Abstract
Eukaryotic initiation factor 4A (eIF4A) is a DEAD-box helicase that stimulates translation initiation by unwinding mRNA secondary structure. The accessory proteins, eIF4G, eIF4B, and eIF4H enhance the duplex unwinding activity of eIF4A, but the extent to which they modulate eIF4A activity is poorly understood. Here, we use real time fluorescence assays to determine the kinetic parameters of duplex unwinding and ATP hydrolysis by these initiation factors. To ensure efficient duplex unwinding, eIF4B and eIF4G cooperatively activate the duplex unwinding activity of eIF4A. Our data reveal that eIF4H is much less efficient at stimulating eIF4A unwinding activity than eIF4B, implying that eIF4H is not able to completely substitute for eIF4B in duplex unwinding. By monitoring unwinding and ATPase assays using identical conditions, we demonstrate that eIF4B couples the ATP hydrolysis cycle of eIF4A with strand separation, thereby minimizing non-productive unwinding events. Using duplex substrates with altered GC contents, but with similar predicted thermal stabilities, we further show that the rate of formation of productive unwinding complexes is strongly influenced by the local stability per base pair in addition to the stability of the entire duplex. This finding explains how a change in the GC content of a hairpin while maintaining overall predicted thermal stability is able to influence translation initiation.
Keywords: Translation, Initiation, Protein synthesis, Ribosome
Introduction
Recruitment and positioning of an mRNA on the small ribosomal subunit is inhibited by secondary structure. In eukaryotes, initiation factors have evolved to unwind mRNA secondary structure so that an unstructured region can be positioned directly in the decoding site of the 40S ribosomal subunit. Eukaryotic initiation factor 4A (eIF4A) is a DEAD-box helicase that has been implicated in mRNA duplex unwinding. Consistent with this function, eIF4A is an RNA dependent ATPase that bidirectionally unwinds short RNA duplexes 1; 2; 3. It belongs to the helicase superfamily 2 (SF2) with a structure possessing characteristic RecA homology domains (Fig. 1A and ref. 4; 5). Binding of ATP and RNA triggers the RecA domains to adopt a compact closed structure that stabilizes substrate binding 6; 7; 8; 9. Subsequent hydrolysis and release of phosphate promotes an opening in the cleft between the RecA domains, resulting in dissociation of RNA6; 7; 8; 10 A current model suggests that duplex binding by a DEAD-box helicase results in local strand separation11; 12. A detailed molecular mechanism of duplex unwinding by eIF4A is not known, however. Purified eIF4A functions as a less efficient helicase compared to other DEAD-box proteins, even though they share considerable structural homology2; 10; 13 To overcome this limitation, the in vitro duplex unwinding activity of eIF4A is enhanced by eIF4B, eIF4H, and incorporation into the eIF4F complex.
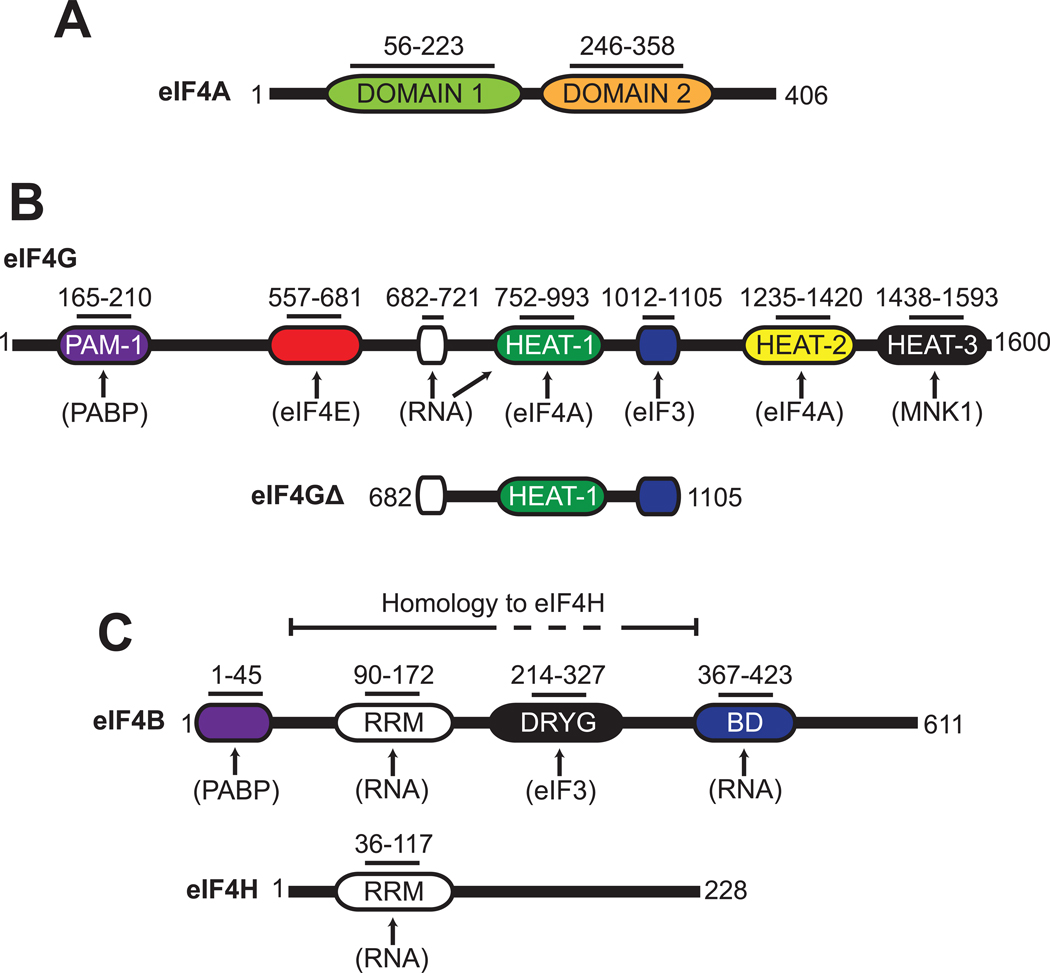
Fig. 1. Schematic representation of eIF4A, eIF4G, eIF4B and eIF4H.
Organization of protein domains found in human eIF4A (A), eIF4G (B), eIF4B and eIF4H (C). The eIF4G truncation construct used in this study (eIF4GΔ) is also depicted (B). Functional domains in each protein are shown with their corresponding amino acid regions. Known protein and RNA interaction sites are indicated by arrows. Abbreviations: PAM-1, PABP-binding motif; RRM, RNA recognition motif; DRYG, aspartic acid, arginine, tyrosine and glycine domain; BD, RNA binding domain.
eIF4F is a stable complex that can be purified from mammalian cells and is composed of eIF4E, eIF4G and eIF4A14; 15 The eIF4E subunit functions to tether the complex to the mRNA by binding directly to the 5′ 7-methyl guanosine cap structure. eIF4G possesses binding domains for eIF4E and eIF4A in addition to eIF3 and poly(A) binding protein (Fig. 1B and ref16). A single eIF4A protein makes specific contacts with the HEAT-1 and HEAT-2 domains in eIF4G (Fig. 1B and ref.7). With regard to eIF4A activity, incorporation into the eIF4F complex increases the RNA binding affinity and the rate of ATP hydrolysis7; 8; 17; 18; 19. Stoichiometric comparisons indicate that native eIF4F is 20-fold more efficient than eIF4A alone in duplex unwinding, as determined by an indirect nuclease sensitivity assay20. A straightforward kinetic analysis using a gel-shift unwinding assay, however, has indicated that the observed initial rate of unwinding is only increased 2-fold by incorporation into the eIF4F complex21. The interpretation of in vitro experiments using eIF4F is complicated by the fact that purified native eIF4F typically contains variable amounts of eIF4A and eIF4E proteins. To circumvent this problem, recent work identified a truncation of eIF4G that is able to recruit an mRNA to the ribosome in vitro and in vivo without the need for eIF4E22; 23. Unlike full-length eIF4G, this truncation can be readily purified and increases the RNA binding affinity of eIF4A and its rate of ATP hydrolysis to a similar extent to that found for native eIF4F18.
eIF4B functions as an accessory protein that enhances the duplex unwinding activity of eIF4A alone or in the eIF4F complex1; 2; 21; 24. Human eIF4B is an RNA binding protein that contains an RNA recognition motif (RRM) in its amino terminus and a separate RNA binding domain in its carboxyl terminus (Fig. 1C and ref.25; 26). An additional region rich in aspartic acid, arginine, tyrosine and glycine (DRYG) has been shown to mediate the interaction between eIF4B and eIF327. eIF4B increases the affinity of eIF4A for RNA and ATP, which may explain its ability to enhance duplex unwinding17; 24; 26.
The unwinding activity of eIF4A is also enhanced by eIF4H, which was first identified as a protein that stimulates an in vitro globin synthesis assay28. This protein possesses sequence homology to the N-terminal region of eIF4B, which includes the RRM motif, but not the DRYG or C-terminal RNA binding domains (Fig. 1C and ref28). Similar to eIF4B, eIF4H has been shown to increase the affinity of eIF4A for RNA, albeit to a lesser extent, which may account for its role in duplex unwinding2; 28; 29.
Despite extensive prior studies showing that eIF4F, eIF4B and eIF4H enhance duplex unwinding by eIF4A, the extent to which each accessory protein contributes to duplex unwinding has not been determined. Moreover, insight into how these accessory proteins couple the ATPase activity of eIF4A with duplex unwinding can only come from using identical reactions. In this work, we have used real time fluorescence methods to determine the kinetic parameters of duplex unwinding and ATP hydrolysis for these components under identical assay conditions. Our kinetic data reveal how eIF4B and the eIF4G subunit of eIF4F function to coordinate the duplex unwinding and ATPase activities of eIF4A to promote strand separation.
Results
eIF4G and eIF4B cooperatively activate eIF4A duplex unwinding activity
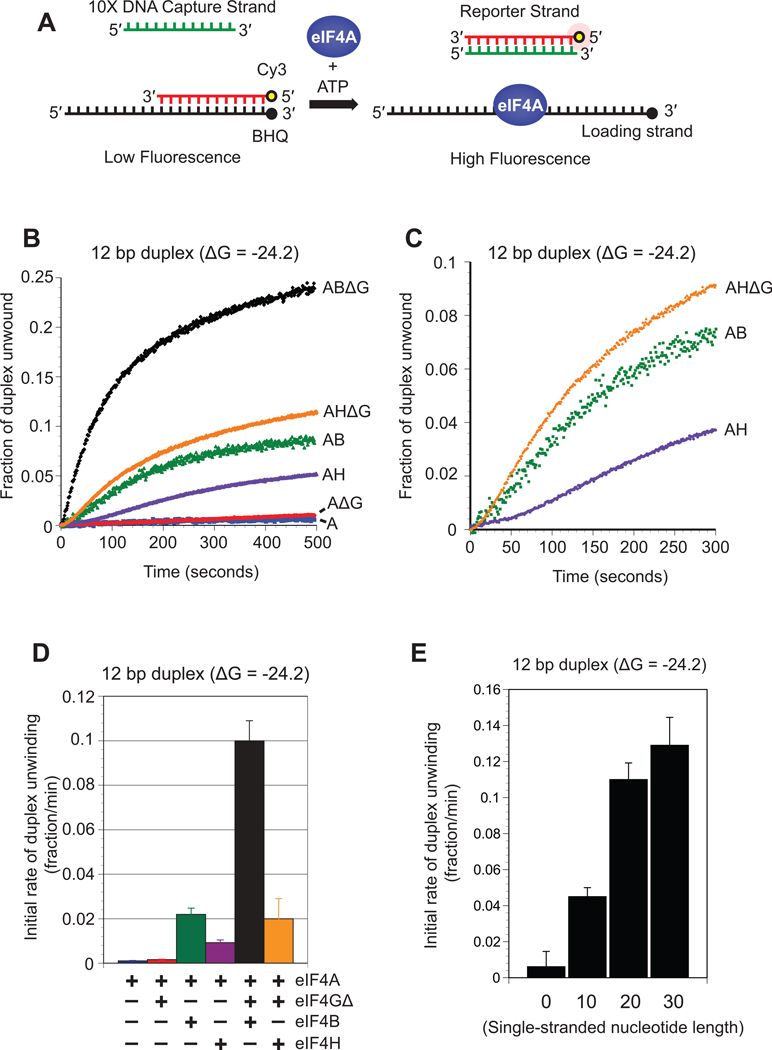
We have established an in vitro fluorescence assay capable of detecting RNA strand separation in real time using highly purified recombinant components. Full-length human eIF4A and eIF4B are used in addition to the previously characterized functional “core” region of eIF4GI, which spans amino acids 682–1105 (eIF4G Δ, Fig. 1B and ref 18; 23). In this assay, the reporter strand of the duplex is modified on its 5′ end with a cyanine 3 (Cy3) fluorescent dye while the loading strand is modified on its 3′ end with a spectrally paired quencher dye (BHQ). Annealing of the strands brings the two dyes close together, resulting in efficient fluorescence quenching. Duplex unwinding is monitored by observing an increase in Cy3 fluorescence following strand separation (Fig. 2A). Reannealing of the reporter strand to the loading strand is suppressed by the presence of a 10-fold molar excess of a DNA “capture strand” that is complementary to the reporter strand. A 10-fold molar excess of DNA was found to largely prevent reannealing and result in the maximum rate of unwinding (Fig. S1). For each unwinding reaction, the observed fluorescence increase is calibrated to the fraction of duplex unwound, as described in the Materials and Methods. To establish a suitable substrate to use in this assay, we initially selected a well-characterized 12bp duplex that has previously been used to monitor eIF4A unwinding activity 2; 3 Stoichiometric amounts of each protein component are used in our reactions. With the exception of eIF4A, saturating concentrations of each protein is used in each unwinding reaction, as described in the Materials and Methods (Fig. S2).
Fig. 2. Unwinding of an RNA duplex by eIF4A and its accessory proteins.
(A) Cartoon depiction of the fluorescent duplex unwinding assay. A reporter RNA strand is modified on its 5′ end with cyanine 3 (Cy3) and annealed to a complimentary loading strand that is modified on its 3′ end with a spectrally paired black hole quencher (BHQ). The loading strand also possesses a 20nt 5′ extension. Addition of eIF4A and ATP to the reaction results in strand separation, which is visualized as an increase in Cy3 fluorescence. Reannealing of the strands is reduced by the presence of a 10-fold molar excess of a DNA capture strand that is complementary to the reporter strand. (B) Representative unwinding time courses of a 12bp duplex substrate (duplex 1; see Materials and Methods) by eIF4A in the absence (blue line) or presence of eIF4GΔ (red line), eIF4H (purple line), eIF4B (green line), eIF4H/4GΔ (orange line) and eIF4B/4GΔ (black line). The zero time point represents the reaction before ATP addition. The change in total fluorescence is converted to the fraction of duplex unwound over time, as described in Materials and Methods. (C) A magnified view of a portion of the unwinding time course from panel B to show the sigmoidal traces for eIF4A in the presence of eIF4H (purple line), eIF4B (green line) and eIF4H/4GΔ (orange line). (D) Initial rates during the initial linear portion of the unwinding time course are determined by linear fits. The initial rate of duplex unwinding by each protein combination is shown as indicated. Error bars represent the standard error from at least three separate experiments. (E) Initial rates of unwinding for a 12bp duplex (duplex 1) possessing either 0, 10, 20 and 30 single-stranded nucleotides added to the 5′ end of the loading strand (see Materials and Methods for sequences). Each assay is carried out in the presence of 50 nM RNA substrate and 1 µM eIF4A/4GΔ/4B. The change in total fluorescence is converted to the fraction of duplex unwound and the initial rate of unwinding during the linear portion of the unwinding time course is shown. Error bars represent the standard error from at least three separate unwinding reactions.
To accurately quantitate the kinetic activity of each initiation factor, we monitored a change in Cy3 fluorescence for each protein combination (Fig. 2B). The observed initial rate of each unwinding reaction was calculated by using the initial linear portion of the time course, as described in Materials and Methods. As expected, eIF4A does not promote a high degree of duplex unwinding under these conditions, with an observed initial fraction of duplex unwound per minute of only 1.1×10−3 ± 0.2×10−3 (Fig. 2D and Table 1). In the presence of eIF4A and eIF4GΔ the fraction of duplex unwound per minute is relatively unchanged at 1.6×10−3 ± 0.3×10−3 (Fig. 2D and Table 1). In contrast, the fraction of duplex unwound per minute by eIF4A in the presence of eIF4B increases roughly 20-fold to 23×10−3 ± 3×10−3 (Fig. 2D and Table 1). We then tested the activity of eIF4A in the presence of both eIF4GΔ and eIF4B. Unexpectedly, a very strong coupled enhancement of unwinding activity is observed, increasing the fraction of duplex unwound per minute by 100-fold to 100×10−3 ± 9×10−3 (Fig. 2D and Table 1). This coupled increase shows that these proteins must coordinate their activities to obtain maximum duplex unwinding and reveals a much larger increase in activity than suggested by an indirect nuclease sensitivity assay20 However, it is difficult to compare these results to previous data that were obtained with much lower protein concentrations than used here20 We find that replacing ATP with the same concentration of the non-hydrolyzable analog, AMPPNP, results in no change in fluorescence (data not shown). This suggests that the fast phase observed in the unwinding kinetics does not appear to be due to protein binding to the duplex substrate. It is worth noting that, consistent with previous work, we found it necessary to add a 20nt single-stranded 5′ extension to the loading strand of this duplex to ensure efficient protein loading (Fig. 2E and ref.21). A 3′ extension was not tested, but would likely perform in a similar way to the 5′ extension based on previous experiments from the Merrick lab21. Importantly, the kinetic data of our fluorescence assay correspond well with a traditional gel shift assay, where an eIF4A/4GΔ/4B complex generates an observed initial fraction of duplex unwound per minute of 128×10−3 ± 20×10−3 (Fig. S3).
Table 1.
Initial rates of duplex unwinding for RNA duplex substrates.
| Proteins | 12bp ΔG* = −24.2 %GC = 58 |
18bp ΔG* = −23.9 %GC = 22 |
24bp ΔG* = −24.7 %GC = 8 |
24bp ΔG* = −29.3 %GC = 16.6 |
|---|---|---|---|---|
| IR† (×10−3) | IR† (×10−3) | IR† (×10−3) | IR† (×10−3) | |
| A | 1.1±0.2 | 1.7±0.2 | 9±1 | 1.3±0.2 |
| AGΔ | 1.6±0.3 | 7±1 | 20±1 | 0.7±0.2 |
| AB | 23±3 | 52±1 | 48±2 | 11±1 |
| ABGΔ | 100±9 | 254±15 | 576±5 | 158±8 |
| AH | 9±1 | |||
| AHGΔ | 20±9 | |||
Stabilities of duplex regions are calculated as described in Materials and Methods.
Initial rate (IR) of unwinding is the fraction of duplex unwound min−1. Mean values and standard errors are from at least three independent experiments.
Duplex unwinding in the presence of only eIF4A and eIF4B reveals a significant lag phase, indicative of a rate-limiting intermediate step during duplex unwinding (Fig. 2B and magnified in Fig. 2C). This lag phase is completely removed by addition of eIF4GΔ, indicating that eIF4GΔ alters the rate-limiting step of duplex unwinding. The observed increase in the rate of duplex unwinding may reflect an enhanced association rate of a productive unwinding complex on the duplex substrate. We use the term productive unwinding complex here to describe the formation of an eIF4A•ATP•RNA complex in the absence or presence of accessory proteins. Alternatively, it is possible that there is an increase in the intrinsic unwinding activity of eIF4A. We use intrinsic unwinding activity here to describe the rate of strand separation by eIF4A following RNA binding. Interestingly, there appears to be a much slower second “inhibitory” component of each unwinding reaction that predominates over time, resulting in biphasic kinetics. The inability to obtain complete duplex unwinding within the experimental timeframe is surprising since each reaction contains a 20-fold molar excess of protein components to duplex substrate. This observation, however, is in good agreement with a previous study that used a 100-fold excess of protein components to substrate, yet only promoted partial unwinding2. The reason for this inhibition is not clear, but previous work has reported that it does not appear to be due to protein inactivation or inhibition by ADP2. It is also possible that the generation of free loading strand following reporter strand removal may inhibit unwinding events due to sequestering eIF4A. This is unlikely to be true since it is possible to double the concentration of substrate RNA (100 nM) and still observe similar unwinding kinetics (Fig. S1). It is possible that the inhibitory effect is due to non-productive, damaged, complexes in the reaction. It has not been possible, however, to detect the formation of such slow unwinding complexes that may be responsible for this inhibitory effect2. It is worth noting, however, that duplex unwinding by some DNA helicases using single turnover reaction conditions also reveals biphasic unwinding kinetics for unknown reasons30; 31; 32.
eIF4H does not completely substitute for eIF4B in duplex unwinding
To determine if eIF4H can substitute for eIF4B in unwinding reactions, we determined its kinetic activity using analogous experiments to those described above. The fraction of duplex unwound per minute by eIF4A in the presence of eIF4H is increased roughly 9-fold to 9.2×10−3 ± 1.3×10−3 (Fig. 2B, 2D and Table 1). This rate is half that observed for eIF4A in the presence of eIF4B and comparable to the 5-fold increase in observed initial rate of unwinding by eIF4H for the same duplex in a previous study21; 29 Further addition of eIF4GΔ increases the fraction of duplex unwound per minute by an additional two-fold to 20×10−3 ± 9.0×10−3 This unexpected result indicates that eIF4H is less effective than eIF4B in coordinating its activity with eIF4GΔ to maximize duplex unwinding by eIF4A. As we discovered for an eIF4A/4B complex, duplex unwinding by eIF4A in the presence of eIF4H also reveals a lag phase early in the duplex unwinding time course (Fig. 2B,C). In contrast to eIF4B, however, this lag phase is still evident following the addition of eIF4GΔ (Fig. 2B,C). This indicates that in the presence of eIF4H, eIF4GΔ does not appear to increase the reaction rate enough to completely remove the lag phase. These data strongly indicate that while eIF4H does enhance duplex unwinding by eIF4A, it is not completely interchangeable with eIF4B in duplex unwinding. This may suggest that the C-terminal RNA binding region of eIF4B is important for coordinating the duplex unwinding activity of eIF4A in the presence of eIF4GΔ.
eIF4B increases the efficiency of strand separation by eIF4A
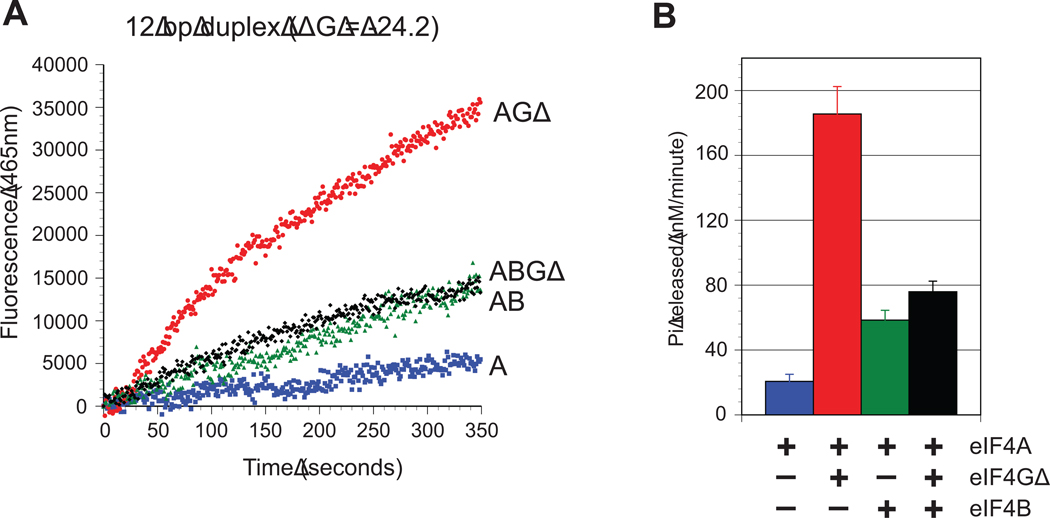
To measure the coupling between ATP hydrolysis and duplex unwinding by eIF4A, both activities were analyzed using identical reaction conditions. To accurately probe the rate of ATP hydrolysis in our unwinding reactions, we employed a highly sensitive assay that utilizes a well-characterized fluorescently labeled recombinant phosphate binding protein from E. coli, PBP-MDCC33; 34. Using the same 12bp duplex substrate already described, the rate of Pi release for each combination of proteins was determined by monitoring the change in total fluorescence of PBP-MDCC over time (Fig. 3A). To ensure that only the RNA-dependent ATPase rate is monitored, the change in fluorescence from each combination of proteins in the absence of RNA is subtracted, as described in the Materials and Methods and shown in Figure S4. The initial rate of Pi release was calculated by using the initial linear portion of the time course for each trace. In the case of the eIF4A/4B complex, the initial rate of phosphate release was measured following the lag phase in duplex unwinding. Consistent with the unwinding reactions, eIF4A alone possesses the slowest rate of Pi release, with only 21 ± 4 nM Pi released min−1 (Fig. 3B and Table 2). There is a dramatic increase in the rate of Pi released by the addition of eIF4GΔ, increasing the observed initial rate by almost 10 fold to 186 ± 17 nM Pi min−1 (Fig. 3B and Table 2). This is consistent with the finding that eIF4G with a similar truncation lowers the KmRNA by 10-fold and increases the kcat of ATP hydrolysis by almost 4-fold18. A reaction containing eIF4A and eIF4B also increases the rate of Pi released by roughly 3-fold compared to eIF4A alone, with an observed initial rate of 58 ± 6 nM Pi min−1 (Fig. 3B and Table 2). Surprisingly, in the presence of eIF4A/4GA/4B, the rate of Pi release is very similar to that observed for the eIF4A/4B complex, with an initial rate of 76 ± 7 nM Pi min−1 (Fig. 3B and Table 2).
Fig. 3. ATP hydrolysis by eIF4A and its accessory proteins.
(A) Representative time courses of phosphate release by eIF4A on a 12bp duplex (duplex 1) in the absence (blue line) or presence of eIF4GΔ (red line), eIF4B (green line) and eIF4B/4GΔ (black line). The change in RNA-dependent total fluorescence intensity (arbitrary units) of PBP-MDCC at 465 nm against time is shown for each protein combination. The zero time point represents the reaction before ATP addition. (B) Quantitation of the initial rate of RNA-dependent Pi release by eIF4A in the absence or presence of eIF4GΔ, eIF4B and eIF4B/4GΔ, as indicated. The initial change in total fluorescence is converted to the concentration of Pi released (nM min−1). Error bars represent the standard error from at least three separate experiments.
Table 2.
Coupling efficiency of duplex unwinding to ATP hydrolysis.
| Proteins | Duplex | ATP hydrolysis (nM min−1)* |
Duplex unwinding (nM min−1)† |
ATP hydrolyzed per duplex unwound |
|---|---|---|---|---|
| A | 12bp | 21±4 | 0.05±0.01 | 420±16 |
| AGΔ | 12bp | 186±17 | 0.08±0.01 | 2325±360 |
| AB | 12bp | 58±6 | 1.2±0.14 | 48±8 |
| ABGΔ | 12bp | 76±7 | 5.3±0.50 | 14±2 |
RNA dependent ATP hydrolysis rates are obtained by subtracting ATP hydrolysis rates in the absence of RNA, as described in Materials and Methods.
The initial rate of duplex unwinding is obtained from the fraction of duplex unwound (Table 1) and assumes that 100% of the duplex is annealed before the addition of ATP to initiate the unwinding reaction. All mean values and standard errors are from at least three independent experiments.
The apparent efficiency of coupling between ATP hydrolysis and duplex unwinding can be calculated for each combination of proteins by dividing the initial unwinding rate by the initial rate of ATP hydrolysis. To simplify the comparisons between different protein combinations, the relative efficiency for each protein combination is presented as moles of ATP hydrolyzed per mole of duplex unwound (Table 2). The results show that an eIF4A/4B complex is nearly ten fold more efficient at coupling unwinding with ATP hydrolysis than eIF4A alone (Table 2). This is in contrast to the efficiency of unwinding for an eIF4A/4GΔ complex, which is found to be roughly 6-fold lower than for eIF4A alone (Table 2). This decrease reflects the large increase in the rate of ATP hydrolysis by the addition of eIF4GΔ without an appreciable increase in duplex unwinding. Strikingly, the eIF4A/4GΔ/4B complex is 30-fold more efficient than eIF4A and 160-fold more efficient than the eIF4A/4GΔ complex at coupling unwinding with ATP hydrolysis (Table 2). These results show that unwinding events become considerably more productive when eIF4B is present, inferring that eIF4B functions to increase the likelihood of strand separation for each ATP hydrolysis cycle of eIF4A.
It is important to note that the rate of Pi release by an eIF4A/4GΔ complex on the 32nt loading strand is comparable to that observed for the 32nt loading strand that includes the annealed 12nt RNA to form a duplex region (178±19 nM Pi min−1; Fig. S4). This is consistent with the notion that eIF4A binds to single stranded RNA regions, previously shown in experiments that employed poly(A) and poly(U) as substrates to stimulate the ATPase activity of eIF4A in the absence and presence of accessory proteins17; 18; 35. This makes it very difficult to precisely determine exactly how much of the observed ATP hydrolysis is attributed to duplex unwinding compared to side reactions that are non-productive. This indicates that the values obtained here likely represent a maximum amount of ATP that can be hydrolyzed per duplex unwound for each combination of proteins. To this end, it is interesting to note that an eIF4A/4GΔ/4B complex hydrolyzes roughly 10-fold more ATP per strand separation compared to that previously shown for the DEAD-box helicase CYT-19 on an 11bp duplex36. Nevertheless, these data strongly indicate that the presence of eIF4B is required to dramatically increase the efficiency of strand separation with each cycle of ATP hydrolysis.
Strand separation by eIF4A is sensitive to the duplex energy landscape
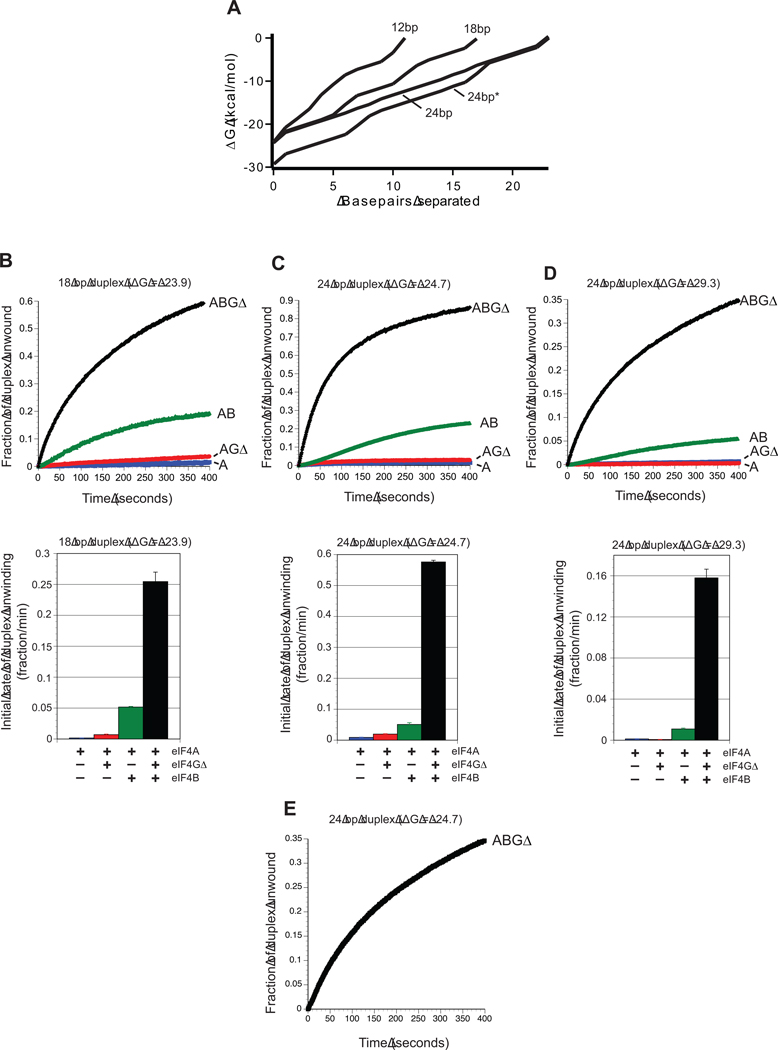
Stability of a duplex region is typically determined by calculating the Gibbs free energy of the entire duplex region. Two duplex regions of different length but equal ΔG values must therefore possess different numbers of GC base pairs. Plotting the change in ΔG per base pair of the duplex unwound generates a depiction of the duplex energy landscape (Fig. 4A). The presence of more GC base pairs therefore increases the steepness of energy landscape per base pair of a duplex. Previous work has shown that duplex unwinding by eIF4A is sensitive to the total ΔG of a duplex rather than the steepness of the energy landscape3. In contrast, translation efficiency increases as the steepness of the energy landscape of a duplex region located four nucleotides from the mRNA cap structure is reduced while maintaining the overall duplex ΔG37. To resolve these apparently conflicting data, we designed two additional duplex constructs with greatly altered GC content compared to that of the 12bp duplex (58% GC). To maintain a thermal stability close to −24 kcal/mol, these duplexes were increased in length by half helical turns, resulting in 18bp (22% GC) and 24bp (8% GC) duplexes with identical 20nt single-stranded regions for protein loading. This creates predicted free energy landscapes with progressively less steep gradients per base pair across the duplex region (Fig. 4A). We monitored the unwinding time courses for these duplex substrates using each combination of protein factors (Fig. 4B, C). Noticeably, reducing the % GC content of the duplex region results in an increase in observed initial duplex unwinding rates for all combinations of proteins compared with the 12 bp duplex (Fig. 4B, 4C and Table 1). Specifically, reducing the GC content from 58% (12bp duplex) to 22% (18bp duplex) increases the fraction of duplex unwound per minute by the eIF4A/4GΔ/4B complex roughly 2.4-fold from 100×10−3±9×10−3 to 254×10−3±15×10−3 (Fig 4B and Table 1). A further decrease in the GC content to 8% (24bp duplex) increases the fraction of duplex unwound per minute by the eIF4A/4GΔ/4B complex an additional 2.3-fold to 576×10−3±5.4×10−3 (Fig. 4C and Table 1). Similar increases in the initial rate of duplex unwinding are observed for eIF4A, eIF4A/4GΔ and eIF4A/4B when the GC content is reduced from 58% (12bp duplex) to 22% (18bp duplex) (Fig. 4B and Table 1). Further reduction in the GC content to 8% (24bp duplex) increases the initial rate of duplex unwinding by eIF4A and eIF4A/4GΔ (Fig. 4C and Table 1). Curiously, this reduction in the GC content to 8% (24bp duplex) does not further increase the initial rate of duplex unwinding by an eIF4A/4B complex (Fig. 4C). The reason for this is not clear, but may reflect a change in the number of protein binding sites between different duplex lengths. As expected, each increase in the initial rate of unwinding in response to lowering the GC content is also observed in the maximum amount of duplex unwound (Fig. 4B, C).
Fig. 4. Effect of energy landscape on duplex unwinding by eIF4A.
(A) The energy landscape of each duplex substrate used is plotted as the change in Gibbs free energy (kcal/mol) per base pair of the duplex unwound. Each trace starts with the 3′ most base pair of the reporter strand and is labeled according to the duplex length. The 24bp* duplex represents the more stable 24bp duplex, as described in the Materials and Methods. (B–D) Upper panels show representative unwinding time courses for duplex substrates possessing different stabilities by eIF4A in the absence (blue line) or presence of eIF4GΔ (red line), eIF4B (green line) and eIF4B/4GΔ (black line). For each time course, the zero time point represents the reaction before ATP addition. The change in total fluorescence is converted to the fraction of duplex unwound over time, as described in Materials and Methods. Unwinding reactions are presented for each protein combination for an 18bp duplex substrate (duplex 2; B), a 24bp duplex (duplex 3, C), and a more stable 24bp duplex (duplex 4; D). Each duplex possesses identical 20 nucleotide 5′ extensions to the loading strand, as described in Materials and Methods. Lower panels in each case show the quantitation of the initial rate of duplex unwinding by eIF4A in the absence (blue bar) or presence of eIF4GΔ (red bar), eIF4B (green bar) and eIF4B/4GΔ (black bar). Initial rates from unwinding reactions during the initial linear portion of each unwinding time course are shown for the 18bp (duplex 2; A), 24bp (duplex 3; B) and more stable 24bp (duplex 4; C) duplexes. Each data set is from at least three separate experiments and error bars represent the standard error. (E) A representative unwinding reaction by an eIF4A/4GΔ/4B complex for a blunt-ended 24bp duplex is shown (duplex 5).
To determine if duplex unwinding by eIF4A is also sensitive to the overall stability of the duplex, we generated a more stable 24bp duplex with an increased GC content (16.6%) to compare directly to the 24bp duplex with lower GC content (8%). In agreement with previous data, the observed initial rate of duplex unwinding by all combinations of proteins is strongly reduced in response to increased overall stability while maintaining duplex length (Fig. 4D, Table 1 and ref3; 21). It should be noted, however, that the increase in GC content also has the effect of increasing the steepness of the energy landscape per base pair of the duplex (Fig. 4A). These data show that duplex unwinding by eIF4A and associated proteins is sensitive to the steepness of the energy landscape of the duplex per base pair, and help to explain why translation initiation is influenced by the energy landscape of hairpin regions in the 5′ UTR.
The duplex energy landscape influences productive complex formation
To gain mechanistic insight into how the energy landscape of a duplex regulates eIF4A activity, we have determined the rate constants of unwinding by an eIF4A/4GΔ/4B complex for each duplex. As mentioned above, the time courses of duplex unwinding by eIF4A/4GΔ/4B are biphasic, implying a rapid unwinding phase (k1) and a slower unwinding phase (k2) (Fig. S4). Each time course can be fitted well using a double exponential equation (Equation 1) to generate rate constants and amplitudes of unwinding for each phase (Table 3). Interestingly, we only observe relatively small 2- to 3-fold changes in the rate constant of the rapid unwinding phase (k1) between the duplexes tested (Table 3). This is in spite of observing an almost 6-fold increase in the apparent initial rate of unwinding between the 12bp (58% GC) and 24bp (8% GC) duplexes (Table 1). This implies that the duplex energy landscape predominantly influences the rate of productive complex formation rather than changing the intrinsic rate of unwinding by eIF4A. Moreover, the rapid unwinding rate constant (k1) of the 24bp duplex (8% GC) is not greatly influenced by the absence of an overhang region, inferring that the overhang region only serves to increase the rate of productive complex formation. This is in good agreement with previous work elegantly showing that single stranded regions only serve to promote DEAD-box protein loading to a duplex region38 Interpretation of the slow unwinding phase (k2) is more complicated. There is ~5-fold increase in the rate of the slow unwinding phase (k2) when the duplex length is increased from 12bp to 18bp (Table 3). A further increase in the duplex length, however, does not result in an additional increase in the rate constant, which is an order of magnitude less than the rapid unwinding rate constant (k1). The reason for the increase in the rate constant of the slow unwinding phase (k2) between the 12bp and 18bp duplexes is not clear, but may indicate the existence of multiple unwinding complexes associated with the duplex region on longer duplexes. Indeed, native eIF4F cooperatively binds to long RNAs that are more than ~60 nucleotides in length, but it is not clear whether there is a similar length dependence for an eIF4A/GΔ complex39. As mentioned above, the slow unwinding phase may reflect the presence of slow, perhaps damaged, complexes in the unwinding reactions. Nevertheless, our data strongly suggest that the duplex energy landscape influences the rate of productive complex formation on the substrate rather than intrinsic unwinding activity.
Table 3.
Kinetic parameters for duplex unwinding by eIF4A/4GΔ/4B.
| Factors | Duplex Length |
Overhang Length |
Duplex Stability (ΔG) |
k1 | A1 (Fraction unwound) |
k2 | A2 (Fraction unwound) |
|---|---|---|---|---|---|---|---|
| AGΔB | 12 | 20 | −24.2 | 1.2×10−2 ± 4×10−4 | 0.17 ± 0.01 | 7.0×10−4 ± 1×10−4 | 0.26 ± 0.03 |
| AGΔB | 18 | 20 | −23.9 | 3.7×10−2 ± 8×10−3 | 0.17 ± 0.01 | 3.8×10−3 ± 5×10−4 | 0.61 ± 0.03 |
| AGΔB | 24 | 20 | −24.7 | 2.7×10−2 ± 4×10−3 | 0.36 ± 0.06 | 6.1×10−3 ± 1×10−3 | 0.47 ± 0.01 |
| AGΔB | 24 | 20 | −29.3 | 2.6×10−2 ± 2×10−3 | 0.08 ± 0.01 | 3.3×10−3 ± 1×10−4 | 0.37 ± 0.01 |
| AGΔB | 24 | 0 | −24.7 | 1.3×10−2 ± 7×10−4 | 0.10 ± 0.01 | 2.0×10−3 ± 2×10−4 | 0.44 ± 0.01 |
Kinetic constants were obtained from at least three separate unwinding reactions for each duplex substrate similar to that shown in Fig. 2 and Fig. S3. The data are fit to a double exponential equation (Equation 1), yielding rate constants for rapid (k1) and slow (k2) unwinding phases. The corresponding amplitude of unwinding for each phase is shown as the fraction of duplex unwound (A1 and A2). All mean values and standard errors are from at least three independent experiments.
Discussion
Using a real-time fluorescent duplex unwinding assay, we have established that eIF4G and eIF4B function to cooperatively activate the duplex unwinding activity of eIF4A. Our data reveal that an eIF4A/4GΔ/4B complex possesses an observed initial rate of duplex unwinding that is 100-fold faster than that of eIF4A alone. Efficient strand separation by eIF4A therefore requires both eIF4G and eIF4B, explaining why knockdown of eIF4B in vivo inhibits the translation of highly structured mRNAs40. Our data also reveal that eIF4H is much less efficient at stimulating eIF4A unwinding activity than eIF4B. This implies that the C-terminal RNA binding domain of eIF4B may be responsible for stimulating unwinding since it is entirely absent in eIF4H. This finding raises the question of what the role of eIF4H is in protein synthesis initiation. It will be important in the future to determine if eIF4H regulates translation initiation on a subset of mRNAs in vivo.
Combining the kinetic unwinding data and ATPase assays performed under identical conditions has enabled us to determine the relative coupling efficiency between ATP hydrolysis and duplex unwinding. Our data show that eIF4B functions to dramatically increase the probability of strand separation by eIF4A per ATP hydrolyzed. Moreover, the presence of eIF4B further ensures that the eIF4G–dependent increase in the rate of eIF4A ATP hydrolysis is used productively to maximize duplex unwinding. As noted above, the moles of ATP hydrolyzed per duplex unwound is likely to be an overestimate since the rate of ATP hydrolysis on the single stranded loading strand is comparable to that of the duplex substrate (Fig. S3). Nevertheless, eIF4B clearly regulates the rate of ATP hydrolysis by eIF4A and directs this energy to promote duplex unwinding. How eIF4B increases the likelihood of strand separation by eIF4A remains an open question. Structures of eIF4AIII bound to RNA in the exon junction complex show that the resulting RNA conformation is incompatible with duplex formation9; 41. It is possible that such an eIF4A–induced conformation may be stabilized by eIF4B, and possibly eIF4G, to increase the probability of strand separation. Since eIF4B enhances the RNA binding affinity of eIF4A, it is possible that it stabilizes a productive unwinding conformation. Previous data, however, have shown that both eIF4B and eIF4G increase the binding affinity of eIF4A to RNA and ATP17; 18; 24 It is therefore unlikely that the function of eIF4B in unwinding can be explained entirely by increasing substrate affinity since an eIF4A/4GΔ complex possesses little duplex unwinding activity. The stimulation of eIF4A duplex unwinding by eIF4B increases with greater duplex stability, implying that eIF4B may actually function at a kinetic step following substrate binding21. The data presented here using two 24bp duplexes with different stabilities is in good agreement with this finding. Unfortunately, our kinetic assay can only monitor complete strand separation, preventing us from detecting intermediates in the unwinding process. It is therefore entirely possible that eIF4B may function to stabilize an unwinding intermediate that prevents strand reannealing. It will be useful to investigate the affinity of eIF4B and eIF4G to unwinding intermediates in future experiments to help answer this question.
In this study, we also investigated how the steepness of the duplex energy landscape influences unwinding by eIF4A and its accessory proteins. A reduction in the GC content of a duplex while maintaining the overall stability strongly increases the rate of unwinding by an eIF4A/4GΔ/4B complex, even though the duplex is longer. By comparing rate constants, our data indicate that the energy landscape may directly influence the rate at which a productive complex (eIF4A/4GΔ/4B + ATP) forms on a duplex substrate rather than change in the intrinsic unwinding activity. It is important to account for the changes observed in the amplitude of unwinding between the duplexes tested. To this end, we propose that there must be a rapid kinetic competition between the formation of a productive unwinding complex and the inactivation process that is observed in our data. These data therefore help to explain why translation efficiency increases as the GC content of a hairpin stem placed close to the mRNA cap structure is reduced without changing the overall ΔG37 It will be important in the future to determine if the location of GC base pairs within a duplex region further influences duplex unwinding by eIF4A. It also will be important to define the exact contributions that the steepness of the energy landscape of a duplex makes to regulate unwinding and initiation efficiency.
On the basis of these findings and those of other studies, we propose a model in which eIF4A and its accessory factors use a mechanism of local unwinding to promote strand separation12; 42. The rate of productive complex formation is influenced by the local stability per base pair in addition to the stability of the entire duplex region being separated. To ensure efficient duplex unwinding, eIF4A requires eIF4B and eIF4G to couple the ATP hydrolysis cycle of eIF4A with duplex unwinding. Further investigation will be required to completely understand the complex mechanism by which eIF4B and eIF4G stimulate eIF4A duplex unwinding activity. This includes the need to determine the order of binding for each protein to a duplex substrate. The relative affinities and kinetic parameters for the interaction of each accessory protein with RNA and eIF4A must also be determined for each step in the ATP hydrolysis cycle. Additional crystal structures of these components bound to RNA will undoubtedly help to reveal how these proteins associate with RNA to coordinate duplex unwinding and ATP hydrolysis. Finally, high-resolution single molecule techniques will likely demonstrate if eIF4G and eIF4B function to alter the unwinding step size and possibly the processivity of eIF4A. It is important to stress here that we have employed a truncation of eIF4G that was previously identified to constitute the functional core region of human eIF4G that is able to recruit an active ribosome in a tethered assay 23. In particular, the data presented here indicate how the activity of the evolutionarily conserved HEAT-1 domain regulates eIF4A helicase activity. It will clearly be important in the future to determine if other domains of human eIF4G regulate eIF4A activity.
There is growing evidence that additional helicases may further synergize with eIF4A to unwind mRNA secondary structure43. The real-time fluorescent duplex unwinding assay described here will provide a valuable tool with which to carry out kinetic unwinding assays using other initiation components, including the 40S ribosomal subunit. By introducing fluorescent reporter and quench strands at different positions within a natural 5′ UTR, it should be possible to also determine how each initiation component functions in unwinding mRNA secondary structure during different stages of translation initiation.
Materials and Methods
Materials
Human eIF4AI (406 amino acids), eIF4H isoform 2 (228 amino acids) and eIF4GΔ (amino acids 682–1166 from eIF4GI) are expressed in E. coli as maltose-binding protein (MBP) fusion constructs using a previously modified pET28c expression vector using NdeI and XhoI sites (Novagen)44. Human eIF4B (611 amino acids) is is inserted into a modified His-tagged pFASTBAC1 vector (Invitrogen) and expressed in insect cells (sf9), as described previously for human eIF3j44. Expression and initial purification of each protein using Ni-NTA Superflow resin (Qiagen) is carried out using a standard protocol for eukaryotic initiation factors 44. Each protein is cleaved using recombinant TEV protease, and purified by ion-exchange chromatography (Heparin HP, GE Healthcare) and size exclusion chromatography (Superose 6, GE Healthcare) to yield untagged proteins. Following purification, eIF4A, eIF4B and eIF4GΔ proteins are stored at −80 °C in storage buffer (20 mM Hepes, pH 7.5, 200 mM KCl, 1 mM DTT) supplemented with 10% glycerol. To maintain stability, purified eIF4H is stored at −80 °C in storage buffer supplemented with 20% glycerol. Recombinant eIF4B purified from E.coli poorly substitutes for native eIF4B in the assembly of 48S pre-initiation complexes 45 Our kinetic unwinding assays show that human eIF4B expressed and purified using a baculovirus system as described here is remarkably stable over many months of storage at −80°C. Purified PBP-MDCC is obtained by using a previously described protocol 34
All RNA oligonucleotides are chemically synthesized, modified and HPLC purified by Integrated DNA Technologies. Reporter strands are modified with cyanine 3 (Cy3) on their 5′ end and loading strands are modified with a spectrally paired black hole quencher (BHQ) on their 3′ end. RNA substrate sequences are as follows: (underlined regions correspond to the duplex region): 12bp duplex with 20nt 5′ overhang (duplex 1), 5′-GAACAACAACAACAACCAUGGCACCGUAAAGC-BHQ-3′ (loading strand 1) annealed to 5′-Cy3-GCUUUACGGUGC- 3′ (reporter strand 1); 12bp blunt end duplex, 5′-GCACCGUAAAGC-BHQ-3′ (loading strand 2) annealed to reporter strand 1; 12bp duplex with 10nt 5′ overhang, 5′- GAACACCAUGGCACCGUAAAGC-BHQ-3′ (loading strand 3) annealed to reporter strand 1; 12bp duplex with 30nt 5′ overhang, 5′-GAACAACAACAACAACAACAACAAACCAUGGCACCGUAAAGC-BHQ-3′ (loading strand 4) annealed to reporter strand 1; 18bp duplex with 20nt 5′ overhang (duplex 2), 5′-GAACAACAACAACAACCAUGGAAAAUCAAAACUAAAAC-BHQ-3′ (loading strand 5) and 5′-Cy3-GUUUUAGUUUUGAUUUUC-3′ (reporter strand 2); 24bp duplex with 20nt 5′ overhang (duplex 3), 5′-GAACAACAACAACAACCAUGGAAAAAAUUAAAAAAUUAAAAAAC-BHQ-3′ (loading strand 6) and 5′-Cy3-GUUUUUUAAUUUUUUAAUUUUUUC-3′ (reporter strand 3); 24bp blunt end duplex, 5′-GAAAAAAUUAAAAAAUUAAAAAAC-BHQ-3′ (loading strand 7) and reporter strand 3; 24bp duplex with 20nt 5′ overhang (duplex 4), 5′-GAACAACAACAACAACCAUGGAAAAAAGUAAAAAAUUGAAAAAC-BHQ-3′ (loading strand 8) and 5′-Cy3-GUUUUUCAAUUUUUUACUUUUUUC- 3′ (reporter strand 4). DNA capture strand sequences were as follows: 12bp, 5′-GCACCGUAAAGC-3′ (capture 1); 18bp, 5′-GAAAAUCAAAACUAAAAC-3′ (capture 2); 24bp, 5′-GAAAAAAUUAAAAAAUUAAAAAAC-3′ (capture 3); 24bp, 5′-GAAAAAAGUAAAAAAUUGAAAAAC-3′ (capture 4).
The stability of individual duplex regions are determined by the nearest neighbor method, which involves the connection of the duplex strands by a tetra loop sequence for computation purposes 46. The absence of a tetra loop connecting the duplexes in our experiments is accounted for by addition of the tetra loop positive energy in the final stability calculation. This accounts for the slightly more positive stability compared to a previous calculation of the 12bp duplex region3.
ATP was purchased from Sigma and N-[2-1-maleimidyl)ethyl]-7-diethylamino coumarin-3-carboxamide (MDCC) was from Invitrogen.
Unwinding reactions
Duplex annealing: For each duplex substrate, a 10X concentrated stock is generated by annealing loading and reporter strands to a final concentration of 500 nM in unwinding buffer (20 mM Tris acetate (pH 7.5), 2 mM magnesium acetate, 100 mM KCl and 0.2 mM DTT). The annealing reaction is placed into a heating block at 70 °C and then cooled to room temperature by allowing the block to cool on the bench. A 10-fold molar excess of competitor DNA is then added, resulting in a 5X stock of duplex substrate in unwinding buffer.
Unwinding reactions: Each unwinding reaction (70 µl) is prepared in 1X unwinding buffer and contains 50 nM duplex RNA, 500 nM competitor DNA and 1 µM of each protein (unless otherwise noted). The reaction is mixed and added to a 50 µl quartz sub-micro cell (Starna) and placed into a Fluorolog-3 spectrofluorometer (Horiba). Reactions are incubated at 25 °C for the duration of the unwinding experiment. An initial fluorescent measurement is made to obtain a stable baseline of the annealed duplex using excitation and emission wavelengths of 550 nm and 570 nm respectively. A final concentration of 2 mM ATP•Mg acetate is added to initiate unwinding. The change in emission fluorescence is monitored as a function of time, recording data points at one-second intervals.
Data analysis: The change in fluorescence is calibrated to the fraction of duplex unwound over time by determining the maximum fluorescence of the Cy3-modified reporter strand (50 nM) in the absence of the loading strand. The maximum fluorescence is monitored prior to the start of each experiment and typically varies by ~10% from day to day. The maximum fluorescence of the Cy3 reporter strand is not altered by the incubation of protein components and ATP•Mg. Any change in the maximum fluorescence by the DNA chase strand is taken into consideration when calibrating the amount of unwinding. The fluorescence baseline before ATP addition is subtracted from the maximum fluorescence to reveal the total change in fluorescence. The dilution factor of the added ATP•Mg is taken into account so that an accurate change in fluorescence is obtained. Quenching of the Cy3 fluorescence is typically found to be in the range of 70–90% compared to free reporter. The total change in fluorescence is converted to the fraction of quenched duplex unwound and plotted against time following the addition of ATP•Mg. All data is plotted using Delta Graph software (Redrock software).
The initial rate of unwinding is determined by measuring the fraction of duplex unwinding in the initial linear portion of the unwinding reaction. This is typically collected over the first 60 seconds of the reaction. Linear regression fits are applied, and the initial fraction of duplex unwound per minute is taken from the slope of the line. In the case of unwinding time courses that possess a lag phase (eIF4H and eIF4B), the initial rate of unwinding is calculated from the linear portion of the unwinding reaction immediately following the lag phase.
The kinetic unwinding time course for each eIF4A/4GΔ/4B reaction is fit to a double exponential equation using Deltagraph software:
| (Eq. 1) |
Where RNAss represents the fraction of single stranded RNA released as a function of time; A1 is the total unwinding amplitude of the rapid unwinding phase; k1 is the unwinding rate constant of the rapid phase; A2 is the total unwinding amplitude of the slow unwinding phase; and k2 is the unwinding rate constant of the slow unwinding phase.
Phosphate release assay
Measurement of inorganic phosphate (Pi) release was determined by an established method using a fluorescently modified phosphate binding protein from E.coli 33; 34; 47 with minor modifications. Briefly, an identical duplex unwinding reaction to that described above is prepared with minor modifications. Reactions (70 µl) are supplemented with 10 µM PBP-MDCC and 2 mM ATP•Mg is added prior to the addition of protein initiation factors. Importantly, the addition of 10 µM PBP-MDCC does not influence the rate of duplex unwinding. Total PBP-MDCC fluorescence is determined by excitation at 430 nm and monitoring the emission wavelength at 465 nm in a Fluorolog-3 spectrofluorometer. Reactions are incubated at 25 °C for 2 minutes to obtain a constant baseline. The reaction is initiated by the addition of 1 µM protein components and the change in total fluorescence is monitored. The change in fluorescence due to RNA binding is determined by subtracting a control reaction in the absence of duplex substrate. The addition of a phosphate mop (0.01 U ml−1 purine nucleoside phosphorylase and 200 µM 7-methylguanosine) was not found to be necessary due to low levels of contaminating Pi in the components used. The change in total fluorescence is calibrated as described 47. The initial rate of Pi release by combinations of initiation factors is determined by measuring the change in total fluorescence in the initial linear portion of each reaction. This is typically collected over the first 60 seconds of the reaction, recording data points every second. Linear regression fits are applied, and the concentration of Pi released per minute is determined by calibration using the slope of the linear fit of the Pi calibration curve. PBP-MDCC fluorescence is determined by excitation at 430 nm and monitoring the emission wavelength at 465 nm.
Supplementary Material
Acknowledgments
We thank Behzad Rad, Stephen Kowalczykowski and members of the Kowalczykowski lab for their expert help and advice with real-time fluorescence assays. We thank John Hershey, Cuauhtémoc García-García and Enoch Baldwin for their insightful comments and critical reading of the manuscript. We gratefully acknowledge Martin Webb for his generous gift of PBP-MDCC and Angelie Do for excellent technical assistance. This work was supported by grant R01GM092927 from the National Institute of General Medical Sciences.
Abbreviations
- eIF
eukaryotic initiation factor
- 5’ UTR
5’ untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rozen F, Edery I, Meerovitch K, Dever TE, Merrick WC, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rogers GW, Jr, Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 3.Rogers GW, Jr, Lima WF, Merrick WC. Further characterization of the helicase activity of eIF4A. Substrate specificity. J Biol Chem. 2001;276:12598–12608. doi: 10.1074/jbc.M007560200. [DOI] [PubMed] [Google Scholar]
- 4.Caruthers JM, Johnson ER, McKay DB. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc Natl Acad Sci U S A. 2000;97:13080–13085. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schutz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, Baumann U. Crystal structure of the yeast eIF4A–eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci U S A. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lorsch JR, Herschlag D. The DEAD box protein eIF4A. 2. A cycle of nucleotide and RNA-dependent conformational changes. Biochemistry. 1998;37:2194–2206. doi: 10.1021/bi9724319. [DOI] [PubMed] [Google Scholar]
- 7.Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilbert M, Kebbel F, Gubaev A, Klostermeier D. eIF4G stimulates the activity of the DEAD box protein eIF4A by a conformational guidance mechanism. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Seraphin B, Le Hir H, Andersen GR. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 10.Lorsch JR, Herschlag D. The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry. 1998;37:2180–2193. doi: 10.1021/bi972430g. [DOI] [PubMed] [Google Scholar]
- 11.Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nature reviews. Molecular cell biology. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 12.Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28:253–263. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 13.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci U S A. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grifo JA, Tahara SM, Morgan MA, Shatkin AJ, Merrick WC. New initiation factor activity required for globin mRNA translation. J Biol Chem. 1983;258:5804–5810. [PubMed] [Google Scholar]
- 15.Muckenthaler M, Gray NK, Hentze MW. IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Mol Cell. 1998;2:383–388. doi: 10.1016/s1097-2765(00)80282-8. [DOI] [PubMed] [Google Scholar]
- 16.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 17.Abramson RD, Dever TE, Merrick WC. Biochemical evidence supporting a mechanism for cap-independent and internal initiation of eukaryotic mRNA. J Biol Chem. 1988;263:6016–6019. [PubMed] [Google Scholar]
- 18.Korneeva NL, First EA, Benoit CA, Rhoads RE. Interaction between the NH2-terminal domain of eIF4A and the central domain of eIF4G modulates RNA-stimulated ATPase activity. J Biol Chem. 2005;280:1872–1881. doi: 10.1074/jbc.M406168200. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen KH, Behrens MA, He Y, Oliveira CL, Sottrup Jensen L, Hoffmann SV, Pedersen JS, Andersen GR. Synergistic activation of eIF4A by eIF4B and eIF4G. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ray BK, Lawson TG, Kramer JC, Cladaras MH, Grifo JA, Abramson RD, Merrick WC, Thach RE. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J Biol Chem. 1985;260:7651–7658. [PubMed] [Google Scholar]
- 21.Rogers GW, Jr, Richter NJ, Lima WF, Merrick WC. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem. 2001;276:30914–30922. doi: 10.1074/jbc.M100157200. [DOI] [PubMed] [Google Scholar]
- 22.Morino S, Imataka H, Svitkin YV, Pestova TV, Sonenberg N. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol Cell Biol. 2000;20:468–477. doi: 10.1128/mcb.20.2.468-477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Gregorio E, Preiss T, Hentze MW. Translation driven by an eIF4G core domain in vivo. EMBO J. 1999;18:4865–4874. doi: 10.1093/emboj/18.17.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bi X, Ren J, Goss DJ. Wheat germ translation initiation factor eIF4B affects eIF4A and eIFiso4F helicase activity by increasing the ATP binding affinity of eIF4A. Biochemistry. 2000;39:5758–5765. doi: 10.1021/bi992322p. [DOI] [PubMed] [Google Scholar]
- 25.Milburn SC, Hershey JW, Davies MV, Kelleher K, Kaufman RJ. Cloning and expression of eukaryotic initiation factor 4B cDNA: sequence determination identifies a common RNA recognition motif. Embo J. 1990;9:2783–2790. doi: 10.1002/j.1460-2075.1990.tb07466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Methot N, Pause A, Hershey JW, Sonenberg N. The translation initiation factor eIF-4B contains an RNA-binding region that is distinct and independent from its ribonucleoprotein consensus sequence. Mol Cell Biol. 1994;14:2307–2316. doi: 10.1128/mcb.14.4.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Methot N, Song MS, Sonenberg N. A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol Cell Biol. 1996;16:5328–5334. doi: 10.1128/mcb.16.10.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richter-Cook NJ, Dever TE, Hensold JO, Merrick WC. Purification and characterization of a new eukaryotic protein translation factor. Eukaryotic initiation factor 4H. J Biol Chem. 1998;273:7579–7587. doi: 10.1074/jbc.273.13.7579. [DOI] [PubMed] [Google Scholar]
- 29.Richter NJ, Rogers GW, Jr, Hensold JO, Merrick WC. Further biochemical and kinetic characterization of human eukaryotic initiation factor 4H. J Biol Chem. 1999;274:35415–35424. doi: 10.1074/jbc.274.50.35415. [DOI] [PubMed] [Google Scholar]
- 30.Ali JA, Lohman TM. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science. 1997;275:377–380. doi: 10.1126/science.275.5298.377. [DOI] [PubMed] [Google Scholar]
- 31.Bjornson KP, Amaratunga M, Moore KJ, Lohman TM. Single-turnover kinetics of helicase-catalyzed DNA unwinding monitored continuously by fluorescence energy transfer. Biochemistry. 1994;33:14306–14316. doi: 10.1021/bi00251a044. [DOI] [PubMed] [Google Scholar]
- 32.Sikora B, Eoff RL, Matson SW, Raney KD. DNA unwinding by Escherichia coli DNA helicase I (TraI) provides evidence for a processive monomeric molecular motor. J Biol Chem. 2006;281:36110–36116. doi: 10.1074/jbc.M604412200. [DOI] [PubMed] [Google Scholar]
- 33.Brune M, Hunter JL, Corrie JE, Webb MR. Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry. 1994;33:8262–8271. doi: 10.1021/bi00193a013. [DOI] [PubMed] [Google Scholar]
- 34.Brune M, Hunter JL, Howell SA, Martin SR, Hazlett TL, Corrie JE, Webb MR. Mechanism of inorganic phosphate interaction with phosphate binding protein from Escherichia coli. Biochemistry. 1998;37:10370–10380. doi: 10.1021/bi9804277. [DOI] [PubMed] [Google Scholar]
- 35.Grifo JA, Abramson RD, Satler CA, Merrick WC. RNA-stimulated ATPase activity of eukaryotic initiation factors. J Biol Chem. 1984;259:8648–8654. [PubMed] [Google Scholar]
- 36.Chen Y, Potratz JP, Tijerina P, Del Campo M, Lambowitz AM, Russell R. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc Natl Acad Sci U S A. 2008;105:20203–20208. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Babendure JR, Babendure JL, Ding JH, Tsien RY. Control of mammalian translation by mRNA structure near caps. RNA. 2006;12:851–861. doi: 10.1261/rna.2309906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 39.Kaye NM, Emmett KJ, Merrick WC, Jankowsky E. Intrinsic RNA binding by the eukaryotic initiation factor 4F depends on a minimal RNA length but not on the m7G cap. J Biol Chem. 2009;284:17742–17750. doi: 10.1074/jbc.M109.009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shahbazian D, Parsyan A, Petroulakis E, Topisirovic I, Martineau Y, Gibbs BF, Svitkin Y, Sonenberg N. Control of cell survival and proliferation by mammalian eukaryotic initiation factor 4B. Mol Cell Biol. 2010;30:1478–1485. doi: 10.1128/MCB.01218-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 42.Del Campo M, Mohr S, Jiang Y, Jia H, Jankowsky E, Lambowitz AM. Unwinding by local strand separation is critical for the function of DEAD-box proteins as RNA chaperones. J Mol Biol. 2009;389:674–693. doi: 10.1016/j.jmb.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, Merrick WC, Sonenberg N. mRNA helicases: the tacticians of translational control. Nat Rev Mol Cell Biol. 2011;12:235–245. doi: 10.1038/nrm3083. [DOI] [PubMed] [Google Scholar]
- 44.Fraser CS, Berry KE, Hershey JW, Doudna JA. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol Cell. 2007;26:811–819. doi: 10.1016/j.molcel.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 45.Dmitriev SE, Terenin IM, Dunaevsky YE, Merrick WC, Shatsky IN. Assembly of 48S translation initiation complexes from purified components with mRNAs that have some base pairing within their 5' untranslated regions. Mol Cell Biol. 2003;23:8925–8933. doi: 10.1128/MCB.23.24.8925-8933.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic acids research. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toseland CP, Webb MR. Fluorescence tools to measure helicase activity in real time. Methods. 2010;51:259–268. doi: 10.1016/j.ymeth.2010.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.