Abstract
High-risk HPV infection leads to aberrant expression of cellular oncogenic and tumor suppressive miRNAs. A large number of these miRNA genes are downstream targets of the transcription factors c-Myc, p53, and E2F and their expression can therefore be modulated by oncogenic HPV E6 and E7. Cervical cancer represents an unique tumor model for understanding how viral E6 and E7 oncoproteins deregulate the expression of the miR-15/16 cluster, miR-17-92 family, miR-21, miR-23b, miR-34a, and miR-106b/93/25 cluster via the E6–p53 and E7–pRb pathways. Moreover, miRNAs may influence the expression of papillomavirus genes in a differentiation-dependent manner by targeting viral RNA transcripts. Cellular miRNAs affecting HPV DNA replication are of great interest and will be a future focus. We are entering an era focusing on miRNA and noncoding RNA, and the study of HPV and host miRNA interactions will continue to shed more light on our understanding of the HPV life cycle and the mechanistic underpinnings of HPV-induced oncogenesis.
Keywords: human papillomaviruses, microRNAs, oncogenes, tumor suppressor genes, cervical cancer, gene expression
Introduction
Human papillomaviruses (HPVs) are a group of small DNA tumor viruses ~55 nm in diameter. HPVs contain a small, double-stranded circular genome of ~8 kb and encode six viral early proteins (E6, E7, E1, E2, E4, and E5) that have regulatory functions and two viral late proteins (L2 and L1) for viral capsid formation [1]. Infection through sexual intercourse is initiated when a viral particle gains entry into a basal epithelial cell of the cervix. Viral gene expression and multiplication occur exclusively in the nuclei of the infected cells and are tightly linked to the state of differentiation of the cells. Viral early genes are expressed in the undifferentiated basal and parabasal layers, and expression of viral late genes and viral DNA replication occur in the upper spinous and more differentiated granular or cornified layers of the infected cervical epithelium (Fig. 1). Some HPVs, such as HPV16 and HPV18, are associated with oncogenesis and are therefore considered “high risk (HR)”. Viral E6 and E7 of HR HPVs are viral oncoproteins and respectively inactivate p53 and pRB, two major cellular tumor suppressors, thereby contributing to cervical carcinogenesis [2-4].
Fig. 1.
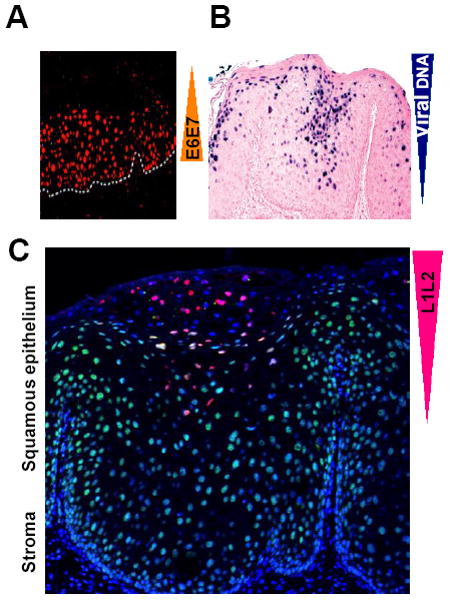
Keratinocyte differentiation–dependent HPV16 life cycle. In general, HPV16 infects the cervical basal cells through microtrauma during sexual intercourse and initiates viral E6 and E7 expression in the infected cells (orange color in A for MCM7 as a surrogate for viral E6 and E7, modified with permission from John Doorbar [104]). Viral DNA replication takes place in the spinous and granular keratinocytes under intermediate or high differentiation (navy blue color in B for viral DNA, modified with permission from Ming Guo[119]). However, viral L1 and L2 (red color for L1 in C) become detectable only in the granular and cornified keratinocytes under terminal differentiation (Jia R and Zheng ZM, unpublished observation) [120].
MicroRNAs (miRNAs) are noncoding regulatory RNAs 18-25 nucleotides in size that are derived from RNA polymerase II (pol II) transcripts of coding or noncoding genes. Many miRNAs are tissue- or differentiation-specific, and their temporal or short-lived expression modulates gene expression at the posttranscriptional level by base-pairing with complementary nucleotide sequences (seed matching) of target mRNAs [5;6]. Depending on the degree of sequence complimentarity, the binding of miRNA(s) to a target mRNA inhibits protein translation, degrades the target mRNA, or both. As of September 2010, the miRBase database (http://www.mirbase.org/) had collected 15,172 entries representing hairpin precursor pre-miRNAs expressing 17,341 mature miRNAs in 143 species. Human genome contains ~416 miRNA genes encoding 1048 distinct mature miRNAs from every chromosome except Y (Fig. 2). Approximately, ~113 miRNA genes encode a cluster of miRNAs and produce ~390 miRNA sequences. Bioinformatics prediction shows that each miRNA targets ~200 RNA transcripts directly or indirectly, and up to one-third of the total number of human mRNAs are targets of more than one miRNA [7;8]. Thus, the actions of miRNAs exert profound effects on gene expression at the posttranscriptional level in almost every biological process. However, miRNA expression itself, similar to any other transcription mediated by pol II, is regulated both at the transcriptional and posttranscriptional levels. Many cellular transcription factors, including c-Myc, p53, and E2F, have been described to regulate miRNA transcription. Other factors involved in miRNA maturation and processing after transcription are Drosha (an RNase-III endonuclease that produces pre-miRNA from pri-miRNA), DGCR8 (DiGeorge syndrome critical region gene 8, a double-stranded RNA-binding protein needed for Drosha activity), exportin 5 (for pre-miRNA export), Dicer (an RNase-III enzyme that produces mature miRNA from pre-miRNA), TRBP (a Dicer partner), and Ago2 (a major component of RISC) [9;10]. Because oncogenic HPV E6 induces degradation of p53 and E7 mediates degradation of pRB to release E2F from the pRB-E2F complex, it is conceivable that oncogenic HPV infection causes aberrant expression of cellular miRNAs.
Fig. 2.
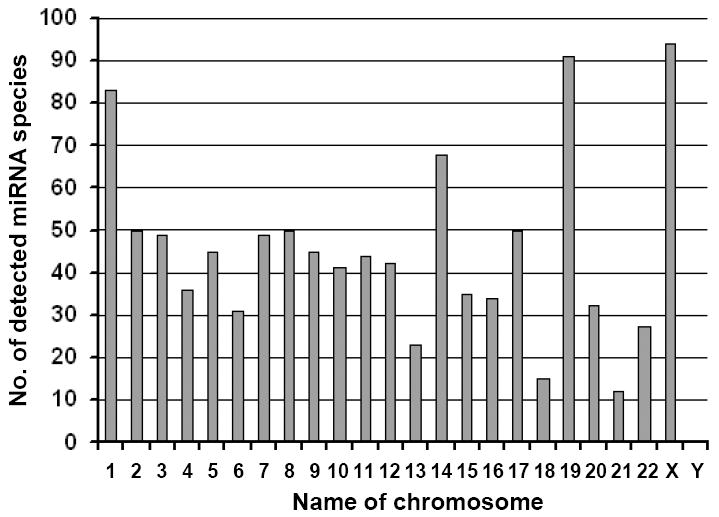
miRNA expression and human chromosomes. Data were obtained from the miRbase sequence database, release 16, September 2010 (http://www.mirbase.org/).
Modulation of cellular miRNA expression by c-Myc, p53, and E2F, which are regulated by oncogenic HPV E6 and E7
High-risk E6 interacts with c-Myc to enhance c-Myc binding to the hTERT (human telomerase reverse transcriptase) promoter and induces hTERT mRNA transcription [11-13]. Viral E7 induces the expression of c-Myc by binding pRB protein, thereby releasing E2F to activate c-Myc [14-16]. High-risk E7 also interacts directly with c-Myc [17]. Myc is a basic helix–loop–helix leucine zipper protein that dimerizes with Max to bind the DNA sequence CACGTG, known as an E box, and thereby activates gene transcription [18;19]. c-Myc is a potent transcriptional regulator of miRNA expression [20;21]. c-Myc induces the expression of the miR-17-92 family on chromosome 13 and of E2F1, but miR-17-5p and miR-20a in the family target the E2F1 3′ UTR to prevent its efficient translation [20;21]. However, c-Myc overexpression widely represses miRNA expression. Chromatin immunoprecipitation reveals that much of this repression is likely to be a direct result of c-Myc binding to miRNA promoters, including those of let-7a-1/f-1/d, miR-15a/16-1, miR-22, miR-26a-2, miR-26b, miR-29a/b-1, miR-29b-2/c, miR-30e/30c-1, miR-34a, and miR-146a [21].
Oncogenic E6 targets p53 for degradation via the E6AP ubiquitin proteolytic pathway [22;23]. The tumor suppressor p53 is a transcription factor that binds to a promoter=s p53 binding site (a palindrome DNA sequence of RRRCWWGYYY [R=A,G;W=A,T;Y=C,T]) [24]. It regulates many cellular miRNAs by increasing the expression of miR-23a, miR-26a, and miR-34a [25] via direct transactivation of these miRNA genes. Other studies show that p53 decreases expression of miRNA clusters, including miR-106b/miR-93/miR-25, miR-17-5p/18a/19a/20a/19b-1/92-1, and miR-106a/18b/20b/19b-2/92-2 [26]. Interestingly, p53 decreases expression of the miRNA clusters by an indirect mechanism through repression of E2F1 [26]. It has been reported that p53 transactivates expression of the BTG3 (B-cell translocation gene 3) gene, which directly binds E2F1 and inhibits its activity [27]. p53-induced miR-34a also targets the E2F1 3′ UTR and inhibits its expression [28]. Moreover, p53 interacts with the Drosha/p68 complex to facilitate Drosha-mediated pri-miRNA processing, consequently promoting expression of miR-15a/16-1, miR-103/107, miR-143/145, miR-203, and miR-206 at the posttranscriptional level [25].
As noted above, E7-mediated degradation of tumor suppressor protein pRB frees E2F from the pRB-E2F complex. The promoter regions of many miRNA genes contain an E2F binding site (TTTSSCGC, S= C or G) [29], and binding of E2F to an E2F binding site in the promoter region transactivates the expression of miRNA genes including miR-17-92, let-7a-d, let-7i, miR-15b/16-2, and miR-106b-25 [20;30;31]. In addition, E2F1 directly activates the transcription of coding genes by binding to their promoter regions, and thus promotes miRNA expression from these transcripts. For example, E2F1 promotes MCM7 expression from chromosome 7q22 and production of miR-106b/93/25 from its intron 13 [32].
miRNA signatures in cervical cancer
Cervical cancer ranks as the most common cancer in women in the developing world, with an estimated global incidence of 493,243 cases and approximately 273,505 deaths per year (www.who.int/hpvcentre). Cervical cancer is the fourth most common type of cancers in women in the U.S., with an estimated 12,200 new cases in 2010, and is the eighth leading cause of cancer mortality in women, accounting for an estimated 4,210 deaths in 2010 [33]. Sexual infection with oncogenic HPVs is widely recognized as a leading cause of cervical, penile, and anal cancers. Among over 120 genotypes isolated from humans [34], oncogenic HPVs, such as HPV16, HPV18, and HPV31, have been detected in up to 99.7% of cervical squamous cell carcinomas and 94%-100% of cervical adeno- and adenosquamous carcinomas [35;36].
Genome-wide profiling of miRNA signatures have indicated that aberrant (increased or decreased) miRNA expression is common in most human tumors [10;37]. Besides regulating cell cycle progression, proliferation/differentiation, apoptosis, and senescence, miRNAs have been linked to cancer etiology, progression, metastasis, and prognosis and could function as oncogenes (e.g., miR-17-92, miR-21, miR155, miR-372/373) or tumor suppressors (e.g., let-7, miR-15a/16-1, miR-34a, miR-143/145) by modulating oncogenic or tumor suppressive pathways, including the Ras, Myc, p53, and pRB pathways. Some miRNAs, such as the miR-221/222 and miR-17-92 clusters, may be oncogenic in one cell or tissue type but tumor suppressive in another, depending on the tissue context and target genes [37]. Moreover, the tumor suppressive miR-200 family (miR-200a, miR-200b, miR-200c, miR-141, and miR-149) and tumor suppressive miR-205 have been identified as controlling cancer metastasis [38-40].
In the course of understanding how HPV16 develops resistance to E7-specific siRNA, in 2004 we isolated and cloned E7-specific siRNAs derived from shRNA precursors [41] and 174 cellular miRNAs from HPV16+ CaSki cells [42]. In combination with Northern blot and miRNA array analyses of 11 cervical cancer cell lines and cervical cancer tissues, as well as of HPV16-induced pre-neoplastic lesions in raft tissues derived from primary human vaginal keratinocytes, we concluded that a substantial number of cellular miRNAs exhibited altered expression due to HPV infection (Table 1) [42]. A recent study by Li et al. [43] independently analyzed six paired normal and cervical cancer tissues by using the same miRNA array platform and showed a very similar result. Increased expression of 12 miRNAs (miR-15b, miR-16, miR-17-5p, miR-20a, miR-20b, miR-21, miR-93, miR-106a, miR-155, miR-182, miR-185, and miR-224) and decreased expression of 9 miRNAs (miR-29a, miR-34a, miR-126, miR-127, miR-145, miR-218, miR-424, miR-450, and miR-455) were found in cervical cancer tissues in both laboratories (Table 1) [42-45]. Among the altered miRNAs reported by both laboratories, miR-126, miR-143/145, miR-155, miR-424/450 were also confirmed by deep sequencing (Table 1)[46]; increased expression of miR-20a, miR-20b, miR-93, and miR-224 and decreased expression of miR-127, miR-143/145, and miR-218 were confirmed in cervical cancer tissues when compared with adjacent normal cervical tissues (2 cm away from the cancerous tissues) using a different miRNA array platform [47]. Other studies with customized miRNA arrays [48-50] or different assay platforms [51] revealed more variable results. However, one study [51] found increased expression of miR-21 in cervical cancer, and two others [48;51] found decreased expression of both miR-143 and miR-145. Together, the data indicate that cervical cancer expresses no or very little of the miR-143/145 cluster. However, reduced miR-143/145 expression is common in other tumor types unrelated to HPV infection [52;53]. The miR-143/145 cluster is a class II miRNA whose expression is unrelated to any E2F factor and is greatly increased during G1 and maintained or further increased in proliferating cells [30]. A recent study indicates that downregulation of the miR-143/145 cluster requires the Ras-responsive element-binding protein (RREB1) to repress its promoter, but K-Ras and RREB1 are themselves targets of miR-143/145, arguing for a feed-forward mechanism that potentiates Ras signaling [54].
Table 1.
Summary of miRNA expression profiling studies in cervical cancer
| miRNA | Chr. Location | Wang et ala | Li et ala | Witten et alb |
|---|---|---|---|---|
| miR-15a | 13q14.2 | Up | ||
| miR-15b | 3q25.33 | Up | Up | |
| miR-16 | 13q14.2 | Up | Up | |
| miR-17-5p | 13q31.3 | Up | Up | |
| miR-20a | 13q31.3 | Up | Up | |
| miR-20b | Xq26.2 | Up | Up | |
| miR-21 | 17q23.1 | Up | Up | miR-21* Up |
| miR-93 | 7q22.1 | Up | Up | |
| miR-106a | Xq26.2 | Up | Up | |
| miR-146a | 5q34 | Up | ||
| miR-148a | 7p15.2 | Up | ||
| miR-155 | 21q21.3 | Up | Up | Up |
| miR-181c | 19p13.13 | Up | ||
| miR-182 | 7q32.2 | Up | Up | |
| miR-183 | 7q32.2 | Up | miR-183* Up | |
| miR-185 | 22q11.21 | Up | Up | |
| miR-223 | Xq12 | Up | ||
| miR-224 | Xq28 | Up | Up | |
| miR-324-5p | 17p13.1 | Up | ||
| miR-10b | 2q31.1 | Down | Down | |
| miR-29a | 7q32.3 | Down | Down | |
| miR-30b | 8q24.22 | Down | ||
| miR-34ac | 1p36.22 | Down | Down | |
| miR-125a | 19q13.41 | Down | ||
| miR-125b | 11q24.1 | Down | Down | |
| miR-126 | 9q34.3 | Down | Down | Down |
| miR-127 | 14q32.2 | Down | Down | |
| miR-133a | 18q11.2 | Down | ||
| miR-133b | 6p12.2 | Down | ||
| miR-143 | 5q32 | Down | Down | |
| miR-145 | 5q32 | Down | Down | Down |
| miR-191* | 3p21.31 | Down | ||
| miR-218 | 4p15.31;5q34 | Down | Down | |
| miR-378(422b) | 5q32 | Down | ||
| miR-422a | 15q22.31 | Down | ||
| miR-424 | Xq26.3 | Down | Down | Down |
| miR-450 | Xq26.3 | Down | Down | Down |
| miR-455 | 9q32 | Down | Down | |
| miR-574 | 4p14 | Down |
Using a PCR-based miRNA assay to analyze 102 cervical cancer samples, Hu et al. identified miR-200a and miR-9 as two promising miRNAs that could be used to predict cervical cancer survival [50]. However, in other studies neither miR-9 nor miR-200a appeared to be expressed at significantly different levels in cervical cancer tissues than in age-matched normal cervical tissues [42;43;46]. In one report, the altered miR-200a expression showed no correlation with cervical cancer invasion and metastasis [47]. Thus, further independent analyses are needed to validate the reported observation [50].
HPV E6 regulates the expression of miR-23b, miR-34a, and miR-218
The major function of oncogenic HPV E6 is to target p53 for degradation. Structural and functional analyses of HPV16 E6 indicate that an E6 F47R mutant is defective for polyubiquitination and degradation of p53 [55;56]. As a transcription factor, p53 plays an important role in the transcription of numerous coding and noncoding genes [25;57;58]. Conceivably, oncogenic HPV E6 is capable of regulating the expression of many cellular miRNAs via p53. We initially observed this in our laboratory when we were investigating how high-risk HPV infection leads to reduction of miR-34a expression [44]. Because the miR-34a gene is a direct transcriptional target of p53, and its expression can be transactivated by the binding of p53 to a consensus p53 binding site in the miR-34a promoter region [59-61], we and others found that E6-mediated degradation of p53, and not E7-mediated reduction of pRB, leads to the reduction of miR-34a that is mediated by high-risk HPV infection in raft cultures, cervical intraepithelial neoplasia (CIN), and cervical cancer tissues. Conversely, knockdown of viral E6 expression in HPV16+ and HPV18+ cervical cancer cell lines by siRNAs increases the expression of p53 and also of miR-34a [44;45]. A direct link between p53 and miR-34a expression was convincingly achieved in C33A cells, an HPV-negative cervical cancer cell line expressing a mutant p53 and producing no miR-34a. Ectopic expression of wild-type p53 in C33A cells induces production of miR-34a [44]. Because miR-34a affects the expression of cell cycle regulators, including cyclin E2, cyclin D1, CDK4, CDK6, E2F1, E2F3, E2F5, Bcl-2, SIRT1, and p18Ink4c [28;59-65], viral E6–mediated reduction of p53 and miR-34a quickly relieves the multi-step controls on cell cycle progression, senescence, and apoptosis, resulting in cell proliferation and transformation (Fig. 3). However, recent studies indicate that p53-independent upregulation of miR-34a can be triggered in cells undergoing terminal differentiation [44] or senescence [66]. In contrast, cancer cells may inactivate miR-34a expression by aberrant CpG methylation [67] independent of HPV E6.
Fig. 3.
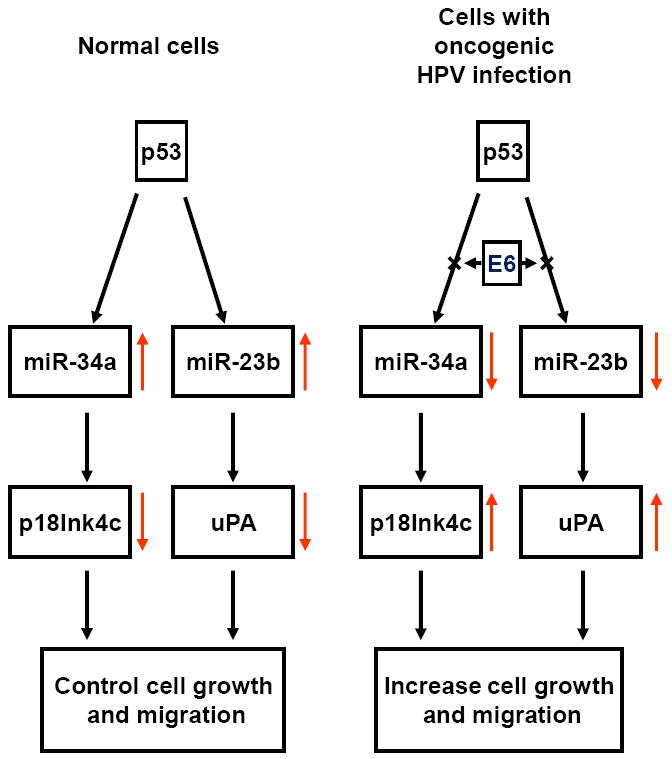
Oncogenic HPV regulates p53 expression and its downstream targets miR-34a and miR-23b. Arrows in color indicate expression level (increase or decrease).
Viral E6 also regulates expression of miR-218 and miR-23b. The reduction of miR-218 by oncogenic HPV E6 leads to increased LAMB3 (Laminin subunit beta-3) production and appears to be unrelated to p53, because miR-218 is highly expressed in the HPV-negative C33A cells containing a mutant p53 gene and producing no miR-34a [68]. Although how E6 reduces miR-218 expression remains unknown, HPV16 E6 reduction of miR-23b has been linked to p53. Au Yeung and colleagues found that downregulation of miR-23b expression by HPV16 E6 results in an increased expression of urokinase-type plasminogen activator (uPA) [69;70] and is related to E6-mediated p53 reduction [71]. Similar to miR-34a, the gene encoding miR-23b on chromosome 9 contains a promoter region with a p53 binding site [71], and in the presence of p53, expresses miR-23b as a cluster of miR-23b/27b/24-1. The data provide miR-23b as another example that p53 mediates the HPV16 E6 downregulation of cellular miRNAs (Fig. 3).
HPV E7 regulates the expression of miR-15a/miR-16-1 and miR-203
Tumor suppressive miR-15a and miR-16-1 control cell proliferation, survival, and invasion [72-74] and are expressed as an miRNA cluster from an intron region of the DLEU2 (deleted in lymphocytic leukemia 2) transcript (Fig. 4). The gene encoding the miR-15a/16-1 cluster is a noncoding DLEU2 gene and is positioned in the antisense orientation in the 13q14.3 locus, which is frequently deleted in chronic lymphocytic leukemia (CLL) [75;76]. In mouse, a deletion of DLEU2/miR-15a/16-1 from the 14qC3 region, a conserved region equivalent to human 13q14, also accelerates the proliferation of mouse B cells by modulating the expression of genes controlling cell-cycle progression, further defining the role of 13q14 deletions in the pathogenesis of CLL [77]. Although both miR-15a and miR-16-1 are abundantly expressed in normal tissues, studies of cancer-related miRNA signatures indicate that the miR-15a/16-1 cluster is downregulated in multiple types of human cancer [78]. However, our initial study using miRNA array analyses showed higher levels of miR-15a and miR-16-1 expression in cervical cancer tissues than in normal cervical tissue [42]. Despite increased expression in cervical cancer, miR-15a and miR-16-1 appear not to function efficiently in controlling the growth of cervical cancer cells.
Fig. 4.

Production of the miR-15a/16-1 cluster from a DLEU-2-001 transcript containing 4 exons (boxes) and 3 introns (lines). Numbers below exons and introns indicate the size in nucleotides.
The underlines are miRNA stem-loop regions, with the miR-15a and miR-16-1 sequences in color.
To investigate whether increased expression of the miR-15a/16-1 cluster in cervical cancer tissues is related to high-risk HPV infection, we used Northern blotting to compare the expression levels of miR-16-1 in raft tissues derived from human foreskin keratinocytes with and without HPV18 infection and observed a more than two-fold increase of miR-16-1 expression in the rafts with HPV18 infection over the expression in control rafts without HPV18 infection (Fig. 5A). When rafts infected with a retrovirus expressing HPV18 E6, E7, or E6E7 were examined, the production of miR-16-1 was increased only in the rafts expressing viral E7, not viral E6 (Fig. 5B), indicating that viral E7 is responsible for the increased expression of miR-16-1. The reverse was true in cervical cancer cell lines with decreased expression of viral E7: by knocking down viral E7 expression with RNAi in HPV16+ CaSki cells or HPV18+ HeLa cells, we demonstrated decreased expression of miR-16-1 in the cells treated with an E7-specific siRNA compared to the control cells treated with a non-specific siRNA (Fig. 5C). Together, these data lead to the conclusion that viral E7 regulates miR-15/16 cluster expression in cells with HPV16 or HPV18 infection. The expression of miR-15a/16-1 is controlled by binding of c-Myc, c-Myb, or PPAR (peroxisome proliferator-activated receptor δ) to the DLEU2 promoter region to positively (c-Myb and PPAR) or negatively (c-Myc) regulate DLEU2 transcription [21;79;80]. E2F1 transactivates c-Myb expression [81;82] but represses c-Myc expression [83]. Given these relationships, it is understandable that the increased expression of miR-15a and miR-16-1 in cervical cancer tissues could be attributed to viral E7–mediated degradation of the tumor suppressor pRB. Viral E7–mediated degradation of pRB [4] frees E2F from the pRB-E2F complex to interact with the promoter regions of c-Myb and c-Myc, consequently regulating DLEU2 transcription and promoting miR-15a/16-1 cluster expression. In addition to transcriptional regulation of DLEU2 gene expression, however, posttranscriptional regulation may also play an important role in the expression of miR-15a and miR-16-1. Because miR-15a and miR-16-1 are derived from the intron 3 region of the DLEU2-001 transcript (Fig. 4), it remains to be understood how RNA splicing contributes to the Drosha-dependent production of miR-15a and miR-16-1 from the DLEU2-001 transcripts during high-risk HPV infection of cervical tissues.
Fig. 5.
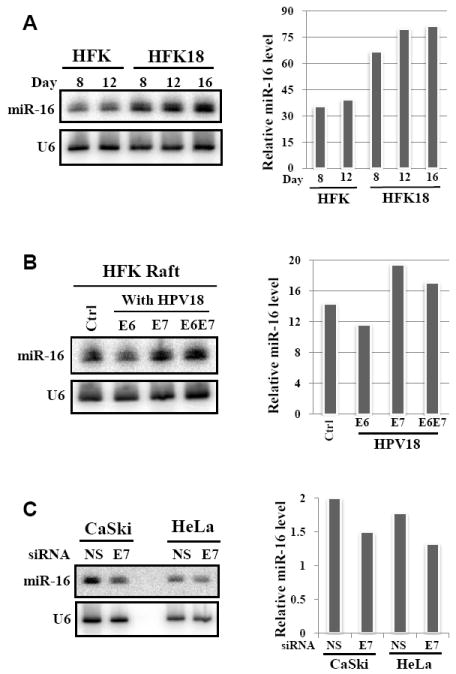
Oncogenic HPV infection increases miR-16 expression through viral E7. Northern blotting was used to detect miR-16 from total RNA (40 μg) isolated from day-8 to day-16 raft tissues derived from human foreskin keratinocytes with (HFK18) or without (HFK) HPV18 infection (A), or from day-10 rafts with or without HPV18 E6, E7, or E6E7 retrovirus infection (B). Northern blotting was also conducted to detect miR-16 expression in CaSki cells treated for 48 h with HPV16 E7–specific siRNA198 (40 nM) or in HeLa cells treated for 48 h with HPV18 E7-specific siRNA220 (40 nM) [121]). A nonspecific (NS) siRNA at the same dose was used as a control for both cells (C). U6 RNA was probed as internal loading control. Bar graphs on the right show relative miR-16 levels detected by Northern blot in each sample from a representative gel on the left after being normalized to U6 for sample loading.
High-risk HPV E7 in human keratinocytes downregulates the expression of miR-203 [84] which controls the shift of keratinocytes in differentiating epithelia from a proliferative state to a nonproliferative state by repressing stemness [85;86]. Although primarily expressed in superbasal layers of stratified epithelia, miR-203 expression can be induced in vitro in primary keratinocytes in parallel with differentiation. In normal human foreskin keratinocytes with stable expression of HPV31 episomes or HPV31 E6 or E7, differentiation-dependent miR-203 expression is severely blocked only by viral E7, with a corresponding increase in expression of ΔNp63, a member of the p53 family that is highly expressed in proliferative undifferentiated basal keratinocytes, but poorly expressed in differentiated nonproliferative cells [86;87]. Viral E7 perhaps blocks the MAPK/PKC pathway–dependent activation of miR-203 expression [84]. Other studies have shown that the primary role of miR-203 is to inhibit the proliferative capacity of epithelial cells upon differentiation by targeting the 3′ UTR regions of ΔNp63 [86;87].
Cellular miRNAs regulate HPV gene expression and replication
Oncogenic HPVs regulate the expression of many cellular miRNAs; conversely, it is now emerging that cellular miRNAs modulate the expression of HPV genes. As every viral gene transcript derived from the HPV genome is in a bicistronic or polycistronic form, with a long 5′ UTR or 3′ UTR, we have hypothesized that HPVs express their genes under the control of multiple cellular miRNAs [1]. In fact, ectopic expression of any given HPV gene has been a challenge; cervical tissues with high-risk HPV infection express very little viral protein, which in general practice is scarcely detectable. Codon optimization to remove “the suppressive nucleotides” (presumably the nucleotides in miRNA seed matches) of a given viral transcript was introduced to enhance the expression of viral genes and has been a common practice in papillomavirology. We recently analyzed the genome-wide distribution of miRNA binding sites of ~450 hsa-miRNAs (homo sapiens miRNAs) in the HPV6, HPV11, HPV16, and HPV18 genomes and identified several dozen potential miRNA binding sites in each HPV gene transcript. Our preliminary results indicate that expression of both HPV early and late genes is subject to miRNA-mediated regulation at the post-transcriptional level in cervical cancer cell lines and in reporter assays (unpublished observations).
Interestingly, Melar-New and Laimins [84] recently found that a differentiation-dependent miR-203 increase in CIN612 cells containing HPV31 episomes promotes viral genome amplification in the short term, but a high level of miR-203 expression interferes with viral genome amplification in the long term [84]. Although the mechanism by which miR-203 is involved in viral DNA replication remains to be investigated, this study provides the first evidence that cellular miRNAs may also play an important role in regulation of viral gene expression and DNA replication. In a separate study, HPV16 DNA replication was suppressed by ectopic miR-125b, presumably through the sequence homology of HPV16 L2 and miR-125b. Conversely, reduced miR-125b expression was found in cervical pre-cancerous lesions during early HPV infection and may also be associated with viral DNA replication [88]. Together, the current data are not yet sufficient to conclude that HPV gene expression is under massive regulation by cellular miRNAs, but this presents a valuable direction for understanding HPV biology and pathogenesis.
Conclusions and Remarks
Cervical cancer, like many other cancer types [10;37], displays notably increased or decreased expression of a large number of cellular oncogenic or tumor suppressive miRNAs (Table 1). Because tumor suppressive miRNAs with decreased expression are more numerous in cervical cancer tissues than oncogenic miRNAs with increased expression (Table 2), the altered miRNA expression appears unlikely to be attributable to Drosha overexpression from the chromosome 5p gain in cervical cancer [89;90]. It is clear that oncogenic HPVs, despite producing no viral miRNAs [42;91], are responsible for the aberrant expression of oncogenic or tumor suppressive miRNAs. Although these observations shed more light on the mechanistic underpinnings of HPV-induced oncogenesis (Fig. 6), whether the altered miRNA expression would sensitize the HPV genome to integration or infected cells to unrestricted proliferation remains to be discovered.
Table 2.
Alterations of cancer-associated microRNAs in cervical cancer
| Oncogenic miRNAs | Tumor suppressive miRNAs |
|---|---|
| miR17-92 family | let-7 family |
| miR-21 | miR-1 |
| miR-26a | miR-9 |
| miR-146a | miR-15a/16-1 cluster |
| miR-155 | miR-26a |
| miR-205 | miR-29 |
| miR-221/222 cluster | miR-34a/b/c |
| miR-372/373 cluster | miR-101 |
| miR-124a | |
| miR-127 | |
| miR-128 | |
| miR-143/145 cluster | |
| miR-200/141 family | |
| miR-203 | |
| miR-205 | |
| miR-206/133b cluster | |
| miR-223 |
Fig. 6.
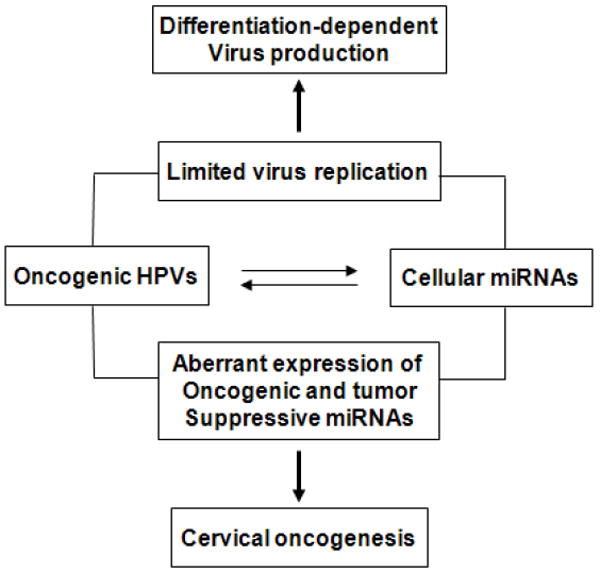
Outcomes of interactions between oncogenic HPVs and cellular miRNAs. These intimate interplays may lead to limited virus gene expression or uncontrolled cell proliferation by altering miRNA expression.
To date, mounting evidence indicates that HPV regulation of cellular miRNA expression most likely occurs through viral E6 and E7, although other viral protein(s) might be involved. In this regard, E6 degradation of p53 contributes to reduced expression of miR-34a [42;45] and miR-23b [71] at the transcriptional level. However, the mechanism by which E6 reduces miR-218 expression and how E7 regulates the expression of miR-203 and the miR-15/16 cluster remain elusive. High-risk E6 and E7 interact separately with several dozens or even hundreds of cellular factors [92-94], and these interactions could lead to increased or decreased expression of cellular miRNAs. Some of the altered miRNA expression could be a result from both E6 and E7. The matter is to discover what these miRNAs are and how E6 and E7 regulate them.
Increased miR-21 expression in cervical cancer was noticed in several studies [42;43;51] and may be attributable to both E6 and E7. Expression of miR-21 in human and mouse cells depends on STAT3 and p65 NF-κB binding to the miR-21 promoter region [95;96]. Consistent with this, cervical cancer tissues display an increased amount of STAT3 [97-99] and high-risk HPV E6 and E7 increase p65 NF-κB expression [100]. Moreover, miR-21 targets the PTEN tumor suppressor, also leading to increased NF-κB activity [96] and inhibits negative regulators of the Ras/MeK/ERK pathway to block apoptosis [101], feeding forward to viral E6 and E7 functions. Together, these reports help to solve a long-standing puzzle of how E7 could operate with activated Ras to fully transform primary rat cells [102;103].
Considering that each miRNA subtly affects hundreds of different gene transcripts for protein expression, HPV transcripts could be targeted in turn by these altered miRNAs in a miRNA-directed negative feedback loop. Notably, inefficient translation of HPV16 transcripts has been thought to be due to rare codon usage in the viral open reading frames; therefore, codon optimization, which may disrupt the miRNA binding sites, has been introduced to improve the production of various HPV16 proteins [1]. Thus, future studies should elucidate the effects of such miRNAs on HPV protein expression (Fig. 6). The altered miRNAs may also deregulate downstream targets of viral E6 or E7. For example, high-risk E6 and E7 increase MCM7 expression through E2F1-dependent and -independent pathways [104;105]. Production of the MCM7 transcripts that produce the miR-106b/93/25 cluster decreases along with the increase of p53 expression, because p53 induces transcriptional repression of E2F1 [26]. Because viral E6 degrades p53 and E7 interacts with pRB to release E2F1 from the pRB-E2F complex, increased expression of the miR-106b/93/25 cluster is expected in cervical cancer tissues with high-risk HPV infection. However, both miR-106b and miR-93 target and negatively regulate the expression of p21 and E2F1 [32]. The finding of miR-93 upregulation in cervical cancer tissues in three separate studies [42;43;47] indicates that the circuit favors the transactivation of MCM7 expression and miR-106b/93/25 cluster production by E2F1 due to viral E6 and E7.
Although the viral oncoproteins E6 and E7 play important roles in regulating cellular miRNA expression, their function can be reduced due to epigenetic modification of miRNA genes. Because miRNA genes are pol II genes [106], their transcription is driven by a pol II promoter, similar to all other eukaryotic coding genes. Therefore, aberrant CpG methylation and histone modification, which affect expression of coding genes, will also influence the expression of miRNA genes [107-109] including those under regulation by viral E6 and/or E7. DNA methylation–mediated silencing of miR-34a expression has been reported in multiple tumor types [67]. CpG island hypermethylation is a hallmark of cervical cancer [110-112]. Consistent with this, a recent report indicates that miR-124 reduction in cervical cancer correlates with a high frequency of DNA methylation in the miR-124 locus [113].
There is no doubt that biomedicine is entering an era of miRNAs and non-coding RNAs. The widespread research in laboratories and clinical settings today relies heavily on microarray screening and deep sequencing of biological samples and qRT-PCR. As different miRNA array platforms are now commercially available, with variable qualities and sensitivities [114;115], special care should be taken when making a conclusion based on array data from a single platform. Dreher and colleagues have compared three different miRNA array platforms (Affymetrix, Invitrogen, and Exiqon) in profiling hsa-miRNA expression from HaCaT cells transfected with the full-length HPV-11 genome and found inconsistent results among the three platforms. They also show that TaqMan qRT-PCR validation is of limited use for less abundant miRNAs [116], which argues that qRT-PCR is not a “gold-standard” for miRNA validation [114] and cannot be used to replace the Northern blotting method [117]. Despite these technical issues, the study of miRNAs shows great promise and will be a prospective focus in coming years to further understand HPV life cycle and pathogenesis.
Research Highlights.
Oncogenic HPV infection leads to development of cervical cancer and deregulates the expression of oncogenic and tumor suppresive miRNAs via E6-p53 and E7-pRb pathways. Cellular miRNAs may also influence the expression of papillomavirus genes in a differentiation-dependent manner by targeting viral RNA transcripts. Cervical cancer provides a unique cancer model for understanding the interplays between HPV oncogenes and host miRNAs and their roles in cervical carcinogenesis.
Acknowledgments
We thank Louise Chow, Hsu-Kun Wang, and Nilam Banerjee for providing the raft tissues used in the experiments shown in Fig. 5A and 5B. We also thank John Doorbar and Ming Guo for their permission to use the modified images shown in Fig. 1A and 1B. This work was supported by the intramural research program of the National Institutes of Health, the National Cancer Institute, and the Center for Cancer Research.
Abbreviations
- HPV
human papillomavirus
- miRNA
microRNA
- RISC
RNA-induced silencing complex
- hTERT
human telomerase reverse transcriptase
- BTG3
B-cell translocation gene 3
- CIN
cervical intraepithelial neoplasia
- uPA
urokinase-type plasminogen activator
- DLEU2
deleted in lymphocytic leukemia 2
- CLL
chronic lymphocytic leukemia
- HFK
human foreskin keratinocytes
- RREB1
Ras-responsive element-binding protein 1
- PPAR
peroxisome proliferator-activated receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zheng ZM, Baker CC. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front Biosci. 2006;11:2286–2302. doi: 10.2741/1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 3.Dyson N, Howley PM, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez SL, Stremlau M, He X, Basile JR, Munger K. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J Virol. 2001;75:7583–7591. doi: 10.1128/JVI.75.16.7583-7591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 6.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 8.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen BR. Viruses and microRNAs. Nat Genet. 2006;38(Suppl):S25–S30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 10.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McMurray HR, McCance DJ. Human papillomavirus type 16 E6 activates TERT gene transcription through induction of c-Myc and release of USF-mediated repression. J Virol. 2003;77:9852–9861. doi: 10.1128/JVI.77.18.9852-9861.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veldman T, Liu X, Yuan H, Schlegel R. Human papillomavirus E6 and Myc proteins associate in vivo and bind to and cooperatively activate the telomerase reverse transcriptase promoter. Proc Natl Acad Sci USA. 2003;100:8211–8216. doi: 10.1073/pnas.1435900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, Dakic A, Chen R, Disbrow GL, Zhang Y, Dai Y, Schlegel R. Cell-restricted immortalization by human papillomavirus correlates with telomerase activation and engagement of the hTERT promoter by Myc. J Virol. 2008;82:11568–11576. doi: 10.1128/JVI.01318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gewin L, Galloway DA. E box-dependent activation of telomerase by human papillomavirus type 16 E6 does not require induction of c-myc. J Virol. 2001;75:7198–7201. doi: 10.1128/JVI.75.15.7198-7201.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh ST, Kyo S, Laimins LA. Telomerase activation by human papillomavirus type 16 E6 protein: induction of human telomerase reverse transcriptase expression through Myc and GC-rich Sp1 binding sites. J Virol. 2001;75:5559–5566. doi: 10.1128/JVI.75.12.5559-5566.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al Moustafa AE, Foulkes WD, Benlimame N, Wong A, Yen L, Bergeron J, Batist G, Alpert L, Alaoui-Jamali MA. E6/E7 proteins of HPV type 16 and ErbB-2 cooperate to induce neoplastic transformation of primary normal oral epithelial cells. Oncogene. 2004;23:350–358. doi: 10.1038/sj.onc.1207148. [DOI] [PubMed] [Google Scholar]
- 17.Wang YW, Chang HS, Lin CH, Yu WC. HPV-18 E7 conjugates to c-Myc and mediates its transcriptional activity. Int J Biochem Cell Biol. 2007;39:402–412. doi: 10.1016/j.biocel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 19.Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, Fu Y, Weng Z, Kuznetsov VA, Sung WK, Ruan Y, Dang CV, Wei CL. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci USA. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 21.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huibregtse JM, Scheffner M, Howley PM. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol Cell Biol. 1993;13:4918–4927. doi: 10.1128/mcb.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 24.Beno I, Rosenthal K, Levitine M, Shaulov L, Haran TE. Sequence-dependent cooperative binding of p53 to DNA targets and its relationship to the structural properties of the DNA targets. Nucleic Acids Res. 2011;39:1919–1932. doi: 10.1093/nar/gkq1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 26.Brosh R, Shalgi R, Liran A, Landan G, Korotayev K, Nguyen GH, Enerly E, Johnsen H, Buganim Y, Solomon H, Goldstein I, Madar S, Goldfinger N, Borresen-Dale AL, Ginsberg D, Harris CC, Pilpel Y, Oren M, Rotter V. p53-Repressed miRNAs are involved with E2F in a feed-forward loop promoting proliferation. Mol Syst Biol. 2008;4:229. doi: 10.1038/msb.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ou YH, Chung PH, Hsu FF, Sun TP, Chang WY, Shieh SY. The candidate tumor suppressor BTG3 is a transcriptional target of p53 that inhibits E2F1. EMBO J. 2007;26:3968–3980. doi: 10.1038/sj.emboj.7601825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao Y, Kassatly RF, Cress WD, Horowitz JM. Subunit composition determines E2F DNA-binding site specificity. Mol Cell Biol. 1997;17:6994–7007. doi: 10.1128/mcb.17.12.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bueno MJ, Gomez dC, Laresgoiti U, Fernandez-Piqueras J, Zubiaga AM, Malumbres M. Multiple E2F-induced microRNAs prevent replicative stress in response to mitogenic signaling. Mol Cell Biol. 2010;30:2983–2995. doi: 10.1128/MCB.01372-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 32.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de M, Iliopoulos ID, Pilozzi E, Liu CG, Negrini M, Cavazzini L, Volinia S, Alder H, Ruco LP, Baldassarre G, Croce CM, Vecchione A. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 33.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 34.Bernard HU, Burk RD, Chen Z, van Doorslaer K, Hausen H, de Villiers EM. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401:70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clifford G, Franceschi S, Diaz M, Munoz N, Villa LL. Chapter 3: HPV type-distribution in women with and without cervical neoplastic diseases. Vaccine. 2006;24(Suppl 3):S3-26–S3/34. doi: 10.1016/j.vaccine.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 36.Parkin DM, Bray F. Chapter 2: The burden of HPV-related cancers. Vaccine. 2006;24(Suppl 3):S3-11–S3/25. doi: 10.1016/j.vaccine.2006.05.111. [DOI] [PubMed] [Google Scholar]
- 37.Di Leva G, Croce CM. Roles of small RNAs in tumor formation. Trends Mol Med. 2010;16:257–267. doi: 10.1016/j.molmed.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 39.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang S, Tao M, McCoy JP, Zheng ZM. Short-term induction and long-term suppression of HPV16 oncogene silencing by RNA interference in cervical cancer cells. Oncogene. 2006;25:2094–2104. doi: 10.1038/sj.onc.1209244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, Zheng ZM. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PloS ONE. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Y, Wang F, Xu J, Ye F, Shen Y, Zhou J, Lu W, Wan X, Ma D, Xie X. Progressive miRNA expression profiles in cervical carcinogenesis and identification of HPV related target genes for miR-29. J Pathol. 2011 doi: 10.1002/path.2873. In press. [DOI] [PubMed] [Google Scholar]
- 44.Wang X, Wang H-K, McCoy JP, Banerjee NS, Rader JS, Broker TR, Meyers C, Chow LT, Zheng ZM. Oncogenic HPV infection interrupts the expression of tumor-suppressive miR-34a through viral oncoprotein E6. RNA. 2009;15:637–647. doi: 10.1261/rna.1442309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li B, Hu Y, Ye F, Li Y, Lv W, Xie X. Reduced miR-34a expression in normal cervical tissues and cervical lesions with high-risk human papillomavirus infection. Int J Gynecol Cancer. 2010;20:597–604. doi: 10.1111/IGC.0b013e3181d63170. [DOI] [PubMed] [Google Scholar]
- 46.Witten D, Tibshirani R, Gu SG, Fire A, Lui WO. Ultra-high throughput sequencing-based small RNA discovery and discrete statistical biomarker analysis in a collection of cervical tumours and matched controls. BMC Biol. 2010;8:58. doi: 10.1186/1741-7007-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rao Q, Zhou H, Peng Y, Li J, Lin Z. Aberrant microRNA expression in human cervical carcinomas. Med Oncol. 2011 doi: 10.1007/s12032-011-9830-2. In press. [DOI] [PubMed] [Google Scholar]
- 48.Pereira PM, Marques JP, Soares AR, Carreto L, Santos MA. MicroRNA expression variability in human cervical tissues. PloS ONE. 2010;5:e11780. doi: 10.1371/journal.pone.0011780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JW, Choi CH, Choi JJ, Park YA, Kim SJ, Hwang SY, Kim WY, Kim TJ, Lee JH, Kim BG, Bae DS. Altered MicroRNA expression in cervical carcinomas. Clin Cancer Res. 2008;14:2535–2542. doi: 10.1158/1078-0432.CCR-07-1231. [DOI] [PubMed] [Google Scholar]
- 50.Hu X, Schwarz JK, Lewis JS, Jr, Huettner PC, Rader JS, Deasy JO, Grigsby PW, Wang X. A microRNA expression signature for cervical cancer prognosis. Cancer Res. 2010;70:1441–1448. doi: 10.1158/0008-5472.CAN-09-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67:6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- 52.Michael MZ, O’Connor SM, Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 53.Akao Y, Nakagawa Y, Kitade Y, Kinoshita T, Naoe T. Downregulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci. 2007;98:1914–1920. doi: 10.1111/j.1349-7006.2007.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kent OA, Chivukula RR, Mullendore M, Wentzel EA, Feldmann G, Lee KH, Liu S, Leach SD, Maitra A, Mendell JT. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev. 2010;24:2754–2759. doi: 10.1101/gad.1950610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nomine Y, Masson M, Charbonnier S, Zanier K, Ristriani T, Deryckere F, Sibler AP, Desplancq D, Atkinson RA, Weiss E, Orfanoudakis G, Kieffer B, Trave G. Structural and functional analysis of E6 oncoprotein: insights in the molecular pathways of human papillomavirus-mediated pathogenesis. Mol Cell. 2006;21:665–678. doi: 10.1016/j.molcel.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 56.Ristriani T, Fournane S, Orfanoudakis G, Trave G, Masson M. A single-codon mutation converts HPV16 E6 oncoprotein into a potential tumor suppressor, which induces p53-dependent senescence of HPV-positive HeLa cervical cancer cells. Oncogene. 2009;28:762–772. doi: 10.1038/onc.2008.422. [DOI] [PubMed] [Google Scholar]
- 57.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 59.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 62.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582:1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- 64.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, MacDougald OA, Cho KR, Fearon ER. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 65.Wang X, Meyers C, Guo M, Zheng ZM. Up-regulation of p18Ink4c expression by oncogenic HPV E6 via p53-miR-34a pathway. Int J Cancer. 2010 doi: 10.1002/ijc.25800. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Christoffersen NR, Shalgi R, Frankel LB, Leucci E, Lees M, Klausen M, Pilpel Y, Nielsen FC, Oren M, Lund AH. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2010;17:236–245. doi: 10.1038/cdd.2009.109. [DOI] [PubMed] [Google Scholar]
- 67.Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Korner H, Knyazev P, Diebold J, Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 68.Martinez I, Gardiner AS, Board KF, Monzon FA, Edwards RP, Khan SA. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene. 2008;27:2575–2582. doi: 10.1038/sj.onc.1210919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Turner MA, Palefsky JM. Urokinase plasminogen activator expression by primary and HPV 16-transformed keratinocytes. Clin Exp Metastasis. 1995;13:260–268. doi: 10.1007/BF00133481. [DOI] [PubMed] [Google Scholar]
- 70.Riethdorf L, Riethdorf S, Petersen S, Bauer M, Herbst H, Janicke F, Loning T. Urokinase gene expression indicates early invasive growth in squamous cell lesions of the uterine cervix. J Pathol. 1999;189:245–250. doi: 10.1002/(SICI)1096-9896(199910)189:2<245::AID-PATH427>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 71.Au Yeung CL, Tsang TY, Yau PL, Kwok TT. Human papillomavirus type 16 E6 induces cervical cancer cell migration through the p53/microRNA-23b/urokinase-type plasminogen activator pathway. Oncogene. 2011 doi: 10.1038/onc.2010.613. In press. [DOI] [PubMed] [Google Scholar]
- 72.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D’Urso L, Pagliuca A, Biffoni M, Labbaye C, Bartucci M, Muto G, Peschle C, De Maria R. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 73.Bandi N, Zbinden S, Gugger M, Arnold M, Kocher V, Hasan L, Kappeler A, Brunner T, Vassella E. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69:5553–5559. doi: 10.1158/0008-5472.CAN-08-4277. [DOI] [PubMed] [Google Scholar]
- 74.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lerner M, Harada M, Loven J, Castro J, Davis Z, Oscier D, Henriksson M, Sangfelt O, Grander D, Corcoran MM. DLEU2, frequently deleted in malignancy, functions as a critical host gene of the cell cycle inhibitory microRNAs miR-15a and miR-16-1. Exp Cell Res. 2009;315:2941–2952. doi: 10.1016/j.yexcr.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 77.Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, Dalla-Favera R. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 78.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17:215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 79.Zhao H, Kalota A, Jin S, Gewirtz AM. The c-myb proto-oncogene and microRNA-15a comprise an active autoregulatory feedback loop in human hematopoietic cells. Blood. 2009;113:505–516. doi: 10.1182/blood-2008-01-136218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yin KJ, Deng Z, Hamblin M, Xiang Y, Huang H, Zhang J, Jiang X, Wang Y, Chen YE. Peroxisome proliferator-activated receptor delta regulation of miR-15a in ischemia-induced cerebral vascular endothelial injury. J Neurosci. 2010;30:6398–6408. doi: 10.1523/JNEUROSCI.0780-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sala A, Nicolaides NC, Engelhard A, Bellon T, Lawe DC, Arnold A, Grana X, Giordano A, Calabretta B. Correlation between E2F-1 requirement in the S phase and E2F-1 transactivation of cell cycle-related genes in human cells. Cancer Res. 1994;54:1402–1406. [PubMed] [Google Scholar]
- 82.Campanero MR, Armstrong M, Flemington E. Distinct cellular factors regulate the c-myb promoter through its E2F element. Mol Cell Biol. 1999;19:8442–8450. doi: 10.1128/mcb.19.12.8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sahin F, Sladek TL. E2F-1 has dual roles depending on the cell cycle. Int J Biol Sci. 2010;6:116–128. doi: 10.7150/ijbs.6.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Melar-New M, Laimins LA. Human papillomaviruses modulate expression of microRNA 203 upon epithelial differentiation to control levels of p63 proteins. J Virol. 2010;84:5212–5221. doi: 10.1128/JVI.00078-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lena AM, Shalom-Feuerstein R, Rivetti d, Aberdam VD, Knight RA, Melino G, Candi E. miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 2008;15:1187–1195. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- 87.Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nuovo GJ, Wu X, Volinia S, Yan F, Di Leva G, Chin N, Nicol AF, Jiang J, Otterson G, Schmittgen TD, Croce C. Strong inverse correlation between microRNA-125b and human papillomavirus DNA in productive infection. Diagn Mol Pathol. 2010;19:135–143. doi: 10.1097/PDM.0b013e3181c4daaa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Scotto L, Narayan G, Nandula SV, Subramaniyam S, Kaufmann AM, Wright JD, Pothuri B, Mansukhani M, chneider AS, Arias-Pulido H, Murty VV. Integrative genomics analysis of chromosome 5p gain in cervical cancer reveals target over-expressed genes, including Drosha. Mol Cancer. 2008;7:58. doi: 10.1186/1476-4598-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Muralidhar B, Goldstein LD, Ng G, Winder DM, Palmer RD, Gooding EL, Barbosa-Morais NL, Mukherjee G, Thorne NP, Roberts I, Pett MR, Coleman N. Global microRNA profiles in cervical squamous cell carcinoma depend on Drosha expression levels. J Pathol. 2007;212:368–377. doi: 10.1002/path.2179. [DOI] [PubMed] [Google Scholar]
- 91.Cai X, Li G, Laimins LA, Cullen BR. Human papillomavirus genotype 31 does not express detectable microRNA levels during latent or productive virus replication. J Virol. 2006;80:10890–10893. doi: 10.1128/JVI.01175-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zheng ZM. Viral oncogenes, noncoding RNAs, and RNA splicing in human tumor viruses. Int J Biol Sci. 2010;6:730–755. doi: 10.7150/ijbs.6.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Narisawa-Saito M, Kiyono T. Basic mechanisms of high-risk human papillomavirus-induced carcinogenesis: roles of E6 and E7 proteins. Cancer Sci. 2007;98:1505–1511. doi: 10.1111/j.1349-7006.2007.00546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McLaughlin-Drubin ME, Munger K. The human papillomavirus E7 oncoprotein. Virology. 2009;384:335–344. doi: 10.1016/j.virol.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang CH, Yue J, Fan M, Pfeffer LM. IFN induces miR-21 through a signal transducer and activator of transcription 3-dependent pathway as a suppressive negative feedback on IFN-induced apoptosis. Cancer Res. 2010;70:8108–8116. doi: 10.1158/0008-5472.CAN-10-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang SF, Yuan SS, Yeh YT, Wu MT, Su JH, Hung SC, Chai CY. The role of p-STAT3 (ser727) revealed by its association with Ki-67 in cervical intraepithelial neoplasia. Gynecol Oncol. 2005;98:446–452. doi: 10.1016/j.ygyno.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 98.Sobti RC, Singh N, Hussain S, Suri V, Bharti AC, Das BC. Overexpression of STAT3 in HPV-mediated cervical cancer in a north Indian population. Mol Cell Biochem. 2009;330:193–199. doi: 10.1007/s11010-009-0133-2. [DOI] [PubMed] [Google Scholar]
- 99.Shukla S, Shishodia G, Mahata S, Hedau S, Pandey A, Bhambhani S, Batra S, Basir SF, Das BC, Bharti AC. Aberrant expression and constitutive activation of STAT3 in cervical carcinogenesis: implications in high-risk human papillomavirus infection. Mol Cancer. 2010;9:282. doi: 10.1186/1476-4598-9-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nees M, Geoghegan JM, Hyman T, Frank S, Miller L, Woodworth CD. Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-kappaB-responsive genes in cervical keratinocytes. J Virol. 2001;75:4283–4296. doi: 10.1128/JVI.75.9.4283-4296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, Olson EN. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Matlashewski G, Schneider J, Banks L, Jones N, Murray A, Crawford L. Human papillomavirus type 16 DNA cooperates with activated ras in transforming primary cells. EMBO J. 1987;6:1741–1746. doi: 10.1002/j.1460-2075.1987.tb02426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Phelps WC, Yee CL, Munger K, Howley PM. The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell. 1988;53:539–547. doi: 10.1016/0092-8674(88)90570-3. [DOI] [PubMed] [Google Scholar]
- 104.Middleton K, Peh W, Southern S, Griffin H, Sotlar K, Nakahara T, El Sherif A, Morris L, Seth R, Hibma M, Jenkins D, Lambert P, Coleman N, Doorbar J. Organization of human papillomavirus productive cycle during neoplastic progression provides a basis for selection of diagnostic markers. J Virol. 2003;77:10186–10201. doi: 10.1128/JVI.77.19.10186-10201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shai A, Brake T, Somoza C, Lambert PF. The human papillomavirus E6 oncogene dysregulates the cell cycle and contributes to cervical carcinogenesis through two independent activities. Cancer Res. 2007;67:1626–1635. doi: 10.1158/0008-5472.CAN-06-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lujambio A, Portela A, Liz J, Melo SA, Rossi S, Spizzo R, Croce CM, Calin GA, Esteller M. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29:6390–6401. doi: 10.1038/onc.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J, Prosper F, Garcia-Foncillas J. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125:2737–2743. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- 109.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 110.Badal V, Chuang LS, Tan EH, Badal S, Villa LL, Wheeler CM, Li BF, Bernard HU. CpG methylation of human papillomavirus type 16 DNA in cervical cancer cell lines and in clinical specimens: genomic hypomethylation correlates with carcinogenic progression. J Virol. 2003;77:6227–6234. doi: 10.1128/JVI.77.11.6227-6234.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wentzensen N, Sherman ME, Schiffman M, Wang M. Utility of methylation markers in cervical cancer early detection: appraisal of the state-of-the-science. Gynecol Oncol. 2009;112:293–299. doi: 10.1016/j.ygyno.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang TH, Lai HC, Liu HW, Lin CJ, Wang KH, Ding DC, Chu TY. Quantitative analysis of methylation status of the PAX1 gene for detection of cervical cancer. Int J Gynecol Cancer. 2010;20:513–519. doi: 10.1111/IGC.0b013e3181c7fe6e. [DOI] [PubMed] [Google Scholar]
- 113.Wilting SM, van Boerdonk RA, Henken FE, Meijer CJ, Diosdado B, Meijer GA, le Sage C, Agami R, Snijders PJ, Steenbergen RD. Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol Cancer. 2010;9:167. doi: 10.1186/1476-4598-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Git A, Dvinge H, Salmon-Divon M, Osborne M, Kutter C, Hadfield J, Bertone P, Caldas C. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA. 2010;16:991–1006. doi: 10.1261/rna.1947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sato F, Tsuchiya S, Terasawa K, Tsujimoto G. Intra-platform repeatability and inter-platform comparability of microRNA microarray technology. PloS ONE. 2009;4:e5540. doi: 10.1371/journal.pone.0005540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dreher A, Rossing M, Kaczkowski B, Nielsen FC, Norrild B. Differential expression of cellular microRNAs in HPV-11 transfected cells. An analysis by three different array platforms and qRT-PCR. Biochem Biophys Res Commun. 2010;403:357–362. doi: 10.1016/j.bbrc.2010.11.035. [DOI] [PubMed] [Google Scholar]
- 117.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, Matzke M, Ruvkun G, Tuschl T. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Friedman JM, Jones PA. MicroRNAs: critical mediators of differentiation, development and disease. Swiss Med Wkly. 2009;139:466–472. doi: 10.4414/smw.2009.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Guo M, Gong Y, Deavers M, Silva EG, Jan YJ, Cogdell DE, Luthra R, Lin E, Lai HC, Zhang W, Sneige N. Evaluation of a commercialized in situ hybridization assay for detecting human papillomavirus DNA in tissue specimens from patients with cervical intraepithelial neoplasia and cervical carcinoma. J Clin Microbiol. 2008;46:274–280. doi: 10.1128/JCM.01299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jia R, Liu X, Tao M, Kruhlak M, Guo M, Meyers C, Baker CC, Zheng ZM. Control of the papillomavirus early-to-late switch by differentially expressed SRp20. J Virol. 2009;83:167–180. doi: 10.1128/JVI.01719-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tang S, Tao M, McCoy JP, Jr, Zheng ZM. The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J Virol. 2006;80:4249–4263. doi: 10.1128/JVI.80.9.4249-4263.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]


