Abstract
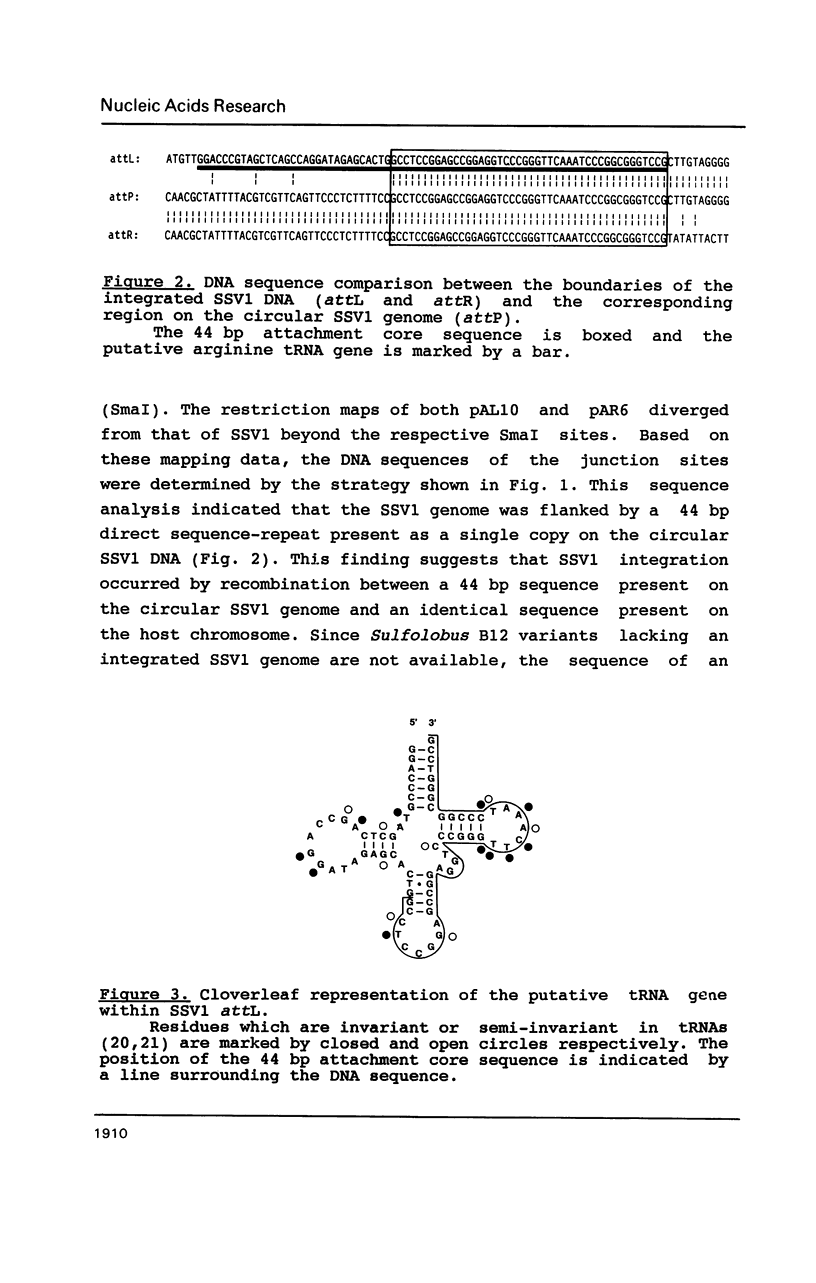
The DNA sequences were determined at the boundaries of the integrated copy of the archaebacterial genetic element SSV1. A 44 bp sequence present as a single copy on the 15.5 kb circular SSV1 DNA flanked the integrated copy as a direct DNA sequence repeat, suggesting that SSV1 integration occurred by recombination between this 44 bp SSV1 sequence and an identical sequence on the bacterial chromosome. At the left attachment site, a region encompassing the 44 bp attachment core sequence and the 31 nucleotides upstream of it displayed all characteristics expected for an arginine tRNA gene. An analysis of published attachment site sequences of other systems revealed that tRNA genes also constitute the bacterial attachment site in the case of three temperate phages and two transmissible plasmids in eubacteria, indicating a widespread occurrence of tRNA genes as integration target sites. This finding may be important for the understanding of mechanisms and evolution of site-specific recombination.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Argos P., Landy A., Abremski K., Egan J. B., Haggard-Ljungquist E., Hoess R. H., Kahn M. L., Kalionis B., Narayana S. V., Pierson L. S., 3rd The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986 Feb;5(2):433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978 Oct;4(2):121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Brody H., Hill C. W. Attachment site of the genetic element e14. J Bacteriol. 1988 May;170(5):2040–2044. doi: 10.1128/jb.170.5.2040-2044.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss D. H., Sprinzl M. Compilation of tRNA sequences. Nucleic Acids Res. 1983 Jan 11;11(1):r1–53. [PMC free article] [PubMed] [Google Scholar]
- Hoess R. H., Ziese M., Sternberg N. P1 site-specific recombination: nucleotide sequence of the recombining sites. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3398–3402. doi: 10.1073/pnas.79.11.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy A., Ross W. Viral integration and excision: structure of the lambda att sites. Science. 1977 Sep 16;197(4309):1147–1160. doi: 10.1126/science.331474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Iandolo J. J. Integration of staphylococcal phage L54a occurs by site-specific recombination: structural analysis of the attachment sites. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5474–5478. doi: 10.1073/pnas.83.15.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Iandolo J. J. Structural analysis of staphylococcal bacteriophage phi 11 attachment sites. J Bacteriol. 1988 May;170(5):2409–2411. doi: 10.1128/jb.170.5.2409-2411.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong J. M., Nunes-Düby S., Lesser C. F., Youderian P., Susskind M. M., Landy A. The phi 80 and P22 attachment sites. Primary structure and interaction with Escherichia coli integration host factor. J Biol Chem. 1985 Apr 10;260(7):4468–4477. [PubMed] [Google Scholar]
- Madon J., Moretti P., Hütter R. Site-specific integration and excision of pMEA100 in Nocardia mediterranei. Mol Gen Genet. 1987 Sep;209(2):257–264. doi: 10.1007/BF00329651. [DOI] [PubMed] [Google Scholar]
- Martin A., Yeats S., Janekovic D., Reiter W. D., Aicher W., Zillig W. SAV 1, a temperate u.v.-inducible DNA virus-like particle from the archaebacterium Sulfolobus acidocaldarius isolate B12. EMBO J. 1984 Sep;3(9):2165–2168. doi: 10.1002/j.1460-2075.1984.tb02107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Omer C. A., Cohen S. N. Structural analysis of plasmid and chromosomal loci involved in site-specific excision and integration of the SLP1 element of Streptomyces coelicolor. J Bacteriol. 1986 Jun;166(3):999–1006. doi: 10.1128/jb.166.3.999-1006.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson L. S., 3rd, Kahn M. L. Integration of satellite bacteriophage P4 in Escherichia coli. DNA sequences of the phage and host regions involved in site-specific recombination. J Mol Biol. 1987 Aug 5;196(3):487–496. doi: 10.1016/0022-2836(87)90026-x. [DOI] [PubMed] [Google Scholar]
- Rich A., RajBhandary U. L. Transfer RNA: molecular structure, sequence, and properties. Annu Rev Biochem. 1976;45:805–860. doi: 10.1146/annurev.bi.45.070176.004105. [DOI] [PubMed] [Google Scholar]
- Ross W., Landy A. Patterns of lambda Int recognition in the regions of strand exchange. Cell. 1983 May;33(1):261–272. doi: 10.1016/0092-8674(83)90355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senecoff J. F., Bruckner R. C., Cox M. M. The FLP recombinase of the yeast 2-micron plasmid: characterization of its recombination site. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7270–7274. doi: 10.1073/pnas.82.21.7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman A. S., Goodman S. D., Scocca J. J. Nucleotide sequences and properties of the sites involved in lysogenic insertion of the bacteriophage HP1c1 genome into the Haemophilus influenzae chromosome. J Bacteriol. 1987 Jan;169(1):238–246. doi: 10.1128/jb.169.1.238-246.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats S., McWilliam P., Zillig W. A plasmid in the archaebacterium Sulfolobus acidocaldarius. EMBO J. 1982;1(9):1035–1038. doi: 10.1002/j.1460-2075.1982.tb01292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


