oncogene. 2010;29(13):1952–62. doi: 10.1038/onc.2009.480. [DOI] [PubMed] [Google Scholar]
- 40.Jin S, White E. Tumor suppression by autophagy through the management of metabolic stress. Autophagy. 2008;4(5):563–6. [PMC free article] [PubMed] [Google Scholar]
- 41.Bates SE. DNA Repair: A Reinvigorated Therapeutic Target. Clin Cancer Res. 2010 doi: 10.1158/1078-0432.CCR-10-2086. [DOI] [PubMed] [Google Scholar]
- 42.Hsiang YH, Hertzberg R, Hecht S, Liu LF. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem. 1985;260(27):14873–8. [PubMed] [Google Scholar]
- 43.Gupta M, Fan S, Zhan Q, Kohn KW, O'Connor PM, Pommier Y. Inactivation of p53 increases the cytotoxicity of camptothecin in human colon HCT116 and breast MCF-7 cancer cells. Clin Cancer Res. 1997;3(9):1653–60. [PubMed] [Google Scholar]
- 44.Liu F, Lee JY, Wei H, et al. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood. 2010 doi: 10.1182/blood-2010-06-288589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Qi H, Taylor R, Xu W, Liu LF, Jin S. The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy. 2007;3(4):337–46. doi: 10.4161/auto.4127. [DOI] [PubMed] [Google Scholar]
- 46.Baldwin EL, Osheroff N. Etoposide, topoisomerase II and cancer. Curr Med Chem Anticancer Agents. 2005;5(4):363–72. doi: 10.2174/1568011054222364. [DOI] [PubMed] [Google Scholar]
- 47.Muslimovic A, Nystrom S, Gao Y, Hammarsten O. Numerical analysis of etoposide induced DNA breaks. PLoS One. 2009;4(6):e5859. doi: 10.1371/journal.pone.0005859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137(6):1001–4. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pankiv S, Lamark T, Bruun JA, Overvatn A, Bjorkoy G, Johansen T. Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies. J Biol Chem. 2010;285(8):5941–53. doi: 10.1074/jbc.M109.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lallemand-Breitenbach V, de The H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2(5):a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.