Abstract
During cell division, chromosomes are segregated to nascent daughter cells by attaching to the microtubules of the mitotic spindle through the kinetochore. Kinetochores are assembled on a specialized chromatin domain, called the centromere that is characterized by the replacement of nucleosomal histone H3 with the histone H3 variant centromere protein A (CENP-A). CENP-A is essential for centromere and kinetochore formation in all eukaryotes but it is unknown how CENP-A chromatin directs centromere and kinetochore assembly 1. Here we generate synthetic CENP-A chromatin that recapitulates essential steps of centromere and kinetochore assembly in vitro. We show that reconstituted CENP-A chromatin when added to cell free extracts is sufficient for the assembly of centromere and kinetochore proteins, microtubule binding and stabilization, and mitotic checkpoint function. Using chromatin assembled from histone H3/CENP-A chimeras, we demonstrate that the conserved C-terminus of CENP-A is necessary and sufficient for centromere and kinetochore protein recruitment and function but that the CENP-A targeting domain (CATD), required for new CENP-A histone assembly 2, is not. These data show that two of the primary requirements for accurate chromosome segregation, the assembly of the kinetochore and the propagation of CENP-A chromatin are specified by different elements in the CENP-A histone. Our unique cell-free system enables complete control and manipulation of the chromatin substrate and thus presents a powerful tool to study centromere and kinetochore assembly in higher eukaryotes.
Metazoan centromeres are specified epigenetically by the presence of CENP-A nucleosomes3. Structural differences between CENP-A and histone H3 nucleosomes 2,4 and/or specific protein recognition elements in CENP-A appear to provide the information that specifies centromere identity and directs kinetochore assembly in a DNA sequence independent manner 5-10 ENREF 4. Moreover, many metazoan centromeres are complex in their organization with interspersed blocks of CENP-A nucleosomes and histone H3 nucleosomes assembled on long arrays of repetitive DNA 11-13. The difficulty in purifying and manipulating complex centromeres has limited our understanding of how centromeric chromatin promotes centromere and kinetochore formation and chromosome segregation.
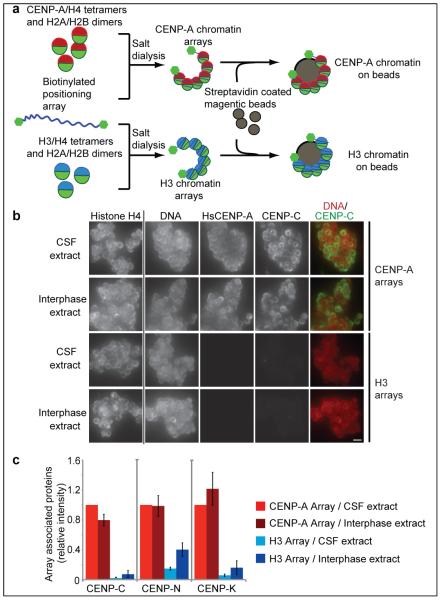
To mimic the arrays of CENP-A nucleosomes present in complex vertebrate centromeres, we reconstituted human CENP-A chromatin from recombinant components (Figure 1a). We generated saturated chromatin arrays by salt dialysis of purified histone proteins H2A, H2B, H4, and either CENP-A or H3 with a biotinylated DNA template containing 19 repeats of a 147 bp high affinity nucleosome positioning sequence (19×601) (Figure S1a,b) 14,15. We bound the biotinylated arrays to streptavidin coated magnetic beads, thereby immobilizing the arrays so that they can be easily added to and recovered from cell extracts (Figure 1a, Figure S1c, d, e).
Figure 1. Reconstituted CENP-A chromatin supports centromere assembly in Xenopus egg extracts.
(a) A schematic showing the reconstitution of CENP-A and H3 chromatin arrays and the attachment of the chromatin to magnetic beads via biotin end-labeled DNA. (b) Representative images comparing CENP-C binding to CENP-A and H3 chromatin arrays in CSF and interphase Xenopus extract. The left column shows the separate histone H4 staining used for normalization of the quantification, followed by staining for DNA, HsCENP-A and CENP-C. A merge image of the DNA (red) and CENP-C (green) channels is shown in the right column. Scale bar, 5 μm (c) Quantification of the array associated centromeric proteins CENP-C, CENP-N and CENP-K in CSF and interphase extracts, normalized to histone H4 levels. The levels are rescaled so that CENP-A arrays in CSF are set at 1. Error bars represent the standard error of the mean (SEM), n = 3 (p < 0.05 between CENP-A and H3 chromatin arrays for CENP-C, CENP-N and CENP-K).
We recently demonstrated that the essential centromere protein CENP-C directly recognizes the C-terminus of CENP-A in mononucleosomes but not in isolated CENP-A2/H42 tetramers 5 (our unpublished observations). Therefore, we tested in vitro translated human and Xenopus laevis CENP-C (Hs- and XlCENP-C respectively) for binding to reconstituted H3 and CENP-A chromatin. Human and Xenopus CENP-A are >50% identical (Figure S2a) and we find that both HsCENP-C and XlCENP-C bind specifically to HsCENP-A chromatin arrays in vitro, when compared to H3 chromatin arrays (Figure S2b).
Xenopus laevis egg extract is a widely used cell free system to study chromosome segregation16. Egg extracts are arrested in metaphase II of meiosis by the activity of cytostatic factor (CSF) and the cell cycle state of the extract can be transitioned into interphase by adding calcium. We developed a quantitative immunofluorescence assay to determine whether centromere proteins bound to CENP-A chromatin arrays when arrays were added to Xenopus egg extracts. CENP-N and CENP-K are centromere proteins that are required for proper centromere and kinetochore assembly in somatic cells, and we have previously shown that CENP-N, similar to CENP-C, directly binds to the CENP-A nucleosome6. We found that CENP-C, CENP-N and CENP-K specifically associated with CENP-A arrays independent of the cell cycle stage of the extract (Figure 1b,c and Figure S2c-f). The centromere protein CENP-T that binds to either H3 nucleosomes or DNA at centromeres did not selectively bind CENP-A chromatin arrays (Figure S3a,b)17. Similarly, the inner centromere protein Incenp and Polo like kinase 1 (Plk1) associated with both types of chromatin arrays (Figure S3c). Xenopus incenp is targeted to chromatin through phosphorylation of both H2A and H3 and thus may have affinity for both CENP-A and H3 chromatin18-20 and Plk1 associates with chromatin in Xenopus egg extract independent of the kinetochore21. Furthermore, reconstituted chromatin segments are unlikely to generate paired sister chromatids with inner centromeres because naked DNA and linear DNA replicates inefficiently in these egg extracts22. The specific recruitment of the centromere proteins CENP-C, CENP-N and CENP-K, however, indicates that reconstituted CENP-A chromatin arrays can support essential steps in the centromere assembly process in vitro.
Functional kinetochores assemble on sperm chromatin in metaphase Xenopus egg extract. At high sperm concentration, microtubule depolymerization causes mitotic checkpoint activation, resulting in the increased association of checkpoint proteins with kinetochores, and cell cycle arrest23. We tested whether reconstituted CENP-A chromatin arrays support kinetochore assembly and checkpoint protein binding after microtubule depolymerization. We added CENP-A or H3 arrays to CSF arrested egg extracts and then cycled the extracts through interphase and back into mitosis, in the presence or absence of nocodazole, as outlined in Figure 2a and demonstrated in Figure S4a. The constitutive centromere protein CENP-C and the microtubule-binding kinetochore protein Ndc80 bound to CENP-A arrays in the presence or absence of nocodazole (Figure 2b,c and S4b). The spindle assembly checkpoint proteins CENP-E, Mad2, Rod and ZW10 associated with CENP-A chromatin at intermediate levels in the absence of nocodazole but upon microtubule depolymerization their binding increased 2-4 fold (Figure 2b). Western blot analysis showed that CENP-C and Ndc80 are precipitated with CENP-A arrays independent of microtubule depolymerization. ZW10 and Rod are enriched on CENP-A arrays upon nocodazole treatment in metaphase, regardless of whether the extract has been cycled through interphase (Figure 2c). These results indicate that CENP-A chromatin arrays respond to microtubule depolymerization by recruiting mitotic checkpoint proteins (Figure 2b,c and S4b).
Figure 2. CENP-A chromatin specifically recruits kinetochore proteins as a response to a mimic of kinetochore detachment from microtubules.
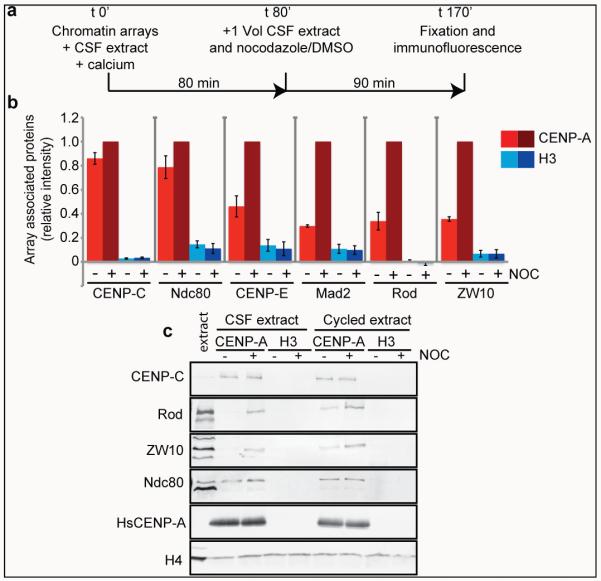
(a) A schematic showing the experimental procedure. (b) Quantification of immunofluorescence analysis of CENP-C, Ndc80, CENP-E, Mad2, Rod or ZW10 recruitment to chromatin arrays with (+) and without (−) nocodazole (NOC). The levels are rescaled so that CENP-A arrays with (+) nocodazole are set at 1. Error bars represent SEM, n = 3 (p < 0.05 between − and + nocodazole for CENP-E, Mad2, Rod and ZW10 binding to CENP-A chromatin arrays). (c) Western blot analysis of CENP-C, Ndc80, Rod and ZW10 recruitment to CENP-A and H3 chromatin arrays with and without (+/−) nocodazole (NOC) in CSF and cycled egg extracts. H4 levels are shown as a loading control.
Microtubule binding is a hallmark of kinetochore function and decondensed sperm chromatin efficiently supports spindle formation in egg extracts (Figure 3a, left panel)24. However, chromatin assembled on naked DNA induces spindle formation in Xenopus egg extracts independent of kinetochores25. When we added CENP-A and H3 chromatin beads into mitotic egg extract we observed microtubule polymerization around the majority of CENP-A arrays but only around a subset of H3 arrays (Figure 3a, left panel). We quantified the amount of microtubule polymer associated with each type of array and found significantly more microtubules associated with CENP-A chromatin beads (Figure 3b and Figure S5a). This indicates that CENP-A chromatin preferentially stabilizes microtubules or promotes their polymerization. We observed heterogeneous microtubule structures around the CENP-A chromatin beads ranging from bipolar spindles to stabilized microtubules or microtubule bundles (Figure 3a, Figure S5a,b). A second property of functional kinetochores is that kinetochore associated microtubule bundles (k-fibers) are stable to cold treatment which depolymerizes non-kinetochore microtubules. We asked whether kinetochores assembled on CENP-A chromatin could stabilize microtubules to cold shock by incubating the microtubule assembly reactions for 10 min at 4°C. We found that kinetochores assembled on CENP-A chromatin arrays stabilized microtubules to cold shock similar to kinetochores assembled on native sperm chromatin while H3 chromatin arrays did not (Figure 3a,c and Figure S5c). When we completely depolymerized microtubules with nocodazole we observed Mad2 recruitment to native sperm centromeres and CENP-A chromatin beads but not H3 chromatin beads (Figure 3a,c and Figure S5c). These results indicate that CENP-A chromatin arrays, similar to native sperm chromatin, assemble functional kinetochores that promote microtubule binding, k-fiber stabilization and spindle checkpoint function (Figure 3a).
Figure 3. Kinetochores assembled on reconstituted CENP-A chromatin bind microtubules and generate a mitotic checkpoint signal.
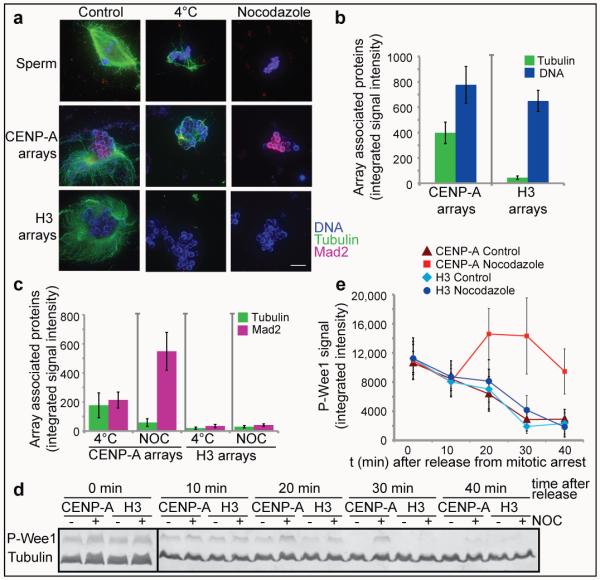
(a) Representative images of microtubule polymerization induced by sperm or reconstituted CENP-A and H3 chromatin. Microtubules (green) and Mad2 (magenta) levels are shown. Scale bar, 10μm (b) Quantification of tubulin and DNA associated with CENP-A and H3 chromatin beads. Error bars represent SEM, n = 5 (c) Quantification of tubulin and Mad2 levels associated with CENP-A and H3 chromatin beads after cold shock (4°C) and nocodazole (NOC) treatment. Error bars represent SEM, n = 5 (d) Western blot showing phospho-Wee1 (P-Wee1) levels as an indicator of the cell cycle stage and tubulin levels as a loading control. Samples from different time points after release from mitotic arrest (t 0′, t 10′, t 20′, t 30′, t 40′) are shown for CENP-A and H3 chromatin arrays, each incubated with nocodazole (+) or with DMSO (−) as a control. (e) Quantification of four independent experiments showing the phospho-Wee1 signal intensity (P-Wee1 signal) over time (min). Error bars represent SEM, n = 4.
In cells, unattached kinetochores activate the mitotic checkpoint and delay mitotic exit until all chromosomes are properly attached and aligned 26,27. We tested whether kinetochores assembled on CENP-A chromatin arrays could generate a mitotic checkpoint response to microtubule depolymerization and delay the cell cycle. We mixed CENP-A and H3 chromatin with CSF extracts, cycled the reactions through interphase and then cycled them back into mitosis in the presence or absence of nocodazole (Figure 2a). We then released the extract from mitosis into interphase a second time and monitored the kinetics of this transition by measuring the mitosis specific phosphorylation of Wee1 (phospho-Wee1) (Figure 3d). Upon release from mitosis, phospho-Wee1 levels rapidly declined and were undetectable after 30 minutes in control extracts containing CENP-A chromatin or H3 chromatin, as well as in extracts containing H3 chromatin in the presence of nocodazole (Figure 3d and 3e). In extracts containing CENP-A chromatin and nocodazole, the phospho-Wee1 signal increased until 20 min after calcium addition and subsequently declined until 40 minutes after calcium addition to a level only slightly lower than the levels prior to release (Figure 3d and 3e). In the presence of CENP-A chromatin and nocodazole, cyclin B levels rapidly declined but then stabilized, similar to the response observed for native sperm chromatin23. However cyclin B was not stabilized in the presence of H3 chromatin and nocodazole (Figure S5d,e). We estimate that the number of CENP-A nucleosomes we are adding to the egg extract exceeds the CENP-A nucleosome concentration required to activate the checkpoint using sperm nuclei23. The lower efficiency of reconstituted arrays for checkpoint signaling may be due to the comparatively short length of our reconstituted CENP-A chromatin to native CENP-A chromatin or the lack of replicated sister chromatids and inner centromeres important for tension dependent checkpoint activation. Despite these differences, our synthetic CENP-A chromatin supports a mitotic checkpoint response that mimics the response of native kinetochores to microtubule depolymerization.
The reconstituted chromatin system we have developed provides a distinct experimental advantage over native metazoan centromeric chromatin because the chromatin template can be easily manipulated to dissect the roles of histone proteins in centromere function. A central question in centromere function is how CENP-A chromatin directs the assembly of the centromere and kinetochore. CENP-N recognizes the CATD region of the CENP-A nucleosome while CENP-C binds the C-terminal tail of CENP-A 5,6. However, the relative importance of these two recognition mechanisms in centromere and kinetochore assembly is incompletely understood.
We generated chromatin arrays containing chimeric CENP-A/H3 proteins to ask how the CENP-A CATD domain and the CENP-A C-terminus influence centromere and kinetochore assembly (Figure 4a). We characterized the level of histone exchange and/or loss from the arrays during incubation in extracts and found that the majority of recombinant HsCENP-A nucleosomes were stable during the incubation indicating low exchange and/or loss rates (Figure S6a,b). We detected a low level of phosphorylated histone H3 on CENP-A chromatin arrays in CSF extract (11.7% ± 7% compared to H3 arrays) and in extract that had been cycled through interphase and back into mitosis (22% ± 13% compared to H3 arrays) (Figure S6c,d). The chimeric arrays containing CENP-A with the histone H3 tail (CENP-A+H3C) exhibited similar levels of exchange (Figure 6c,d). The Xenopus CENP-A present in the extract did not appreciably exchange onto any of the arrays (detection limit ~5-10% exchange) (Figure S6c). The absence of gross rearrangements or bulk histone exchange suggests that chromatin arrays can be used to dissect how individual domains of CENP-A influence kinetochore assembly.
Figure 4. The CENP-A C-terminus is required for centromere and kinetochore assembly in Xenopusegg extract.
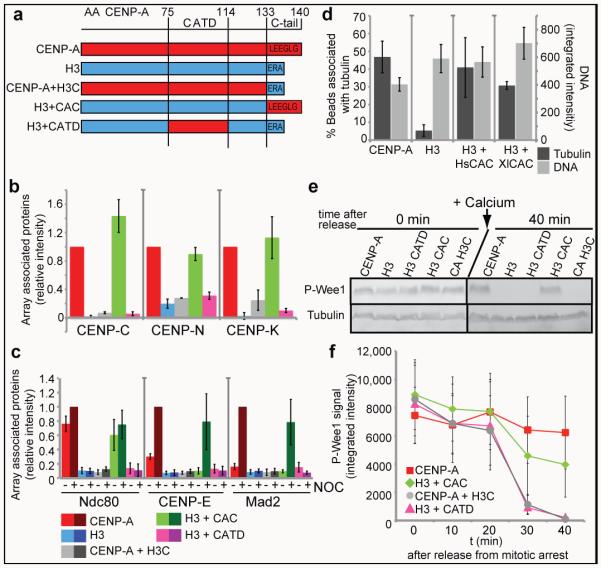
(a) A schematic showing the different CENP-A/H3 chimeras used in this study. The numbers on top represent the amino acid (AA) within HsCENP-A. (b) Quantification of immunofluorescence analysis of CENP-C, CENP-K and CENP-N recruitment to wild type and chimeric arrays. The relative amounts of each centromere protein bound to the arrays are shown relative to CENP-A arrays set to 1. Error bars represent SEM, n = 3 (p ≤ 0.05 for all proteins binding to CENP-A arrays compared to chimeric arrays except for the H3 arrays containing the CENP-A C-terminus). (c) Quantification of immunofluorescence analysis of Ndc80, CENP-E, Mad2 recruitment to chimeric chromatin arrays with (+) and without (−) nocodazole (NOC). Values are displayed relative to CENP-A arrays in the presence of nocodazole set to 1. Error bars represent SEM, n = 4. The efficiencies of recruitment of kinetochore proteins to CENP-A and H3+CAC arrays in nocodazole were not statistically distinguishable (p ≥ 0.26 for Ndc80, CENP-E and Mad2). (d) Quantification of microtubule binding to CENP-A, H3, H3+HsCAC and H3+XlCAC chromatin arrays represented as percentage of beads associated with tubulin levels above threshold (dark grey bars, left y-axis). Average DNA levels on chromatin beads are shown representing the levels of chromatin arrays bound to beads (light grey bars, right y-axis). Error bars represent SEM, n = 4 for CENP-A and H3 arrays, n = 5 for H3+HsCAC arrays and n = 2 for H3+XlCAC arrays. (e) Western blot analysis shows phospho-Wee1 (P-Wee1) levels as an indicator of the cell cycle stage at t 0′ (0 min) and t 40′ (40 min) after mitotic exit. Tubulin levels are shown as a loading control. (f) Quantification of the phospho-Wee1 signal intensity over time (P-Wee1). Error bars represent SEM, n = 5.
Using our in vitro centromere and kinetochore assembly assay, we found that CENP-C bound with equal efficiency to chromatin arrays assembled with either wild type CENP-A or with chimeras of histone H3 with the CENP-A C-terminal 6 amino acids (H3+CAC) but not CENP-A+H3C (Figure 4b and S7a, left panel). This demonstrates that the CENP-A C-terminus is necessary and sufficient for recruiting CENP-C to CENP-A chromatin arrays in egg extracts as it is for CENP-A mononucleosome binding in vitro5.
CENP-K depends on CENP-C for its association with Xenopus sperm centromeres 28 and CENP-K also associated with the wild type and H3+CAC arrays (Figure 4b and Figure S7a). Surprisingly, we found that H3+CAC arrays recruited CENP-N as efficiently as wild-type CENP-A arrays, even though these arrays lack the CATD recognition element for CENP-N6. CENP-N binding to either CENP-A+H3C or H3+CATD arrays was no better than its binding to H3 chromatin arrays, indicating that the CENP-A C-terminus is required for CENP-N association with CENP-A chromatin in Xenopus egg extract (Figure 4b and Figure S7a). The lack of Xenopus CENP-N binding to H3+CATD and CENP-A+H3C chromatin arrays is not due to species differences because XlCENP-N binds HsCENP-A mononucleosomes in vitro in the absence of CENP-C (Figure S7b). The association of CENP-N and -K with chromatin arrays was dependent on CENP-C as CENP-C depletion from the extract (Figure S8a) reduced the binding to background levels (Figure S8b,c). This was not due to depletion of CENP-N or CENP-K by CENP-C as we have previously shown that complementation of CENP-C depleted extracts restores CENP-K binding and CENP-K is known to depend on CENP-N for its centromere localization 6,29,30. The dependence of CENP-N on CENP-C for its localization to CENP-A arrays may reflect a role for CENP-C in altering the geometry of centromeric chromatin to promote CENP-N’s access to CENP-A nucleosomes or may reflect the assembly of CENP-N into the larger CCAN complex recruited to the centromere via CENP-C. Our results demonstrate that CENP-C recognition of the CENP-A C-terminus is necessary and sufficient for CENP-N and CENP-K association with chromatin arrays in Xenopus egg extract.
We analyzed the chromatin requirements for mitotic kinetochore formation using the experimental strategy illustrated in Figure 2a. The kinetochore proteins Ndc80, CENP-E, Mad2, Rod and ZW10 are efficiently recruited to wild type and H3+CAC chromatin arrays, but not to CENP-A+H3C or H3+CATD chromatin arrays (Figure 4c, Figure S9a, Figure S10a). Similar to wild type CENP-A chromatin, only the checkpoint proteins CENP-E, Mad2, ZW10 and Rod increased in their association with H3+CAC after microtubule depolymerization (Figure 4c, Figure S9a, Figure S10a). As with wild type CENP-A arrays, the H3+CAC arrays showed increased associated microtubule polymer indicating that the C-terminus of CENP-A directs the formation of microtubule binding or stabilization activity (Figure 4d). Human and Xenopus CENP-A differ by two amino acids in their C-terminal tail (Figure S2a) and chimeric nucleosome arrays containing the Xenopus C-terminal tail of CENP-A fused to H3 (H3+XlCAC) were equally efficient in CENP-C recruitment and microtubule binding as HsH3+CAC arrays (Figure 4d and Figure S10b); indicating that the mode of interaction between CENP-C and CENP-A is conserved.
We assayed the ability of chimeric nucleosome arrays to promote mitotic checkpoint arrest after microtubule depolymerization and found that H3+CATD and CENP-A+H3C did not delay the exit from mitosis but that H3+CAC did (Figure 4e,f). The delay of mitotic exit caused by H3+CAC arrays was less effective than that of CENP-A chromatin arrays indicating that regions of CENP-A in addition to the C-terminus increase the effectiveness of checkpoint signaling, possibly by stabilizing CCAN and kinetochore protein interactions with chromatin (Figure 4e,f). Taken together, our data demonstrate that the primary chromatin determinant for functional centromere and kinetochore assembly is the C-terminus of CENP-A and its recognition by CENP-C.
Here we have shown that reconstituted CENP-A chromatin, in the absence of native centromeric DNA, is necessary and sufficient for centromere and kinetochore assembly. Our data imply that short domains of CENP-A chromatin are sufficient for assembling core components of the centromere and kinetochore in the absence of higher order organization of centromeric chromatin and interspersed domains of H3 chromatin. Using our in vitro system, we have directly assessed how domains of CENP-A participate in centromere and kinetochore assembly, even when the mutations we analyze would be expected to be lethal in vivo. We find that, the CENP-A C-terminus is both necessary and sufficient for the recruitment of centromere and kinetochore proteins, for microtubule binding and for a checkpoint response to microtubule depolymerization. We suggest that CENP-A performs two functions that can be separated molecularly: 1) the CENP-A CATD provides a recognition mechanism for targeting of CENP-A to centromeres to maintain centromeric chromatin 2,6-8 and 2) the CENP-A C-terminal tail domain recruits the conserved centromere protein CENP-C to promote centromere and kinetochore assembly5. We envision the use of more complex chromatin templates to understand the importance of higher order chromatin organization and regulatory modifications in centromere assembly and function.
Methods Summary
Histone proteins and chimeras were purified as described previously 5,6,15 and assembled onto a biotin end-labeled tandem array of 19 high affinity nucleosome positioning sequences (19×601) by salt dialysis14. Chromatin arrays were bound to streptavidin coated magnetic Dynabeads (Invitrogen). X. laevis extracts were prepared as previously described 16 and centromere protein binding to chromatin arrays was performed in freshly prepared CSF egg extract for 1 hour with or without calcium addition. Arrays were fixed in formaldehyde and stained for centromere proteins by indirect immunofluorescence. Kinetochore and checkpoint protein assembly was assayed by adding arrays to extracts released into interphase with calcium for 80 min followed by readdition of CSF extract in the presence or absence of nocodazole (10 μg/ml) for another 90 min. To analyze microtubule binding, chromatin arrays were incubated in CSF for 90 min. Reactions were sedimented through a glycerol cushion onto a coverslip followed by tubulin immunofluorescence. Chromatin array dependent inhibition of mitotic exit, was assayed as described for kinetochore protein binding, but calcium was added a second time to release extracts into interphase. The cell cycle state monitored by western blotting using the α-phospho-Wee1 antibody generously provided by Jim Ferrell.
Images were collected as 13 axial planes at 2 μm intervals on a Nikon Eclipse-80i microscope using a 60×, 1.4 NA PlanApo oil lens and a CoolSnapHQ CCD camera (Photometrics) with MetaMorph software (MDS Analytical Technologies). Axial stacks were maximum intensity projected and quantified using custom software. For normalization of each experiment, a separate Histone H4 staining was performed to quantify the exact array coupling efficiency.
Supplementary Material
Acknowledgements
The authors like to thank the Straight Lab members for support and helpful comments, James E. Ferrell, Andrew Murray, Rey-Huei Chen, Geert Kops, and P. Todd Stukenberg for providing antibodies. Daniela Rhodes, Philip Robinson, Karolin Luger, Jeff Hansen, Geeta Narlikar and Janet Yang for providing reagents and advice. A.G. was supported by a postdoctoral fellowship from the German Research Foundation (DFG). C.W.C. was supported by a postdoctoral fellowship from the Helen Hay Whitney Foundation and the American Heart Association (AHA). B.M. was supported by T32GM007276, C.J.F by a Stanford Graduate Fellowship and this work was supported by NIH R01GM074728 to A.F.S.
Appendix
Methods
Histone expression
CENP-A/H4 and H3/H4 wild-type and chimeric tetramers, as well as H2A and H2B dimers were expressed and purified as described previously 31-33.
Preparation of biotinylated array DNA
A tandem array of 19 copies of the high affinity nucleosome positioning sequence (19×601) 34,35 was digested with EcoRI, XbaI, DraI and HaeII (NEB) overnight to excise the 19mer array and to digest the remaining backbone DNA to smaller DNA fragments. The array DNA was then purified by PEG precipitation and dialyzed against 10mM Tris-HCl pH 8.0, 0.25mM EDTA as previously described 34. The array DNA was end-labeled with biotin by end filling the EcoRI and XbaI sites using Klenow DNA polymerase for 4 hr at 37°C in a reaction containing 35μM Biotin-14-dATP (Invitrogen), α-thio-dTTP and α-thio-dGTP (Chemcyte) and dCTP. The labeled DNA was then purified using a PCR fragment purification kit (Qiagen). The biotinylation efficiency was determined by adding FITC-Streptavidin (final concentration of 10 μg/ml) to 500ng of purified array DNA and monitoring the fraction of gel shifted DNA after migration in a 0.7% agarose gel.
Chromatin array assembly
To assemble chromatin arrays, biotinylated DNA, CENP-A/H4 or H3/H4 tetramers and H2A/H2B dimers were mixed at a stochiometry of 1:1:2.2 or at 1:0.9:2.2, respectively, in high salt buffer (10mM Tris-HCl pH 7.5, 0.25mM EDTA, 2M NaCl) and then dialyzed into low salt buffer (10mM Tris-HCl pH 7.5, 0.25mM EDTA, 2.5mM NaCl) over 60-70h at 4°C. Final array DNA concentration typically was 0.15mg/ml – 0.2mg/ml. To assess the efficiency of nucleosome assemblies, arrays were digested at room temperature overnight with AvaI in a low magnesium buffer (50mM potassium acetate, 20mM Tris-acetate, 0.5 mM magnesium acetate, 1mM DTT, pH7.9). Digested chromatin arrays were supplemented with glycerol (20% final concentration) and separated on a native 5% acrylamide gel in 0.5x Tris/Borate/EDTA buffer for 80 min at 10mA. Gels were stained with EtBr (1μg/ml) to visualize DNA.
Coupling of biotinylated chromatin arrays to Dynabeads
Biotinylated chromatin arrays were coupled to prewashed Streptavidin coated magnetic Dynabeads (Invitrogen) at a ratio of 10 μg DNA to 1 mg beads in 50mM Tris-HCl pH 8.0, 75mM NaCl, 0.25mM EDTA, 2.5% polyvinyl alcohol (PVA) and 0.05% Triton-X-100 for 1-2h. The beads were then equilibrated in 75mM Tris-HCl pH 8.0, 75mM NaCl, 0.25mM EDTA, 0.05% Triton-X-100 and either used directly or stored at 4°C for later use.
Xenopus laevis egg extracts
X. laevis CSF extracts were prepared as previously described 36. To assess the binding of centromeric proteins to chromatin arrays in CSF and interphase egg extracts, chromatin arrays were mixed with freshly prepared CSF egg extract with or without CaCl2 (final concentration 0.6 mM) at a nucleosome concentration of ~100nM unless stated otherwise. The reactions were incubated for 1h at 4°C or at 16-20°C in a waterbath, the arrays were re-isolated from extracts by exposure to a magnet and then washed three times in 1x CSF-XB buffer (10 mM Hepes 7.7, 2 mM MgCl2, 0.1mM CaCl2, 100 mM KCl, 5 mM EGTA, 50 mM sucrose) supplemented with 0.05% Triton-X-100. Chromatin arrays were fixed in CSF-XB buffer, 0.05% Triton-X-100, 2% Formaldehyde for 5 min. After fixation, chromatin arrays were washed into antibody dilution buffer (20mM Tris-HCl pH7.5, 150mM NaCl, 0.1% Triton-X-100, 2% BSA) and analyzed by immunofluorescence.
Kinetochore and spindle checkpoint protein assembly were analyzed by mixing chromatin arrays with CSF extract and CaCl2 (final concentration 0.6mM). Reactions were incubated at 16-20°C for 80 min to allow extracts to release into interphase and mixed every 15 min. 1 volume fresh CSF extract was added together with nocodazole (or DMSO) at 10 μg/ml and samples were held at 16-20°C for another 90 min. After 170 min total incubation time, samples for immunofluorescence analysis were washed and fixed as described above.
The cell cycle state was verified by loading 2μl extract of all relevant time points onto a SDS-PAGE, followed by western blotting using the α-phospho-Wee1 antibody 37. To assess the ability of chromatin arrays to inhibit mitotic exit, arrays were mixed with CSF extract and CaCl2 (final concentration: 0.6mM). The samples were incubated for 80 min to induce the release into interphase. In the next step, 1 volume fresh CSF extract, supplemented with nocodazole/DMSO, was added to cycle the extract back into a mitotic arrest. After 90 min, CaCl2 was added again to release from mitotic arrest. Western blot samples were taken at all indicated time points and processed as described.
To analyze microtubule binding by CENP-A and H3 chromatin arrays, chromatin arrays were mixed with CSF extract and incubated for 90 min at 18-20°C. During incubation samples were mixed every 15 min. Reactions were fixed for 10 min in 2.5% formaldehyde, sedimented through a glycerol cushion onto coverslips and post-fixed for 5 min in ice cold methanol followed by immunofluorescence analysis 38. To assay for Mad2 levels and microtubule stabilization, reactions were either supplemented with nocodazole at a final concentration of 10μg/ml or shifted to 4°C for 10 min after the 90 min incubation time.
Immunodepletion
Depletion of XlCENP-C from Xenopus egg extracts was performed as described previously 39.
Cloning and Antibody generation
The X. laevis CENP-N cDNA clone (MGC:83847) was purchased from American Type Culture Collection (Manassas, VA). Peptides against XlCENP-N (acetyl-CPHKARNSFKITEKR-amide) were synthesized by Bio-Synthesis (Lewisville, TX) and peptide antibodies were generated as previously described 40.
Immunofluorescence
For immunofluorescence analysis, fixed chromatin arrays were bound to poly-L-lysine-coated acid-washed coverslips. The following primary antibodies were used for immunofluorescence staining and typically incubated at 4°C overnight: α-HsCENP-A 32 was directly coupled to Alexa 647 (Molecular Probes), α-H4 (Abcam), α-XlCENP-C, α-XlCENP-E, α-XlCENP-K and α-XlCENP-N and α-Tubulin (Dm1α, Sigma). Rabbit antibodies were generated against the full length XlPolo kinase made in Sf9 cells and a GST fusion to the first 379 amino acids of XlIncenp made in E. coli. The α-Mad2 antibody was provided by Andrew Murray (Harvard University, Cambridge, MA), and Rey-Huei Chen (Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan), the α-XlZW10 and α-XlRod antibodies were provided by Geert Kops (University Medical Center Utrecht, Utrecht, The Netherlands) and the α-XlNdc80 antibody was provided by P. Todd Stukenberg (University of Virginia, Charlottesville, VA). Alexa-conjugated secondary antibodies were used at 1 μg/ml (Molecular Probes). Propidium iodide at 1μg/ml or Hoechst at 10μg/ml was used to visualize DNA.
Microscopy and Analysis
Images were collected on a Nikon Eclipse 80i microscope using a 60×, 1.4 NA Plan Apo VC oil immersion lens, a Sedat Quad filter set (Chroma Technology Corp.) using MetaMorph software (MDS Analytical Technologies) and a charge-coupled device camera (CoolSnapHQ; Photometrics). 13 axial planes at 2 μm intervals were acquired with an MFC-2000 Z-axis drive (Applied Scientific Instrumentation). Axial stacks were maximum intensity projected and then quantified using custom software (Matlab) to identify beads in each image and to quantify the integrated intensity for each channel after background subtraction. Briefly, the propidium iodide stained (DNA) channel was used to find beads. Bead centroids were found by filtering the image using a structuring element that had a peak at a 17 pixel radial distance from the structuring element center, corresponding to the bright ring seen around the edges of the beads. A 35 pixel diameter circle around the centroid of each bead identified was used as the region of interest for that bead. After beads were identified, regions of interest were transferred automatically to the remaining channels and the integrated signal intensity was calculated for each bead in each channel, normalized to the area of the bead region (which was uniform except in cases of partially overlapping beads), and background corrected using an average of three bead-sized regions manually chosen to be away from any beads. For each experiment, at least three images per coverslip were acquired and 20 - 300 beads were analyzed per image. For the normalization of each experiment, a separate Histone H4 staining was performed to quantify the exact coupling efficiency for each type of chromatin array and for each experiment.
Immunofluorescence microscopy images of the microtubule binding assays that were subjected to deconvolution were acquired with an Olympus IX70 microscope. The microscope was outfitted with a Deltavision Core system (Applied Precision) using an Olympus 60× 1.4NA Plan Apo lens, a Sedat Quad filter set (Semrock, Inc. Rochester, NY) and a CoolSnap HQ CCD Camera (Photometrics, Tucson, AZ). The microscope was controlled via Softworx 4.1.0 software (Applied Precision) and images were deconvolved using softWoRX v4.1.0 (Applied Precision). Microtubule quantification was performed using a modification of the same software used for centromere protein quantification.
Immunoblotting
Western blot samples were separated by SDS-PAGE and transferred onto PVDF membrane (Bio-Rad) in CAPS transfer buffer [10 mM 3-(cyclohexylamino)-1-propanesulfonic acid, pH 11.3, 0.1% SDS, and 20% methanol]. The following primary antibodies were typically incubated overnight at 4°C: α-XlCENP-C 39, α-tubulin (Dm1α, Sigma), α-H4 (Abcam), α-phospho H3 (Ser10) (Millipore), α-phospho-Wee1. The α-phospho-Wee1 antibody was provided by James E. Ferrell (Stanford University, Stanford, CA) 37. For additional primary antibodies, western blot samples were transferred onto PVDF membrane (Bio-Rad) in 20 mM Tris-Base, 200 mM glycine. Alexa fluorophore conjugated anti-rabbit or anti-mouse secondary antibodies (Molecular Probes) were used according to manufacturer’s specification. Fluorescence was detected on a Typhoon 9400 Variable Mode Imager (Amersham Biosciences) and quantified using ImageJ (http://rsb.info.nih.gov/ij/). Actin antibodies were (provided by Julie Theriot, Stanford University, Stanford, USA) and α-CyclinB was purchased from Santa Cruz Biotechnology.
In vitro binding of centromere proteins to chromatin arrays
Human and Xenopus CENP-C was in vitro translated (IVT) in rabbit reticulocyte extracts in the presence of 10 mCi/Ml [35S]methionine (Perkin Elmer) using the TnT Quick-Coupled Transcription/Translation system (Promega) according to the manufacturer’s instructions. For a binding reaction (60μl total volume), 5 μl of each IVT protein were mixed with chromatin arrays in bead buffer (75mM Tris-HCl pH 7.5, 50mM NaCl, 0.25mM EDTA, 0.05% Triton-X-100). The final nucleosome concentration per reaction was 60nM. Reactions were incubated at 4°C for 1 h. The beads were washed three times with bead buffer and resuspended in 4x SDS loading buffer. Samples were separated on a SDS-PAGE, Coomassie stained and after drying scanned using a phosphorimager (Typhoon 4200, Amersham Biosciences) and quantified using ImageJ (http://rsb.info.nih.gov/ij/).
Statistical analysis
In each experiment, the relative levels of proteins associated with the chromatin arrays were normalized to values for wild type CENP-A arrays set to 1. For calculation of P-values each dataset was anchored at 1 and then log transformed followed by calculation of P-values using a Student’s t-test 41.
References
- 1.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 2.Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 3.Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr Opin Cell Biol. 2008;20:91–100. doi: 10.1016/j.ceb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010 doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll CW, Silva MCC, Godek KM, Jansen LET, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunleavy EM, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 8.Foltz DR, et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu H, et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes & development. 2011;25:901–906. doi: 10.1101/gad.2045111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tachiwana H, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011 doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 11.Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci U S A. 2010 Jun 8;107(23):10484–9. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zinkowski RP, Meyne J, Brinkley BR. The centromere-kinetochore complex: a repeat subunit model. J Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huynh V, Robinson P, Rhodes D. A Method for the In Vitro Reconstitution of a Defined. J Mol Biol. 2005;345:957–968. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- 15.Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 16.Desai A, Murray A, Mitchison TJ, Walczak CE. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 1999;61:385–412. doi: 10.1016/s0091-679x(08)61991-3. [DOI] [PubMed] [Google Scholar]
- 17.Hori T, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 18.Kawashima SA, Yamagishi Y, Honda T, Ishiguro K.-i., Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science (New York, NY) 2010;327:172–177. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- 19.Kelly AE, et al. Survivin Reads Phosphorylated Histone H3 Threonine 3 to Activate the Mitotic Kinase Aurora B. Science (New York, NY) 2010 doi: 10.1126/science.1189505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang F, et al. Histone H3 Thr-3 Phosphorylation by Haspin Positions Aurora B at Centromeres in Mitosis. Science (New York, NY) 2010 doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Budde PP, Kumagai A, Dunphy WG, Heald R. Regulation of Op18 during spindle assembly in Xenopus egg extracts. J Cell Biol. 2001;153:149–158. doi: 10.1083/jcb.153.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- 23.Minshull J, Sun H, Tonks NK, Murray AW. A MAP kinase-dependent spindle assembly checkpoint in Xenopus egg extracts. Cell. 1994;79:475–486. doi: 10.1016/0092-8674(94)90256-9. [DOI] [PubMed] [Google Scholar]
- 24.Sawin KE, Mitchison TJ. Mitotic spindle assembly by two different pathways in vitro. The Journal of cell biology. 1991;112:925–940. doi: 10.1083/jcb.112.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heald R, et al. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- 26.Nicklas RB, Ward SC, Gorbsky GJ. Kinetochore chemistry is sensitive to tension and may link mitotic forces to a cell cycle checkpoint. J Cell Biol. 1995;130:929–939. doi: 10.1083/jcb.130.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol. 1995;130:941–948. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milks KJ, Moree B, Straight AF. Dissection of CENP-C-directed centromere and kinetochore assembly. Molecular Biology of the Cell. 2009;20:4246–4255. doi: 10.1091/mbc.E09-05-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foltz DR, et al. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 30.Mcclelland SE, et al. The CENP-A NAC/CAD kinetochore complex controls chromosome congression and spindle bipolarity. EMBO J. 2007;26:5033–5047. doi: 10.1038/sj.emboj.7601927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carroll CW, Silva MCC, Godek KM, Jansen LET, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luger K, Rechsteiner T, Richmond T. Preparation of nucleosome core particle from recombinant histones. Methods in enzymology. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 34.Huynh V, Robinson P, Rhodes D. A Method for the In Vitro Reconstitution of a Defined. J Mol Biol. 2005;345:957–968. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- 35.Lowary PT, Widom J. New DNA Sequence Rules for High Affinity Binding to Histone Octamer and Sequence-directed Nucleosome Positioning. JMB. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 36.Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 37.Kim S, Song E, Lee K, Ferrell J., Jr Multisite M-phase phosphorylation of Xenopus Wee1A. Molecular and cellular biology. 2005;25:10580. doi: 10.1128/MCB.25.23.10580-10590.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hannak E, Heald R. Investigating mitotic spindle assembly and function in vitro using Xenopus laevis egg extracts. Nat Protoc. 2006;1:2305–2314. doi: 10.1038/nprot.2006.396. doi:10.1038/nprot.2006.396. [DOI] [PubMed] [Google Scholar]
- 39.Milks KJ, Moree B, Straight AF. Dissection of CENP-C-directed centromere and kinetochore assembly. Molecular Biology of the Cell. 2009;20:4246–4255. doi: 10.1091/mbc.E09-05-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Field CM, Oegema K, Zheng Y, Mitchison TJ, Walczak CE. Purification of cytoskeletal proteins using peptide antibodies. Methods Enzymol. 1998;298:525–541. doi: 10.1016/s0076-6879(98)98043-0. [DOI] [PubMed] [Google Scholar]
- 41.Osborne JW. Best practices in quantitative methods. Sage Publications; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.