Abstract
Objectives
To determine mRNA expression differences in genes involved in signaling and modulating sensory fatigue, and muscle pain in patients with Chronic Fatigue Syndrome (CFS) and Fibromyalgia Syndrome (FM) at baseline, and following moderate exercise.
Design
Forty eight Patients with CFS-only, or CFS with comorbid FM, 18 Patients with FM that did not meet criteria for CFS, and 49 healthy Controls underwent moderate exercise (25 minutes at 70% maximum age predicted heart-rate). Visual-analogue measures of fatigue and pain were taken before, during, and after exercise. Blood samples were taken before, and 0.5, 8, 24, and 48 hours after exercise. Leukocytes were immediately isolated from blood, number coded for blind processing and analyses, and flash frozen. Using real-time, quantitative PCR, the amount of mRNA for 13 genes (relative to control genes) involved in sensory, adrenergic, and immune functions was compared between groups at baseline, and following exercise. Changes in amounts of mRNA were correlated with behavioral measures, and functional clinical assessments.
Results
No gene expression changes occurred following exercise in Controls. In 71% of CFS patients, moderate exercise increased most sensory and adrenergic receptor’s and one cytokine gene’s transcription for 48 hours. These post-exercise increases correlated with behavioral measures of fatigue and pain. In contrast, for the other 29% of CFS patients, adrenergic α-2A receptor’s transcription was decreased at all time points after exercise; other genes were not altered. History of orthostatic intolerance was significantly more common in the α-2A decrease subgroup. FM only patients showed no post-exercise alterations in gene expression, but their pre-exercise baseline mRNA for two sensory ion channels and one cytokine were significantly higher than Controls.
Conclusions
At least two subgroups of CFS patients can be identified by gene expression changes following exercise. The larger subgroup showed increases in mRNA for sensory and adrenergic receptors and a cytokine. The smaller subgroup contained most of the CFS patients with orthostatic intolerance, showed no post-exercise increases in any gene, and was defined by decreases in mRNA for α-2A. FM only patients can be identified by baseline increases in 3 genes. Post-exercise increases for 4 genes meet published criteria as an objective biomarker for CFS, and could be useful in guiding treatment selection for different subgroups.
Keywords: chronic fatigue syndrome, fatigue, orthostatic symptoms, autonomic dysfunction, gene expression, fibromyalgia
Introduction
Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (CFS), as defined by the Fukuda research criteria [1], is an otherwise unexplained cluster of symptoms lasting at least 6 months that includes profound, remitting/relapsing fatigue that impairs functioning, and is not relieved by rest or recovery. Four or more of the following 8 symptoms must also be present: 1) post-exertional malaise (described as greatly increased fatigue and the feeling of sickness beginning 6–24 hours after exercise and continuing for several days or weeks). 2) unrefreshing sleep 3) muscle pain 4) joint pain 5) new or a change in headaches 6) impairment of memory or concentration, 7) sore throat, 8) tender lymph nodes [1].
The Canadian Clinical Criteria for CFS require a significant degree of new onset physical and mental fatigue that substantially reduces activity levels and also requires (from above criteria) #1, #2, one of #3–#5, neurocognitive symptoms (including impairment of concentration and short term memory, inability to focus vision, ataxia, muscle weakness, photophobia, periods of anxiety), and at least one symptom from each of the following categories: Immune manifestations (tender lymph nodes, sore throat, or new sensitivities to food and medications and/or chemicals), Autonomic manifestations (orthostatic intolerance, POTS, extreme pallor, nausea and irritable bowel syndrome, palpitations, exertional dyspnea), or Neuroendocrine manifestations (subnormal temperature, sweating episodes, intolerance of heat and cold, weight changes, anorexia or abnormal appetite, worsening of symptoms with stress) [2].
Up to 70% of ME/CFS patients (hereafter referred to as CFS) also meet the criteria set by the American College of Rheumatology for Fibromyalgia Syndrome (FM) as a condition of unknown etiology that consists of widespread muscle/connective tissue pain present in 4 quadrants of the body (bilateral, upper and lower body) for 3 months or longer. There is also painful sensitivity to pressure at tender points (with pain reported for at least 11 of 18 tender points) [3]. Recent guidelines published by ACR experts, but not yet validated, have suggested altering this definition to permit easier application in the clinical setting [4]. In both CFS and FM, the major symptoms, and most of the ancillary symptoms, are subjectively determined.
For CFS, the major symptom, “fatigue” is a clinical measure that is only loosely associated with the physiologists’ traditional definition of “fatigue” which is the inability to voluntarily contract skeletal muscle [5]. The clinical “fatigue” includes both the sensation of tiredness, and increased effort in skeletal muscle contraction, and also increased effort in mental functions. For FM, the major symptom is muscle pain, which has been poorly studied in comparison to cutaneous pain.
Recently, our laboratory was able to identify some of the molecular receptors responsible for the sensory neuron signaling of muscle fatigue and muscle pain [6]. In addition, we have discovered that adrenergic receptors can also contribute to enhanced muscle pain [7, 8]. We further discovered that these molecular receptors are expressed by human leukocytes, and that their expression is altered by exercise that worsens muscle pain and muscle fatigue [8]. In patients with CFS, we showed that mRNAs of sensory and adrenergic molecular receptors as well as two cytokines were dramatically increased 30 minutes to at least 48 hours following moderate exercise (25 minutes of combined arm and leg exercise) [9]. These findings are similar to previous reports suggesting that CFS patients have complex dysregulation of several physiological systems including the immune system (cytokines), the central nervous system, cellular energy and transport, and cardiovascular system (Carruthers BM, van de Sande, MI, De Meirleir, KL, Klimas, NG, Mitchell, T, Staines, D, Powles, AC, Bell, DS, Vallings, R, Speight. Myalgic Encephalomyelitis: International Consensus Criteria submitted). Dysregulation of these systems singly or together could contribute to some or all of the symptoms that define CFS and FM, but especially could help explain the important symptom of post-exertional malaise.
Here, we report on an increased sample of patients with CFS, and compare their gene regulation with that of patients with FM. With the larger sample, we confirmed our original finding of increased gene expression of a number of sensory, adrenergic, and cytokine genes, and discovered a major subgroup of CFS that is characterized by a large post exercise decrease in gene expression of the adrenergic alpha 2A receptor (α-2A). This subgroup was also characterized by a predominance of orthostatic intolerance when compared with other CFS patients. We also discovered that patients with FM only (who do not meet criteria for CFS) do not show large gene expression changes following exercise, but, rather have baseline increases in gene expression for two sensory molecular receptors and the immune cytokine IL10.
Materials and Methods
Study Population
All research reported here was approved by the University of Utah Institutional Review Board, and written informed consent was obtained from all subjects before participation. Exclusion criteria included active viral or upper respiratory infections, chronic cardiovascular or pulmonary disorders, or other chronic conditions such as anemia, cancer or multiple sclerosis. All subjects were required to refrain from any other continuous exercise greater than 5 minutes of walking, for 2 days before, and 2 days after the scheduled exercise task.
The sample for the present report included 48 CFS patients (33 females) [(15 females) of these are the same patients as in our previous publication [8]], 49 Control subjects (29 females) [16 (11 females) are the same Controls as in our previous report], and 18 FM only subjects (15 females). This ratio of more females than males is typical in CFS and FM research, and is also consistent with observations from large scale incidence and prevalence studies including the Wichita sample [10]. The patients reflected the local population with 94% being Caucasian, 6% being minority; thus our findings may not apply to minorities with CFS or FM. All CFS patients met the CDC criteria for CFS [1], and 46 (96%) met the Canadian Criteria for ME/CFS as well [2]. Prior screening by an experienced physician (LB) ruled out all other known causes for persistent or relapsing fatigue in these CFS patients. All patients were also screened for FM using the strict ACR research criteria, which includes presence of widespread pain for at least 6 months, and pain reported at 11 or more of 18 sites during tender point examination [3]. Thirty three of the 48 CFS patients (69%) also met ACR criteria for FM, similar to the high co morbidity of these disorders previously reported [11–13]. Eighteen patients met criteria for FM, but did not meet criteria for CFS (principally due to fatigue causing less compromise in normal daily activities), and were classified as FM-only patients (FM).
For primary analyses, all 48 CFS patients were compared to the 49 Controls. All later analyses were done after separating out the CFS subgroup in which the major identifying feature was large decreases in α-2A receptor mRNA at all times following exercise. All 18 FM patients were also compared with this same control group, and with the CFS group.
Both controls and all patient groups included individuals on prescribed antidepressants, and no subjects were withdrawn from these medications because antidepressants require a relatively long washout and patients may be at risk off medications. Also, our prior study [9] indicated that antidepressants do not substantially alter CFS patients’ expression of the genes in our profile. Thus, 11 of 49 control subjects (23%), 30 of 48 CFS patients (63%) and 9 of the FM patients (50%) were tested while continuing their usual antidepressants. In regard to other prescribed medications, for the first 19 CFS and all 18 FM patients, we requested that these patients be withdrawn from any prescribed pain medications and anticonvulsants for 6 days in order to participate, and only 2 of these CFS patients failed to comply fully. For the remaining 29 CFS patients, in order to increase participation by those with more severe symptoms who were unwilling to interrupt their medications, they were allowed to remain on all their usual physician-prescribed medications. This resulted in 15 (31%) CFS patients being tested on opioid pain medications, and 11 (23%) CFS patients being tested on anticonvulsants. This allowed secondary analyses to be performed comparing CFS patients tested on vs. not on these prescribed medications.
Disorder Onset and Functionality/Severity Ratings
For the CFS patients, CFS symptom onset was reported as sudden by 36 patients (70% of the α-2A increase and 87% of the α-2A decrease subgroups). Thirty five (73%) reported onset to be associated with one or several flu-like, mononucleosis or other viral illnesses or infections. Five patients (10%) associated onset with traumatic injuries and fractures. Three patients (6%) associated onset with surgery, and 5 patients associated onset with life stress or no memorable event. In contrast, the FM only group included only 2 patients (11%) who reported sudden onset of their symptoms, linked to a virus in one and to a herniated disk in the other.
Categorical ratings of disorder severity from 1–4 were assigned for all CFS patients by their physician (LB) based on observations and symptom measurements over multiple visits in the clinical setting, while blind to the gene expression and exercise results. The categories were defined by ability to function in normal daily activities, as follows: 4. Able to work full time 30–40 hours but “nothing left over”, high symptom burden and limitations; 3. Able to do part time school/work/other activities 10–30 hours/week, but easily relapses, and frequent rest needed; 2. Only able to do self-care activities of daily living (ADL), sedentary, <10 hrs/wk of light activities, but could live alone with occasional help; 1. House- or bed-bound, with minimal activity tolerance, marked cognitive dysfunction, and dependent on others for ADL. Thus, lower function scores indicated worse disorder severity.
Exercise Protocol
Exercise testing was always performed at the same time of day (starting between 8 and 9:30 am). Venous blood samples were obtained immediately prior to exercise (baseline) and at 0.5, 8, 24, and 48 hours post-exercise. We assessed the severity of pre-existing and exercise-related fatigue and myalgia symptoms at the time of each blood draw, and at the midpoint and immediately after completing the exercise task; the subject provided numerical ratings of mental fatigue, physical fatigue and overall body pain using a 0–100 scale where 100 was defined as the greatest level of fatigue or pain the subject could ever imagine experiencing.
A combined arm-leg cycle ergometer (Schwinn Air-Dyne) was used for the 25-min, moderate exercise test. In the first 5 min of exercise, subjects were asked to increase pedaling rate until 70% age-predicted maximal heart rate was achieved. Thereafter, work rate was adjusted in order to maintain this target heart rate throughout the sub-maximal exercise protocol. Ratings of Perceived Exertion (RPE) were obtained on a scale of 1–10 every 45 min; heart rate was recorded every minute, and blood pressure was measured at baseline, every 10 min during exercise, and upon completion of the exercise. We elected to use a sustained moderate exercise rather than a maximal exercise test (which typically last only 5–9 min in CFS patients) because of closer similarity to the natural exercise experiences reported to exacerbate CFS symptoms in patients’ daily lives. Our 25 min sub-maximal exercise task did elicit consistent worsening of fatigue and pain symptoms from 8–48 hours post-exercise (see Fig. 1). In contrast, after a briefer maximal exercise task, reports of worsening CFS symptoms were inconsistent or absent until 5 days after the challenge [14], a pattern not typically observed in real life. Maximal exercise protocols have demonstrated few differences in cardiorespiratory and perceptual responses; RPE is an exception being consistently higher in CFS patients than controls [15]. However, it is notable that responses to sub-maximal exercise including VO2 do predict peak exercise performance in CFS patients [16].
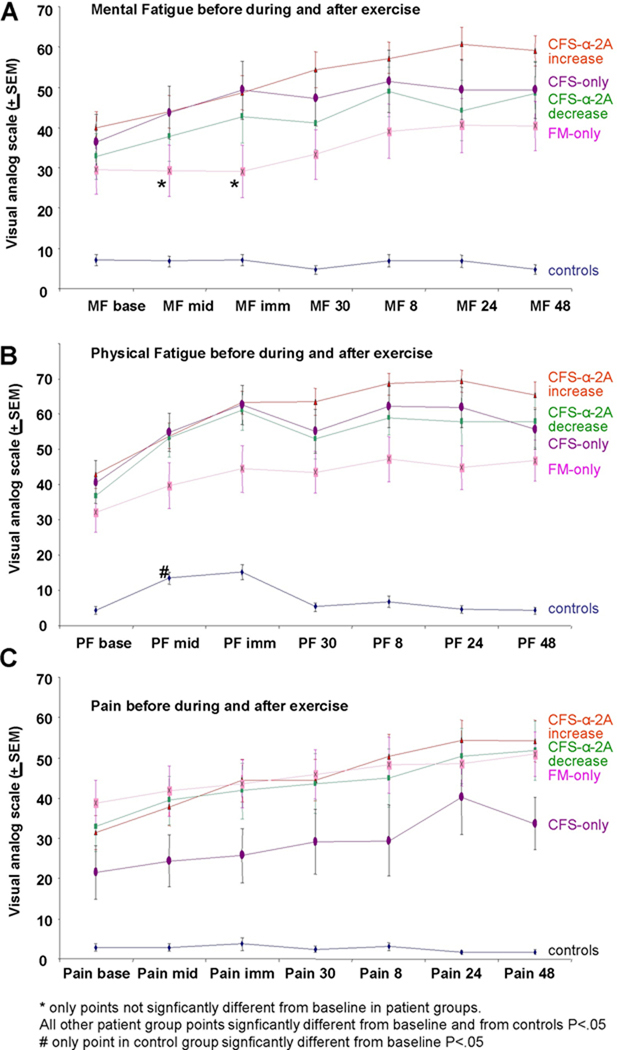
Figure 1.
Behavioral scores for mental and physical fatigue and pain in patients vs. control subjects. A, Average Visual analog scores (maximum 100, minimum 0) for Mental Fatigue for CFS subgroups, FM, and Controls. Data points are at baseline (MF base), half way through the 25 minute exercise period (MF mid), immediately after exercise (MF imm), 30 minutes after exercise (MF 30), and 8 (MF 8), 24 (MF 24), and 48 hours (MF 48) after exercise. Orange line indicates averages of patients identified as showing increases in α-2A adrenergic receptor mRNA following exercise. Purple line indicates patients with CFS that do not meet criteria for FM. Green line indicates patients showing decreases in α-2A adrenergic receptor mRNA following exercise. Pink line indicates patients with FM that do not meet criteria for CFS. Blue line indicates Controls. Asterisks indicate the only two data points for any of the patient groups that were not greater than baseline, all other data points for all groups were significantly greater than baseline. All data points for all patient groups were significantly greater than those of Controls (P<.05).
B, Average Visual analog scores (maximum 100, minimum 0) for Physical Fatigue for CFS subgroups, FM, and Controls. # indicates the only point at which Physical Fatigue was significantly increased from baseline for Controls (P<.05). All data points for all patient groups were significantly greater than controls, and also significantly greater than baseline (P<.05). All other information as in A.
C, Average Visual analog scores (maximum 100, minimum 0) for Pain for CFS subgroups, FM, and Controls. CFS-only patients had overall pain scores that were lower than CFS and FM patients. All patients pain scores were higher at baseline and all time points than controls (P<.05). In addition, all patients showed significant increases from baseline in pain scores during and following exercise (P<.05). All other markings as in A and B.
mRNA extraction and analysis
All blood processing and analyses were performed by personnel blinded to the subject’s group. At each of the 5 blood sampling times, blood was collected in EDTA tubes. Seven minutes after blood collection, the blood was centrifuged at 3200 rpm (1315 × g- Clay Adams Compact II Centrifuge) for 12 minutes, plasma removed, and the white layer carefully collected in RLT+β-ME (Qiagen, Valencia, CA) then quickly frozen using a methanol-dry ice slurry, and stored at −80°. RNA was extracted using RNeasy kits (Qiagen, Valencia, CA), according to manufacture’s directions, and treated with RNase-free DNase-I (Qiagen, Valencia, CA). Immediately following extraction, RNA was converted to a cDNA library using the ABI High Capacity cDNA Archive Kit (Applied Biosystems, Inc., Foster City, CA). The cDNA samples were stored at −2° C until analysis.
RNA integrity was assessed with a Bioanalyzer, and consistently found to have values greater than 9. The cycle counts for the control gene, TF2B) averaged 21.78 ± 1.67 (SD) for control subjects and 22.32 ± 2.09 (SD) for patients. These values and variations indicate consistent, high quality integrity of the RNA, and sufficient amounts of RNA for accurate analysis of the amount of mRNA in these experiments.
The cDNA libraries were analyzed using the ABI quantitative, real-time PCR system on the ABI Prism 7900 Sequence Detection System (Applied Biosystems, Inc., Foster City, CA), using ABI TaqMan Master Mix (Applied Biosystems, Inc., Foster City, CA). Master Mix/primer probe solutions and template solutions were separately loaded onto 96 well pre plates, with robot loading mixing these solutions when placed in the 384 well plates. Plates were centrifuged to remove any air bubbles in the wells. Each sample was run in duplicate with standards being run in quadruplicate. No template control samples were also run. Each 384 well plate contained samples from two subjects/patients, and all genes were analyzed on the same plate. Primer probes (all from TaqMan Gene Expression Assays, Applied Biosystems, Inc., Foster City, CA) were: ASIC3 - Hs00245097_m1; P2X4 - Hs00175706_m1; P2X5 - Hs00175712_m1; TRPV1 - Hs00218912_m1; Adrenergic A2A (α-2A) - Hs00265081_s1; Adrenergic B-1 - Hs02330048_s1; Adrenergic B-2 - Hs00240532_s1; COMT - Hs00241349_m1; IL6 - Hs00174131_m1; IL10 - Hs00174086_m1; TNFβ (Alpha Lymphotoxin or “α-lym”) - Hs00236874_m1; TLR4 - Hs00152937_m1; CD14 - Hs00169122_g1. Control primer probes included TF2B - Hs00155321_m1; β-Actin - Hs99999903_m1; and PSMB6 - Hs00382586_m1. In later experiments, only TF2B was used as the reference gene. All primer probes, except for the adrenergic receptors and CD14 (these genes do not have introns), recognize sequences that cross splice sites, and therefore, make detection of genomic DNA unlikely. In all cases, we quenched the genomic DNA and ran no-template control wells to ensure that genomic DNA did not contaminate the final results. All of these primer probes were designed and tested to be used together, and have similar efficiencies to help eliminate inaccuracy. For the genes that have rarely been described in leukocytes (Adrenergic α-2A, Adrenergic B-1, P2X5, TRPV1, ASIC3) we designed primers that contained 360 to 600 base-pairs, which included the regions ABI indicated the Primer probes listed above spanned. PCR product was generated from our leukocyte samples and sequenced. All of these sequences were 99–100% identical to predicted sequences of these genes. Evaluation of Controls in this and previous experiments indicated that TF2B had less intrinsic variation than other candidates such as β-actin, had a count range that was similar to the genes of interest, and did not increase or decrease due to the exercise protocol. Real-time PCR results were analyzed with SDS 2.1 (Applied Biosystems, Inc., Foster City, CA), and inspected to determine artifacts (loading errors, robot errors, thresholding errors, etc.). Count numbers were exported to an Excel spreadsheet, and analyzed according to the ddCT method described in ABI User Bulletin #2 (Applied Biosystems, Inc., Foster City, CA). Baseline levels for each gene were computed relative to TF2B, and these baselines were used as the comparator for all measures taken after the exercise period (see statistical methods below for further analysis details).
Statistical Analysis
Because the ddCT method used for mRNA analysis necessarily creates a non-normal, rightward skewed distribution, data were log transformed to yield distributions that could be appropriately analyzed with parametric statistics.
Post-exercise values for each gene expression measure were normalized relative to the same subject’s baseline levels (1.00 = baseline). As detailed in our previous report [8], to reduce false positive findings associated with multiple comparisons, we did not examine each sampling time individually. Instead, the relative mRNA values from the 4 post-exercise time points (0.5, 8, 24, and 48 hour) were summed into a single measure labeled Area Under the Curve (AUC) and then log transformed. Initial MANOVAs were performed for the metabolite detecting, adrenergic and immune markers that differed in our prior study [8] yielded significant effects of Groups. These were followed by individual ANOVAs, and when significant, by comparisons of baseline and post-exercise AUC of Controls vs. CFS and FM patients determined with independent two-tailed t-tests. Two tailed rather than 1 tailed tested were used despite the predictions of increases as an additional protection against false positive results. This approach to multiple gene comparisons is similar (but on a much smaller scale) to Pathway Analysis as used to explore the many thousands of gene examined in microarray studies. Secondarily, we compared two CFS subgroups (CFS with increased post-exercise α-2A and CFS with decreased post-exercise α-2A) to Controls using one-way ANOVAs. One-way ANOVAs were used for between group comparisons of cardiovascular, work rate and RPE measures obtained during the exercise task.
To examine whether Group differences were related to differences in age, gender, or body mass index (BMI), we performed one way ANOVAs with each of these included in the model as covariates. In no instance did age, gender, or BMI impact the significance of group differences.
Group by Time, Group, and Time differences in ratings of physical fatigue, mental fatigue, and pain were analyzed using 3 × 7 repeated measures (RM) ANOVAs. When significant Group differences were present, within group Time effects were examined with RM ANOVAs with simple contrasts to determine which time points differed significantly from baseline.
In addition to group comparisons, Pearson r correlations were used to examine relationships between exercise variables, pain and fatigue ratings, and gene expression measures. All data are presented as means and standard errors, with significance set at P < 0.05.
Results
Data were obtained from 48 CFS patients, 18 FM patients and 49 control subjects. Table 1 summarizes the characteristics of these groups, with the CFS patients divided into the two major subgroups suggested by the data in this study. Age and gender were well matched between controls and the CFS patient groups, but less well matched for the FM group; however, neither gender nor age were significant covariates in the FM patient analyses.
Table 1.
Subject Characteristics for Controls, CFS Patients, and FM Patients
| Characteristic | Controls (n=49; 29F) |
All CFS Patients (n=48; 33F) |
α-2A increase CFS Patients (n=34; 25F) |
α-2A decrease CFS Patients (n=14; 8F) |
FM patients (n=18, 15F) |
All CFS P* |
α-2A + CFS Patients P* |
α-2A - CFS Patients P* |
FM P* |
|---|---|---|---|---|---|---|---|---|---|
| Age (years) Median |
42.0 ± 1.9 45 |
41.8 ± 1.9 46 |
41.4 ± 2.2 46 |
42.8 ± 3.5 46 |
50.4 ± 2.6 51 |
.998 | .979 | .971 | .037 |
| Body Mass Index (kg/m2) | 23.9 ± 0.7 | 26.2 ± 0.8 | 26.5 ± 0.9 | 25.6 ± 1.3 | 26.5 ± 1.4 | .055 | .050 | .448 | .135 |
| Resting SBP (mmHg) | 126.4 ± 1.9 | 123.0 ± 2.6 | 120.9 ± 3.0 | 128.3 ± 5.0 | 126.1 ± 3.0 | .550 | .216 | .902 | .994 |
| Resting DBP (mmHg) | 80.5 ± 1.6 | 80.6 ± 1.5 | 80.2 ± 1.8 | 81.7 ± 2.9 | 87.1 ± 2.2 | .996 | .990 | .909 | .048 |
| Exercise SBP (mmHg) | 157.0 ± 3.2 | 148.7± 3.7 | 147.8 ± 4.6 | 150.9 ± 6.4 | 161.1± 5.9 | .199 | .195 | .660 | .766 |
| Exercise DBP (mmHg) | 90.8 ± 1.8 | 88.6 ± 1.6 | 88.2 ± 2.1 | 89.6 ± 2.6 | 95.7 ± 3.1 | .600 | .545 | .931 | .253 |
| Resting HR (baseline) | 76.7 ± 1.8 | 82.4 ± 2.4 | 83.4 ± 2.7 | 80.1 ± 5.0 | 80.2 ± 3.8 | .144 | .087 | .687 | .683 |
| Exercise HR (BPM) | 126.8 ± 1.4 | 119.8 ± 2.2 | 121.6 ± 2.5 | 115.6 ± 4.4 | 122.8 ± 3.2 | .017 | .172 | .067 | .446 |
| Exercise HR (%PMHR) | 71.3 ± 0.5 | 67.3 ± 1.1 | 68.1 ± 1.1 | 65.4 ± 2.5 | 61.7 ± 1.9 | .004 | .031 | .081 | <.001 |
| Exercise WR (Kcal/kg/min) | 7.6 ± 0.7 | 4.3 ± 0.2 | 4.1 ± 0.2 | 4.7 ± 0.4 | 5.1 ± 0.4 | <.001 | <.001 | .002 | .023 |
| Exercise RPE | 3.1 ± 0.1 | 5.0 ± 0.2 | 5.0 ± 0.3 | 5.1 ± 0.5 | 4.5 ± 0.4 | <.001 | <.001 | .004 | .012 |
| # Ortho intolerance | NA | 18 (38%) | 8 (24%) | 10 (71%) | 0 | .0003 | .008 | ||
| Viral onset | NA | 29 (60%) | 19 (56%) | 10 (71%) | 1 (5%) | ||||
| Tested on Pain med | 1 | 15 (31%) | 11 (32%) | 4 (29%) | 3 (16%) | ||||
| On anti-convulsants | 0 | 11 (23%) | 6 (18%) | 5 (36%) | 5 (28%) | ||||
| On anti-depressant | 12 (24%) | 35 (73%) | 24 (71%) | 11 (79%) | 10 (56%) |
Values are mean ± SE
BMI = body mass index; SBP = systolic blood pressure; DBP = diastolic blood pressure; %PMHR = percent of age-predicted maximal heart rate; WR = work rate; RPE = rating of perceived exertion
Bold indicates P<.01
Bold(in italic) indicates .01<P<.05
We attempted to adjust for fitness level mismatches by exercising all patients and controls at the same relative exercise intensity, to 70% of age predicted, maximal heart rate. The majority of CFS and FM patients were able to attain the 70% level, but a few could not, leading to small, but significantly lower percentages of maximal heart rate in all patient groups. In spite of lower relative exercise intensities and lower work rates, all patient groups reported higher perceived exertion (RPE) during the exercise task than controls (see Table 1).
Figure 1 depicts mean ratings of mental fatigue, physical fatigue and pain on our 0–100 scale in the various groups before, during and after the exercise task. In control subjects, exercise did not increase ratings of mental fatigue or pain at any time point; and physical fatigue was increased only at mid-exercise and not at any post-exercise time. In sharp contrast, exercise caused significant increases in all fatigue and pain measures at all time points during and after exercise in CFS only and in CFS+FM patients. Patients with FM reported increases in pain and physical fatigue at all time points, and increases in mental fatigue at all time points except during and immediately after exercise. As can be seen in Figure 1, the two CFS subgroups distinguished by post-exercise adrenergic α-2A increases vs. decreases (described in detail below) did not differ from each other in ratings of pain or fatigue. Not surprisingly (since these patients were defined by having less pain than FM or CFS+FM patients), patients with CFS only had lower pain scores during and immediately after exercise than the CFS+FM or FM groups. Thus, this very moderate level of exercise for 25 min caused post-exertional malaise lasting 48 hours in all CFS and FM patient groups, but not controls.
Gene expression results
CFS patients compared with CFS patients who also have co-morbid FM
Initially, we divided the CFS patients into those that had CFS, but did not meet criteria for FM, and those CFS patients who also met criteria for FM, i.e., they had both CFS and FM (CFS+FM). None of the descriptive variables in Table 1 were significantly different between these groups. Likewise, comparison of these two groups showed very similar gene expression both before and after exercise. Only post-exercise ASIC3 AUC was greater in the CFS+FM vs. the CFS only patients (P < .046). For this reason, in all of the following analyses, CFS only and CFS+FM patients are grouped together.
CFS patients showed no baseline changes in gene expression from Controls; FM patients showed baseline increases in 3 genes
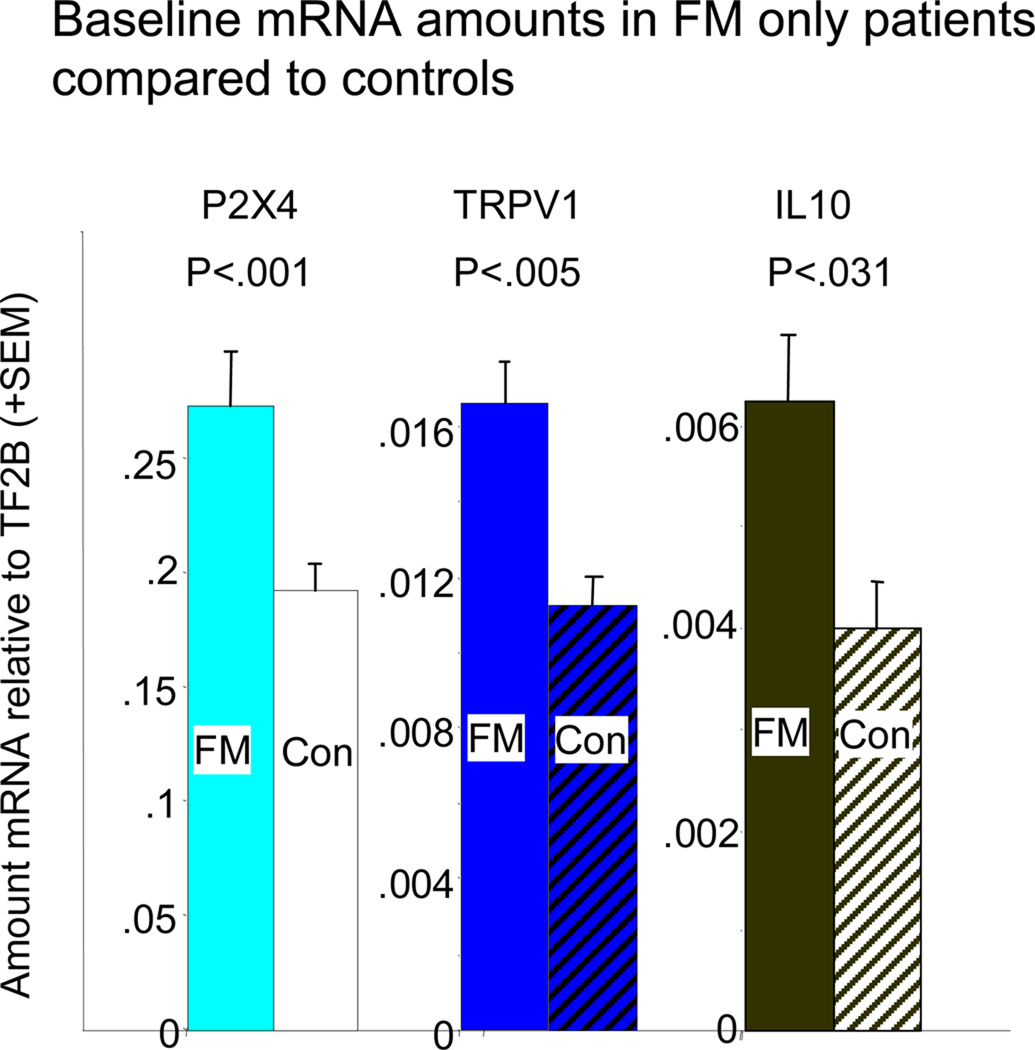
Table 2 contains the average baseline values and significance values for post-exercise AUC (0.5, 8, 24, and 48 hours after exercise) for mRNA increases from baseline levels. None of the CFS subgroups differed from controls in expression of any gene at baseline, either as separate subgroups or combined into a single CFS group (all P values >0.145). In contrast, the FM group showed several differences from controls at baseline before exercise. They had significantly higher baseline quantities of mRNA for sensory receptors P2X4 (P<0.001) and TRPV1 (P<0.005), and for the cytokine IL10 (P<0.031). Figure 2 graphs these differences. These differences were unaltered after covarying for age and gender differences.
Table 2.
MANOVA results, baseline means + SEs and ANOVA results for all mRNAs relative to TF2B, and ANOVA results for post-exercise Area Under Curve (AUC) mRNA differences from Controls in CFS Patients and FM patients.
| Controls Baseline N=49 |
All CFS Patients Baseline N=48 |
CFS α-2A Increase Baseline N=34 |
CFS α-2A Decrease Baseline N=14 |
FM Baseline N=18 |
All CFS Patients Baseline P |
FM Baseline P |
All CFS Patients AUC P N=48 |
CFS α-2A Increase AUC P N=34 |
CFS α-2A Decrease AUC P N=14 |
FM AUC P N=18 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| Metabolite Detecting | MANOVA | .030 | .009 | .022 | .006 | .634 | .576 | ||||
| ASIC3 | 9.02E-03 ± 5.17E-04 |
7.79E-03 ± 7.03E-04 |
8.02E-03 ± 8.56E-04 |
7.26E-03 ± 8.32E-04 |
9.81E-03 ± 1.09E-03 |
.298 | .462 | .200 | .54 | .443 | .925 |
| P2X4 | 2.00E-01 ± 1.05E-02 |
1.86E-01 ± 1.38E-02 |
1.97E-01 ± 1.80E-02 |
1.62E-01 ± 1.50E-02 |
2.74E-01 ± 2.36E-02 |
.693 | .001 | .002 | .002 | .547 | .570 |
| P2X5 | 2.33E-01 ± 2.00E-02 |
2.43E-01 ± 2.47E-02 |
1.97E-01 ± 1.80E-02 |
3.23E-01 ± 4.23E-02 |
2.75E-01 ± 3.35E-02 |
.395 | .282 | .043 | .022 | .888 | .629 |
| TRPV1 | 1.23E-02 ± 7.94E-04 |
1.41E-02 ± 1.00E-03 |
1.35E-02 ± 1.12E-03 |
1.51E-02 ± 1.64E-03 |
1.66E-02 ± 1.09E-03 |
.272 | .005 | .027 | .020 | .645 | .939 |
| Adrenergic | MANOVA | .472 | .304 | .007 | <.0005 | <.0005 | .112 | ||||
| α-2A | 5.42E-03 ± 6.87E-04 |
7.04E-03 ± 1.59E-03 |
7.68E-03 ± 2.10E-03 |
5.75E-03 ± 1.40E-03 |
3.61E-03 ± 6.32E-04 |
.619 | .128 | .023 | .0005 | .019 | .297 |
| β-1 | 3.25E-02 ± 1.03E-02 |
7.27E-02 ± 5.30E-02 |
9.15E-02 ± 7.43E-02 |
3.13E-02 ± 1.80E-02 |
5.92E-02 ± 3.68E-02 |
.648 | .344 | .056 | .012 | .994 | .999 |
| β-2 | 1.13E+00 ± 8.85E-02 |
9.98E-01 ± 7.17E-02 |
1.97E+00 ± 5.87E-01 |
9.09E-01 ± 6.96E-02 |
1.05E+00 ± 2.02E-01 |
.481 | .673 | .002 | .001 | .998 | .594 |
| COMT | 2.33E-01 ± 1.88E-02 |
1.89E-01 ± 1.75E-02 |
1.90E-01 ± 1.91E-02 |
1.89E-01 ± 3.30E-02 |
2.29E-01 ± 1.82E-02 |
.145 | .906 | .009 | .032 | .102 | .845 |
| Cytokine | MANOVA | .506 | .018 | .151 | .093 | .765 | .498 | ||||
| IL6 | 3.69E-03 ± 1.19E-03 |
3.98E-03 ± 4.86E-04 |
4.17E-03 ± 6.07E-04 |
3.55E-03 ± 7.14E-04 |
4.64E-03 ± 1.17E-03 |
.966 | .651 | .253 | .135 | .998 | .701 |
| IL10 | 4.61E-03 ± 3.81E-04 |
2.09E-02 ± 1.45E-02 |
2.85E-02 ± 2.02E-02 |
5.04E-03 ± 7.51E-04 |
6.24E-03 ± 6.47E-04 |
.509 | .031 | .003 | 0.002 | .153 | .799 |
| αLT | 7.95E-02 ± 1.41E-02 |
8.21E-02 ± 1.29E-02 |
8.38E-02 ± 1.64E-02 |
7.84E-02 ± 1.75E-02 |
6.55E-02 ± 7.54E-03 |
.985 | .550 | .307 | .140 | .974 | .980 |
| TLR4 | 4.54E-01 ± 6.13E-02 |
4.34E-01 ± 3.95E-02 |
4.80E-01 ± 4.88E-02 |
3.31E-01 ± 5.10E-02 |
3.64E-01 ± 2.09E-02 |
.939 | .376 | .196 | .110 | .995 | .973 |
| CD14 | 2.14E+00 ± 1.34E-01 |
2.11E+00 ± 1.80E-01 |
2.30E+00 ± 2.27E-01 |
1.73E+00 ± 2.01E-01 |
2.37E+00 ± 2.21E-01 |
.990 | .374 | .658 | .724 | .877 | .840 |
Bold = P<.01, Bold (italics) .01>P>.05 as compared with Controls (all 2 tailed comparisons).
Figure 2.
Comparison of baseline gene expression in FM patients to that of Age and gender matched controls. Because of the large differences in the amount of mRNA for different genes, the average baseline value of each of the significantly increased genes for the FM patients were matched in magnitude for this graph. The scale for each of the genes (in amount relative to the control gene, TF2B) is indicated to the left of each bar. Con = Control subjects. Controls also marked with cross hatching. Colors are the same as for genes in Figure 3.
Post Exercise Gene Expression Changes in patient subgroups vs. controls
Although their gene expression did not differ from controls prior to exercise, after exercise CFS patients showed greater increases in mRNA than controls for 7 of the genes under study. With all CFS subgroups combined, CFS patients showed greater post-exercise AUC increases than controls for P2X4, P2X5, TRPV1, α-2A, β-2, COMT, and IL10 (P <.05 to P<.001;see Table 2).
In our previous study, unusual gene expression patterns in some CFS patients suggested a possible subgroup, which was confirmed clinically by Dr. Bateman. With the additional data from the present study, on the basis of changes in gene expression levels following exercise, we could define two major subgroups of CFS patients. These subgroups were defined by post-exercise increases vs. decreases in mRNA of the adrenergic α-2A receptor. The larger CFS subgroup (71% of all CFS patients) had increases in the α-2A receptor mRNA at one or more time points following exercise (α-2A increase CFS patients). This group showed large increases in all 7 genes listed above as well as increases in β-1 AUC (See Table 2). As can be seen in Figure 3 (compare A with B), increases in expression of the genes listed above compared to controls were observed at 30 minutes following the exercise period, and lasted for the duration of the study, 48 hours after the exercise period. This was confirmed by repeated measures analyses showing significant time effects after exercise indicating increases above baseline that were sustained throughout the post-exercise period.
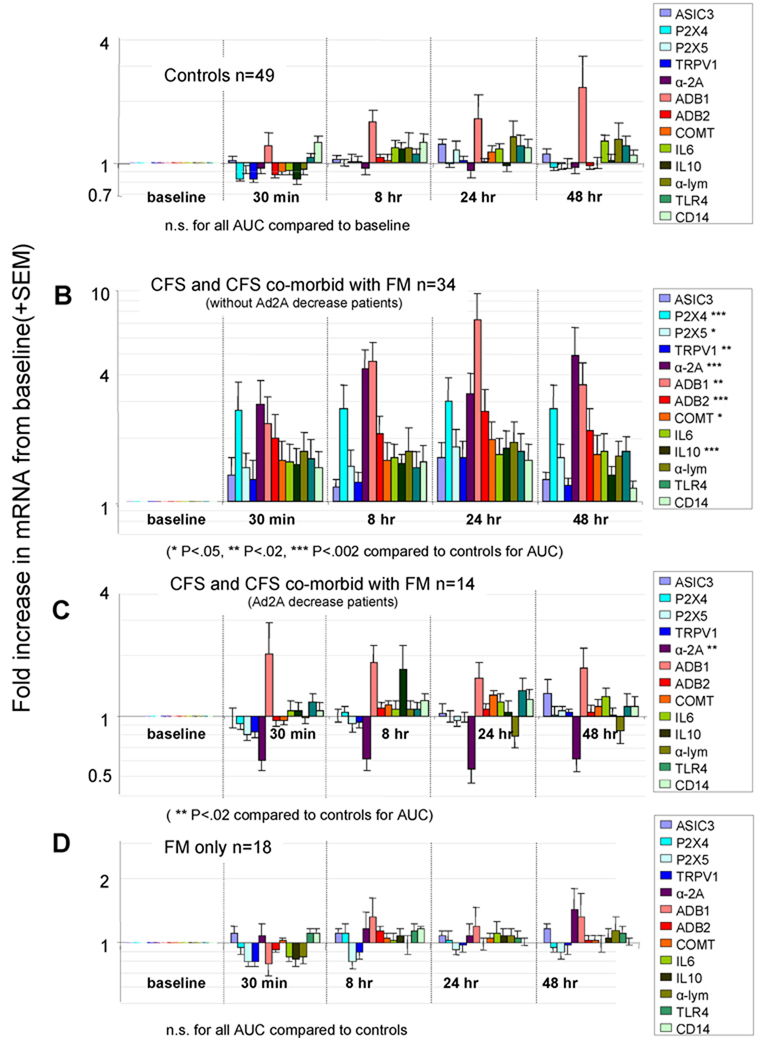
Figure 3.
Graphs comparing gene expression increases following moderate exercise in patients with CFS and FM. All graphs plotted in log10 scale. For all graphs, baselines for all genes were normalized to 1. Scale on graphs are fold changes from baseline in mRNA quantity. Significant differences in the sum of all time points for each gene (AUC) are indicated by * in the legend boxes to the right of each graph. Metabolite detecting receptors are colored in blues, adrenergic receptors and COMT are colored in reds, and cytokine related genes are colored in greens.
A, Averages of mRNA fold changes after exercise for 49 Control subjects. No significant differences from baselines were noted.
B, Averages of mRNA fold changes after exercise in 34 CFS patients and CFS patients that also made criteria for FM that did not show decreases in α-2A adrenergic receptor mRNA following exercise. Significant differences are noted by asterisks in the legend box at right. See key under this graph for P value of asterisks.
C, Averages of mRNA fold changes after exercise for 14 CFS patients and CFS patients that also made criteria for FM that showed decreases in α-2A adrenergic receptor mRNA at all times following exercise. Only α-2A adrenergic receptor mRNA showed significant changes. For this graph, the data for ASIC3 from one extreme outlier was dropped.
D, Averages of mRNA fold changes after exercise for 18 FM only patients. No differences from controls were seen at any time after exercise. This contrasts with baseline changes seen in Figure 2.
As shown in Figure 3C and Table 2, the smaller subgroup (29% of all CFS patients) demonstrated large decreases in α-2A mRNA at all time points following exercise (α-2A decrease group). This group was also distinguished by a clinical history of orthostatic intolerance in most of the patients (see Table 1, #Ortho intolerance). Ten of the 14 α-2A decrease patients had clinical orthostatic intolerance compared with only 8 of 34 α-2A increase CFS patients (71% vs. 18%, Χ2 P< 0.008). In addition to this high rate of orthostatic intolerance, the α-2A decrease patients demonstrated no significant increases in expression of any of the other genes measured in this study compared to the Control group (see Figure 3C and Table 2). CFS only and CFS+FM patients were similarly represented in both subgroups; the α-2A decrease group had 6 CFS only patients, and 8 CFS+FM patients, while the α-2A increase group had 9 CFS only patients and 25 CFS+FM patients.
To ensure that these findings were not solely due to differences in exertion, we performed a secondary analysis after reducing our sample to the CFS and Control subjects who were matched on Ratings of Perceived Exertion (RPE). This analysis confirmed our central findings. This comparison examined 15 controls with the highest RPE (mean 3.89) matched with 27 patients with overlapping RPEs (mean 3.87). Even though this reduced our sample size and statistical power substantially, 6 of the 7 genes’ AUCs were still significantly greater in CFS patients than controls [P2X4 (p<.05), TRPV1 (P<.02), α-2A (P<.03), β-2 (P<.03), COMT (P<.04), and IL10 (P< .01)]. The only gene not reliably different was P2X5 (P=.12).
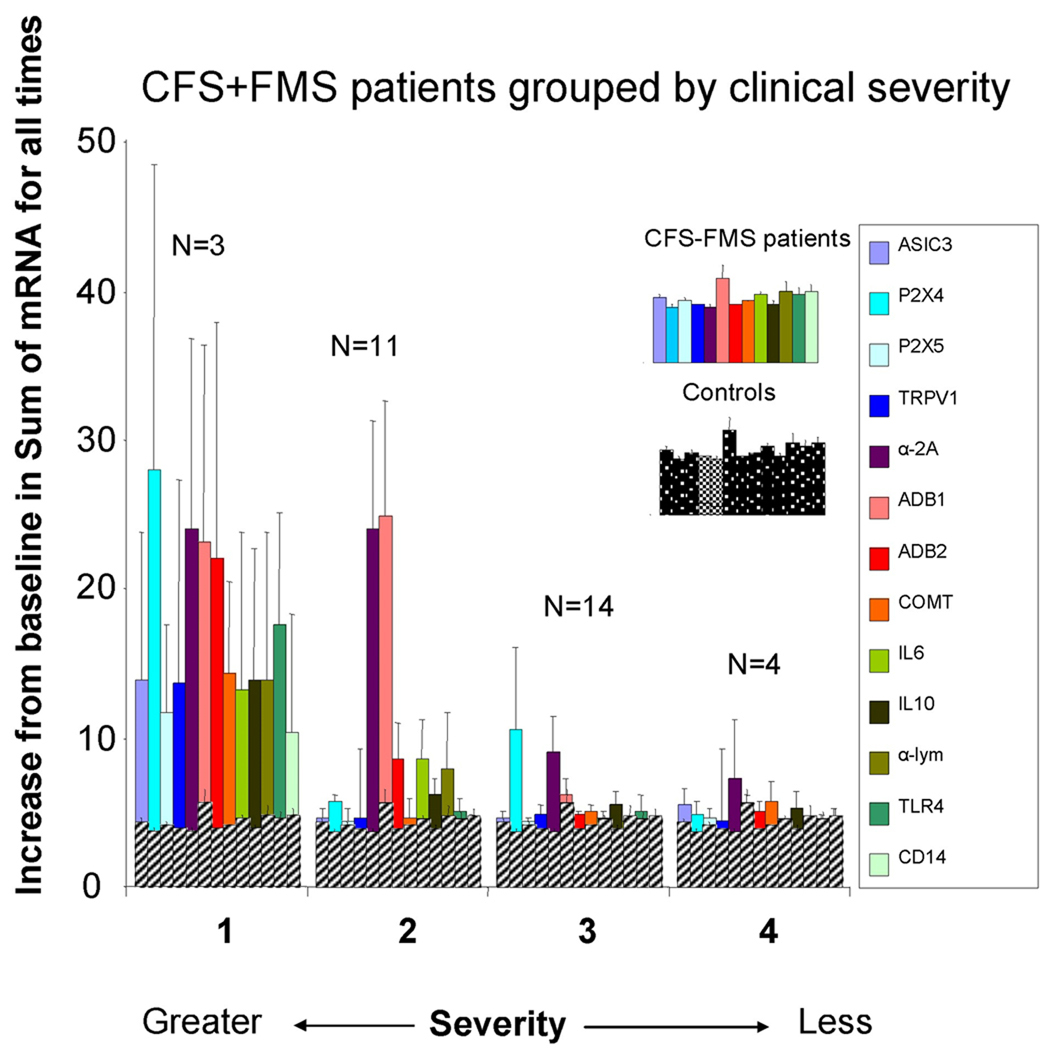
Gene expression varied with the clinical severity of CFS
Figure 4 graphs the 4 severity groups for the α-2A increase CFS patients (n=34). In this subgroup, there were 3 patients with severity 1 (9%), 11 with severity 2 (32%), 14 with severity 3 (41%), and 4 with severity 4 (12%). Two patients were not included for this analysis because they were less severe than 4. This graph indicates that, relative to controls, gene expression was increased most in patients with the highest severity, and least increased in patients with the lowest severity. When the average of all gene AUC was compared, groups 1 and 2 were significantly greater that that of groups 3 and 4 (P< .011).
Figure 4.
Graph of severity groups’ gene expression. For this graph, gene expression increases for all 4 time periods of exercise were summed and a single value shown. Severity scale was: 1. House- or bed-bound, with minimal activity tolerance, marked cognitive dysfunction, and dependent on others for activities of daily living. 2. Only able to do self-care activities of daily living, sedentary, <10 hrs/wk of light activities, but could live alone with occasional help. 3. Able to do part time school/work/other activities 10–30 hours/week, but easily relapses, and frequent rest needed 4. Able to work full time 30–40 hours but “nothing left over”, high symptom burden and limitations. Controls are graphed on top of each of the severity groups in grey cross hatching so that patients can be easily compared with control gene expression levels.
The minor subgroup, the α-2A decrease group (n=14), had 1 patient with severity 1 (7%), 4 patients with severity 2 (29%), 6 patients with severity 3 (43%) and 2 patients with severity 4 (14%). One patient had severity 1. This distribution was very similar to that of the major subgroup (Χ2 P> .85).
Effects of medications
As described previously, 11 CFS patients were on anticonvulsants and 15 were on opioids during testing while the other CFS patients either had not used them or were withdrawn from them prior to testing. To determine if these medications might have affected the gene expression, we compared the total post-exercise AUC gene expression of patients on these medications with the AUC of those not on the medications. There was a trend in the direction of lesser post-exercise increases in our selected genes among CFS patients on anticonvulsants vs. CFS on no medications (P =.055) but even this medicated CFS group still differed from controls (P < .05). CFS patients on opioid pain medications alone did not differ from other CFS patients (P> .245). These findings suggest that anticonvulsant drugs may reduce the post-exercise gene expression increases in some CFS patients, but they also indicate that withdrawal from these medications prior to testing is not critical when attempting to differentiate their responses from those of healthy individuals.
When the 30 CFS patients tested on vs. the 18 not on antidepressants were compared, no statistical difference in expression of mRNA was observed (P> .614). Likewise, when the 11 control subjects also were on antidepressants were compared to the 38 other controls, there were no differences in gene expression.
Correlations
Table 3 shows the correlations between the behavioral scores for fatigue and pain and AUC gene expression measures, as well as inter-correlations between the various genes. These indicate strong positive relationships between post exercise pain and fatigue and increases in P2X4, TRPV1, α-2A, β-2, and IL10. Relationships between the behavioral measures were weaker for the ASIC3, P2X5, and the other cytokine genes measured.
Table 3.
Correlations of Post-exercise AUC Gene Expression Measures with Post-exercise AUC Fatigue and Pain and with Each Other (All α-2A increase CFS patients and controls included in this analysis)
| ASIC3 | P2X4 | P2X5 | TRPV1 | α-2A | β-1 | β-2 | COMT | IL6 | IL10 | TLR4 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mental fatigue | NS | +.51 | +.28 | +.34 | +.60 | +.32 | +.45 | +.39 | +.25 | +.42 | +.27 |
| Physical fatigue | NS | +.51 | +.26 | +.34 | +.60 | +.32 | +.47 | +.36 | NS | +.41 | +.25 |
| Pain | NS | +.43 | NS | +.32 | +.48 | +.30 | +.44 | +.27 | +.25 | +.38 | NS |
| ASIC3 | --- | NS | +.42 | +.35 | NS | −.38 | NS | +.40 | NS | NS | NS |
| P2X4 | --- | +.36 | +.41 | +.57 | +.38 | +.72 | +.59 | +.37 | +.57 | +.36 | |
| P2X5 | --- | +.76 | +.27 | NS | +.30 | +.61 | +.33 | +.24 | NS | ||
| TRPV1 | --- | +.30 | NS | +.31 | +.63 | +.55 | +.55 | +.46 | |||
| α-2A | --- | +.59 | +.76 | +.29 | +.34 | +.49 | +.34 | ||||
| β-1 | --- | +.62 | NS | +.30 | +.45 | NS | |||||
| β-2 | --- | +.37 | +.45 | +.51 | +.36 | ||||||
| COMT | --- | +.33 | +.44 | +.29 | |||||||
| IL6 | --- | +.66 | +.34 | ||||||||
| IL10 | --- | +.50 |
For all listed correlations P < .05. For those correlations > +.30, P < .01. NS = nonsignificant, P> .05
Discussion
1) Confirmation that gene expression changes occur in CFS
The present study confirms our prior findings [9] that moderate exercise in CFS patients leads to increased expression of certain sensory ion channel, adrenergic, and immune genes (P2X4, P2X5, TRPV1, α-2A, β-2, COMT, and IL10) that do not occur in healthy individuals. The functions of these genes and how they contribute to the symptoms of CFS and FM was discussed in our previous report [9]. These mRNA increases in CFS are evident within 30 minutes and are maintained for at least 48 hours. This includes one ion channel receptor, the heat-sensitive capsaicin receptor TRPV1, which was not significantly different between CFS and controls in our prior study with smaller group sizes. However, we did not replicate our prior study in all respects. First, in the present study, post-exercise increases in mRNA for ASIC3 or adrenergic β-1 receptors or in the immune marker TLR4 were not statistically greater in all CFS patients combined vs. controls. Second, we previously reported that baseline mRNA for β-2 and α-2A adrenergic receptors tended to be lower in CFS patients; the present study, however, indicated no differences prior to exercise in expression of any gene in CFS patients compared to controls. These differences are most likely due to the increased sample and a broader range of ages, disease severity, and possibly partly due to the allowed use of pain medications and anticonvulsant medications in the present study. Our present findings indicate that although both milder disorder severity and current use of anticonvulsant medications may be associated with lesser post-exercise increases in expression of these sensory and adrenergic receptors, the CFS patients who are more functional (and all the less functional patients as well) or are tested on medications still differ from healthy controls.
In our previous report [9], we suggested that CFS patients that did not meet criteria for FM were a defined subgroup separate from CFS patients that had co-morbid FM. With the additional sample size in the present study, we found that these two groups had similar gene expression profiles before and after exercise except for ASIC3, which was higher after exercise in the CFS+FM patients than in the CFS only patients. (These two groups also both divided into the α-2A subgroups described below, with similar proportions in each group.) Pre- and post-exercise ratings of mental and physical fatigue severity were likewise similar in these two patient groups, although the CFS only group had lower pain scores at nearly all times before, during and after the exercise task. Whether CFS only vs. CFS+FM belong to one or two different CFS populations cannot be determined from these selected outcome measures. Based on the specific genes in our profile, however, it was clear that these CFS+FM patients were much more similar to CFS alone patients than they were to FM alone patients, who showed greater mRNA for 3 genes at baseline compared to controls, but did not show greater increases in any of these genes after exercise.
2) Discovery of a significant subgroup defined by gene expression
A subgroup (29% of all CFS patients tested here) was defined by consistent decreases in α-2A following exercise. This subgroup (α-2A decrease CFS patients) did not show increases or decreases in other genes relative to Controls. The majority (71%) of this subgroup also had clinical orthostatic intolerance, as opposed to only 24% of the major subgroup (those that showed increases in α-2A following exercise “α-2A increase CFS patients”). The distribution of Clinical Severity was not different between these subgroups, indicating that both groups had similarly debilitating fatigue. Both groups also demonstrated very similar fatigue and pain responses to exercise (see Fig. 1). In the α-2A decrease CFS patients, the very large and consistent decreases in α-2A expression following exercise combined with orthostatic intolerance suggests that different mechanisms cause the debilitating fatigue in this subgroup. The large decreases in α-2A mRNA may reflect a particular type of dysregulation of the sympathetic nervous system. For example, activation of α-2A normally causes a decrease in sympathetic outflow (decreased release of norepinephrine). It is possible that the decrease in transcription of α-2A observed is a response to abnormally low levels of sympathetic outflow (or low levels of vascular and cardiac responses to norepinephrine), and resulting decreased cardiac output and possibly global vasodilatation. These effects would cause inadequate blood flow to working muscles and the brain. Thus, this decrease in α-2A transcription may be an attempt to compensate for a dysfunctional sympathetic response (by increasing norepinephrine release) that does not adequately increase blood flow to working muscles and the brain during and following exercise. This interpretation is supported by previous reports, which found a significant subgroup of CFS patients that were defined by more severe orthostatic intolerance and possibly by cardiac bioenergetic abnormalities that would lead to inadequate increases in blood flow during exercise [17–20]. These investigators also demonstrated that at least some CFS patients showed improper efflux of metabolites from the muscles of CFS patients that also could be a result of abnormally low levels of sympathetic outflow, or reduced vascular and cardiac responses to norepinephrine. One report suggested that postural orthostatic tachycardia syndrome patients (POTS) comprise about 30% of patients with CFS [20]. Another group suggested that while some CFS patients did conform to the POTS diagnosis, other CFS patients without POTS also had autonomic disturbances as well [21]. The relationship between the α-2A decrease CFS patients and POTS and the degree and type of autonomic disturbances needs further study. Given that the α-2A increase CFS patients also have dysregulation of α-2A as well as other adrenergic receptors and COMT, it is likely that dividing the patients by the gene expression measures will lead to better understanding of the specific problems in CFS patients, and lead to better treatment options.
3) Possible transcription factors involved in gene expression increases in the larger subgroup
The larger subgroup of patients (α-2A increase CFS patients) demonstrated large gene expression increases following exercise in many different genes. These include P2X4, P2X5, TRPV1, α-2A, β-1, β-2, COMT, and IL10. The increase of so many genes might indicate that upstream transcription factors common to all of these genes are dysregulated and control these downstream genes in a pathological fashion in CFS. Interestingly, all of the genes measured here are interconnected by the transcription factors CREB, GR-alpha (part of NR3C1, the glucocorticoid receptor), and NF-Kappa B1. Previously, NR3C1 polymorphisms were found to be associated with CFS [22–26]. In addition, Maes et al [27] have shown increased NF-Kappa B production in leukocytes of CFS patients, and Kim et al. [28] have shown increased DNA binding of NF-Kappa B in direct proportion to increases in exercise intensity. Whether these associations predict, or cause future CFS, should be investigated. Interestingly, the genes found to be dysregulated in the present study represent most of the pathways hypothesized by others to be altered in CFS. These include the immune system (IL10, and leukocytes in general) [29–31], cellular energy (P2X4 and 5 that encode ATP levels, and TRPV1 encoding temperature) [32], and the cardiovascular system (adrenergic receptors α-2A, β-1, β-2, and the catecholamine processing gene COMT) [33, 34].
Previously, most groups have used microarrays using RNA collected at a single time-point, from relatively few subjects to examine thousands of genes to attempt to determine genes whose expression differed in CFS patients from Controls or other disease groups e.g. [22, 35–44]. As opposed to these findings of differences, a recent microarray study using identical twins discordant for chronic fatigue found no gene expression differences in CFS [45]. None of the genes that we assayed in the present study were found in these previous studies to be increased in CFS patients. This is not surprising since the gene expression differences reported here were not observed at baseline, but required moderate exercise to expose them. Additionally, previous exercise studies, using a single post exercise time point would also likely have missed the gene increases demonstrated here. This is because the presence of the α-2A decrease group greatly increases the variance of the overall CFS population. With the small sample sizes used for microarray, it is unlikely that the gene expression increases observed in the α-2A increase group could have been discerned, and even more unlikely that the gene expression decreases in the α-2A decrease group would have been found. Dividing these subgroups greatly decreased the variance in gene expression measures in both groups.
4) FM-only patients are defined by increases in gene expression at baseline
Contrary to expectations, FM patients who did not meet criteria for CFS, that is, without chronically diminished function specifically linked to fatigue, did show evidence of post-exertional malaise reflected as increases in self-reported fatigue and pain measures for 48 hours after moderate exercise. However, the FM patients did not show reliable post-exercise increases in mRNA for any gene under study. Although their average work rate was higher than the CFS patients, FM patients had lower average increase in heart rate than CFS patients. This differential in work rate and heart rate increase may be due to less de-conditioning on average in the FM group vs. the CFS group. However, it is unlikely that this explains the post-exercise gene expression differences between the CFS and FM-only patients because a post hoc analysis using only those CFS patients whose work rates were matched to those of the FM patients indicated that these CFS patients still showed greater increases in mRNA than controls (P < .05), unlike the lack of increases observed in FM patients.
Even though they did not show exercise induced increases in gene expression, FM patients were clearly different from healthy controls and from the CFS patients at baseline. Unlike CFS only or CFS+FM, they showed higher baseline mRNA than controls for 2 sensory ion channel genes, P2X4 and TRPV1, and one cytokine, IL10. Given the results for the CFS+FM patients and the similarity of their post-exertional symptoms, this finding was surprising.
What is not surprising is that the affected genes were P2X4, TRPV1 and IL10. In our mouse experiments, genes that seemed most likely to encode noxious levels of muscle produced metabolites were a combination of ASIC3 (and or ASIC1) P2X4, and TRPV1 [6]. The increases in these receptors on leukocytes could make them more sensitive to muscle produced metabolites, and more likely to produce sensitizing cytokines when muscles are activated (see a recent review of how P2X4 on leukocytes could be involved in inflammatory pain [46]). This could lead to more muscle pain at all times. It is also possible that numbers of these same molecular receptors (P2X4, and TRPV1) are increased in sensory neurons in as well as in leukocytes. If increased in sensory neurons, an increased signal for muscle metabolites would occur at all levels of exercise, leading to widespread increases in muscle pain. As Sluka et al [47] have shown, long-term muscle pain can lead to secondary hyperalgesia in skin throughout the body. This secondary hyperalgesia seems to be mediated by ASIC3 [48]. It is important to note that the higher mRNA for P2X4, TRPV1 and IL10 at baseline in FM-only patients did not diminish after exercise—these increases were simply maintained from the pre- to post-exercise time points.
The increased gene expression in FM patients at baseline that is maintained but not increased after moderate exercise is a novel finding that has not been reported previously. One possible explanation for the increased gene expression at baseline in FM patients is that, although all subjects refrained from formal exercise, their general activity level over 24–48 hours prior to their baseline blood draw was high enough to cause the mRNA increases. It is also possible that the CFS+FM patients had lower general activity levels prior to testing, which may explain why they do not show baseline increases. Although this possibility needs to examined more formally in future studies, comments volunteered by many of the CFS+FM patients are consistent with this possibility, reporting many hours spent totally inactive either seated or in bed each week.
Among the array of immune markers that we used for this gene expression profile, only the anti-inflammatory cytokine IL10 differed between patients and controls. It is very intriguing that IL10 was elevated at baseline in the FM-only group and was more increased throughout the post-exercise period in CFS patients. The findings of enhanced post-exercise anti-inflammatory cytokines response in at least a subset of CFS patients has been reported previously using serum levels, which are more definitive measures [49, 50]. Immune activation may also play an important role in post-exertional worsening of fatigue and pain.
5) Gene expression as Biomarkers for CFS and FM
Previously, we suggested that mRNA expression of some of the genes measured here might be useful as objective, blood based biomarkers for CFS [9]. For the major subgroup reported here (the α-2A increase patients) a combination of PX4, α-2A, B-2, and IL10 at all time points after moderate exercise, sensitivity can be as great as 0.93 with a specificity of 0.77, or specificity can be as great as 0.91 with a sensitivity of 0.77. The AUC of the Receiver Operating Curve was 0.91 with a 95% confidence interval of 0.83–0.98. In either case, accuracy was 0.80. This would be considered a Very Good to Excellent diagnostic tool [51]. Reliable diagnostic values for the α-2A decreasing patients could not be accurately computed because of small sample size (n=14).
We have shown in a recently submitted manuscript that these gene markers are not similarly increased in patients with MS who exhibit unexplained increases in fatigue (White, A.T., Light A.R., Hughen R.W., VanHaitsma T.A., and Light K.C. Differences in metabolite-detecting, adrenergic, and immune gene expression following moderate exercise in multiple sclerosis, chronic fatigue syndrome, and healthy controls, submitted). We also have preliminary data indicating that these genes are not increased before or after exercise in patients with unexplained fatigue who have advanced prostate cancer. Furthermore, the observations from this study on the subset of controls tested while on antidepressants prescribed for mild clinical depression suggest that these genes are not increased in medication-responsive depression; we are currently examining patients with moderate to severe medication-refractory depression to reinforce this tentative finding.
For FM patients, using the 3 genes that were significantly different at baseline weighted to compensate for their quantity (P2X4+15×TRPV1+40×IL10), sensitivity could be as great as 0.89 with a specificity of 0.6, or specificity could be as great as 0.89 with a sensitivity of 0.56. The AUC of the ROC was 0.78, with a 95% confidence interval of 0.662– 0.894, and the accuracy was 0.72. This would be less than ideal as a biomarker for diagnostic purposes, but could still identify the majority of FM patients. This test would have the advantage that a single blood draw could be used, and no exercise test would be required.
It is possible that these gene expression tests could be used in combination with behavioral measures before, during and after moderate exercise to more accurately determine the presence of CFS or FM in patients. Other promising candidate biomarkers have been supported by recent studies including other gene expression measures [43, 52], noradrenergic markers like Neuropeptide Y [53], hypothalamic pituitary-adrenal markers including NR3C1 gene expression and polymorphisms [24, 25, 54], immune markers including natural killer cell activity and cytokines [31, 55] mitochondrial dysfunction markers [32], and proteomic markers in cerebrospinal fluid [56]. It is very plausible that some of these other biomarkers could potentially be combined with the gene expression measures reported here to produce a very accurate diagnostic test, and clearly demark different subgroups within the ME/CFS umbrella.
6) Causes of CFS and FM
Genes for this study were selected because they were directly involved in signaling of fatigue by skeletal muscle. Therefore, they were not selected to determine the primary cause of CFS or FM in the subjects tested. Gene expression changes in the patients tested here (both increases and decreases) could be caused by viral infections. Most previous investigations of humans during viral infections have indicated strong Th1 immune responses involving increases in pro-inflammatory cytokines [57], which we did not observe in most of our CFS or FM patients either at baseline or after exercise. On the contrary, the increase in IL10 mRNA we observed at baseline in FM patients and in CFS patients following exercise is more similar to an enhanced Th2 response, which could potentially lead to greater susceptibility to viral infections and tumors [58]. A very recent study of identical twins discordant for CFS found no unique viruses consistently associated with CFS [59], but did find that 9% of twins with CFS had undiagnosed GB type C virus infections while none of the identical twins without CFS had such infections. Until causal factors are determined, studies like this one are helpful in providing objective biological markers indicating neural and immune pathways that become dysregulated in CFS and FM and that are directly associated with severity of symptoms. These pathways also provide new information that may be used to develop and assess treatments in both disorders.
Acknowledgements
Supported by extramural grants from CFIDS, AFSA, and NIH R21 NS057821 from NINDS and NIAMS, and by intramural grants from the University of Utah.
Footnotes
Conflict of Interest Statement
Dr. Bateman has received income from Eli Lilly, Forest, Jazz, Pfizer and Hemispherx Biopharma as a principal investigator for phase III trials of investigational drugs for FM and CFS. She is also a paid speaker and consultant for Eli Lilly, Forest and Pfizer regarding FDA approved drugs for FM. The other authors have no conflicts of interest.
References
- 1.Fukuda K, Straus SE, Hickie I, Sharpe MC, Dobbins JG, Komaroff A. The chronic fatigue syndrome: a comprehensive approach to its definition and study. International Chronic Fatigue Syndrome Study Group. Ann.Intern.Med. 1994;121:953–959. doi: 10.7326/0003-4819-121-12-199412150-00009. [DOI] [PubMed] [Google Scholar]
- 2.Carruthers BMJ, De Meirleir AK, Peterson KL, Klimas DL, Lerner NG, Bested AM, Flor-Henry AC, Joshi P, Powles P, Sherkey AC, van de Sande JA, Myalgic MI. Encephalomyelitis/Chronic Fatigue Syndrome: Clincal Working Case Definition, Diagnostic and Treatment Protocols: A Consensus Document. Journal of Chronic Fatigue Syndrome. 2003;11:7–115. [Google Scholar]
- 3.Wolfe F, Smythe HA, Yunus MB, et al. The American College of Rheumatology 1990 Criteria for the Classification of Fibromyalgia. Report of the Multicenter Criteria Committee. Arthritis Rheum. 1990;33:160–172. doi: 10.1002/art.1780330203. [DOI] [PubMed] [Google Scholar]
- 4.Wolfe F, Clauw DJ, Fitzcharles MA, et al. The American College of Rheumatology preliminary diagnostic criteria for fibromyalgia and measurement of symptom severity. Arthritis Care Res (Hoboken) 2010;62:600–610. doi: 10.1002/acr.20140. [DOI] [PubMed] [Google Scholar]
- 5.Bigland-Ritchie B, Jones DA, Hosking GP, Edwards RH. Central and peripheral fatigue in sustained maximum voluntary contractions of human quadriceps muscle. Clin.Sci.Mol.Med. 1978;54:609–614. doi: 10.1042/cs0540609. [DOI] [PubMed] [Google Scholar]
- 6.Light AR, Hughen RW, Zhang J, Rainier J, Liu Z, Lee J. Dorsal root ganglion neurons innervating skeletal muscle respond to physiological combinations of protons, ATP, and lactate mediated by ASIC, P2X, and TRPV1. J Neurophysiol. 2008;100:1184–1201. doi: 10.1152/jn.01344.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Light KC, Bragdon EE, Grewen KM, Brownley KA, Girdler SS, Maixner W. Adrenergic Dysregulation and Pain With and Without Acute Beta-blockade in Women with Fibromyalgia and Temporomandibular Disorder. J.Pain. 2009;10:542–552. doi: 10.1016/j.jpain.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Light AR, Vierck CJ, Light KC. Myalgia and Fatigue-Translation from Mouse Sensory Neurons to Fibromyalgia and Chronic Fatigue Syndromes. In: Kruger L, Light A, editors. Translational Pain Research - From Mouse to Man. London, New York: CRC Press, Taylor and Francis Group Boca Raton; 2010. pp. 251–282. [PubMed] [Google Scholar]
- 9.Light AR, White AT, Hughen RW, Light KC. Moderate exercise increases expression for sensory, adrenergic, and immune genes in chronic fatigue syndrome patients but not in normal subjects. J Pain. 2009;10:1099–1112. doi: 10.1016/j.jpain.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reyes M, Nisenbaum R, Hoaglin DC, et al. Prevalence and incidence of chronic fatigue syndrome in Wichita, Kansas. Arch Intern Med. 2003;163:1530–1536. doi: 10.1001/archinte.163.13.1530. [DOI] [PubMed] [Google Scholar]
- 11.Kato K, Sullivan PF, Evengard B, Pedersen NL. Premorbid predictors of chronic fatigue. Arch.Gen.Psychiatry. 2006;63:1267–1272. doi: 10.1001/archpsyc.63.11.1267. [DOI] [PubMed] [Google Scholar]
- 12.Ciccone DS, Natelson BH. Comorbid illness in women with chronic fatigue syndrome: a test of the single syndrome hypothesis. Psychosom.Med. 2003;65:268–275. doi: 10.1097/01.psy.0000033125.08272.a9. [DOI] [PubMed] [Google Scholar]
- 13.Aaron LA, Herrell R, Ashton S, Belcourt M, Goldberg J, Buchwald D. Comorbid clinical conditions in chronic fatigue: a co-twin control study. J Gen.Intern.Med. 2001;16:24–31. doi: 10.1111/j.1525-1497.2001.03419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshiuchi K, Cook DB, Ohashi K, Kumano H, Kuboki T, Yamamoto Y, Natelson BH. A real-time assessment of the effect of exercise in chronic fatigue syndrome. Physiol Behav. 2007;92:963–968. doi: 10.1016/j.physbeh.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cook DB, Nagelkirk PR, Poluri A, Mores J, Natelson BH. The influence of aerobic fitness and fibromyalgia on cardiorespiratory and perceptual responses to exercise in patients with chronic fatigue syndrome. Arthritis Rheum. 2006;54:3351–3362. doi: 10.1002/art.22124. [DOI] [PubMed] [Google Scholar]
- 16.Nijs J, Demol S, Wallman K. Can submaximal exercise variables predict peak exercise performance in women with chronic fatigue syndrome? Arch.Med Res. 2007;38:350–353. doi: 10.1016/j.arcmed.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Hollingsworth KG, Jones DE, Taylor R, Blamire AM, Newton JL. Impaired cardiovascular response to standing in chronic fatigue syndrome. Eur J Clin Invest. 2010;40:608–615. doi: 10.1111/j.1365-2362.2010.02310.x. [DOI] [PubMed] [Google Scholar]
- 18.Jones DE, Hollingsworth KG, Taylor R, Blamire AM, Newton JL. Abnormalities in pH handling by peripheral muscle and potential regulation by the autonomic nervous system in chronic fatigue syndrome. J Intern Med. 2010;267:394–401. doi: 10.1111/j.1365-2796.2009.02160.x. [DOI] [PubMed] [Google Scholar]
- 19.Jones DE, Gray J, Frith J, Newton JL. Fatigue severity remains stable over time and independently associated with orthostatic symptoms in chronic fatigue syndrome: a longitudinal study. J Intern Med. 2010;269:182–188. doi: 10.1111/j.1365-2796.2010.02306.x. [DOI] [PubMed] [Google Scholar]
- 20.Hoad A, Spickett G, Elliott J, Newton J. Postural orthostatic tachycardia syndrome is an under-recognized condition in chronic fatigue syndrome. QJM. 2008;101:961–965. doi: 10.1093/qjmed/hcn123. [DOI] [PubMed] [Google Scholar]
- 21.Yoshiuchi K, Quigley KS, Ohashi K, Yamamoto Y, Natelson BH. Use of time-frequency analysis to investigate temporal patterns of cardiac autonomic response during head-up tilt in chronic fatigue syndrome. Auton.Neurosci. 2004;113:55–62. doi: 10.1016/j.autneu.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Lee E, Cho S, Kim K, Park T. An integrated approach to infer causal associations among gene expression, genotype variation, and disease. Genomics. 2009;94:269–277. doi: 10.1016/j.ygeno.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Lin E, Bayesian A. A Bayesian approach to gene-gene and gene-environment interactions in chronic fatigue syndrome. Pharmacogenomics. 2009;10:35–42. doi: 10.2217/14622416.10.1.35. [DOI] [PubMed] [Google Scholar]
- 24.Rajeevan MS, Smith AK, Dimulescu I, Unger ER, Vernon SD, Heim C, Reeves WC. Glucocorticoid receptor polymorphisms and haplotypes associated with chronic fatigue syndrome. Genes Brain Behav. 2007;6:167–176. doi: 10.1111/j.1601-183X.2006.00244.x. [DOI] [PubMed] [Google Scholar]
- 25.Goertzel BN, Pennachin C, de Souza Coelho L, Gurbaxani B, Maloney EM, Jones JF. Combinations of single nucleotide polymorphisms in neuroendocrine effector and receptor genes predict chronic fatigue syndrome. Pharmacogenomics. 2006;7:475–483. doi: 10.2217/14622416.7.3.475. [DOI] [PubMed] [Google Scholar]
- 26.Smith AK, White PD, Aslakson E, Vollmer-Conna U, Rajeevan MS. Polymorphisms in genes regulating the HPA axis associated with empirically delineated classes of unexplained chronic fatigue. Pharmacogenomics. 2006;7:387–394. doi: 10.2217/14622416.7.3.387. [DOI] [PubMed] [Google Scholar]
- 27.Maes M, Mihaylova I, Bosmans E. Not in the mind of neurasthenic lazybones but in the cell nucleus: patients with chronic fatigue syndrome have increased production of nuclear factor kappa beta. Neuro Endocrinol Lett. 2007;28:456–462. [PubMed] [Google Scholar]
- 28.Kim SY, Jun TW, Lee YS, Na HK, Surh YJ, Song W. Effects of exercise on cyclooxygenase-2 expression and nuclear factor-kappaB DNA binding in human peripheral blood mononuclear cells. Ann N Y Acad Sci. 2009;1171:464–471. doi: 10.1111/j.1749-6632.2009.04915.x. [DOI] [PubMed] [Google Scholar]
- 29.Broderick G, Fuite J, Kreitz A, Vernon SD, Klimas N, Fletcher MA. A formal analysis of cytokine networks in chronic fatigue syndrome. Brain Behav Immun. 2010;24:1209–1217. doi: 10.1016/j.bbi.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorusso L, Mikhaylova SV, Capelli E, Ferrari D, Ngonga GK, Ricevuti G. Immunological aspects of chronic fatigue syndrome. Autoimmun Rev. 2009;8:287–291. doi: 10.1016/j.autrev.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Fletcher MA, Zeng XR, Maher K, et al. Biomarkers in chronic fatigue syndrome: evaluation of natural killer cell function and dipeptidyl peptidase IV/CD26. PLoS One. 2010;5:e10817. doi: 10.1371/journal.pone.0010817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myhill S, Booth NE, McLaren-Howard J. Chronic fatigue syndrome and mitochondrial dysfunction. Int J Clin Exp Med. 2009;2:1–16. [PMC free article] [PubMed] [Google Scholar]
- 33.Streeten DH, Thomas D, Bell DS. The roles of orthostatic hypotension, orthostatic tachycardia, and subnormal erythrocyte volume in the pathogenesis of the chronic fatigue syndrome. Am J Med Sci. 2000;320:1–8. doi: 10.1097/00000441-200007000-00001. [DOI] [PubMed] [Google Scholar]
- 34.Peckerman A, LaManca JJ, Dahl KA, Chemitiganti R, Qureishi B, Natelson BH. Abnormal impedance cardiography predicts symptom severity in chronic fatigue syndrome. Am J Med Sci. 2003;326:55–60. doi: 10.1097/00000441-200308000-00001. [DOI] [PubMed] [Google Scholar]
- 35.Vernon SD, Unger ER, Dimulescu IM, Rajeevan M, Reeves WC. Utility of the blood for gene expression profiling and biomarker discovery in chronic fatigue syndrome. Dis Markers. 2002;18:193–199. doi: 10.1155/2002/892374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whistler T, Unger ER, Nisenbaum R, Vernon SD. Integration of gene expression, clinical, and epidemiologic data to characterize Chronic Fatigue Syndrome. J Transl Med. 2003;1:10. doi: 10.1186/1479-5876-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grans H, Nilsson P, Evengard B. Gene expression profiling in the chronic fatigue syndrome. J Intern.Med. 2005;258:388–390. doi: 10.1111/j.1365-2796.2005.01548.x. [DOI] [PubMed] [Google Scholar]
- 38.Kaushik N, Fear D, Richards SC, et al. Gene expression in peripheral blood mononuclear cells from patients with chronic fatigue syndrome. J Clin Pathol. 2005;58:826–832. doi: 10.1136/jcp.2005.025718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carmel L, Efroni S, White PD, Aslakson E, Vollmer-Conna U, Rajeevan MS. Gene expression profile of empirically delineated classes of unexplained chronic fatigue. Pharmacogenomics. 2006;7:375–386. doi: 10.2217/14622416.7.3.375. [DOI] [PubMed] [Google Scholar]
- 40.Whistler T, Taylor R, Craddock RC, Broderick G, Klimas N, Unger ER. Gene expression correlates of unexplained fatigue. Pharmacogenomics. 2006;7:395–405. doi: 10.2217/14622416.7.3.395. [DOI] [PubMed] [Google Scholar]
- 41.Aspler AL, Bolshin C, Vernon SD, Broderick G. Evidence of inflammatory immune signaling in chronic fatigue syndrome: A pilot study of gene expression in peripheral blood. Behav Brain Funct. 2008;4:44. doi: 10.1186/1744-9081-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kerr JR, Petty R, Burke B, et al. Gene expression subtypes in patients with chronic fatigue syndrome/myalgic encephalomyelitis. J Infect Dis. 2008;197:1171–1184. doi: 10.1086/533453. [DOI] [PubMed] [Google Scholar]
- 43.Saiki T, Kawai T, Morita K, Ohta M, Saito T, Rokutan K, Ban N. Identification of marker genes for differential diagnosis of chronic fatigue syndrome. Mol Med. 2008;14:599–607. doi: 10.2119/2007-00059.Saiki. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gow JW, Hagan S, Herzyk P, Cannon C, Behan PO, Chaudhuri A. A gene signature for post-infectious chronic fatigue syndrome. BMC Med Genomics. 2009;2:38. doi: 10.1186/1755-8794-2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Byrnes A, Jacks A, Dahlman-Wright K, Evengard B, Wright FA, Pedersen NL, Sullivan PF. Gene expression in peripheral blood leukocytes in monozygotic twins discordant for chronic fatigue: no evidence of a biomarker. PLoS One. 2009;4:e5805. doi: 10.1371/journal.pone.0005805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jakobsson PJ. Pain: how macrophages mediate inflammatory pain via ATP signaling. Nat Rev Rheumatol. 2010;6:679–681. doi: 10.1038/nrrheum.2010.175. [DOI] [PubMed] [Google Scholar]
- 47.Radhakrishnan R, Moore SA, Sluka KA. Unilateral carrageenan injection into muscle or joint induces chronic bilateral hyperalgesia in rats. Pain. 2003;104:567–577. doi: 10.1016/s0304-3959(03)00114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sluka KA, Price MP, Breese NM, Stucky CL, Wemmie JA, Welsh MJ. Chronic hyperalgesia induced by repeated acid injections in muscle is abolished by the loss of ASIC3, but not ASIC1. Pain. 2003;106:229–239. doi: 10.1016/S0304-3959(03)00269-0. [DOI] [PubMed] [Google Scholar]
- 49.Borish L, Schmaling K, DiClementi JD, Streib J, Negri J, Jones JF. Chronic fatigue syndrome: identification of distinct subgroups on the basis of allergy and psychologic variables. J Allergy Clin Immunol. 1998;102:222–230. doi: 10.1016/s0091-6749(98)70090-9. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Q, Zhou XD, Denny T, et al. Changes in immune parameters seen in Gulf War veterans but not in civilians with chronic fatigue syndrome. Clin Diagn Lab Immunol. 1999;6:6–13. doi: 10.1128/cdli.6.1.6-13.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ray P, Le Manach Y, Riou B, Houle TT. Statistical Evaluation of a Biomarker. Anesthesiology. 2010 doi: 10.1097/ALN.0b013e3181d47604. [DOI] [PubMed] [Google Scholar]
- 52.Kerr JR, Burke B, Petty R, et al. Seven genomic subtypes of chronic fatigue syndrome/myalgic encephalomyelitis: a detailed analysis of gene networks and clinical phenotypes. J Clin Pathol. 2008;61:730–739. doi: 10.1136/jcp.2007.053553. [DOI] [PubMed] [Google Scholar]
- 53.Fletcher MA, Rosenthal M, Antoni M, et al. Plasma neuropeptide Y: a biomarker for symptom severity in chronic fatigue syndrome. Behav Brain Funct. 6:76. doi: 10.1186/1744-9081-6-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin E, Hsu SY. A Bayesian approach to gene-gene and gene-environment interactions in chronic fatigue syndrome. Pharmacogenomics. 2009;10:35–42. doi: 10.2217/14622416.10.1.35. [DOI] [PubMed] [Google Scholar]
- 55.Fletcher MA, Zeng XR, Barnes Z, Levis S, Klimas NG. Plasma cytokines in women with chronic fatigue syndrome. J Transl Med. 2009;7:96. doi: 10.1186/1479-5876-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schutzer SE, Angel TE, Liu T, et al. Distinct cerebrospinal fluid proteomes differentiate post-treatment lyme disease from chronic fatigue syndrome. PLoS One. 2011;6:e17287. doi: 10.1371/journal.pone.0017287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carpentier A, Conti F, Carriere M, et al. Analysis of gene transcription in sera during chronic hepatitis C infection. J Med Virol. 2009;81:473–480. doi: 10.1002/jmv.21398. [DOI] [PubMed] [Google Scholar]
- 58.O'Garra A, Barrat FJ, Castro AG, Vicari A, Hawrylowicz C. Strategies for use of IL-10 or its antagonists in human disease. Immunol Rev. 2008;223:114–131. doi: 10.1111/j.1600-065X.2008.00635.x. [DOI] [PubMed] [Google Scholar]
- 59.Sullivan PF, Allander T, Lysholm F, et al. An unbiased metagenomic search for infectious agents using monozygotic twins discordant for chronic fatigue. BMC Microbiol. 2011;11:2. doi: 10.1186/1471-2180-11-2. [DOI] [PMC free article] [PubMed] [Google Scholar]