Abstract
The Nile tilapia (Oreochromis niloticus) is an important species in aquaculture and an excellent model system for laboratory studies. Functional genetic analysis using this species has been difficult because existing methods for producing transgenics are inefficient. Here we show that the Tol2 transposon system can be used to create transgenic tilapia with high efficiency. We constructed a line that is transgenic for GFP under control of a Xenopus elongation factor 1α (EF1α) promoter. The germline transmission rate of the Tol2 construct to the first generation was about 30%, which is much higher than conventional methods. GFP expression was strong and ubiquitous in the embryos. Application of the Tol2 system for constructing transgenics in tilapia and related species will promote research in many areas, but will be especially useful for studies of evolutionary developmental biology in the cichlid fishes of East Africa.
Keywords: Nile tilapia Oreochromis niloticus, transgenic, EF1α, GFP, Tol2
1. Introduction
The Nile tilapia Oreochromis niloticus (Pisces: Cichlidae) is a globally important aquaculture species (FAO, 2008). It is also an excellent laboratory model for questions ranging from physiology (McCormick et al., 1992; Farrell and Campana, 1996) and endocrinology (Parhar et al., 2000; Strüssmann and Nakamura, 2002), to genomic biology and molecular genetics (Majumdar and McAndrew, 1986; Kocher et al., 1998; Oliveira and Wright, 1998; Lee et al., 2005; Katagiri et al., 2005; Santini and Bernardi, 2005). Tilapia breed year-round (2–3 week spawning cycle), are highly fecund (hundreds of eggs in a clutch), and have a short generation period (six months) (Fujimura and Okada, 2007). They are externally fertilized and it is easy to obtain one-cell stage eggs by artificial fertilization (Fujimura and Okada, 2007).
Transgenic tilapias have been established, especially for the purpose of enhancing the growth rate of tilapia in aquaculture, for more than 20 years (Brem et al., 1988). The usual method is the random integration of plasmid DNA which has been introduced into fertilized eggs by microinjection. However, in the conventional method, the frequency of germline transmission of the injected DNA has been low. Maclean et al. (2002) summarize traditional approaches in which the rate of germline transmission in tilapia is 0.1 – 10%.
Transposable elements have been an invaluable tool for transgenesis and mutagenesis in many organisms (Ivics et al., 2009). The Tol2 transposable element has been particularly useful for generating transgenic zebrafish (Kawakami, 2007). Tol2 was originally identified in the genome of the Japanese medaka fish, Oryzias latipes (Koga et al., 1996). An autonomous member of the Tol2 element, which encodes a gene for a fully functional transposase that is capable of catalyzing transposition, has been identified (Kawakami et al., 1998; Kawakami and Shima, 1999). Furthermore, Kawakami et al. (2000, 2004) developed a two-component transposition system in zebrafish, in which a transposon donor plasmid containing a construct with Tol2 elements, and in vitro synthesized transposase mRNA, are co-injected into the fertilized eggs. In the egg, the construct is excised from the donor plasmid and integrated into the genome of germ cells. The germline transmission frequency is typically very high, and the Tol2 system is now widely used for transgenesis of not only zebrafish, but also several model animals, from frogs (Johnson Hamlet et al., 2006) to flies (Urasaki et al., 2008).
Despite this broad success, there is concern that endogenous transposase activity might limit the utility of the Tol2 system in other fish species. Here we show that the Tol2 system can be used to create transgenic tilapia, by demonstrating germline transmission of transgenes with strong and ubiquitous expression of GFP under the control of a Xenopus EF1α promoter.
2. Materials and methods
2.1. Animals
The parental Nile tilapia used in this work were transferred from the Tokyo Institute of Technology (Yokohama, Japan) to the University of Maryland (College Park MD, USA). Each of the adult fish was maintained under constant conditions (28 ± 1°C, 10h dark/14h light cycle) in a 40-liter tank system, through which fresh water was circulated. The fish were fed three times daily with granulated commercial foods.
Developmental stages were determined according to Fujimura and Okada (2007). We also used hours post-fertilization (hpf), and days post-fertilization (dpf) to supply more detailed staging information.
2.2. Microinjection
The pT2AL200R150G plasmid (Urasaki et al., 2006) was used in this work. The plasmid contains minimal elements for Tol2 transposition as well as the green fluorescent protein (GFP) expression cassette, namely the Xenopus elongation factor 1α (EF1α) enhancer-promoter, the rabbit β-globin intron, the enhanced GFP gene, and the SV40 poly(A) signal. The plasmid was electroporated into E. coli strain DH10B (Invitrogen, Carlsbad CA, USA), cultured, and prepared using the Qiagen Plasmid mini kit (Qiagen, Duesseldorf, Germany). The plasmid solution was further purified using the QIAquick PCR purification kit (Qiagen), and diluted to a stock concentration of 125 ng/μl. The transposase mRNA was synthesized using pCS-TP and mMessage mMachine Sp6 Kit (Ambion, Austin TX, USA) following Kawakami et al. (2004). The RNA solution was purified using Qiagen RNeasy kit (Qiagen), and diluted to a stock concentration of 175 ng/μl. Phenol red (2% stock in H2O, Sigma-Aldrich, St. Louis MO, USA) was used to visualize the solution during injecting. Five μl of injection solution was prepared by mixing each stock solution and RNase-free water. For transposase (+) we used 25 ng/μl plasmid, 35 ng/μl RNA, 0.2% phenol red. For transposase (−) we used 25 ng/ul plasmid, 0.2% phenol red. We filled pipettes from the back-end using microloader pipette tips (Eppendorf, Hamburg, Germany).
The transgenic tilapia were produced by microinjecting eggs at the one-cell stage. Artificially fertilized eggs were obtained as described in Fujimura and Okada (2007). The eggs were kept in a Petri dish in embryo medium (14.97 mM NaCl, 0.50mM KCl, 1.31mM CaCl2, 0.99mM MgSO4, 0.15mM KH2PO4, 0.05mM Na2HPO4, 0.07mM NaHCO3; Fisher et al., 2006). Each egg was held with sharp forceps while injecting. Needles (GD-1; Narishige, Tokyo, Japan) were pulled in a P-97 puller (Sutter Instrument, Novato CA, USA). Before injection, they were carefully broken using forceps while viewing under a microscope. Several nanoliters of the solution were injected into the cell through the micropyle (Brem et al., 1988; Rahman and Maclean, 1992), using a PLI-100 injector (Harvard Apparatus, Holliston MA, USA) and an HI-7 pipette holder (Narishige).
The injected eggs were cultured in round-bottom 200ml-flasks into which recirculating system water was gently supplied. The three embryos that had the strongest GFP expression at 5 dpf were chosen as the first generation transgenic fish (G0). Around 8 dpf, the three G0 larvae were each moved to a separate 40-liter tank in the system, and kept under the same condition as described above for the parental fish. Each of the three was female, and they started to spawn eggs after 6 months. Eggs from the three G0 transgenic females and sperm from a non-transgenic male were used for production of the second generation (G1). The resultant G1 embryos were segregated in two groups according to their GFP expression; GFP (+) and GFP (−).
2.3. Excision assay of Tol2
The excision assay was performed as described for zebrafish (Kawakami, 2004) with some modifications. The DNA samples were prepared from the injected embryos of Stage 9 (2 dpf, 24 hpf), Stage 13 (3 dpf, 48 hpf), and Stage 15 (4 dpf, 72 hpf), and isolated by phenol-chloroform extraction. The PCR product was amplified using the primers T2AexL-2 (5′-ACC CTC ACT AAA GGG AAC AAA AG-3′) and T2AexR-2 (5′-GTG CGG GCC TCT TCG CTA TTA C-3′), as described in Urasaki et al. (2006).
2.4. GFP observation
GFP expression in embryos was examined using a fluorescence-equipped dissecting microscope MZ16FA (Leica Microsystems, Wetzlar, Germany) with GFP filter sets (Leica GFP3, excitation 470/40 nm band-pass, emission 525/50 nm band-pass). Images were captured using a digital camera DFC420 (Leica Microsystems).
3. Results
3.1. Tol2 system in Nile tilapia
We tested whether the Tol2 transposon system is active in Nile tilapia by performing a PCR-based excision assay. We injected circular DNA plasmids of pT2AL200R150G, with or without in vitro synthesized mRNA of medaka transposase into tilapia eggs. DNA was isolated from the injected embryos and assayed by PCR. If the Tol2 transposase excised the Tol2 constructs from the plasmid, a PCR product of ~272 bp would be amplified from the remnant plasmid. The PCR product was not detected when only the plasmid was injected (0/7 embryos), indicating that tilapia did not express an endogenous transposase that might be a concern for this method. The PCR product was detected in six of 18 embryos injected with both of the plasmid and transposase mRNA, demonstrating that a transposase-dependent Tol2 excision did occur. After cloning and sequencing the PCR products, we confirmed that most clones had DNA sequences consistent with clean excision of the Tol2 element.
We also tested whether the Tol2 construct containing EF1α promoter and GFP was integrated into the genomic DNA after excision. We observed GFP expression during development of the injected embryos (Stage 15; Fig. 1). In all of the embryos injected with only the plasmid (27 survivors; Fig. 1a, b), no GFP signal was observed. However, in the embryos injected with both the plasmid and transposase mRNA (Fig. 1c, d), mosaic GFP expression was observed in the entire body. Although the survival rate of the injected embryos was low (44.4%), the GFP-positive rate in 138 survivors was 67.4%. These results imply that the Tol2 transposase promotes the genomic integration of the Tol2 construct at an early developmental stage.
Fig.1.
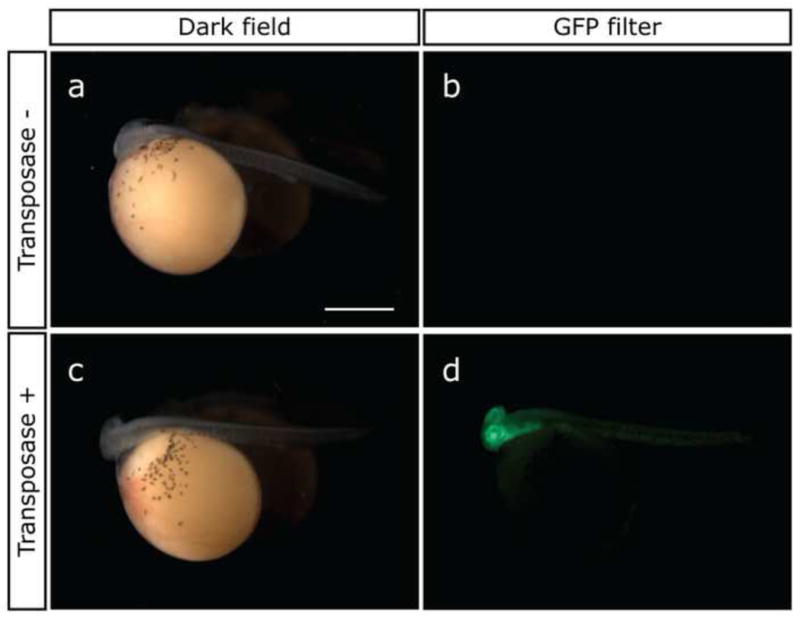
GFP expression in the G0 embryos, into which the plasmid was injected without (a, b) and with (c, d) transposase mRNA, at Stage 15 (4 dpf, 72 hpf). Lateral views of embryos. Left side (a, c), views on dark field. Right side (b, d), views through GFP filter. (a, b) No GFP expression was observed in the transposase (−) embryos. (c, d) Mosaic GPF expression was observed along the entire body in the transposase (+) embryos. Scale bar, 1 mm.
3.2. Germline transmission of Tol2 in Nile tilapia
We further tested whether Tol2 can transpose into the germ lineage of Nile tilapia. In this study, we chose the three G0 embryos (TG14-1, 2, 3 in Table 1) that had strong GFP signals. The three were raised to adult in six months, and all three were female. We examined GFP expression in 2,815 G1 embryos obtained by crosses between a non-transgenic male and the three transgenic females. GFP-positive expression was observed in 29.1% of the G1 embryos (Table 1).
Table 1.
GFP positive rate in G1 clutches
| Mother | Clutch ID | GFP+ | GFP− | GFP Rate |
|---|---|---|---|---|
| TG14-1 | #307 | 38 | 93 | 29.0% |
| #312 | 126 | 381 | 24.9% | |
| #327 | 158 | 423 | 27.2% | |
| #332 | 111 | 340 | 24.6% | |
| TG14-2 | #329 | 96 | 239 | 28.7% |
| TG14-3 | #311 | 132 | 318 | 29.3% |
| #333 | 159 | 201 | 44.2% | |
|
| ||||
| Total | 820 | 1995 | 29.1% | |
The number of embryos with and without GFP signals was counted in each clutch from crossing eggs from G0 transgenic females and sperm from wild type males.
3.3. GFP observation of transgenic green tilapia
GFP expression in the G1 embryos varied in intensity, but there was no indication that this was related to the founder lineage (Fig. 2). Strong and ubiquitous GFP expression (GFP ++) was observed in 43.9% of the GFP-positive embryos (Fig. 2a, b). Weak or mosaic GFP expression (GFP +) was observed in 56.1% of the GFP-positive embryos (Fig. 2c–f).
Fig.2.

Variation in intensity of GFP expression in the G1 embryos. Stage 17 (5 dpf, 98 hpf). Lateral views of embryos. Left side (a, c, e, g), views on dark field. Right side (b, d, f, h), views through GFP filter. (a, b) Example of strong and ubiquitous GFP expression (GFP ++). (c, d, e, f) Examples of weak or mosaic GFP expression (GFP +). (g, h) Example of no GFP expression (GFP −). Scale bar, 1 mm.
In the GFP ++ G1 embryos, GFP signals were observed from the one-cell stage onward (Stage 1; Fig. 3a, b). As a result of convergence and extension movements over the yolk, epiboly began to spread the blastoderm across the yolk at Stage 9. The GFP signals were observed in the cells over the yolk, and the developing embryonic shield was visible with slightly stronger GFP intensity (Stage 9; Fig. 3c, d). At Stage 12, strong GFP expression was observed in the entire body of embryo (Stage 12; Fig. 3e, f). Somitogenesis can be observed along the body axis.
Fig.3.

GFP expression in the GFP ++ of G1 embryos during development from 1 to 3 days post-fertilization. Left side (a, c, e), views on dark field. Right side (b, d, f), views through GFP filter. (a, b) Stage 1 (1 dpf, 1 hpf). Lateral views of one-cell egg. Animal pole at the top. GFP expression was observed in the cell. (c, d) Stage 9 (2 dpf, 27 hpf). Dorsal views of the embryo. GFP expression was observed in the cells of 40% epiboly, especially strong in the cells forming embryonic shield. (e, f) Stage 12 (3 dpf, 46 hpf). Dorsal views of the embryo. GFP expression was observed in the entire body of embryo. About ten somites form along the body axis. Scale bar, 1 mm.
The GFP expression continued in the entire body at later stages of development (Fig. 4). At Stage 15 (Fig. 4a) and Stage 17 (Fig. 4b), strong expression was found in the eyes, optic tectum, and pharyngeal region. Expression was also observed at the myoseptum and pectoral fin buds. After Stage 18 (Fig. 4c), strong expression was found in the developing muscle of the body region. At Stage 19 (Fig. 4d) and Stage 21 (Fig. 4e), strong expression was found in the ‘undifferentiated white tissue’ (Fujimura and Okada, 2007; arrows in Fig. 4d, e) between the yolk and body. Expression in this tissue could be also observed in the earlier stages (arrows in Fig. 4a, b, c). At Stage 21, the signals became weaker as the pigment cells covered the head region and body wall and obscure the signal (Fig. 4e).
Fig.4.
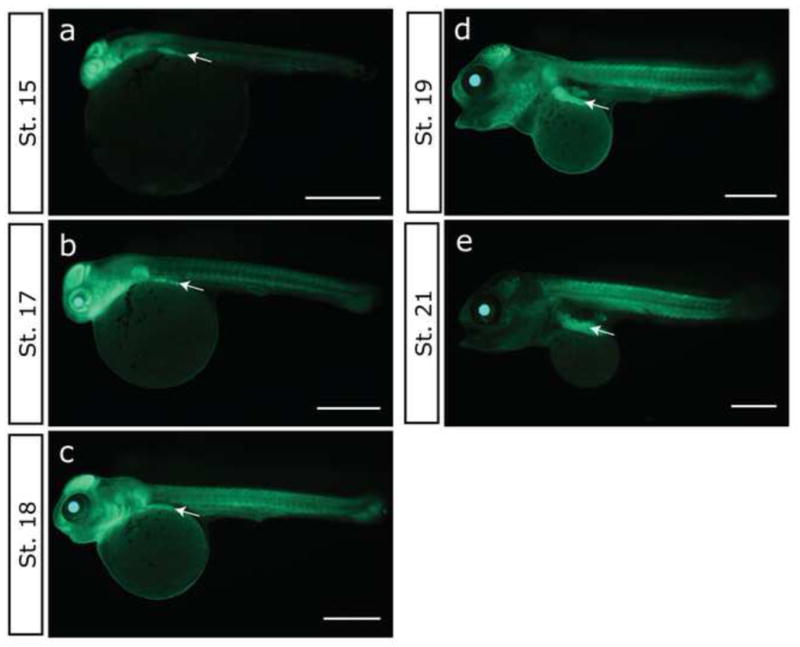
GFP expression in the GFP ++ of G1 embryos during development from 4 to 8 days post-fertilization. Lateral views of embryos through GFP filter. (a) Stage 15 (4 dpf, 74 hpf). (b) Stage 17 (5 dpf, 100 hpf). (c) Stage 18 (6 dpf, 122 hpf). (d) Stage 19 (7 dpf, 143 hpf). (e) Stage 21 (8 dpf, 171hpf). Arrows indicate ‘undifferentiated white tissue’ (Fujimura and Okada 2007). Scale bar, 1 mm.
4. Discussion
In this study, we demonstrated that the Tol2 system is an effective tool for generating transgenic Nile tilapia. We observed a germline transmission rate of the Tol2 construct to the G1 of about 30%. This rate compares favorably with the rates from injection of conventional plasmids, which generally vary from 0.1 – 10% (for review, Maclean et al., 2002; also see Alam et al., 1996; Rahman et al., 1997, 1998; Kobayashi et al., 2007; Farlora et al., 2009). Under the conditions of our experiment, no integration was observed when circular DNA plasmids were injected without transposase mRNA, as evidenced by the lack of GFP signal (Fig. 1). The rate of integration without transposase might be higher if linearized plasmids were used, because linearized form have several times more integration rates than circular one (Chourrout et al. 1986), but would still be much lower than the transposase-mediated integration.
We observed some variation in the intensity of GFP expression (Fig. 2). This variation might be due to position effects at the integration site (Wilson et al., 1990) or variation in copy number (Rahman et al., 2000). Similar variation was found in transgenic Xenopus made using the Tol2 system (Johnson Hamlet et al., 2006).
The Xenopus EF1α promoter was used in this study. EF1α protein promotes guanosine triphosphate (GTP)-dependent binding of aminoacyl-tRNA to ribosome during eukaryotic protein synthesis, and it is ubiquitously expressed in all types of cells (Negrutskii and Elskaya, 1998). Therefore, the EF1α promoter has been used to generate animals with ubiquitous expression of a transgene (Udvadia and Linney, 2003). Our results show that the Xenopus EF1α promoter drives ubiquitous expression in Nile tilapia, as it does in zebrafish (Urasaki et al., 2006).
As shown in Figure 3, the GFP expression was first observed at the one-cell stage. Kinoshita et al. (2000) clearly showed that the expression pattern of GFP depends on the sex of the transgenic parent in EF1α/GFP transgenic medaka. In their study, GFP expression was first observed at the early gastrula stage when crossing transgenic males with non-transgenic females, indicating that the expression is zygotic. On the other hand, GFP expression was first observed at the one-cell stage when crossing transgenic females with non-transgenic males, indicating that the expression is maternal. Consistent with these expectations, the one-cell stage embryos produce by the three G0 tilapia females in this study showed GFP signals that were probably due to maternal expression.
As shown in Figures 3 and 4, these transgenic lines under the control of the Xenopus EF1α have strong and ubiquitous GFP expression in living embryos. This improves visualization of early development in tilapia, which is sometimes difficult due to the large translucent yolk. In this regard, our GFP transgene is an improvement over previous constructs using lacZ expression, which is usually visualized after fixation and staining (Alam et al., 1996). Transgenic tilapia expressing GFP under the control of a β-actin promoter showed weak and mosaic expression during development (Farlora et al., 2009). In contrast, the Xenopus EF1α/eGFP construct used in our work gave strong and ubiquitous GFP signals. We are currently developing a G2 generation from the GFP ++ G1 progenies for use in developmental biology experiments, including cell transplantation.
The transgenic technology we have demonstrated could be applied in aquaculture to produce fish with improved genetic traits. For production of genetically modified fish in aquaculture, it is desired that gene constructs are designed with using sequences from the same or related fish species, and avoiding as much as possible the use of sequences of viral origin (Maclean et al., 2002). Therefore, there is a great potential for transgenic tilapia using the Tol2 system, which is originally derived from the medaka fish.
However, we believe this technology will be best used for basic biological research. The ability to modulate gene expression will enhance the analysis of gene networks controlling growth rate, disease resistance, salinity tolerance, temperature tolerance, sex determination, and reproduction. Several gene promoters of Nile tilapia have already been characterized (Maclean et al., 2002), and many more will be available with the completion of the tilapia genome sequence (www.broadinstitute.org/models/tilapia). It should be possible now to use the Tol2 system to construct a variety of transgenic tilapia for basic research.
Moreover, we hope our work will promote studies of evolutionary developmental biology in African cichlids. Cichlids in the East African Great Lakes are famous as spectacular examples of explosive adaptive radiation (Fryer and Iles, 1972; Kocher, 2004), particularly in their trophic morphologies (Albertson and Kocher, 2006), pigmentation (Roberts et al., 2009) and visual sensitivities (Hofmann et al., 2009). Recent studies suggest that changes in gene expression during development are more important than changes in gene coding sequence (Albertson et al., 2005; Kijimoto et al., 2005). In the context of a well-characterized developmental staging system (Fujimura and Okada, 2007, 2008a, 2008b), a highly efficient transgenic technology will facilitate investigations of the regulatory variants responsible for the dramatic evolutionary radiation of these fishes.
Acknowledgments
The authors thank Drs. Norihiro Okada and Masato Nikaido of Tokyo Institute of Technology for the O. niloticus parental stocks. Dr. Koichi Kawakami of National Institute of Genetics graciously supplied the plasmid stocks. This work was supported by grant R01HD058635 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alam MS, Popplewell A, Maclean N. Germ line transmission and expression of a lacZ containing transgene in tilapia (Oreochromis niloticus) Transgenic Res. 1996;5:87–95. [Google Scholar]
- Albertson RC, Streelman JT, Kocher TD, Yelick PC. Integration and evolution of the cichlid mandible: the molecular basis of alternate feeding strategies. Proc Natl Acad Sci USA. 2005;102:16287–16292. doi: 10.1073/pnas.0506649102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson RC, Kocher TD. Genetic and developmental basis of cichlid trophic diversity. Heredity. 2006;97:211–221. doi: 10.1038/sj.hdy.6800864. [DOI] [PubMed] [Google Scholar]
- Brem G, Brenig B, Hörstgen-Schwark G, Winnacker E. Gene transfer in tilapia (Oreochromis niloticus) Aquaculture. 1988;68:209–219. [Google Scholar]
- Chourrout D, Guyomard R, Houdebine LM. High efficiency gene transfer in rainbow trout (Salmo gairdneri Rich.) by microinjection into egg cytoplasm. Aquaculture. 1986;51:143–150. [Google Scholar]
- Farlora R, Kobayashi S, França LR, Batlouni SR, Lacerda SMSN, Yoshizaki G. Expression of GFP in transgenic tilapia under the control of the medaka β-actin promoter: establishment of a model system for germ cell transplantation. Anim Reprod. 2009;6:450–459. [Google Scholar]
- Farrell JF, Campana SE. Regulation of Calcium and Strontium deposition on the otoliths of juvenile tilapia, Oreochromis niloticus. Comp Biochem Physiol. 1996;115A:103–109. [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, McCallion AS. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgensis in zebrafish. Nature Prot. 2006;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organization (FAO) The State of World Fisheries and Aquaculture. Rome, Italy: 2008. [Accessed 24 January 2011]. Available from URL: http://www.fao.org/fishery/sofia/en. [Google Scholar]
- Fryer G, Iles T. The Cichlid Fishes of the Great Lakes of Africa. Oliver & Boyd; Edinburgh: 1972. [Google Scholar]
- Fujimura K, Okada N. Development of the embryo, larva and early juvenile of Nile tilapia Oreochromis niloticus (Pisces: Cichlidae). Developmental staging system. Dev Growth Differ. 2007;49:301–324. doi: 10.1111/j.1440-169X.2007.00926.x. [DOI] [PubMed] [Google Scholar]
- Fujimura K, Okada N. Bone development in the jaw of Nile tilapia Oreochromis niloticus (Pisces: Cichlidae) Dev Growth Differ. 2008a;50:339–355. doi: 10.1111/j.1440-169X.2008.01032.x. [DOI] [PubMed] [Google Scholar]
- Fujimura K, Okada N. Shaping of the lower jaw bone during growth of Nile tilapia Oreochromis niloticus and a Lake Victoria cichlid Haplochromis chilotes: a geometric morphometric approach. Dev Growth Differ. 2008b;50:653–663. doi: 10.1111/j.1440-169X.2008.01063.x. [DOI] [PubMed] [Google Scholar]
- Hofmann CM, O’Quin KE, Marshall NJ, Cronin TW, Seehausen O, Carleton KL. The eyes have it: regulatory and structural changes both underlie cichlid visual pigment diversity. PLoS Biol. 2009;7:e1000266. doi: 10.1371/journal.pbio.1000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z, Li MA, Mates L, Boeke JD, Nagy A, Bradley A, Izsvak Z. Transposon-mediated genome manipulation in vertebrates. Nature Methods. 2009;6:415–422. doi: 10.1038/nmeth.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson Hamlet MR, Yergeau DA, Kuliyev E, Takeda M, Taira M, Kawakami K, Mead PE. Tol2 transposon-mediated transgenesis in Xenopus tropicalis. Genesis. 2006;44:438–445. doi: 10.1002/dvg.20234. [DOI] [PubMed] [Google Scholar]
- Katagiri T, Kidd C, Tomasino E, Davis JT, Wishon C, Stern JE, Carleton KL, Howe AE, Kocher TD. A BAC-based physical map of the Nile tilapia genome. BMC Genomics. 2005;6:89. doi: 10.1186/1471-2164-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K. Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Meth Cell Biol. 2004;77:201–222. doi: 10.1016/s0091-679x(04)77011-9. [DOI] [PubMed] [Google Scholar]
- Kawakami K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 2007;8:S7. doi: 10.1186/gb-2007-8-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Koga A, Hori H, Shima A. Excision of the Tol2 transposable element of the medaka fish, Oryzias latipes, in zebrafish, Danio rerio. Gene. 1998;225:17–22. doi: 10.1016/s0378-1119(98)00537-x. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Shima A. Identification of the Tol2 transposase of the medaka fish Oryzias latipes that catalyzes excision of a nonautonomous Tol2 element in zebrafish Danio rerio. Gene. 1999;240:239–244. doi: 10.1016/s0378-1119(99)00444-8. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci USA. 2000;97:11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Kijimoto T, Watanabe M, Fujimura K, Nakazawa M, Murakami Y, Kuratani S, Kohara Y, Gojobori T, Okada N. cimp1, a novel astacin family metalloproteinase gene from East African cichlids, is differentially expressed between species during growth. Mol Biol Evol. 2005;22:1649–1660. doi: 10.1093/molbev/msi159. [DOI] [PubMed] [Google Scholar]
- Kinoshita M, Kani S, Ozato K, Wakamatsu Y. Activity of the medaka translation elongation factor 1alpha-A promoter examined using the GFP gene as a reporter. Dev Growth Differ. 2000;42:469–478. doi: 10.1046/j.1440-169x.2000.00530.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Alimuddin, Morita T, Miwa M, Lu L, Endo M, Takeuchi T, Yoshizaki G. Transgenic Nile tilapia (Oreochromis niloticus) over-expressing growth hormone show reduced ammonia excretion. Aquaculture. 2007;270:427–435. [Google Scholar]
- Kocher TD. Adaptive evolution and explosive speciation: the cichlid fish model. Nature Rev Genet. 2004;5:288–298. doi: 10.1038/nrg1316. [DOI] [PubMed] [Google Scholar]
- Kocher TD, Lee WJ, Sobolewska H, Penman D, McAndrew B. A genetic linkage map of a cichlid fish, the Tilapia (Oreochromis niloticus) Genetics. 1998;148:1225–1232. doi: 10.1093/genetics/148.3.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga A, Suzuki M, Inagaki H, Bessho Y, Hori H. Transposable element in fish. Nature. 1996;383:30. doi: 10.1038/383030a0. [DOI] [PubMed] [Google Scholar]
- Lee BY, Lee WJ, Streelman JT, Carleton KL, Howe AE, Hulata G, Slettan A, Stern JE, Terai Y, Kocher TD. A second-generation genetic linkage map of tilapia (Oreochromis spp.) Genetics. 2005;170:237–244. doi: 10.1534/genetics.104.035022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean N, Rahman MA, Sohm F, Hwang G, Iyengar A, Ayad H, Smith A, Farahmand H. Transgenic tilapia and the tilapia genome. Gene. 2002;295:265–277. doi: 10.1016/s0378-1119(02)00735-7. [DOI] [PubMed] [Google Scholar]
- Majumdar KC, McAndrew BJ. Relative DNA content of somatic nuclei and chromosomal studies in three genera, Tilapia, Sarotherodon, and Oreochromis of the tribe Tilapiini (Pisces, Cichlidae) Genetica. 1986;68:175–188. [Google Scholar]
- McCormick SD, Hasegawa S, Hirano T. Calcium uptake in the skin of a freshwater teleost. Proc Natl Acad Sci USA. 1992;89:3635–3638. doi: 10.1073/pnas.89.8.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrutskii BS, Elskaya AV. Eukaryotic translation elongation factor 1: Structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Prog Nucleic Acids Res. 1998;60:47–78. doi: 10.1016/s0079-6603(08)60889-2. [DOI] [PubMed] [Google Scholar]
- Oliveira C, Wright JM. Molecular cytogenetic analysis of heterochromatin in the chromosomes of tilapia, Oreochromis niloticus (Teleostei: Cichlidae) Chromosome Res. 1998;6:205–211. doi: 10.1023/a:1009211701829. [DOI] [PubMed] [Google Scholar]
- Parhar IS, Soga T, Sakuma Y. Thyroid hormone and estrogen regulate brain region- specific messenger ribonucleic acids encoding three gonadotropin-releasing hormone genes in sexually immature male fish, Oreochromis niloticus. Endocrinology. 2000;141:1618–1625. doi: 10.1210/endo.141.5.7460. [DOI] [PubMed] [Google Scholar]
- Rahman MA, Maclean N. Production of transgenic tilapia (Oreochromis niloticus) by one-cell-stage microinjection. Aquaculture. 1992;105:219–232. [Google Scholar]
- Rahman MA, Iyengar A, Maclean N. Co-injection strategy improves integration efficiency of a growth hormone gene construct, resulting in lines of transgenic tilapia (Oreochromis niloticus) expressing an exogenous growth hormone gene. Transgenic Res. 1997;6:369–378. [Google Scholar]
- Rahman MA, Mak R, Ayad H, Smith A, Maclean N. Expression of a novel piscine growth hormone gene results in growth enhancement in transgenic tilapia (Oreochromis niloticus) Transgenic Res. 1998;7:357–369. doi: 10.1023/a:1008837105299. [DOI] [PubMed] [Google Scholar]
- Rahman MA, Hwang GL, Razak SA, Sohm F, Maclean N. Copy number related transgene expression and mosaic somatic expression in hemizygous and homozygous transgenic tilapia (Oreochromis niloticus) Transgenic Res. 2000;9:417–427. doi: 10.1023/a:1026517212807. [DOI] [PubMed] [Google Scholar]
- Roberts RB, Ser J, Kocher TD. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlids. Science. 2009;326:998–1001. doi: 10.1126/science.1174705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini S, Bernardi G. Organization and base composition of tilapia Hox genes: implications for the evolution of Hox clusters in fish. Gene. 2005;346:51–61. doi: 10.1016/j.gene.2004.10.027. [DOI] [PubMed] [Google Scholar]
- Strüssmann CA, Nakamura M. Morphology, endocrinology, and environmental modulation of gonadal sex differentiation in teleost fishes. Fish Physiol Biochem. 2002;26:13–29. [Google Scholar]
- Udvadia AJ, Linney E. Windows into development: historic, current, and future perspectives on transgenic zebrafish. Dev Biol. 2003;256:1–17. doi: 10.1016/s0012-1606(02)00083-0. [DOI] [PubMed] [Google Scholar]
- Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174:639–649. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urasaki A, Mito T, Noji S, Ueda R, Kawakami K. Transposition of the vertebrate Tol2 transposable element in Drosophila melanogaster. Gene. 2008;425:64–68. doi: 10.1016/j.gene.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Wilson C, Bellen HJ, Gehring WJ. Position effects on eukaryotic gene expression. Ann Rev Cell Biol. 1990;6:679–714. doi: 10.1146/annurev.cb.06.110190.003335. [DOI] [PubMed] [Google Scholar]


