Abstract
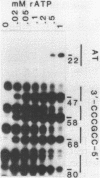
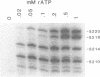
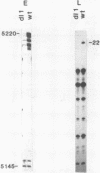
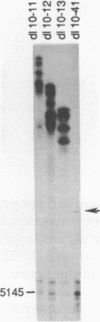
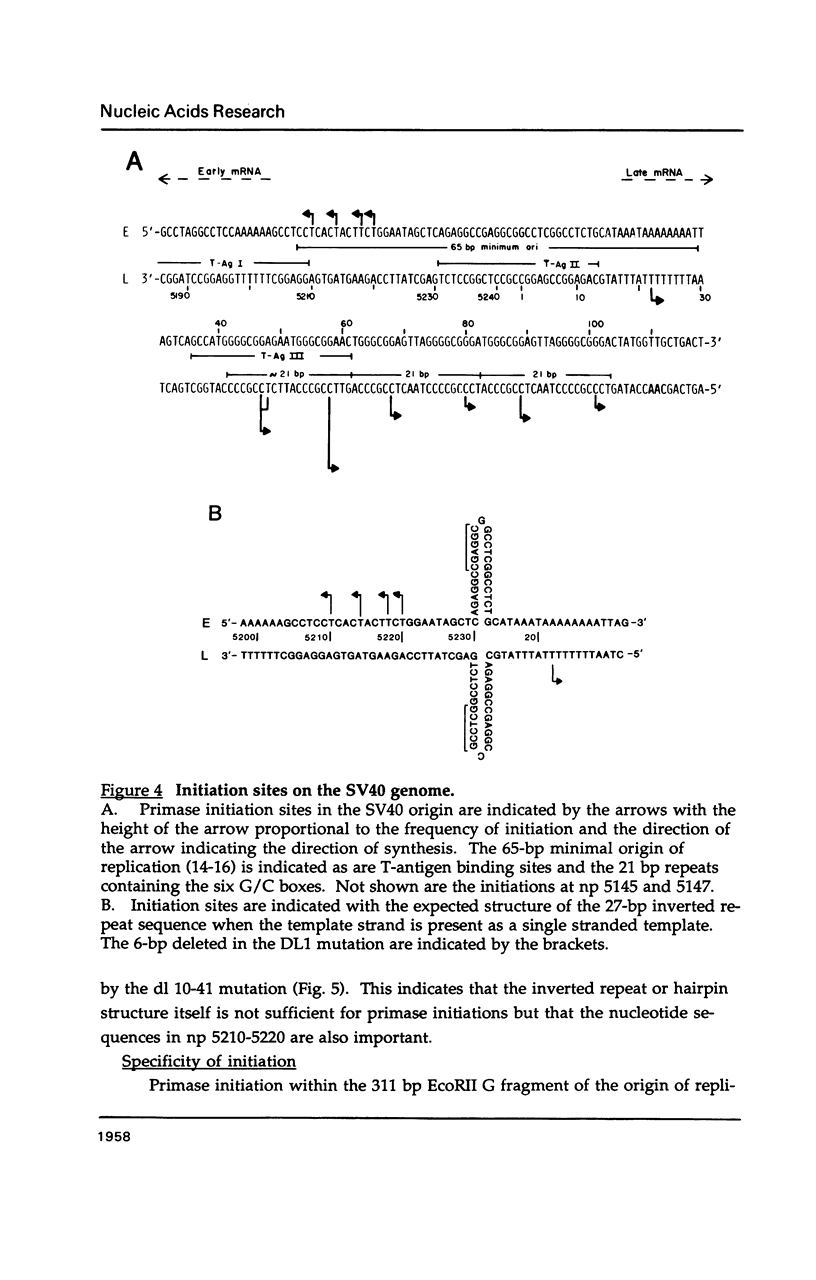
Primase synthesizes decaribonucleotides for priming of lagging and possibly leading strand synthesis at a replication fork. The sites of initiation by purified mouse primase were shown to be highly specific within the SV40 origin of replication. This study further examines the role of the 27-bp inverted repeat in the origin for initiation. A site is observed on the L-strand template at nucleotide position (np) 22 positioned a similar distance from the 27-bp inverted repeat as sites previously reported on the E-strand. The initiations adjacent to the 27-bp repeat have a higher Km for rATP than other sites. A deletion within the inverted repeat eliminated initiation at sites proximal to the hairpin on both E and L strands but had no effect at more distant sites. A deletion mutant which left the inverted repeat intact but deleted the initiation sites at np 5210-5220 on the E-strand was not active as a template for proximal sites. These results indicate that primase has two modes of recognition, one that requires the SV40 inverted repeat structure and a specific sequence and another that requires sequence alone. Additional regions of the SV40 genome have also been examined and of approximately 2000 nucleotides of single stranded template examined, only one additional site was observed at np 2412 on the E-strand. This indicates that primase initiations are highly specific for the SV40 origin and their potential functional role is discussed.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bergsma D. J., Olive D. M., Hartzell S. W., Subramanian K. N. Territorial limits and functional anatomy of the simian virus 40 replication origin. Proc Natl Acad Sci U S A. 1982 Jan;79(2):381–385. doi: 10.1073/pnas.79.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway R. C., Lehman I. R. Synthesis by the DNA primase of Drosophila melanogaster of a primer with a unique chain length. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4585–4588. doi: 10.1073/pnas.79.15.4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean F. B., Borowiec J. A., Ishimi Y., Deb S., Tegtmeyer P., Hurwitz J. Simian virus 40 large tumor antigen requires three core replication origin domains for DNA unwinding and replication in vitro. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8267–8271. doi: 10.1073/pnas.84.23.8267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb S., DeLucia A. L., Baur C. P., Koff A., Tegtmeyer P. Domain structure of the simian virus 40 core origin of replication. Mol Cell Biol. 1986 May;6(5):1663–1670. doi: 10.1128/mcb.6.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio D., Nathans D. Cold-sensitive regulatory mutants of simian virus 40. J Mol Biol. 1980 Jun 15;140(1):129–142. doi: 10.1016/0022-2836(80)90359-9. [DOI] [PubMed] [Google Scholar]
- Eliasson R., Reichard P. Primase initiates Okazaki pieces during polyoma DNA synthesis. Nature. 1978 Mar 9;272(5649):184–185. doi: 10.1038/272184a0. [DOI] [PubMed] [Google Scholar]
- Hay R. T., DePamphilis M. L. Initiation of SV40 DNA replication in vivo: location and structure of 5' ends of DNA synthesized in the ori region. Cell. 1982 Apr;28(4):767–779. doi: 10.1016/0092-8674(82)90056-3. [DOI] [PubMed] [Google Scholar]
- Legerski R. J., Hodnett J. L., Gray H. B., Jr Extracellular nucleases of pseudomonas BAL 31. III. Use of the double-strand deoxyriboexonuclease activity as the basis of a convenient method for the mapping of fragments of DNA produced by cleavage with restriction enzymes. Nucleic Acids Res. 1978 May;5(5):1445–1464. doi: 10.1093/nar/5.5.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Tjian R. Construction and analysis of simian virus 40 origins defective in tumor antigen binding and DNA replication. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6491–6495. doi: 10.1073/pnas.77.11.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prussak C. E., Tseng B. Y. Isolation of the DNA polymerase alpha core enzyme from mouse cells. J Biol Chem. 1987 May 5;262(13):6018–6022. [PubMed] [Google Scholar]
- Reichard P., Eliasson R., Söderman G. Initiator RNA in discontinuous polyoma DNA synthesis. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4901–4905. doi: 10.1073/pnas.71.12.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tack L. C., Proctor G. N. Two major replicating simian virus 40 chromosome classes. Synchronous replication fork movement is associated with bound large T antigen during elongation. J Biol Chem. 1987 May 5;262(13):6339–6349. [PubMed] [Google Scholar]
- Tseng B. Y., Ahlem C. N. A DNA primase from mouse cells. Purification and partial characterization. J Biol Chem. 1983 Aug 25;258(16):9845–9849. [PubMed] [Google Scholar]
- Tseng B. Y., Ahlem C. N. DNA primase activity from human lymphocytes. Synthesis of oligoribonucleotides that prime DNA synthesis. J Biol Chem. 1982 Jul 10;257(13):7280–7283. [PubMed] [Google Scholar]
- Tseng B. Y., Ahlem C. N. Mouse primase initiation sites in the origin region of simian virus 40. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2342–2346. doi: 10.1073/pnas.81.8.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng B. Y., Erickson J. M., Goulian M. Initiator RNA of nascent DNA from animal cells. J Mol Biol. 1979 Apr 25;129(4):531–545. doi: 10.1016/0022-2836(79)90467-4. [DOI] [PubMed] [Google Scholar]
- Tseng B. Y., Goulian M. Initiator RNA of discontinuous DNA synthesis in human lymphocytes. Cell. 1977 Oct;12(2):483–489. doi: 10.1016/0092-8674(77)90124-6. [DOI] [PubMed] [Google Scholar]
- Tseng B. Y., Goulian M. Initiator RNA synthesis upon ribonucleotide depletion. Evidence for base substitutions. J Biol Chem. 1980 Mar 10;255(5):2062–2066. [PubMed] [Google Scholar]
- Vishwanatha J. K., Yamaguchi M., DePamphilis M. L., Baril E. F. Selection of template initiation sites and the lengths of RNA primers synthesized by DNA primase are strongly affected by its organization in a multiprotein DNA polymerase alpha complex. Nucleic Acids Res. 1986 Sep 25;14(18):7305–7323. doi: 10.1093/nar/14.18.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Hendrickson E. A., DePamphilis M. L. DNA primase-DNA polymerase alpha from simian cells: sequence specificity of initiation sites on simian virus 40 DNA. Mol Cell Biol. 1985 May;5(5):1170–1183. doi: 10.1128/mcb.5.5.1170. [DOI] [PMC free article] [PubMed] [Google Scholar]