Abstract
While thousands of large intergenic non-coding RNAs (lincRNAs) have been identified in mammals, few have been functionally characterized, leading to debate about their biological role. To address this, we performed loss-of-function studies on most lincRNAs expressed in mouse embryonic stem cells (ESC) and characterized the effects on gene expression. Here we show that knockdown of lincRNAs has major consequences on gene expression patterns, comparable to knockdown of well-known ESC regulators. Notably, lincRNAs primarily affect gene expression in trans. Knockdown of dozens of lincRNAs causes either exit from the pluripotent state or upregulation of lineage commitment programs. We integrate lincRNAs into the molecular circuitry of ESCs and show that lincRNA genes are regulated by key transcription factors and that lincRNA transcripts bind to multiple chromatin regulatory proteins to affect shared gene expression programs. Together, the results demonstrate that lincRNAs have key roles in the circuitry controlling ESC state.
INTRODUCTION
The mammalian genome encodes many thousands of large non-coding transcripts1 including a class of ~3500 large intergenic ncRNAs (lincRNAs) identified using a chromatin signature of actively transcribed genes2–4. These lincRNA genes have been shown to have interesting properties, including clear evolutionary conservation2–5, expression patterns correlated with various cellular processes2,6 and binding of key transcription factors to their promoters2,6, and the lincRNAs themselves physically associate with chromatin regulatory proteins4,7. Yet, it remains unclear whether the RNA transcripts themselves have biological functions8–10. Few have been demonstrated to have phenotypic consequences by loss-of-function experiments6. As a result, the functional role of lincRNA genes has been widely debated. Various proposals include that lincRNA genes act as enhancer regions, with the RNA transcript simply being an incidental by-product8,9, that lincRNA transcripts act in cis to activate transcription11, and that lincRNA transcripts can act in trans to repress transcription12,13.
We therefore sought to undertake systematic loss-of-function experiments on all lincRNAs known to be expressed in mouse embryonic stem cells (ESCs)2,3. ESCs are pluripotent cells that can self-renew in culture and can give rise to cells of any of the three primary germ layers including the germline14. The signalling14, transcriptional15–17, and chromatin15,18–21 regulatory networks controlling pluripotency have been well characterized providing an ideal system to determine how lincRNAs may integrate into these processes.
Here we show that knockdown of the vast majority of ESC-expressed lincRNAs has a strong effect on gene expression patterns in ESCs, of comparable magnitude to that seen for the well-known ESC regulatory proteins. We identify dozens of lincRNAs that upon loss-of-function cause an exit from the pluripotent state and dozens of additional lincRNAs that, while not essential for the maintenance of pluripotency, act to repress lineage-specific gene expression programs in ESCs. We integrate the lincRNAs into the molecular circuitry of ESCs by demonstrating that most lincRNAs are directly regulated by critical pluripotency-associated transcription factors and ~30% of lincRNAs physically interact with specific chromatin regulatory proteins to affect gene expression. Together, these results demonstrate a regulatory network in ESCs whereby transcription factors directly regulate the expression of lincRNA genes, many of which can physically interact with chromatin proteins, affect gene expression programs, and maintain the ESC state.
RESULTS
lincRNAs affect global gene expression
To perform loss-of-function experiments, we generated five lentiviral-based short hairpin RNAs (shRNAs)22 targeting each of the 226 lincRNAs previously identified in ESCs2,3 (see Methods, Supplemental Table 1). These shRNAs successfully targeted 147 lincRNAs and reduced their expression by an average of ~75% compared to endogenous levels in ESCs (see Methods, Figure 1a, Supplemental Figure 1, Supplemental Table 2). As positive controls, we generated shRNAs targeting ~50 genes encoding regulatory proteins, including both transcription and chromatin factors that have been shown to play critical roles in ESC regulation17,20,23; validated hairpins were obtained against 40 of these genes (Supplemental Table 2). As negative controls, we performed independent infections with lentiviruses containing 27 different shRNAs with no known cellular target RNA.
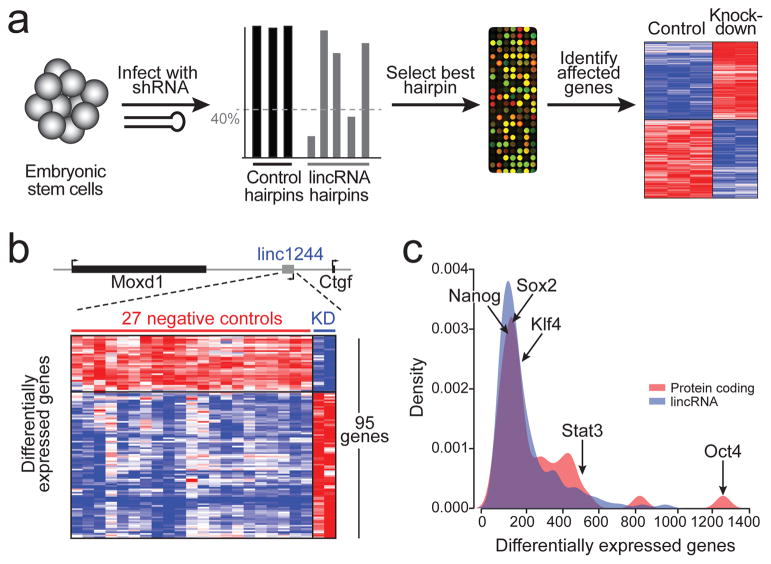
Figure 1. Functional effects of lincRNAs.
(a) A schematic of lincRNA perturbation experiments. ESCs are infected with shRNAs, knockdown level is computed, the best hairpin is selected and profiled on expression arrays, and differential gene expression is computed relative to negative control hairpins. (b) Example of a lincRNA knockdown. Top: Genomic locus containing the lincRNA. Bottom: Heatmap of the 95 genes affected by knockdown of the lincRNA, expression for control hairpins (red line) and expression for lincRNA hairpins (blue line) are shown. (c) Distribution of number of affected genes upon knockdown of 147 lincRNAs (blue) and 40 well-known ESC regulatory proteins (red). Points corresponding to five specific ESC regulatory proteins are marked.
We infected each shRNA into ESCs, isolated RNA after 4 days, and profiled their effects on global transcription by hybridization to genome-wide microarrays (Figure 1a, see Methods). We employed a stringent procedure to control for non-specific effects due to viral infection, generic RNAi responses, or ‘off-target’ effects. Expression changes were deemed significant only if they exceeded the maximum levels observed in any of the negative controls, showed a two-fold change in expression compared to the negative controls, and had a low false discovery rate (FDR) assessed across all genes based on permutation tests (Figure 1b, see Methods). This approach controls for the overall rate of non-specific effects by estimating the number and magnitude of observed effects in the negative control hairpins, where all effects are non-specific.
For 137 of the 147 lincRNAs (93%), knockdown caused a significant impact on gene expression (Supplemental Table 3), with an average of 175 protein-coding transcripts affected (range: 20–936) (Figure 1c, Supplemental Figure 2, Supplemental Table 4). These results were similar to those obtained upon knockdown of the 40 well-studied ESC regulatory proteins: 38 (95%) showed significant effects on gene expression, with an average of 207 genes affected (range: 28 (for DNMT3L) to 1187 (for Oct4)) (Figure 1c, Supplemental Figure 2, Supplemental Table 4). Although some individual lincRNAs have been found to lead primarily to gene repression12,13, we find that knockdown of the lincRNAs studied here largely led to comparable numbers of activated and repressed genes (Supplemental Figure 2, Supplemental Table 4). To further assess ‘off-target’ effects, we also profiled the effects of the second-best validated shRNA targeting 10 randomly selected lincRNA genes. In all cases, second shRNAs against the same target produced significantly similar expression changes (see Methods, Supplemental Table 5). These results indicate that the vast majority of lincRNAs have functional consequences on overall gene expression of comparable magnitude (in terms of number of affected genes and impact on levels) to the known transcriptional regulators in ESCs.
lincRNAs affect gene expression in trans
Following the observation that a few lincRNAs act in cis 24,25, some recent papers have claimed that most lincRNAs act primarily in cis8,11,26. We found no evidence to support this latter notion: knockdown of only 2 lincRNAs showed effects on a neighbouring gene, only 13 showed effects within a window of ten genes on either side, and only 8 showed effects on genes within 300 kb; these proportions are no greater than observed for protein-coding genes (Supplemental Figure 3, Supplemental Table 6). In short, lincRNAs appear to affect expression largely in trans.
Our results contrast with a recent study that concluded that lincRNAs act in cis, based on the observation that knockdown of 7 out of 12 lincRNAs affected expression of a gene within 300 kb11. The explanation appears to be that the threshold in the previous study failed to account for multiple hypothesis testing within the local region. Accounting for this, the effects on neighbouring genes are no greater than expected by chance and are consistent with our observations here (see Methods).
While some lincRNAs can regulate gene expression in cis11,24,25, determining the precise proportion of cis regulators requires more direct experimental approaches. We note that our results are consistent with observed correlations between lincRNAs and neighbouring genes2,26, which may represent shared upstream regulation2,12 or local transcriptional effects10,27. In addition, the lincRNAs studied here should be distinguished from transcripts that are produced at enhancer sites8,9, the function of which has yet to be determined.
lincRNAs maintain the pluripotent state
We next sought to investigate whether lincRNAs play a role in regulating the ESC state. Regulation of the ESC state involves two components, maintaining the pluripotency program and repressing differentiation programs15. To determine whether lincRNAs play a role in the maintenance of the pluripotency program, we studied their effects on the expression of Nanog, a key transcription factor that is required to establish28 and uniquely marks the pluripotent state29,30. We infected ESCs carrying a luciferase reporter gene expressed from the endogenous Nanog promoter31 with shRNAs targeting lincRNAs or protein-coding genes. We monitored loss of reporter activity after 8 days relative to 25 negative control hairpins across biological replicates (see Methods). To ensure that the observed effects were not simply due to a reduction in cell viability, we excluded shRNAs that caused a reduction in cell numbers (see Methods, Supplemental Figure 4, Supplemental Table 7). Altogether, we identified 26 lincRNAs that had major effects on endogenous Nanog levels with many at comparable levels to the knockdown of the known protein-coding regulators of pluripotency such as Oct4 and Nanog (Figure 2a, Supplemental Table 7). This establishes that these lincRNAs have a role in maintaining the pluripotent state.
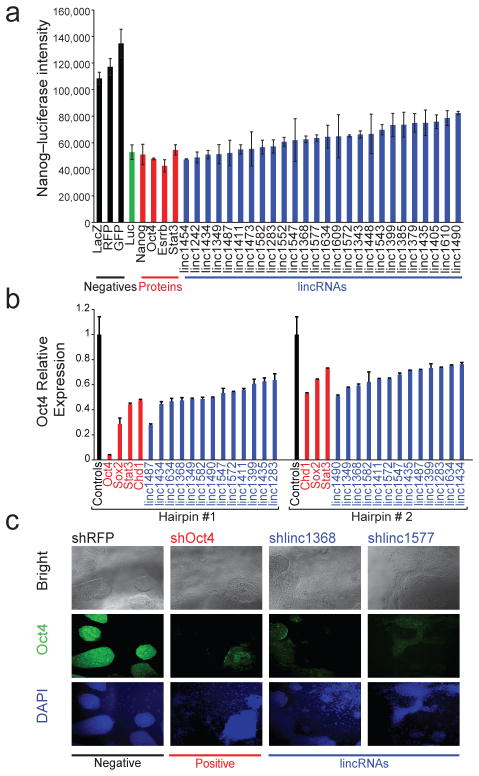
Figure 2. lincRNAs are critical for the maintenance of pluripotency.
(a) Activity from a Nanog promoter driving luciferase, following treatment with control hairpins (black) or hairpins targeting luciferase (green), selected protein-coding regulators (red), and lincRNAs (blue). (b) Relative mRNA expression levels of Oct4 following knockdown of selected protein-coding (red) and lincRNA (blue) genes affecting Nanog–luciferase levels. The best hairpin (black line) and second best hairpin (grey line) are shown. All knockdowns are significant with a p-value<0.01. Error bars represent standard error (n=4). (c) Morphology of ESCs and immunofluorescence staining of Oct4 for a negative control hairpin (black line), and hairpins targeting Oct4 (red line), and two lincRNAs (blue line). The first row shows bright field images, the second row shows immunofluorescence staining of the Oct4 protein, and the third row shows DAPI staining of the nuclei.
To further validate the role of these 26 lincRNAs in regulating the pluripotent state, we knocked down these lincRNAs in wild-type ESCs and measured mRNA levels of pluripotency marker genes Oct4, Sox2, Nanog, Klf4, and Zfp42 after 8 days. In all cases we observe a significant reduction in the expression of multiple pluripotency markers with >90% showing a significant decrease in both Oct4 and Nanog levels (Supplemental Figure 5, Supplemental Tables 8,9). To control for ‘off-target’ effects, we studied additional hairpins targeting these lincRNAs. For 15 lincRNAs we had an effective second hairpin. In all 15 cases, the second hairpin produced comparable reductions in Oct4 expression levels, showing that the observations were not due to ‘off-target’ effects (Figure 2b, Supplemental Table 10). Notably, >90% of lincRNA knockdowns affecting Nanog reporter levels led to loss of ESC morphology (Figure 2c, Supplemental Figures 6, 7). Thus, inhibition of these 26 lincRNAs lead to an increased exit from the pluripotent state.
lincRNAs repress lineage programs
To determine if lincRNAs act in repressing differentiation programs we compared the overall gene expression patterns resulting from knockdown of the lincRNAs to published gene expression patterns resulting from induced differentiation of ESCs32,33 and assessed significance using a permutation-derived FDR34 (see Methods). These states include differentiation into endoderm, ectoderm, neuroectoderm, mesoderm, and trophectoderm lineages. As a positive control for our analytical method, we confirmed the expected results that the expression pattern caused by Oct4 knockdown was strongly associated with the trophoectoderm lineage35 and the pattern caused by Nanog knockdown was strongly associated with endoderm differentiation30 (Figure 3a).
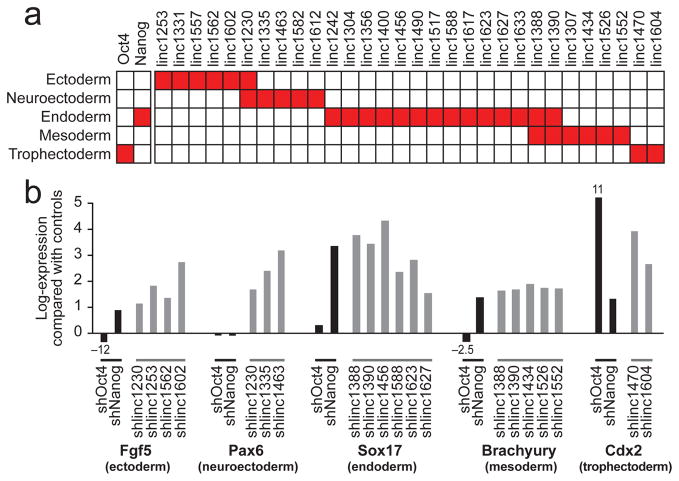
Figure 3. lincRNAs repress specific differentiation lineages.
(a) Expression changes for each lincRNA compared to gene expression of five differentiation patterns. Each box shows significant positive association (red, FDR<0.01) for Oct4 and Nanog (left) and for lincRNAs (right). (b) Expression changes upon knockdown of Oct4 and Nanog (black bars) and representative lincRNAs (grey bars) for five lineage marker genes. The expression changes (FDR<0.05) are displayed on a log scale as the t-statistic compared to a panel of negative control hairpins.
Using this approach, we identified 30 lincRNAs whose knockdown produced expression patterns similar to differentiation into specific lineages (Supplemental Table 11). Amongst these lincRNAs, 13 are associated with endoderm differentiation, 7 with ectoderm differentiation, 5 with neuroectoderm differentiation, 7 with mesoderm differentiation, and 2 with the trophectoderm lineage (Figure 3a). Consistent with these functional assignments, we observe that the majority (>85%) of the 30 lincRNAs associated with specific differentiation lineages showed upregulation of the well-known marker genes for the identified states17,32 upon knockdown (such as Sox17 (endoderm), Fgf5 (ectoderm), Pax6 (neuroectoderm), Brachyury (mesoderm), and Cdx2 (trophectoderm)) (Figure 3b, Supplemental Figures 8, 9, Supplemental Tables 12, 13).
The fact that knockdown of these 30 lincRNAs induces gene expression programs associated with specific early differentiation lineages suggests that these lincRNAs normally are a barrier to such differentiation. Interestingly, most of the lincRNA knockdowns (~85%) that induce gene expression patterns associated with these lineages did not cause the cells to differentiate as determined by Nanog reporter levels (Supplemental Table 7) and Oct4 expression (Supplemental Figure 10). This is consistent with observations for several critical ESC chromatin regulators, such as the polycomb complex; loss-of-function of these regulators similarly induce lineage-specific markers without causing differentiation18,36,37.
Together, these data indicate that many lincRNAs play important roles in regulating the ESC state, including maintaining the pluripotent state and repressing specific differentiation lineages.
lincRNAs are direct targets of ESC TFs
Having demonstrated a functional role for lincRNAs in ESCs, we sought to integrate the lincRNAs into the molecular circuitry controlling the pluripotent state. First, we explored how lincRNA expression is regulated in ESCs. Toward this end, we utilized published genome-wide maps of 9 pluripotency-associated transcription factors (TFs)16,38 and determined whether they bind to the promoters of lincRNA genes. Of the 226 lincRNA promoters ~75% are bound by at least one of 9 pluripotency-associated TFs (including Oct4, Sox2, Nanog, cMyc, nMyc, Klf4, Zfx, Smad, and Tcf3) with a median of 3 factors bound to each promoter (Figure 4a, Supplemental Figure 11, Supplemental Table 14), comparable to the proportion reported for protein-coding genes16. Interestingly, the 3 core factors (Oct4, Sox2, and Nanog) bind to the promoters of ~12% of all ESC lincRNAs and ~50% of lincRNAs involved in the regulation of the pluripotent state.
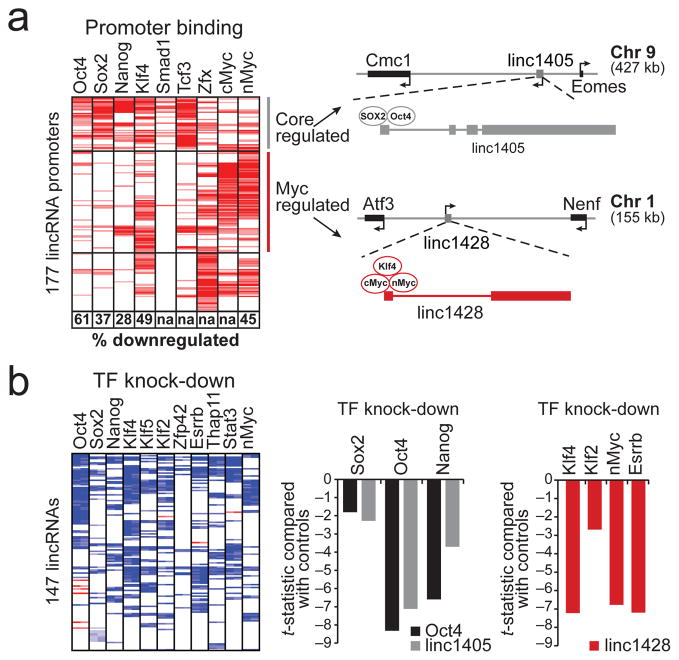
Figure 4. lincRNAs are direct regulatory targets of the ESC transcriptional circuitry.
(a) A heatmap representing ChIP-Seq enrichments for 9 transcription factors (columns) at lincRNA promoters (rows). The percentage of bound lincRNAs downregulated upon knock-down of the TF are indicated in boxes (‘na’ were not measured). Right: Examples of lincRNAs from two clusters (‘core regulated’ and ‘myc regulated’) showing their genomic neighbourhood and TF binding. (b) A heatmap representing changes in lincRNA expression (rows) following knockdown of 11 TFs (columns). Middle: Effect of knockdown of Sox2, Oct4 and Nanog on expression levels of linc1405 (gray) and Oct4 (black). Right: Effect of knockdown of Klf2, Klf4, nMyc, and Esrrb on expression levels of linc1428.
To determine if lincRNA expression is functionally regulated by the pluripotency-associated TFs, we used shRNAs to knock down the expression of 5 of the 9 pluripotency-associated TF genes for which we could obtain validated hairpins and profiled the resulting changes in lincRNA expression after 4 days. Upon knockdown of a TF, ~50% of lincRNAs genes whose promoters are bound by the TF exhibit expression changes (Figure 4a); this proportion is comparable to that seen for protein-coding genes whose promoters are bound by the TF (Supplemental Figure 12). The strong but imperfect correlation between TF-binding and effect of TF-knockdown is consistent with previous observations39 and may reflect regulatory redundancy in the pluripotency network40. In addition, we profiled the knockdown of an additional 7 pluripotency-associated transcription factors (including Esrrb, Zfp42, and Stat3). Altogether, for ~60% of the ESC lincRNAs, we identified a significant downregulation upon knockdown of one of these 11 TFs (Figure 4b, Supplemental Table 15).
After retinoic-acid-induced differentiation of ESCs, the ESC lincRNAs show temporal changes across the time course with ~75% showing a decrease in expression compared to untreated ESCs (Supplemental Figure 13, Supplemental Table 16). Notably, all of the lincRNAs shown to regulate pluripotency are down-regulated upon retinoic acid treatment (Supplemental Figure 13). Our results establish that lincRNAs are direct transcriptional targets of pluripotency-associated TFs and are dynamically expressed across differentiation. Collectively, these results demonstrate that lincRNAs are an important regulatory component within the ESC circuitry.
lincRNAs bind diverse chromatin proteins
To explore how lincRNAs carry out their regulatory roles, we studied whether lincRNAs physically associate with chromatin regulatory proteins in ESCs. We previously showed that many human lincRNAs can interact with the polycomb repressive complex4, a complex that plays a critical functional role in the regulation of ESCs18,19. To determine whether the ESC lincRNAs physically associate with the polycomb complex, we crosslinked RNA-protein complexes using formaldehyde, immunoprecipitated the complex using antibodies specific to both the Suz12 and Ezh2 components of Polycomb, and profiled the co-precipitated lincRNAs using a direct RNA quantification method41 (see Methods). We performed immunoprecipitation of the Polycomb complex across 5 biological replicates and 8 mock-IgG controls, and we assessed significance using a permutation test (see Methods, Supplemental Figure 16). Altogether, we identified 24 lincRNAs (~10% of the ESC lincRNAs) that were strongly enriched for both Polycomb components (Figure 5b, Supplemental Table 17).
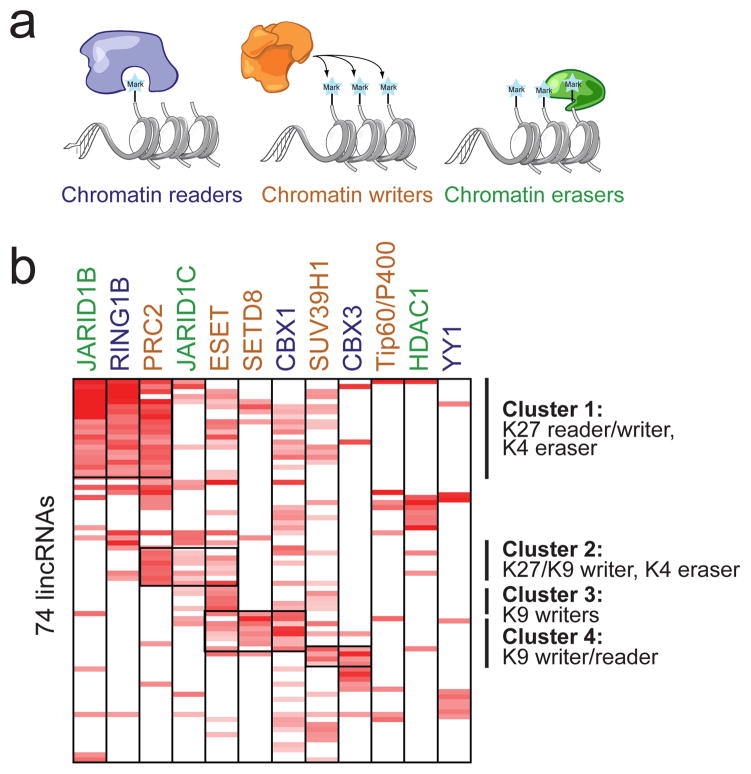
Figure 5. lincRNAs physically interact with chromatin regulatory proteins.
(a) A schematic of the classes of chromatin regulators profiled: ‘readers’ (blue), ‘writers’ (orange), and ‘erasers’ (green). (b) A heatmap showing the enrichment of 74 lincRNAs (rows) for one of 12 chromatin regulatory complexes (columns). The names are color-coded by chromatin-regulatory mechanism. Major clusters are indicated by vertical lines with a description of the chromatin components.
To determine if lincRNAs interact with additional chromatin proteins, we systematically analysed chromatin-modifying proteins that have been shown to play critical roles in ESCs18–21,42. Specifically, we screened antibodies against 28 chromatin complexes (see Methods, Supplemental Figure 14, Supplemental Table 18) and identified 11 additional chromatin complexes that are strongly and reproducibly associated with lincRNAs (see Methods, Supplemental Figure 15, 16). These chromatin complexes are involved in ‘reading’ (PRC1, Cbx1, and Cbx3), ‘writing’ (Tip60/P400, PRC2, Setd8, ESET, and Suv39h1), and ‘erasing’ (Jarid1b, Jarid1c, and HDAC1) histone modifications, as well as a chromatin-associated DNA binding protein (YY1) (Figure 5a). Altogether, we found that 74 (~30%) of the ESC lincRNAs are associated with at least one of these 12 chromatin complexes (Figure 5b, Supplemental Table 17). While most of the identified interactions are with repressive chromatin regulators, this is likely due to limitations of our selection criteria and available antibodies.
Many lincRNAs are strongly associated with multiple chromatin complexes (Figure 5b). For example, we identified 8 lincRNAs that bind to the PRC2 H3K27 and ESET H3K9 methyltransferase complexes (‘writers’ of repressive marks) and the Jarid1c H3K4 demethylase complex (an ‘eraser’ of activating marks). Consistent with this, the PRC2 and ESET complexes have been reported to bind at many of the same ‘bivalent’ domains21 and to functionally associate with the Jarid1c complex43. Similarly, we identified a distinct set of 17 lincRNAs that bind to the PRC2 complex (‘writer’ of K27 repressive marks), PRC1 complex (‘reader’ of K27 repressive marks), and Jarid1b complex (‘eraser’ of K4 activating marks) (Figure 5b), as well as other functionally consistent ‘reader’, ‘writer’, and ‘eraser’ combinations (Supplemental Figure 17). One of several potential models consistent with this data is that lincRNAs may bind to multiple distinct protein complexes, perhaps serving as ‘flexible scaffolds’ to bridge functionally related complexes as previously described for telomerase RNA44.
To determine if the identified lincRNA-protein interactions have a functional role, we examined the effects on gene expression resulting from knockdown of individual lincRNAs that are physically associated with particular chromatin complexes and from knockdown of genes encoding the associated complex itself (see Methods). For >40% of these lincRNA-protein interactions, we identified a highly significant overlap in affected gene expression programs compared to just ~6% for random lincRNA-protein pairs (see Methods, Supplemental Table 19). Other cases may reflect the limited power to detect the overlaps, because specific lincRNA-protein complexes may be related to only a fraction of the overall expression pattern mediated by the chromatin complex.
Together, these data demonstrate that many ESC lincRNAs physically associate with multiple different chromatin regulatory proteins and these interactions are likely to be important for the regulation of gene expression programs.
DISCUSSION
While the mammalian genome encodes thousands of lincRNA genes, few have been functionally characterized. We performed an unbiased loss-of-function analysis of lincRNAs expressed in ESCs and show that lincRNAs are clearly functional and primarily act in trans to affect global gene expression. We establish that lincRNAs are key components of the ESC transcriptional network that are functionally important for maintaining the pluripotent state, and that many are down-regulated upon differentiation. The ESC lincRNAs physically interact with chromatin proteins, many of which have been previously implicated in the maintenance of the pluripotent state18,20,21. In addition to chromatin proteins, lincRNAs interact with other protein complexes including many RNA-binding proteins (data not shown).
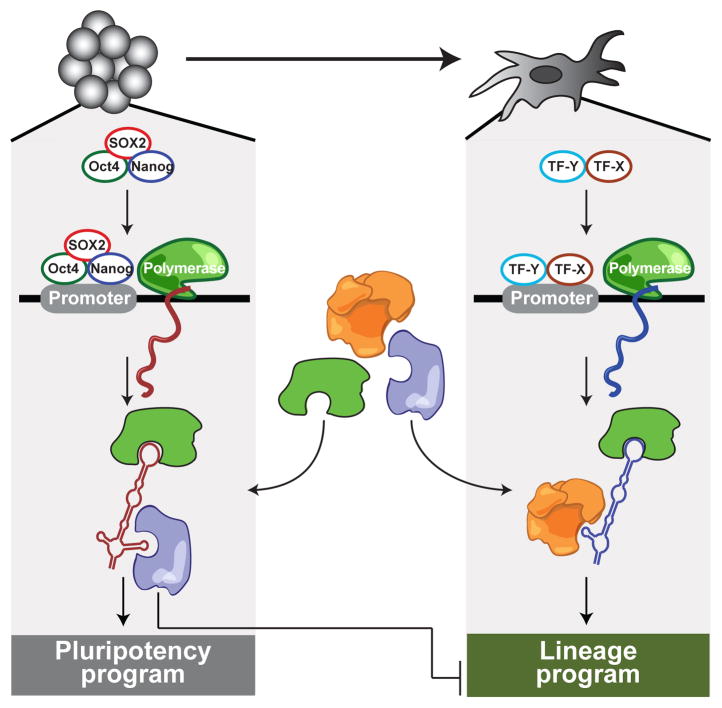
Our data suggest a model whereby a distinct set of lincRNAs is transcribed in a given cell type and interacts with ubiquitous regulatory protein complexes to form cell-type-specific RNA-protein complexes that coordinate cell-type specific gene expression programs (Figure 6). Because many of the lincRNAs studied here interact with multiple different protein complexes, they may act as cell-type specific ‘flexible scaffolds’44 to bring together protein complexes into larger functional units (Figure 6). This model has been previously demonstrated for the yeast telomerase RNA44 and suggested for the XIST45 and HOTAIR46 lincRNAs. The hypothesis that lincRNAs serve as flexible scaffolds could explain the uneven patterns of evolutionary conservation seen across the length of lincRNA genes3: the more highly conserved patches could correspond to regions of interaction with protein complexes.
Figure 6. A model for lincRNA integration into the molecular circuitry of the cell.
ESC-specific transcription factors (such as Oct4, Sox2, and Nanog) bind to the promoter of a lincRNA gene and drive its transcription. The lincRNA binds to ubiquitous regulatory proteins, giving rise to cell-type specific RNA–protein complexes. Through different combinations of protein interactions, the lincRNA–protein complex can give rise to unique transcriptional programs. Right: A similar process may also work in other cell types with specific transcription factors regulating lincRNAs, creating cell-type–specific RNA–protein complexes and regulating cell-type–specific expression programs.
While a model of lincRNAs acting as ‘flexible scaffolds’ is attractive, it is far from proven. Testing the hypothesis for lincRNAs will require systematic studies, including defining all protein-complexes with which lincRNAs interact, determining where these protein interactions assemble on RNA, and ascertaining whether they bind simultaneously or alternatively. Moreover, understanding how lincRNA-protein interactions give rise to specific patterns of gene expression will require determination of the functional contribution of each interaction and possible localization of the complex to its genomic targets.
Methods Summary
RNAi expression affects
We cloned 5 shRNAs targeting each lincRNA into a puromycin-resistant lentiviral vector22. ESCs were plated on pre-gelatinized 96-well plates and infected with lentivirus prior to addition of irradiated DR4 MEFs. Media containing 1ug/mL puromycin was added 24 hr after infection. On-target knockdown was assessed after 4 days and the best hairpin showing a knockdown >60% was selected. RNA from 147 lincRNAs, 40 protein-coding genes, and 27 negative controls were hybridized to Agilent microarrays. Differentially expressed genes were defined as having an FDR<5% and fold-change >2-fold compared to controls.
Screening for pluripotency affects
Nanog-Luciferase ESCs31 were infected and measured after 8 days. Hits were identified if they reduced luciferase levels (z<−6) across all replicates and did not reduce AlamarBlue levels. Hits were validated in wild-type ESCs by measuring mRNA levels of Oct4, Nanog, Sox2, Klf4, and Zfp42. Oct4 expression was assessed using immunofluorescence staining and morphology was visually assessed.
Lineage expression affects
Lineage expression programs were defined using published datasets (GSE12982, GSE11523, and GSE4082) and curated gene expression signatures32,33. Overlaps in gene expression affects were assessed using a modified GSEA34. Expression changes in lineage markers were determined using qPCR.
TF binding and regulation
ChIP-Seq data was downloaded (GSE11724 and GSE11431), aligned, and analysed. lincRNA promoters were previously defined using H3K4me3 peaks3. Changes in expression of the lincRNAs upon knockdown of the TFs were analyzed using Agilent microarrays.
Chromatin binding and overlap in expression
ESCs were crosslinked with formaldehyde, lysed, immunoprecipitated, washed, and reverse crosslinked. RNA was hybridized to the Nanostring codeset. We tested antibodies for 28 chromatin complexes and selected successful antibodies that (i) had >10 lincRNAs exceeding a 5-fold change and (ii) had significant enrichments across 3 replicates. We compared the overlap in gene expression using a modified GSEA34.
METHODS
ES Cell Culture
V6.5 (genotype 129SvJae × C57BL/6) and Nanog-Luciferase31 ES cells were co-cultured with irradiated C57BL/6 MEFs (GlobalStem; GSC-6002C) on pre-gelatinized plates as previously described47. Briefly, cells were cultured in mES media consisting of knock-out DMEM (Invitrogen; 10829018) supplemented with 10% FBS (GlobalStem; GSM-6002), 1% penicillin-streptomycin (Invitrogen; 15140-163), 1% L-glutamine (Invitrogen; 25030-164), 0.001% Beta-mercaptoethanol (Sigma; M3148-100ML) and 0.01% ESGRO (Millipore; ESG1106).
Picking lincRNA gene candidates
Using our previous catalogue of K4-K36 defined lincRNAs2 along with the reconstructed full-length sequences we determined using RNA-Seq3, we designed shRNA hairpins targeting each lincRNA identified in both sets. Specifically, we used the conservative K4-K36 definitions from our previous work2 that were expressed in mouse ES cells. We further filtered the list to include only multi-exonic lincRNAs that were reconstructed in mouse ES cells3. Together, this yielded 226 lincRNA genes.
Picking protein-coding gene candidates
We selected protein coding gene controls consisting of both transcription factors and chromatin proteins. These proteins were selected based on their well-characterized role in regulating mouse ES cells and include Pou5f1 (Oct4)35,48, Sox217,49, Nanog29,30, Stat350, Klf451, and Zfp42 (Rex1)52. In addition, we selected additional transcriptional and chromatin regulators that were identified by RNAi screens as regulators of pluripotency17,20,23 and/or were found in smaller focused studies to have critical roles in the maintenance of the pluripotent state (such as Carm153, Chd154, Thap1155, Suz1218,19,36, and Setdb121,56). A full list is provided in Supplemental Table 2.
shRNA Design Rules
For each lincRNA we designed 5 hairpins by extending the previously described design rules22 accounting for the sequence content of the hairpin, miRNA seed matches, uniqueness to the target compared to the transcriptome and the genome, and number of lincRNA isoforms covered.
For each lincRNA we enumerated all 21-mer sub-sequences and scored them as follows: (i) A “clamp score” was computed by looking at the nucleotides at positions 18, 19, and 20. If all three positions contained an A/T it was assigned a score of 4, if two positions were A/T it was assigned a score of 1.5 and if one was A/T it was assigned a score of 0.8. We then looked at positions 16, 17, and 21 if all 3 were A/T it was assigned a score of 1.25, if 2 were A/T it was assigned a score of 1.1, and if 1 was A/T is was assigned a score of 0.8. The clamp score was computed as the product of these two scores. (ii) A “GC score” was computed by looking at the total GC percentage of the 21-mer sequence. If the sequence was <25% GC it was assigned a score of 0.01 if it was <55% it was assigned a score of 3, if it was <60% it was assigned a score of 1, and if >60% it was assigned a score of 0.01. (iii) A “4-mer penalty” of 0.01 was assigned for any hairpin containing the same nucleotide in 4 subsequent nucleotides. (iv) A “7 GC penalty” of 0.01 was assigned to any hairpin containing any 7 consecutive G/C nucleotides. (v) We removed all hairpins containing an A in either position 1 or position 2 of the hairpin. (vi) We removed all hairpins containing a repeat masked nucleotide. (vii) Finally, we computed a “miRNA-seed penalty” by looking at the forward positions 11–17, 12–20, and 13–19 of the hairpin as well as the reverse complement of positions 14–20, 15–21, or 16–21 plus a 3′ C. We then looked up whether these positions matched known miRNA seeds and with what frequency. We computed the scores for the forward and reverse positions and defined the score as the product of the forward and reverse scores. The final score for each hairpin sequence is defined as the product of all seven scores.
We then sorted the candidate hairpin sequences by score, breaking high scoring ties by the total number of lincRNA isoforms that are covered by the hairpin. We then aligned each hairpin sequence against both the genome and the RefSeq-defined transcriptome (NCBI Release 39), and filtered any hairpin with fewer than three mismatches to any other gene or position in the genome. Candidate sequences were chosen for shRNA production by first picking the highest scoring candidate and then proceeding to successively lower scores. As each hairpin was selected, all other hairpins overlapping this hairpin were removed. We repeated this process until we identified 5 hairpins that covered each lincRNA.
shRNA cloning and virus prep
We designed 1,143 hairpins targeting 226 lincRNA genes. Of these, we successfully cloned 1,010 hairpins targeting 214 lincRNAs. These hairpins were cloned into a vector containing a puromycin resistance gene and incorporated into a lentiviral vector as previously described22. Briefly, synthetic double stranded oligos that represent a stem-loop hairpin structure were cloned into the second-generation TRC (the RNAi Consortium) lentiviral vector, pLKO.5; the expression of a given hairpin produces a shRNA that targets the gene of interest. Lentivirus was prepared as previously described22. Briefly, 100ng of shRNA plasmid, 100ng of packaging plasmid (psPAX2) and 10ng of envelope plasmid (VSV-G) were used to transfect packaging cells (293T) with TransIT-LT1 (Mirus Bio). Virus was harvested 48 and 70 hours post-transfection. Two harvests were combined. Virus titers were measured as previously described22. Briefly, we measured virus titers by infecting A549 cells with appropriately diluted viruses. 24 hours post infection, puromycin was added to a final concentration of 5ug/ml and the selection proceeded for 48 hours. The number of surviving cells, which is correlated to virus titer, was measured by alamarBlue (BioSource) staining utilizing the Envision 2103 Multilabel plate reader (PerkinElmer)
Infection and selection protocol
V6.5 ESCs or Nanog-Luciferase ESCs were plated at a density of 5000 cells/well (8-day time point) or 25,000 cells/well (4-day time point) in 100ul mES media onto pre-gelatinized 96-well dishes (VWR; BD356689). Cells were infected with 5ul of a lentiviral shRNA stock and incubated at 37°C for 30 minutes. Puromycin resistant DR4 MEFs (GlobalStem; GSC-6004G) were then added to the plates at a density of ~6000 cells/well and incubated overnight at 37°C, 5% CO2. After 24 hours, all media was removed from the cells and replaced with media containing 1ug/mL puromycin. Media was then changed every other day with fresh media containing 1ug/mL puromycin. The end-point depended on the assay and was either 4-days post infection (knockdown validation and microarrays) or 8-days (reporters and qPCR of marker genes).
RNA Extraction
ES cells were infected and lysed at day 4 with 150ul of Qiagen’s RLT buffer and 3 replicates of each virus plate were pooled for RNA extraction using Qiagen’s RNeasy 96-well columns (74181). RNA extraction was completed following Qiagen’s RNeasy 96-well protocol with the following modifications: 450ul of 70% ethanol was added to 450ul total lysate prior to the first spin. An additional RPE wash was added to the protocol, for a total of 3 RPE washes.
lincRNA primer design and prescreen
lincRNA primers were designed using primer3 (http://frodo.wi.mit.edu/primer3/). Specifically, we designed primers spanning exon-exon junctions by specifying each of the regions as preferred inclusion regions in the primer3 program. When a low scoring primer pair (primer penalty <1) was available it was used. If none was available, we then identified all primers that contained amplicons that spanned an exon-exon junction. In a few cases, when we could not identify a primer pair spanning an exon-exon junction, we designed primers within an exon of the lincRNA. For each primer pair, we tested the specificity against the transcriptome57 (Ref Seq NCBI Release 39) and the genome (Mouse MM9) using the isPCR (http://genome.ucsc.edu/cgi-bin/hgPcr) program. Specifically, we required that the primer pair amplify the lincRNA gene and no other genomic of gene amplicon.
For each primer pair, we validated the quantification and specificity prior to use. Specifically, we tested primers in qPCR reactions using a dilution series of mouse ES cDNA including a no reverse transcriptase (RT) sample. We excluded any primer that did not have robust quantification across a 64-fold dilution curve, had high signal in the no RT sample, or had low detectable expression in the undiluted sample (cycle number >34). For primers that failed this validation we redesigned and tested new primers.
Knockdown validation using qPCR
To determine if lincRNA hairpins were effective at knocking down the lincRNA of interest, we infected each hairpin into mouse embryonic stem cells, selected for lentiviral integration, and measured changes in the targeted lincRNA expression level. We isolated total cellular RNA after 4 days; this time-point was chosen to allow for identification of robust changes while minimizing secondary effects due to differentiation of the ES cells. We reasoned that this would allow us to determine more direct effects due to RNAi rather than to differentiation.
Gene panels were constructed that contained all 5 hairpins targeting a gene along with an empty vector control pLKO.5-nullT and the GFP-targeting hairpin clonetechGfp_437s1c1. cDNA was generated using 10ul of RNA and 10ul of 2x cDNA master mix containing 5x Transcriptor RT Reaction Buffer (Roche), DTT, MMLV-RT (Roche), dNTPs (Agilent; 200415-51), Random 9-mer oligos (IDT), Oligo-dT (IDT) and water. cDNA was diluted 1:9 and quantitative PCR was performed using 250 nM of each primer in 2x Sybr green master mix (Roche) and run on a Roche Light-Cycler 480. Target lincRNA expression and GAPDH levels were computed for each panel. lincRNA expression levels were normalized by GAPDH levels and this normalized value was compared to the reference control hairpins within the panel. Knockdown levels were computed as the average of the fold decrease compared to the two control hairpins. Hairpins showing a knockdown greater than 60% of the endogenous level were considered validated and the best validated hairpin from a lincRNA panel was selected for microarray studies.
Picking candidates for microarray analysis
To assess the effects of a lincRNA on gene expression, we profiled the changes in gene expression after knocking down each lincRNA gene. Specifically, for each lincRNA with at least 1 validated hairpin we profiled the genome-wide expression level changes after knockdown across 2 independent infections (see above). To control for expression changes due to viral infection, we performed five independent infections containing no RNAi hairpin (pLKO.5-nullT). This control hairpin was embedded in each RNA prep plate. To control for effects due to an off-target RNAi effect, we profiled 27 distinct negative control hairpins which do not have a known target in the cell. These hairpins included 6 RFP hairpins, 10 GFP hairpins, 6 Luciferase hairpins, and 5 LacZ hairpins. These hairpins provide a measurement of the variability of the RNAi response triggered due to non-specific effects. Furthermore, we profiled hairpins targeting 147 lincRNAs, including 10 with a second best hairpin, and 40 protein-coding genes in biological replicate. The hairpins and their replicates were randomly distributed across 7 96-well plates and prepared in batches. Each RNA preparation batch contained 1 pLKO hairpin and 1 clonetechGfp_437s1c1 hairpin in a random location on the plate. To minimize batch effects, the plate locations of the biological replicates were scrambled and the positions within the plates were scrambled for all hairpins and replicates.
Agilent Microarray hybridization
Using Agilent’s One-Color Quick Amp Labelling kit (5190-0442), we amplified and labelled total RNA for hybridization to prototype mouse lincRNA arrays (G4140-90040) according to manufacturer’s instructions with a few variations. The custom Agilent SurePrint G3 8×60K mouse array design used for this study (G4102A, AMADID 025725 G4852A) has probes to 21,503 Entrez genes and 2,230 lincRNA genes. A new updated version of this mouse design is commercially available that contains probes to 34,017 Entrez gene targets as well as 2,230 lincRNA genes (G4825A). The cRNA samples were prepared by diluting 200ng of RNA in 8.3ul water and adding positive control one-color RNA spike-in mix (Agilent, 5188-5282) that was diluted serially 1:20, then 1:25 and finally 1:10. We annealed the T7 promoter primer from the kit by incubating at 65°C for 10 minutes. We prepared the cDNA master mix and added it to the annealed RNA and incubated at 40°C for 2 hours, followed by 65°C for 15 minutes. We prepared the cRNA transcription master mix and added it to the cDNA and incubated at 40°C for 2 hours protected from light. We purified the labeled cRNA using Qiagen’s RNeasy 96-well columns (Qiagen, 74181) by adding 350ul of Qiagen RLT (without BME) to the cRNA followed by the addition of 250ul of 95% ethanol before applying to the plate column. After a 4 minute spin at 6000RPM, we washed the columns 3 times with 800ul buffer RPE. We dried the columns by spinning for 10 minutes and eluted the cRNA with 50ul of water. We measured the cRNA yield and dye incorporation using the Nanodrop 8000 Microarray measurement setting. We mixed 600ng of cRNA with blocking agent and fragmentation buffer (Agilent, 5190-0404) and fragmented for 30 minutes in the dark at 60°C. We added 2x Hybridization Buffer to each sample and loaded 40ul onto an 8-pack Hybridization gasket. We placed the microarray slides on top, sealed in the Hybridization Chamber, and incubated for 18 hours at 65°C. We washed the slides for 1 minute in room temperature GE Wash Buffer 1 and then for 1 minute in 37°C GE Wash Buffer 2 (Agilent 5188-5327, no triton addition). We scanned the microarrays using an Agilent Scanner C (G2565CA) using the following settings: Dye Channel = Red&Green, Scan Region = ScanArea (61×21.6mm), Scan Resolution = 3 μm. We prepared all of the samples simultaneously using homogenous master mixes to limit variability. Fragmentation and hybridization was staggered over time in batches of 3 to 4 slides (24 to 32 samples).
Array filtering, Normalization, and Probe filtering
Each array was processed and data extracted using the Agilent feature extraction software (G4462AA, Version 10.7.3). Samples were retained if they passed all the following quality control statistics:
AnyColorPrcntFeatNonUnifOL<1,
eQCOneColorSpikeDetectionLimit >0.01 and <2.0,
Metric_absGE1E1aSlope between 0.9 and
Metric_gE1aMedCVProcSignal <8, 1.2,
gNegCtrlAveBGSubSig >−10 and <5,
Metric_gNegCtrlAveNetSig <40,
gNegCtrlSDevBGSubSig <10,
Metric_gNonCntrlMedCVProcSignal <8,
Metric_gSpatialDetrendRMSFilterMinusFit <15,
SpotAnalysis_PixelSkewCookiePct >0.8 and <1.2
Gene expression values were determined using the gProcessedSignal intensity values. Probes were flagged if they were not detectable well above background or had an expression level lower than the lowest detectable spike-in control value. The values were floored across all samples by taking the maximum of the minimum non-flagged values across all experiments. Any value less than this maximum value were set to the maximum. This conservatively eliminates any detection variability across the samples due to stringency or other array variables.
The result of this is a single value for each probe per array. To normalize expression values across arrays, we performed quantile normalization as previously described58. Briefly, we ranked each array from lowest to highest expression. For each rank, we computed the average expression and each experiment with this value at the associated rank. For each probe, we computed the difference between the second smallest expression value and the second largest expression value. If this difference was less than 2, we filtered the probe. This metric was chosen to eliminate bias due to single sample outliers.
Identifying significant gene expression hits from RNAi KDs
To control for effects due to non-specific effects of shRNAs, we profiled 27 distinct negative control hairpins which do not have a known target in the cell. These hairpins provide a measurement of the variability of the expression profiles due to random variability or triggered by ‘off-target’ effects of the shRNA lentiviruses. Assuming that any observed effects in the negative control hairpins are due to ‘off-target’ effects and observed effects in the targeting hairpins include a mix of both ‘off-target’ effects and ‘on-target’ effects, we use permutations of the negative controls to assign a false discovery rate (FDR) confidence level for being an ‘on-target’ hit to each gene. As such, a gene would only reach genome-wide significance if the number of genes and scale of the effect was much larger than would be observed randomly among all of the expression changes found for the negative control hairpin.
Specifically, for each gene we computed a t-statistic between shRNAs targeting the lincRNA and control shRNA samples. To assess the significance of each gene we permuted the sample and control groups retaining the relative sizes of the groups and computing the same t-statistic. We then assigned an FDR value to each gene by computing the average number of values in the permuted t-statistics that were greater than the observed value of interest and divided this by the number of all observed t-statistics that were greater than the observed value. We defined genes as significantly differentially expressed if the FDR was <5% and the fold-change compared to the negative controls was >2-fold. Using this approach, an effect would only reach a significant FDR if the scale is significantly larger than would be observed in the negative controls. Knockdown of a lincRNA was considered to have a significant effect of gene expression if we identified at least 10 genes that had an effect that passed all of the criteria.
Gene-Neighbour analysis
We identified neighbouring genes based on the RefSeq genome annotation57 (NCBI Release 39). We excluded from analysis all RefSeq genes that corresponded to our lincRNA of interest but included all other coding and non-coding transcripts. We identified a significant hit as any lincRNA affecting a neighbour within 10 genes on either side with an FDR<.05 and 2-fold expression change. To compute the closest affected neighbour, we classified all genes affected upon knockdown of the lincRNAs using the same criteria above. We computed the distance between each affected gene and the locus of the lincRNA gene (and protein-coding gene) that was perturbed and took the minimum absolute distance across all affected genes.
Analysis of expected number of neighbouring genes that will change by chance
To determine the expected number of differentially expressed “neighbouring” genes occurring by chance assuming that the knockdown has no effect on gene expression, we calculated the average number of genes in a 300Kb window around a randomly selected gene in the human and mouse genome. We calculated this to be 11.2 (human) and 11.8 (mouse). For simplicity, we will conservatively round this down to 11. Assuming that no genes are changing between the knockdown and control, using a nominal p-value, which has a uniform distribution under the null hypothesis (nothing effected), we would expect to see a difference called in 5% of cases at a p-value of 0.05. If we test one locus, which has on average 11 neighbours we would expect to identify 0.55 hits by chance (11 × 0.05=0.55). However, if we now test 12 loci we would expect to see 6.6 (12 × 0.55) knockdowns which appear to have an effect under the null hypothesis.
Luciferase analysis of Nanog ES lines
ES cells containing a Nanog-Luciferase construct31 were infected in biological duplicate and monitored after 7 days. Luciferase activity was measured using Bright-Glo (Promega). All reagents and cells were equilibrated to room temperature. 100ul Bright-Glo solution was added to each plate well. Plates were incubated in the dark at room temperature for 10 minutes and luciferase was measured on a plate reader. The luciferase units were normalized to the control hairpins and a z-score compared to the negative controls (excluding luciferase hairpins) was computed. For each hairpin, we computed a Z-Score relative to the negative control hairpins and identified hits reducing Luciferase levels more than 6 standard deviations (Z<−6) for both independent replicates. In all cases we were able to identify a significant reduction in luciferase levels when using distinct hairpins targeting luciferase. To exclude hits that were due to an overall reduction in proliferation (which would also cause a reduction of Nanog positive cells in this read-out) we excluded all hairpins that caused a reduction in proliferation as measured by AlamarBlue incorporation (described below). AlamarBlue incorporation was measured in the same cells immediately before reading out Nanog-Luciferase levels.
AlamarBlue analysis of ES lines
After a 7 day infection, Nanog-Luciferase cell viability was measured using AlamarBlue (Invitrogen; DAL1025). AlamarBlue was mixed with mES media in a 1:10 ratio, added to the cells and incubated at 37°C for 1 hour. Absorbance readings at 570nm were taken. To control for possible effects due to virus titer, we measured AlamarBlue incorporation on both puromycin treated and non-puromycin treated samples for each infection.
mRNA Analysis of pluripotency markers
V6.5 ESCs were infected with shRNAs targeting lincRNAs, protein-coding genes, and 21 negative controls. After 8-days, RNA was extracted and mRNA levels of the Oct4, Nanog, Sox2, Klf4, and Zfp42 pluripotency markers were analysed using qPCR. Primer sequences are listed in Supplemental Table 9. Each sample was normalized to Gapdh levels. Significance was assessed compared to the negative control hairpins using a one-tailed t-test.
To control for ‘off-target’ effects, we analysed additional hairpins against the 26 lincRNAs affecting Nanog-Luciferase levels. Of the 26 lincRNAs, we identified 15 lincRNAs that contained an additional hairpin that reduced lincRNA expression by >50%. V6.5 ESCs were infected with the best and additional hairpin across biological replicates for these 15 lincRNAs and 21 negative control hairpins. RNA was extracted after 8 days and Oct4 expression levels were determined using qPCR. Significance was assessed relative to the negative controls using a one-tailed t-test.
Immunofluorescence
We crosslinked cells in 4% paraformaldehyde for 15 minutes, and washed in 1x PBS three times. To permeabilize the cells, we washed with 1x PBS + 0.1% Triton and then blocked in 1x PBS + 0.1% Triton + 1% BSA for 45 minutes at room temperature. We incubated cells with α-Pou5f1 antibody (Santa Cruz: SC-9081) at 1:100 dilution in blocking solution for 1.5 hours at room temperature and then washed in blocking solution three times. Next, we incubated cells in α-rabbit secondary antibody coupled to GFP (Jackson ImmunoResearch: 111-486-152) at a dilution of 1:1000 in blocking solution for 45 minutes. Finally, we thoroughly washed cells in blocking solution three times, and added vectashield containing DAPI (VWR: 101098-044) to each well.
Public Dataset curation
Traditionally, lineage markers are used to identify changes in phenotypic states. While these markers can be good indicators of differentiation potential, there are two major limitations with this approach. First, there are multiple genes that are associated with each lineage so simply looking at one can often be misleading. Second, this approach only works for classifying states with well-characterized marker genes but would not work for a comprehensive characterization of the function in the cell. Therefore, we decided to take a different approach and look at the entire gene expression profile of each lincRNA knockdown to determine what cell state each lincRNA resembles.
We curated a set of ES perturbations and differentiation states from publicly available sources. Specifically, we utilized the NCBI e-utils (http://eutils.ncbi.nlm.nih.gov/) to programmatically identify all published datasets containing keywords associated with embryonic stem cells. We filtered the list to only include mouse data sets that were generated across one of three commercial array platforms (Affymetrix, Agilent, and Illumina). Following this approach, we manually curated the list to include datasets associated with ESC perturbations (genetic deletions, RNAi, or chemical perturbations) and differentiation or induced differentiation profiles. This curation yielded 41 GEO datasets corresponding to >150 samples.
Specifically, we defined differentiation lineage states using the following datasets.
Neuro-ectoderm. We downloaded a dataset (GSE12982) corresponding to mouse ES cells containing a Sox1-GFP reporter construct. Upon differentiation of Sox1-GFP ES cells into Embryoid bodies (EBs), Sox1-GFP positive cells were collected and their global expression was profiled59. In addition, we downloaded a dataset (GSE4082)60 corresponding to direct neuroectoderm differentiation61.
Mesoderm. We downloaded the same dataset (GSE12982) as above, where the authors differentiated Brachyury-GFP reporter ES cells into EBs and sorted and profiled Brachyury-GFP positive cells59.
Endoderm. We downloaded a dataset (GSE11523) corresponding to mouse ES cells which were engineered to overexpress GATA633. GATA6 overexpression has been shown to drive ES cells into a primitive endoderm-like state62.
Ectoderm. We downloaded a dataset (GSE4082)60 corresponding to mouse ES cells differentiated into primitive ectoderm like cells with defined media61.
Trophectoderm. We downloaded a dataset (GSE11523)33 corresponding to mouse ES cells which were engineered to deplete Oct435. These cells have been shown to enter a trophectoderm-like state35. To ensure specificity to the trophectoderm state, we also compared the expression effects to trophoblast stem cells33. For all lincRNAs identified, we required a significant enrichment for both induced Oct4 knock-out and trophoblast stem cell programs.
In addition, for all lineage states we utilized a curated discrete gene expression signature of differentiation which was previously functionally tested and shown to correspond specifically to differentiation into the associated states63.
Continuous enrichment analysis and Phenotype-projection analysis
To determine relationships between lincRNA knockdowns and functional states, we employ a modified Gene Set Enrichment Analysis34 approach that accounts for the continuous nature of the two datasets, similar to previously described extensions34,64,65. For each lincRNA knockdown by functional pair we compute a continuous enrichment score. Specifically, (i) for each lincRNA knockdown we compute a normalized score matrix compared to a panel of negative control hairpins by computing a t-statistic for each gene between the replicate lincRNA knockdown expression values and the control knockdown values. (ii) For each experiment, we sort the matrix by the normalized score such that the most differentially expressed upregulated gene is first and the most differentially expressed downregulated genes is last. Using this ordering we sort the functional dataset such that the ordering corresponds to the differential rank of the lincRNA knockdown set. (iii) We compute a score Si as the running average of values from the first position to position i. We then define the enrichment score E as the maximum of the absolute value of Si for all values of i>10. We require i>10 to avoid small fluctuations in the beginning of the ranked list causing fluctuations in the enrichment score. This score is computed for each lincRNA knockdown by functional set. Since we have many lincRNA knockdowns and functional sets, in reality we have a matrix of scores and we will refer to the enrichment score of the ith knockdown and jth functional set as Eij.
To assess the significance of these scores, we compute a permutation derived false discovery rate and assign a confidence value for each projection. Specifically, to assess the significance of Eij, we permute the lincRNA knockdown samples and control samples and compute the enrichment score for each pair across all permutations. To account for the false discovery rate associated with many lincRNAs and functional sets, we use the values of all permutations directly to assess the FDR level of Eij. Specifically, to assess the FDR for each enrichment value Eij, we accumulate all the permutation values for all lincRNA knockdowns and functional sets and compute the number of values greater than Eij as well as a vector of values greater than Eij corresponding to each permutation. The FDR is computed as the average number of permuted values greater than Eij divided by the observed number greater than Eij. Using this approach, we assign an FDR value to each lincRNA knockdown by functional set and identify significant hits as those with an FDR<0.01.
To highlight the accuracy of this approach, we observed that for publicly available gene perturbations for which we also perturbed the gene we were able to identify a significant association of target genes in ~75% of cases. While the remaining few did not pass our conservative significance criteria, they also showed increased enrichments consistent with their common effects. In addition, the projected effects are highly reproducible across distinct experiments originating from many groups and across multiple expression platforms. Highlighting the specificity of this approach, we note that there are many profiles for which no lincRNA had a similar effect.
Analysis of gene-expression overlaps between independent hairpin knockdowns
To determine whether independent hairpins targeting the same lincRNA gene share common gene targets, we computed a continuous enrichment score described above. Briefly, we computed a t-statistic for both hairpins against the negative controls. We then took the second best hairpin and sorted the genes. We scored the best hairpin affected genes based on this ranked order. We assessed the significance of this enrichment by permuting the samples and controls and assigned an FDR of the overlap of the expression effect (as described above).
Discrete gene set analysis
Discrete gene sets were analysed using the Gene Set Enrichment Analysis with a slight modification to the scoring procedure to be more analogous to our continuous scoring procedure (described above). Specifically, we computed the average of the expression changes (defined by the t-statistic) for all genes within the discrete gene set upon knockdown63. Significance was assessed by permuting the control and sample labels and recomputing the average statistic for each permutation. The FDR was assessed off of these values as described above.
Lineage marker gene analysis
We curated lineage marker gene sets from published work and publicly available sources17,32,63. We identified lineage marker genes as significantly upregulated using the differential expression criteria outlined above. We validated the expression of these lineage marker genes for a selected set of lineage marker genes using qPCR (as described above) after a 4-day infection. Specifically, we looked at the expression of FGF5 (ectoderm), Sox1 (neuroectoderm), Sox17 (endoderm), Brachyury (mesoderm), and Cdx2 (trophectoderm). Primer sequences are listed in Supplemental Table 9. Expression estimates were normalized to Gapdh and compared to a panel of 25 negative control hairpins.
Identifying bound lincRNA promoters
We obtained genome-wide transcription factor binding data in mouse ES cells from 2 sources. The transcription factors Oct4, Sox2, Nanog, and Tcf3 were downloaded from the Gene Expression Omnibus (GSE11724) and the cMyc, nMyc, Zfx, Stat3, Smad1, Klf4, and Esrrb from GEO (GSE11431). For each ChIP-Seq dataset, the raw reads were obtained from the SRA (http://www.ncbi.nlm.nih.gov/sra) and processed as follows. (i) The reads were all aligned to the mouse genome assembly (build MM9) using the Bowtie aligner66, requiring a single best placement of each read. All reads with multiple acceptable placements were removed from the analysis. (ii) Binding sites were determined from the aligned reads using the MACS67 (http://liulab.dfci.harvard.edu/MACS/) algorithm using the default parameters with –mfold 8 to account for varying read counts in the libraries. (iii) lincRNA promoter regions were defined as previously described2,3 using the location of the K4me3 peaks overlapping or within 5Kb of the transcriptional start site determined by RNA-Seq reconstruction. (iv) The transcription factor binding locations and lincRNA promoter locations were intersected and the enrichment level of the peak overlapping a lincRNA promoter was assigned transcription factor binding enrichment for each lincRNA. We defined transcription factor binding locations for protein-coding genes in a comparable way. (v) To exclude the possibility that some of this binding might be due to transcription factor binding at distal enhancers, we excluded all binding events that showed evidence of P300, a protein associated with active enhancers68, localization. Altogether, we only identified ~5% of promoters overlapping with any P300 enrichment signal, a slightly lower percentage than identified for protein-coding gene promoters with detectable P300 signal.
Identifying TF-regulated lincRNA genes
lincRNA probes on the Agilent microarray were analysed using the differential expression methodology described above after knockdown of the transcription factor and comparison to the negative control hairpins. To confirm the expression changes of these lincRNAs, we hybridized 12 transcription factor knockdowns on a custom lincRNA codeset using the Nanostring nCounter assay41 (LIN-MES1-96). The knockdowns were profiled in biological duplicate along with 15 negative controls. Regulated lincRNAs were identified using the differential expression approach described above.
Nanostring probeset design
Nanostring probes against lincRNA genes were designed following the standard nanostring design principles with the following modifications specifically for the lincRNA probes. (i) To exclude possible cross-hybridization, probes were screened for cross-hybridization against both the standard mouse transcriptome as well as a background database constructed from all the lincRNA sequences. (ii) To account for isoform coverage, a first pass design attempted to select a probe that would target as many isoforms as possible for each lincRNA. In cases where it was not possible to target all isoforms for a given lincRNA, the probe that targeted the largest number was selected, and additional probes were chosen when possible to target the remaining isoforms. (iii) The standard restrictions on Tm and sequence composition were relaxed to include probes for as many lincRNAs as possible.
Retinoic Acid differentiation
V6.5 cells were cultured on gelatin-coated dishes in mES media in the absence of LIF. 5μM of retinoic acid was added daily and cell samples were taken daily for 6 days. RNA was extracted using Qiagen’s RNeasy spin columns following the manufacturer’s protocol.
Western blots
30ug of mESC nuclear protein extracts were run on 10% Bis-Tris gels (Invitrogen NP0316BOX) in MOPS buffer (Invitrogen NP0001) at 75 volts for 20 minutes followed by 120 volts for 1 hour. Gels were incubated for 30 minutes in 20% methanol transfer buffer (Invitrogen NP0006-1) and transferred onto PVDF membranes (Invitrogen 831605) at 20 volts for 1 hour using the Bio-Rad semi-dry transfer system (170–3940). Membranes were blocked in Blotto (Pierce, 37530) at room temperature for 1 hour. Antibodies were diluted in Blotto and membranes were incubated overnight at 4°C. Antibodies were diluted in using the following concentrations. Ezh2 1:2000, Suz12 1:5000, hnRNPH 1:1000, Ruvbl2 1:1000, Jarid1b 1:500, HDAC1 1:250, Cbx6 1:500, YY1 1:500. All antibodies tested were raised in rabbit. The next day, membranes were washed 3x in 0.1% TBST for 5 minutes each. The membranes were probed with anti-Rabbit-horse radish peroxidase (GE Healthcare; NA9340V) at a 1:10,000 dilution, washed 3x in 0.1% TBST, incubated in ECL reagent (GE Healthcare RPN2132), and exposed.
Crosslinked RNA immunoprecipitation
V6.5 mES cells were fixed with 1% formaldehyde for 10 minutes at room temperature, quenched with 2.5M glycine, washed with 1x PBS (3x) harvested by scraping, pelleting, and resuspended in modified RIPA lysis buffer (150mM NaCl, 50mM Tris, 0.5% Sodium deoxycholate, 0.2% SDS, 1% NP-40) supplemented with RNase inhibitors (Ambion, AM2694) and protease inhibitors. For UV crosslinking experiments, cells were irradiated with 254nm UV light. Cells were kept on ice and crosslinked in 1x PBS using 400,000 μjoules/cm2.
Cell suspension was sonicated using Branson 250 Sonifier for 3 × 20 s cycles at 20% amplitude. 10ul of Turbo DNase (Ambion, AM2238) was added to sonicated material, incubated at 37°C for 10 minutes, and spun down at max speed for 10 minutes at 4°C. Protein-G beads were washed and pre-incubated with antibodies for 30 minutes at room temperature. Lysate and beads were incubated at 4°C for 2 hours. Beads were washed 3x using the following wash buffer (1x PBS, 0.1% SDS, 0.5% NP-40) followed by 2x using a high salt wash buffer (5x PBS, 0.1% SDS, 0.5% NP-40) and crosslinks were reversed and proteins were digested with 5ul proteinase-K (NEB, P8102S) at 65° for 2–4 hours. RNA was purified using phenol/chloroform/isoamyl alcohol and RNA was precipitated in isopropanol.
Nanostring hybridization
500ng of total RNA was hybridized for 17 hours using the lincRNA codeset. The hybridized material was loaded into the nCounter prep station followed by quantification on the nCounter Digital Analyzer following the manufacturer’s protocol. For RNA immunoprecipitation experiments, we used a modified protocol. After reverse crosslinking, RNA was extracted using phenol/chloroform and ethanol precipitation methods and resuspended in 10ul of H2O. 5ul of the eluted material was hybridized for 17 hours using the lincRNA codeset.
Nanostring analysis
Probe values were normalized to negative control probes by dividing the value of the probe by the maximum negative control probe. Probe values were floored to a normalized value of 3 (3-fold higher than maximum negative control). Probes with no value greater than this floor across all samples were removed from the analysis. The values were log transformed. To control for variability between runs and different input material amounts, we normalized all samples simultaneously using the quantile normalization approach described above. The result is a set of normalized log-expression values for each probe normalized across all experiments.
Validation of RNA immunoprecipitation methods
To validate our formaldehyde based RNA immunoprecipitation method we immunoprecipitated the RNA binding protein hnRNPH, which plays a role in mRNA splicing69 and identified the associated RNAs. Consistent with known interactions, we identified a strong enrichment for its binding to intronic regions of mRNA genes. We validated these observed results in mouse ES cells by performing UV-crosslinking experiments70–72 and identified nearly identical results. We identified a similar correlation between the UV and formaldehyde crosslinked samples as for biological replicates of UV crosslinked samples and formaldehyde crosslinked samples and highly comparable enrichments (data not shown).
Antibody Selection
We selected chromatin proteins that have been implicated in regulation of the pluripotent state along with their known associated ‘reader’, ‘writer’, and ‘eraser’ complexes. Specifically, we tested antibodies against 40 chromatin proteins, corresponding to 28 chromatin complexes. In many cases, we tested multiple antibodies against the same target protein to try and identify an antibody that worked well for immunoprecipitation. A full list of tested complexes and their associated antibodies are listed in Supplemental Table 18.
Determining significant chromatin-lincRNA enrichments
We tested each antibody using formaldehyde crosslinked cells and had a two-step procedure for considering an antibody successful. (i) We tested all selected antibodies in batches, with each batch containing a mock-IGG (Santa Cruz) negative control and hnRNPH (Bethyl) positive control. Batches with variability in either the mock-IGG or hnRNPH controls were excluded and retested. For each successful batch, we computed enrichment for each lincRNA between the tested antibody and mock-IGG. We considered an antibody successful in the first step if the highest enrichment level exceeded a 5-fold change compared to the mock-IGG control and more than 10 lincRNAs exceeded this threshold. While this approach can yield false positives (antibodies that pass but are not efficient) it significantly reduced the number of antibodies to be tested in the next step. (ii) For all antibodies that successfully passed the first criteria, we performed immunoprecipitation on two additional biological replicates along with 4 mock-IGG controls. We computed a t-statistic for each lincRNA compared to the controls and assessed the significance using a permutation test, by permuting the samples and IGG samples (as above). Hits were considered significant if they exceed a t-statistic cutoff of 2 (log scale) compared to the controls and had an FDR<0.2. We allowed a slightly higher FDR cutoff since the number of permutations was far smaller yielding lower power to estimate the FDR. Only antibodies yielding significant lincRNAs were considered successful. In total, we identified 12 of the 28 complexes (55 antibodies) with at least one successful antibody.
Determining significant overlaps between lincRNA and chromatin protein knockdown effects
To determine the functional overlap between the lincRNA and the chromatin complexes it physically interacts with, we compared the effects on gene expression upon knockdown of the lincRNA and the associated protein complex. To do this, we utilized the gene expression profiles determined for each lincRNA knockdown and knockdowns of 9 of the 12 identified chromatin complexes for which we had good hairpins. We defined each interaction between a lincRNA and protein, and compute a continuous enrichment score, generated all permutations of the control hairpins and sample hairpins and assigned a false discovery rate to the scores (as described above). At an FDR<0.05 we identified 43% of the interactions to be significant. For 69% of the interactions, we were able to identify an overlap at an FDR<0.1.
Supplementary Material
Acknowledgments
We thank Dianali Rivera, Thomas Green, Tashfeen Bhimdi, Griet Verstappen, Christine Surka, Serena Silver, Adam Brown, Daniel Lam, and Oren Ram for technical help, Casey Gifford, Stella Markoulaki, and Rudolph Jaenisch for providing cell lines used in this study, Peter Tsang, Bo Curry, Anya Tsalenko, and Agilent Technologies for microarray and technical help, Bart Challis and Active Motif for antibodies, Gary Geiss, Rich Boykin, and Nanostring technologies for technical help, Eric Wang and Chris Burge for help with RNA immunoprecipitation experiments and helpful discussions, Piyush Gupta, Andreas Gnirke, John Cassady, Erez Lieberman-Aiden, Moran Cabili, and Matt Thompson for discussions and ideas, and Leslie Gaffney for assistance with figures. M. Guttman is a Vertex scholar. This work was funded by NHGRI, a Center for Excellence for Genomic Science, the Merkin Foundation for Stem Cell Research, and funds from the Broad Institute of MIT and Harvard.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions
M. Guttman and ESL conceived and designed the overall project with help from AM, AR, JLR, and DER, M. Guttman and JD designed experiments with help from JKG, XY (RNAi), BWC (pluripotency assays), and IA (RNA IP), M. Guttman, JD, GM, ABL, RA, GY performed experiments, M. Guttman, JD, M. Garber analysed data, LB, AM, DER provided reagents, M. Guttman and ESL wrote the manuscript.
Microarray data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE30245.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 2.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guttman M, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koziol MJ, Rinn JL. RNA traffic control of chromatin complexes. Curr Opin Genet Dev. 2010;20:142–148. doi: 10.1016/j.gde.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Santa F, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebisuya M, Yamamoto T, Nakajima M, Nishida E. Ripples from neighbouring transcription. Nat Cell Biol. 2008;10:1106–1113. doi: 10.1038/ncb1771. [DOI] [PubMed] [Google Scholar]
- 11.Orom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith AG. Embryo-derived stem cells: of mice and men. Annu Rev Cell Dev Biol. 2001;17:435–462. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- 15.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 17.Ivanova N, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 18.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 20.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 2009;23:2484–2489. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 23.Hu G, et al. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet. 2002;36:233–278. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- 25.Koerner MV, Pauler FM, Huang R, Barlow DP. The function of non-coding RNAs in genomic imprinting. Development. 2009;136:1771–1783. doi: 10.1242/dev.030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5:e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sproul D, Gilbert N, Bickmore WA. The role of chromatin structure in regulating the expression of clustered genes. Nat Rev Genet. 2005;6:775–781. doi: 10.1038/nrg1688. [DOI] [PubMed] [Google Scholar]
- 28.Silva J, et al. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chambers I, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 30.Mitsui K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 31.Brambrink T, et al. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherwood RI, et al. Prospective isolation and global gene expression analysis of definitive and visceral endoderm. Dev Biol. 2007;304:541–555. doi: 10.1016/j.ydbio.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 33.Aiba K, et al. Defining developmental potency and cell lineage trajectories by expression profiling of differentiating mouse embryonic stem cells. DNA Res. 2009;16:73–80. doi: 10.1093/dnares/dsn035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 36.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang H, et al. Role for Dpy-30 in ES Cell-Fate Specification by Regulation of H3K4 Methylation within Bivalent Domains. Cell. 2011;144:513–525. doi: 10.1016/j.cell.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marson A, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kunarso G, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 40.Jiang J, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 41.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 42.Dey BK, et al. The histone demethylase KDM5b/JARID1b plays a role in cell fate decisions by blocking terminal differentiation. Mol Cell Biol. 2008;28:5312–5327. doi: 10.1128/MCB.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zappulla DC, Cech TR. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc Natl Acad Sci U S A. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 46.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meissner A, Eminli S, Jaenisch R. Derivation and manipulation of murine embryonic stem cells. Methods Mol Biol. 2009;482:3–19. doi: 10.1007/978-1-59745-060-7_1. [DOI] [PubMed] [Google Scholar]
- 48.Nichols J, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 49.Avilion AA, et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakatake Y, et al. Klf4 cooperates with Oct3/4 and Sox2 to activate the Lefty1 core promoter in embryonic stem cells. Mol Cell Biol. 2006;26:7772–7782. doi: 10.1128/MCB.00468-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brons IG, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 53.Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–218. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaspar-Maia A, et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dejosez M, et al. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell. 2008;133:1162–1174. doi: 10.1016/j.cell.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan P, et al. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev. 2009;23:2507–2520. doi: 10.1101/gad.1831909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang YH, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aiba K, et al. Defining a developmental path to neural fate by global expression profiling of mouse embryonic stem cells and adult neural stem/progenitor cells. Stem Cells. 2006;24:889–895. doi: 10.1634/stemcells.2005-0332. [DOI] [PubMed] [Google Scholar]
- 61.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 62.Morrisey EE, et al. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 1998;12:3579–3590. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bock C, et al. Reference Maps of Human ES and iPS Cell Variation Enable High-Throughput Characterization of Pluripotent Cell Lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 66.Langmead B, Hansen KD, Leek JT. Cloud-scale RNA-sequencing differential expression analysis with Myrna. Genome Biol. 2010;11:R83. doi: 10.1186/gb-2010-11-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ule J, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 72.Wang Z, Tollervey J, Briese M, Turner D, Ule J. CLIP: construction of cDNA libraries for high-throughput sequencing from RNAs cross-linked to proteins in vivo. Methods. 2009;48:287–293. doi: 10.1016/j.ymeth.2009.02.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.