Abstract
Background
There are few data in the literature on viral sequence variation between host generations/successive transmission events. Relatively little is known about the sequence heterogeneity of the influenza viruses transmitted within families.
Objectives
To study the molecular epidemiology of influenza virus and to determine the sequence variation within an individual, a household and a community during the first wave of influenza pandemic in 2009.
Study design
A prospective study of household transmission of influenza A in Hong Kong was conducted during the pandemic in 2009. The HA and NA sequences of pandemic and seasonal influenza A viral isolates identified in this household transmission study were sequences and analyzed.
Results
Our results indicated that there were multiple introductions of influenza viruses into Hong Kong. Sequence analysis of these isolates suggested that members of these family clusters acquired the infection by household transmissions. Interestingly, unlike those concluded from previous household transmission studies, we observed sequence variations between sequential samples from the same person and also within the same household.
Conclusions
Family clusters of influenza A viral infection are predominantly the result of secondary transmission within a household. Our results also suggested that the intra-host viral sequence variation might be more common that than previously thought.
Background
The emergence of pandemic H1N1/2009 virus provided an unique opportunity to study the dynamics of influenza virus transmission in humans 1, 2 and comparative studies of pandemic and seasonal viruses within households may provide important understanding in this regard.3 We previously conducted a prospective household transmission study and concluded that pandemic and seasonal viruses have broadly similar viral-load dynamics, disease severities, and transmissibilities in the household setting.4 This study cohort had sequential specimens collected from index cases with confirmed influenza and their family contacts. The clinical material provides an opportunity to compare the virus genetic variability within a host and/or during successive transmissions.
Objectives
The aim of this study is to elucidate the genetic sequence variation in HA and NA virus gene segments in these sequential specimens from index cases and secondary cases of infection. In addition, the epidemiological links of these household samples were also studied.
Study design
Three hundred forty-eight index patients with acute respiratory illness from a local network of 14 outpatient clinics were recruited from June to August 2009. We followed the household contacts of 99 index patients who tested positive for influenza A virus on rapid diagnostic testing in an outpatient setting and collected nasal and throat swabs (NTS) from these index cases as well as all household members regardless of illness at three subsequent home visits within 7 days. NTS were sampled and tested by RT-PCR for influenza viral matrix gene as described.5 The study design and epidemiological findings were reported elsewhere.4 NTS specimens that were RT-PCR positive for influenza virus were cultured for virus isolation in MDCK cells as described.5 The full-length surface glycoprotein genes of these virus isolates were determined. Briefly, viral RNA extraction, complementary DNA synthesis, PCR and sequencing were carried out as described.6, 7 Sequencing reactions were performed using the Big Dye-Terminator v3.1 Cycle Sequencing Reaction Kit on an ABI PRISM 3700 DNA Analyser (Applied Biosystems) following the manufacturer’s instructions. All sequences were assembled and edited with Bioedit version 7.0.9 (http://www.mbio.ncsu.edu/bioedit/bioedit.html).
Results and discussion
A total of 119 influenza viruses were isolated from 98 individuals in 78 households (pandemic H1N1 (pH1N1)= 61; seasonal H1N1 (sH1N1)= 4; seasonal H3N2 (sH3N2)= 54). Of these 119 viral isolates, 42 were sampled from 21 individuals (pH1N1= 14; sH3N2= 7) who were positive in two consecutive visits. Twenty pH1N1 and 15 sH3N2 viruses were isolated from households with confirmed secondary transmission (pH1N1=8; sH3N2=8). On one occasion, a pH1N1 and a sH3N2 viruses were isolated from different members of the same household. The phylogenetic trees of HA and NA sequences of each subtype are shown (Figs. 1–2; Supplementary Figs. 1–3). As the HA phylogeny were found to be more informative than that derived from the NA gene, all the subsequent phylogenetic analyses are based on the HA sequences. Four sH1N1 viruses (A/Brisbane/59/2007-like) were detected only once in different households and they were not studied further (Supplementary Fig. 3). All viral sequences reported in this study have been deposited in GenBank (Accession numbers JN256681-JN256918).
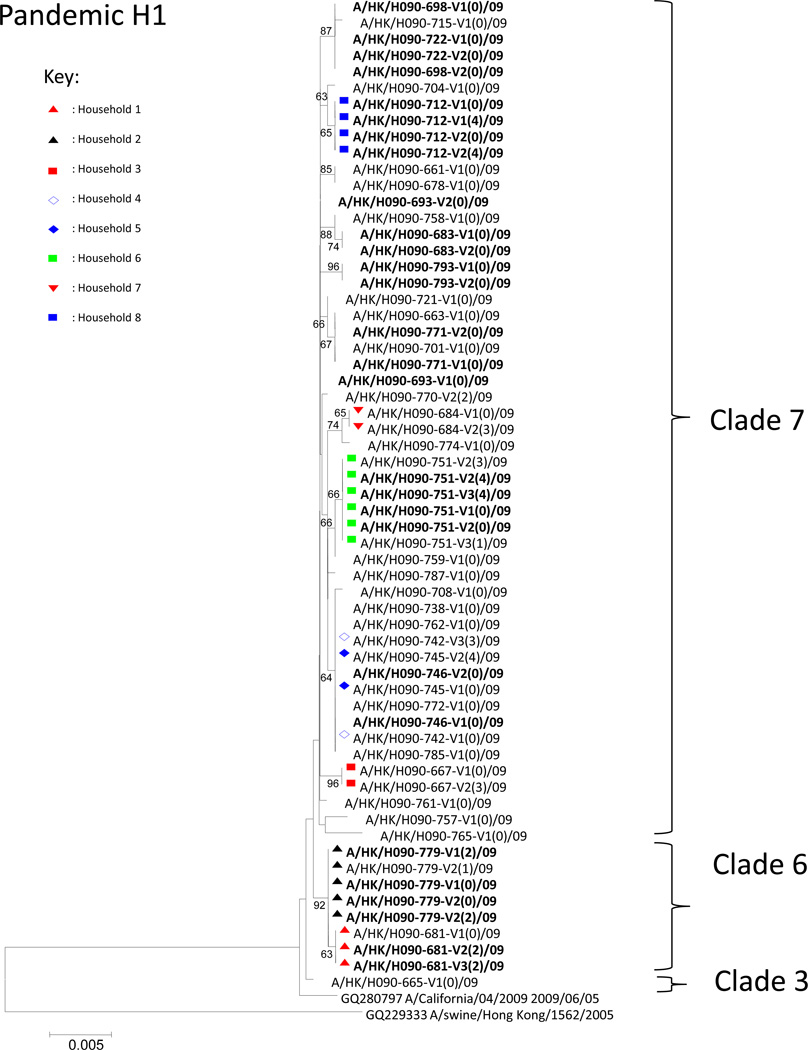
Figure 1.
Phylogenetic analysis of pH1N1 HA. Viruses isolated from households with household transmissions (Household 1–8) were marked by colored symbols. Within the name of each virus, the number after “V” and the number in the brackets denotes the sampling occasion (Visits 1 to 3) and household member identity (Index case=0), respectively. Viruses isolated from the same individuals, but at different sampling occasions, were highlighted in bold. The phylogenetic tree was constructed by neighbor-joining method with default parameters in MEGA4. Bootstrap values of the phylogenetic tree constructed were generated by doing 1,000 replicates. Bootstrap values ≥ 70% were considered well-supported. The scale bar represents genetic distance between homologues (nucleotide substitutions per site). The GenBank accession numbers of reference sequences are shown.
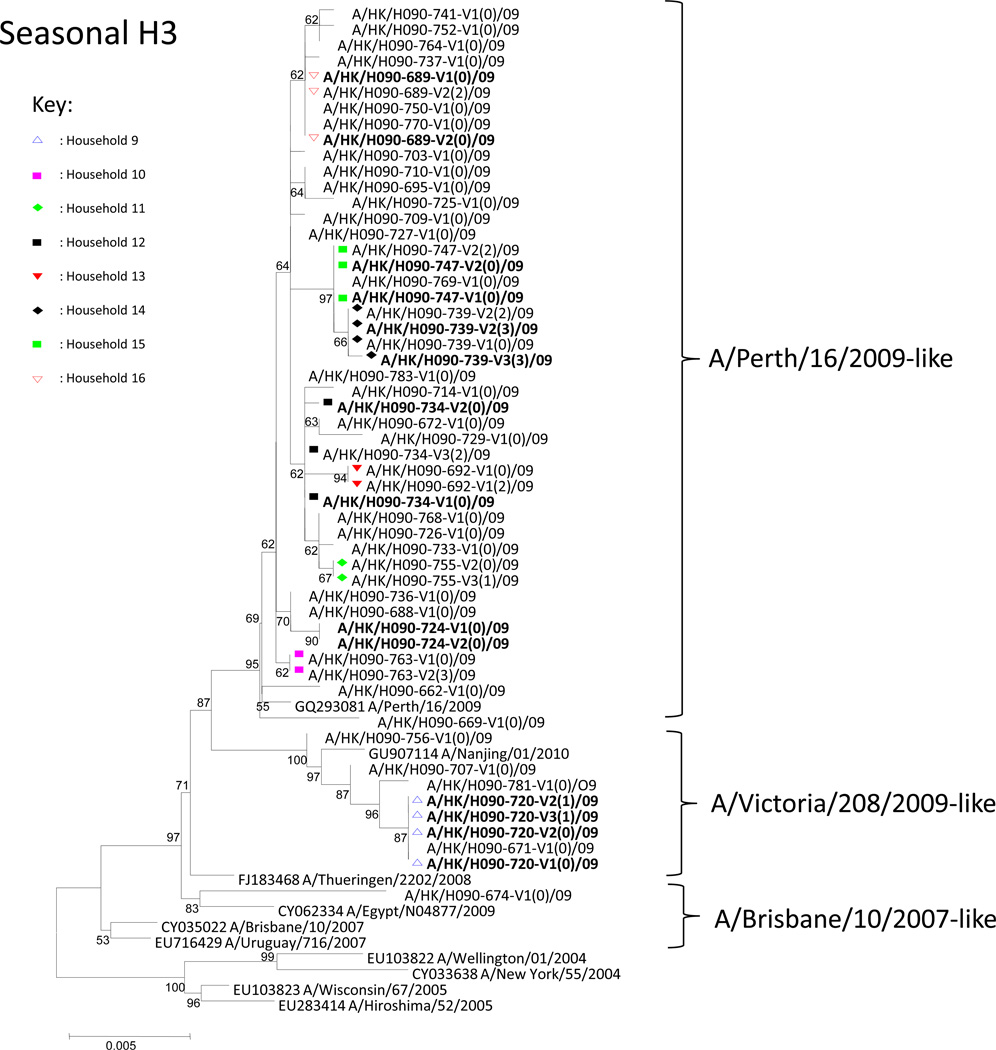
Figure 2.
Phylogenetic analysis of sH3N2 HA. Viruses isolated from households with household transmissions (Household 9–18) were marked by colored symbols. Within the name of each virus, the number after “V” and the number in the brackets denotes the sampling occasion (Visits 1 to 3) and household member identity (Index case=0), respectively. Viruses isolated from the same individuals, but at different sampling occasions, were highlighted in bold. The phylogenetic tree was constructed by neighbor-joining method with default parameters in MEGA4. Bootstrap values of the phylogenetic tree constructed were generated by doing 1,000 replicates. Bootstrap values ≥ 70% were considered well-supported. The scale bar represents genetic distance between homologues (nucleotide substitutions per site). The GenBank accession numbers of reference sequences are shown.
Global genetic sequence analysis of pH1N1 has revealed the presence of at least 7 distinct clades of virus.8 Phylogenetic analysis revealed that clades 3, 6 and 7 viruses were detected in this study (Fig. 1). None of these viruses were found to be genetically similar to the first laboratory confirmed case in Hong Kong (Supplementary Fig. 4, yellow dots).6 Similar to findings concluded in some Asian countries8, 9 the majority of our isolates belong to clade 7. These results also indicated that there were multiple introductions of pH1N1 viruses into Hong Kong. To further illustrate this point, all the publicly available pH1N1 sequences sampled before this study were included for further analysis. Many of our clade 7 viruses were found to be clustered with those isolated from other areas (Supplementary Fig. 4, red dots). Interestingly, some of our clade 7 viruses formed a unique branch in the tree (Supplementary Fig. 4, green), suggesting that this cluster was associated with a single introduction. A dating analysis on these viruses was performed 10 and this particular subclade was estimated to have been introduced into Hong Kong on the 3rd June (95% CI: 11th May to 25th June; Supplementary Fig. 5, highlighted in green). This agreed with our previous estimate that community transmission of pandemic H1N1 in Hong Kong began in early June.11
For the sH3N2 isolates, 3 viral lineages were detected in 39 households (A/Brisbane/10/2007-like =1, A/Victoria/208/2009-like =5 and A/Perth/16/2009-like =33; Fig. 2).12 Using other sH3N2 viruses sampled from 1st January 2008 to 20th June 209 as references, multiple introductions of A/Perth/16/2009-like viruses into Hong Kong were detected (Supplementary Fig. 6). In particular, a large number of our A/Perth/16/2009-like viruses were found to be associated with a single introduction (Supplementary Fig. 6, highlighted in green). Subsequent dating analysis revealed that this introduction occurred around the 19th April 2009 (95% CI: 24th Feb 2009 to 25th May, 2009).
Previous studies on sequence variation in human influenza virus primarily relied on studying viral sequences available from public databases or viruses sampled at a community level with little accompanying epidemiological data. By contrast, there is little information on viral sequence variation between host generations/successive transmission events.13, 14 Gubareva et al studied the HA sequence variation during seasonal influenza virus transmission in 19 informative families.14 In their study, no viral sequence variation within an individual/family was found, suggesting that there is a substantial genetic conservation during virus transmission within households. Our data indicated that the average sequence identity between viruses sampled from the same individual/household was high and was much greater than the one deduced from viruses that had no direct epidemiological links (Table 1; p<0.001). In addition, we found that the intra-household and intra-host sequence variations were not statistically significantly different from each other (p>0.05). In contrast with previous findings,14 we found evidence of intra-household or intra-host sequence variations in both HA and NA genes of pH1N1 and sH3N2 viruses. Two of these nucleotide variations (N=6) resulted in non-silent mutations (Supplementary Table 1). These mutations were also detected in subsequent confirmed secondary infections of household members and/or confirmed by repeated sequencing.
Table 1.
Average sequence identity of viruses sampled within an individual, a household and a community.
| Sequence Identity (%) |
||||
|---|---|---|---|---|
| Viral source | Pandemic H1 | Pandemic N1 | Seasonal H3 | Seasonal N2 |
| Within an individual | 0.999914 | 0.999957 | 0.999829 | 1.000000 |
| (SD) | (0.000218) | (0.000160) | (0.000293) | (0.000000) |
| Within a household* | 0.999865 | 1.000000 | 0.999900 | 0.999933 |
| (SD) | (0.000255) | (0.000000) | (0.000230) | (0.000194) |
| Non-related cases | 0.996654 | 0.998322 | 0.994036 | 0.993669 |
| (SD) | (0.001244) | (0.001390) | (0.004485) | (0.005621) |
Consecutive samples collected from the same individual within a household were excluded in the calculation.
Using the intra-host HA sequence variations and sampling dates as references (Table 1), the nucleotide mutation rates in the pandemic H1 and seasonal H3 genes were estimated to be 1.032 x10-2 and 2.052 x10-2 nucleotide substitutions/site per year, respectively. These data agree with previous findings that pH1N1 and sH3N2 have comparable evolutionary rates,8, 15, 16 but our estimates were about 2.6–4.5 times higher than those estimated from viruses that were sampled at a global level.15, 16 This might be partly explained by the hypothesis that not of all these intra-host variants are fit for sustainable transmission at a community level. As the sample size of specimens from the same host is small, the implication of intra-host sequence variation on the evolution of human influenza virus might require further large-scale analyses on intra- and inter-host viral samples. Nonetheless, our data suggested that both pH1N1 and sH3N2 have similar intrinsic properties to generate infectious variants.
Conclusion
Our results demonstrated that there were multiple introductions of pH1N1 and sH3N2 viruses into Hong Kong in 2009. Although multiple clades of pH1N1 and sH3N2 viruses co-circulated at the community level, viruses isolated from the same household were usually derived from the same viral lineage. Natural sequence variations could be detected in some sequential samples from the same person and also within the same household, suggesting influenza viral sequence variations might be more dynamic that than previously appreciated.
Supplementary Material
Acknowledgements
This study was supported by the Area of Excellence Scheme of the University Grants Committee Hong Kong (AoE/M-12/06), the Research Fund for the Control of Infectious Disease Commissioned Project from Food and Health Bureau, the NIH (NIAID contracts HHS-N266200700005C and N01-AI-70005), and the Harvard Center for Communicable Disease Dynamics from the National Institute of General Medical Sciences (grant number U54 GM088558). The funding bodies were not involved in the collection, analysis and interpretation of data, the writing of the manuscript, or the decision to submit for publication. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
We thank Chan Kit Man, Rita Fung, Lai-Ming Ho, Ho Yuk Ling, Lam Yiu Pong, Lincoln Lau, Tom Lui, Tong Hok Leung, Loretta Mak, Gloria Ng, Teresa So, Alfred Yeung, Eileen Yeung and Jenny Yuen for research support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors have no commercial or other associations that may pose a conflict of interest.
Ethical approval
The procedures described in this study was reviewed and approved by the Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster Ref Number: UW 06-350 T/1375)
References
- 1.Rambaut A, Holmes E. The early molecular epidemiology of the swine-origin A/H1N1 human influenza pandemic. PLoS Curr. 2009;1:RRN1003. doi: 10.1371/currents.RRN1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 3.Uyeki TM. 2009 H1N1 virus transmission and outbreaks. N Engl J Med. 2010;362:2221–2223. doi: 10.1056/NEJMe1004468. [DOI] [PubMed] [Google Scholar]
- 4.Cowling BJ, Chan KH, Fang VJ, Lau LL, So HC, Fung RO, et al. Comparative epidemiology of pandemic and seasonal influenza A in households. N Engl J Med. 362:2175–2184. doi: 10.1056/NEJMoa0911530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan KH, Peiris JS, Lim W, Nicholls JM, Chiu SS. Comparison of nasopharyngeal flocked swabs and aspirates for rapid diagnosis of respiratory viruses in children. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2008;42:65–69. doi: 10.1016/j.jcv.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Poon LL, Chan KH, Smith GJ, Leung CS, Guan Y, Yuen KY, et al. Molecular detection of a novel human influenza (H1N1) of pandemic potential by conventional and real-time quantitative RT-PCR assays. Clin Chem. 2009;55:1555–1558. doi: 10.1373/clinchem.2009.130229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vijaykrishna D, Poon LL, Zhu HC, Ma SK, Li OT, Cheung CL, et al. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science. 2010;328:1529. doi: 10.1126/science.1189132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson M, Spiro D, Wentworth D, Beck E, Fan J, Ghedin E, et al. The early diversification of influenza A/H1N1pdm. PLoS Curr. 2009;1:RRN1126. doi: 10.1371/currents.RRN1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Potdar VA, Chadha MS, Jadhav SM, Mullick J, Cherian SS, Mishra AC. Genetic characterization of the influenza A pandemic (H1N1) 2009 virus isolates from India. PLoS One. 5:e9693. doi: 10.1371/journal.pone.0009693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu JT, Cowling BJ, Lau EH, Ip DK, Ho LM, Tsang T, et al. School closure and mitigation of pandemic (H1N1) 2009, Hong Kong. Emerg Infect Dis. 16:538–541. doi: 10.3201/eid1603.091216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World_Health_Organization. Recommended viruses for influenza vaccines for use in the 2010–2011 northern hemisphere influenza season. 2010 [PubMed]
- 13.Teo SS, Ellis JS, Aitken C, Booy R. Transmission of influenza A in families. Pediatr Infect Dis J. 2007;26:645–647. doi: 10.1097/INF.0b013e3180616cfb. [DOI] [PubMed] [Google Scholar]
- 14.Gubareva LV, Novikov DV, Hayden FG. Assessment of hemagglutinin sequence heterogeneity during influenza virus transmission in families. J Infect Dis. 2002;186:1575–1581. doi: 10.1086/345372. [DOI] [PubMed] [Google Scholar]
- 15.Shiino T, Okabe N, Yasui Y, Sunagawa T, Ujike M, Obuchi M, et al. Molecular evolutionary analysis of the influenza A(H1N1)pdm, May–September, 2009: temporal and spatial spreading profile of the viruses in Japan. PLoS One. 5:e11057. doi: 10.1371/journal.pone.0011057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaraket H, Saito R, Sato I, Suzuki Y, Li D, Dapat C, et al. Molecular evolution of human influenza A viruses in a local area during eight influenza epidemics from 2000 to 2007. Arch Virol. 2009;154:285–295. doi: 10.1007/s00705-009-0309-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.