Abstract
Trinucleotide expansion underlies several human diseases. Expansion occurs during multiple stages of human development in different cell types, and is sensitive to the gender of the parent who transmits the repeats. Repair and replication models for expansions have been described, but we do not know whether the pathway involved is the same under all conditions and for all repeat tract lengths, which differ among diseases. Currently, researchers rely on bacteria, yeast and mice to study expansion, but these models differ substantially from humans. We need now to connect the dots among human genetics, pathway biochemistry and the appropriate model systems to understand the mechanism of expansion as it occurs in human disease.
Expansions in simple DNA repeats underlie ~20 severe neuromuscular and neurodegenerative disorders1,2. Our understanding of the pathogenic mechanisms for trinucleotide repeat (TNR) expansion diseases has advanced substantially in recent years (recently reviewed in refs 3-6), but many aspects of the mutational mechanism remain enigmatic.
Repetitive sequences constitute 30% of the human genome and, in most species, alterations in the lengths of repetitive DNA during evolution create diversity7. However, the rapid alteration in TNR length observed in human expansion diseases is surprising. Mammals have developed systems for resisting rapid changes that could be deleterious. However, when longer than a crucial threshold length, these simple TNRs over-ride genomic safeguards and expand during most parent–child transmissions and during development of the progeny.
The changes in TNR length can be substantial. For TNR repeats in coding sequences, the repeats become unstable at ~29–35 units in length, and the changes in tract size are modest, typically ≤10 repeats per generation1,2 (TABLE 1). By contrast, unstable parent–child transmissions of TNRs in non-coding regions initiate from pre-mutation alleles of ~55–200 units and increase by 100–10,000 units per generation1,2 (TABLE 1). For both coding and non-coding alleles, as the repeat length grows beyond a threshold length, the size of the successive expansions and the likelihood of another unstable event increase. Also, the disease becomes more severe and has an earlier age of onset with each successive generation, a phenomenon known as anticipation1,2.
Table 1. Features of trinucleotide expansion in humans.
| Disease | Sequence | Location | Parent of origin of expansion |
Repeat number (normal) |
Repeat number (pre-mutation) |
Repeat number (disease) |
Somatic instability |
|---|---|---|---|---|---|---|---|
| Diseases with coding TNRs | |||||||
| DRPLA | CAG | ATN1 (exon 5) | P | 6–35 | 35–48 | 49–88 | Yes |
| HD | CAG | HTT (exon 1) | P | 6–29 | 29–37 | 38–180 | Yes |
| OPMD | GCN | PABPN1 (exon 1) | P and M | 10 | 12–17 | >11 | None found in tissue tested (hypothalamus) |
| SCA1 | CAG | ATXN1 (exon 8) | P | 6–39 | 40 | 41–83 | Yes |
| SCA2 | CAG | ATXN2 (exon 1) | P | <31 | 31–32 | 32–200 | Unknown |
| SCA3 (Machado– Joseph disease) |
CAG | ATXN3 (exon 8) | P | 12–40 | 41–85 | 52–86 | Unknown |
| SCA6 | CAG | CACNA1A (exon 47) | P | <18 | 19 | 20–33 | None found |
| SCA7 | CAG | ATXN7 (exon 3) | P | 4–17 | 28–33 | >36 to >460 | Yes |
| SCA17 | CAG | TBP (exon 3) | P > M | 25–42 | 43–48 | 45–66 | Yes |
| SMBA | CAG | AR (exon 1) | P | 13–31 | 32–39 | 40 | None found |
| Diseases with non-coding TNRs | |||||||
| DM1 | CTG | DMPK (3′ UTR) | M | 5–37 | 37–50 | >50 | Yes |
| DM2 | CCTG | CNBP (intron 1) | Uncertain | <30 | 31–74 | 75–11,000 | Yes |
| FRAX-E | GCC | AFF2 (5′ UTR) | M | 4–39 | 40–200 | >200 | Unknown |
| FRDA | GAA | FXN (intron 1) | Recessive | 5–30 | 31–100 | 70–1,000 | Yes |
| FXS | CGG | FMR1 (5′ UTR ) | M | 6–50 | 55–200 | 200–4,000 | Yes |
| HDL2 | CTG | JPH3 (exon 2A) | M | 6–27 | 29–35 | 36–57 | Unknown |
| SCA8 | CTG | ATXN8OS (3′ UTR) | M | 15–34 | 34–89 | 89–250 | Unknown |
| SCA10 | ATTCT | ATXN10 (intron 9) | M and P (smaller changes with M) |
10–29 | 29–400 | 400–4,500 | Yes |
| SCA12 | CAG | PPP2R2B (5′ UTR) | M and P (more unstable with P) |
7–28 | 28–66 | 66–78 | None found |
AFF2, AF4/FMR2 family, member 2; AR, androgen receptor; ATN1, atrophin 1; ATXN, ataxin; ATXN8OS, ATXN8 opposite strand (non-protein coding); CACNA1A, calcium channel, voltage-dependent, P/Q type, alpha 1A subunit; CNBP, CCHC-type zinc finger nucleic acid binding protein; DM, myotonic dystrophy; DMPK, dystrophia myotonica-protein kinase; DRPLA, dentatorubral-pallidoluysian atrophy; FMR1, fragile X mental retardation 1; FRAX-E, mental retardation, X-linked, associated with FRAXE; FRDA, Friedreich’s ataxia; FXN, frataxin; FXS, fragile X syndrome; FXTAS, fragile X-associated tremor/ataxia syndrome; HD, Huntington’s disease; HDL2, Huntington’s disease-like 2; HTT, huntingtin; JPH3, junctophilin 3; M, maternal; OPMD, oculopharyngeal muscular dystrophy; P, paternal; PABPN1, poly(A) binding protein nuclear 1; PPP2R2B, protein phosphatase 2, regulatory subunit B; SCA, spinocerebellar ataxia; SMBA, spinomuscular bulbar atrophy; TBP, TATA-box binding protein; TNR, trinucleotide repeat.
Why some repeats expand more than others remains an important unresolved question. Most models for repeat expansion agree that expansion occurs through the formation of looped intermediates1,2, which are incorporated into DNA. However, thermodynamically, the differences in the physical properties of looped intermediates formed from different TNRs are subtle. For example, although the complementary sequences CAG and CTG are associated with short and long expansions, respectively, the free energy difference between CAG and CTG is only 1–2 kcal/mole for hairpins comprising a 30-repeat tract8,9. Neither the sequences nor the hairpin structure accounts for the differences in the expansion size. We now know that DNA repair and/or replication promote expansion in some manner. What we do not know is whether there are mechanistic differences between expansions of long and short repeats or whether the same pathways for expansion are used under all conditions — that is, in different cell types — during distinct developmental stages and in maternal and paternal transmission. These are important issues to address.
Here I focus on two long-standing questions: do long and short TNR expansions occur by the same mechanisms, and do they occur differently during human development? To address these questions, this Review is organized into two parts. In the first part, I examine the human genetic data of four TNR expansion diseases in which the TNR tract differs in length and position (in coding or non-coding regions) and discuss instability during development. This section lays a foundation for understanding the impact of cell type and parent-of-origin on long or short expansions in humans. The DNA replication and repair pathways that are thought to be relevant to expansion have been reviewed recently1,2. Thus, in the second part of this Review, I integrate genetic and biochemical results to discuss which replication and repair mechanisms fit the properties of expansion observed in humans during development. Finally, I consider directions for future research.
Repeat dynamics during development
Any mechanism for expansion must explain the features of human disease. However, many of our concepts of expansion have been defined by studies of microsatellite instability in bacteria, yeast and mice. These systems differ substantially from humans and from each other in terms of DNA replication rate, the cell type in which expansion occurs and chromatin structure. How well an experimental system reproduces the features of human disease is an important consideration. If our ultimate aim is to understand the mutational mechanism in humans, the logical starting point is to assess the TNR dynamics in human disease.
Non-coding TNRs
In fragile X syndrome (FXS, also known as FMR1), CGG repeats in the non-coding region of fragile X mental retardation 1 (FMR1) expand from the pre-mutation allele length (55–200 units) to a full mutation (200–4,000 units) almost exclusively through maternal transmission (TABLE 1). In females, the probability of expansion increases with the length of the TNR, but expansion is almost certain if the pre-mutation allele is 90 units or longer10. Normal length and intermediate length FXS alleles are more prone to expand into the pre-mutation range during paternal transmission than during maternal transmission11. However, paternal transmission of a fully expanded disease allele (TABLE 1) results either in no change or in contraction in the CGG tract10, and the contraction frequency increases with the length of the transmitted allele10. Some somatic heterogeneity occurs in both males and females. For example, in one family, monozygotic twin brothers with FXS had CGG tracts that differed by ~100–500 units12.
The developmental timing and the cell type in which instability occurs provide insights into whether the expansion mechanism is replication or repair dependent. For FXS, the timing of expansion is not known precisely but is likely to occur in the maternal oocyte during arrest in meiotic prophase I13-16 (FIG. 1). In female carriers of a pre-mutation allele, expansion is already present in seven-cell pre-implantation embryos14,15. Developing oocytes arrest during the first meiotic division (which starts during embryogenesis) (FIG 1), suggesting that the TNR alterations occur in quiescent cells and do not require replication (although some genetic selection cannot be excluded). Mice are good models for developmental aspects of TNR instability, and several mouse models for CGG expansion have been generated17,18. Although no animals exhibit large expansions (that is, expansions of thousands of units) through female transmission, the developmental timing of instability in a recent knock-in model is consistent with the timing observed in humans18.
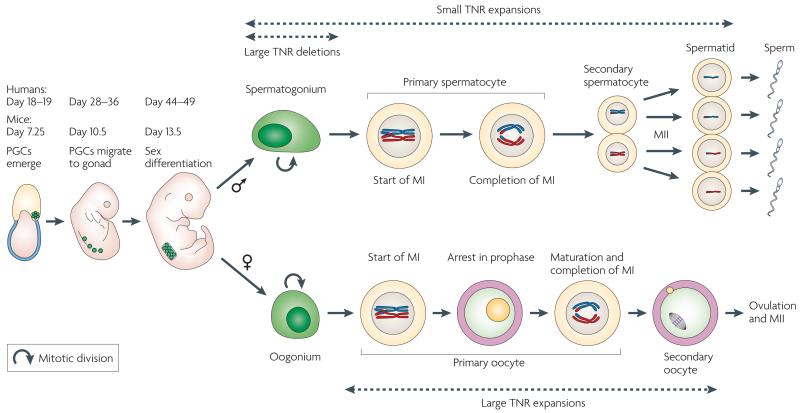
Figure 1. Human germ cell development.
Primordial germ cell (PGC) precursors emerge during embryogenesis at day 18–19 in humans (day 7.25 in mice) as a cluster of about 20 cells. They migrate to the gonad between day 28 and 36 (or at day 10.5 in mice). Sex differentiation to spermatogonium in males and oogonium in females occurs around day 44–49 (day 13.5 in mice). In the testes of the male (top panels), the spermatogonia proliferate (curved arrow). They enter meiosis I (MI), differentiate to primary spermatocytes and undergo two meiotic reductions (MI and MII): first to secondary spermatocytes and then to haploid spermatids. The germ cells differentiate to mature sperm in the epididymus (not shown). Small expansions of pre-mutation alleles in coding and non-coding trinucleotide repeats (TNRs) are observed in both primary spermatocytes and in haploid germ cells. Large deletions of full-mutation-length alleles of non-coding TNRs are observed in dividing spermatogonia. In females (lower panels), PGCs undergo a limited number of divisions (~20) (curved arrow) and differentiate into primary oocytes that arrest during MI. The primary oocytes remain arrested in MI for years in humans. Primary oocytes complete MI at sexual maturity and form a secondary oocyte, which initiates MII but does not complete it until fertilization. Large non-coding TNR expansions occur in primary oocytes that are arrested during MI.
Males who inherit a full mutation allele from their mother harbour the expanded allele in their somatic cells but do not transmit the expanded allele to their progeny10,19,20. This is because large CGG repeat tracts in their spermatogonia are shortened around weeks 13 to 17 of fetal development20 (FIG. 1). Some genetic selection among spermatogonia for cells with smaller TNRs cannot be ruled out19. However, during development in sons of mothers with pre-mutation alleles, only the full mutation is observed in the spermatogonia at 13 weeks of gestation, but pre-mutation alleles are evident at week 17 (ref. 20). These observations suggest that at least some of the large expanded alleles are actively shortened during cell proliferation between 13 and 17 weeks of human development20 (FIG. 1). Thus, large expansions during female transmission appear to be repair-dependent, whereas the shortening of long repeat tracts during male transmission appears to depend on replication.
The patterns of mutation in patients with myotonic dystrophy type 1 (DM1) are similar to those of FXS (TABLE 1). Full mutations are almost exclusively transmitted maternally21,22. Expansion can be observed as early as the two-cell stage in pre-implantation embryos, and the length of expansion correlates with maternal age, which implies that expansion occurs in quiescent oocytes before transmission21,22. In germ cells of DM1 males, the TNR tract contracts if a disease-length TNR tract is large23-26, and the contraction frequency during male transmission increases with allele length23-26. Smaller expansions of CTG tracts from the normal to pre-mutation allele length (55–200 units) are observed in the male germ cells (TABLE 1). A study of mice that harbour ~300 CTG repeats in a human dystrophia myotonica-protein kinase (DMPK) transgene pinpointed the timing of this expansion to spermatogonia, the premeiotic, proliferation phase of male germ cell development27 (FIG. 1). TNR tracts in patients with DM1 (ref. 25) and in mouse models28,29 display a high degree of somatic instability, which increases with age in terminally differentiated somatic cells.
Overall, shared patterns of mutation emerge in patients with DM1 and FXS. Surprisingly, the large expansions of these non-coding TNRs arise in non-dividing oocytes and in terminally differentiated somatic cells; the full mutation alleles tend to contract in the dividing male spermatogonia, and small gains and losses in the pre-mutation range can be observed in both spermatogonia and in somatic cells.
Coding TNRs
Do TNRs in coding sequences expand differently from those in non-coding regions during development? For Huntington’s disease (HD), growth from a pre-mutation repeat length allele (29–35 CAGs) to the disease range (>35 CAGs) and large expansions (>7 CAGs) occur almost exclusively through paternal transmission30-32 (TABLE 1). Offspring of affected mothers are more likely to show no change or to show contractions in CAG repeat size, even if the maternal allele is of a similar size to one that expands through paternal transmission33. In males with pre-mutation alleles, small gains or losses occur over a series of generations, but after the inherited allele approaches the disease threshold, expansion is favoured three- to 175-fold over contraction30-33. Similar patterns have been observed in an HD mouse model34.
The timing of expansion in the HD male germ line is not clear but seems to occur at multiple stages of germ cell development both in humans and mice35-38. The variations in CAG tract sizes of four patients with HD were significant in their mature sperm, but tract size variations in the testes (comprising dividing spermatogonia, differentiated spermatocytes and haploid spermatids) of three patients with HD were minimal35. Therefore, some expansions apparently occur late in germ cell development. Indeed, in the R6/1 mouse model for HD, expansion is most obvious in postmeiotic haploid cells36. In patients with HD31,35 and in transgenic mouse models for HD34, instability does not correlate with the age of the father, as would be expected if expansion occurred in spermatogonia that renew germ cells throughout adult life. However, in laser-captured testicular cells from two patients with HD, the majority of disease-length expansions were found in human primordial spermatogonia, and fewer disease-length expansions were detected in the postmeiotic cell populations (but the expansions in the latter population contained the longest TNRs)37. As suggested by Yoon et al.37, expansions apparently occur before meiosis in dividing spermatogonia37,38 or after meiosis is complete in differentiating germ cells.
Somatic heterogeneity is extensive in humans with HD30,31,39,40 and in HD mouse models41-44. Expansion in humans cannot be monitored with age, but in mice the size of the allele is stable from the eight-cell-stage embryo until roughly 6–11 weeks postnatal41,43, when expansion resumes in somatic tissues and continues throughout the lifetime of the animal. In mice, laser capture44 and fluorescence activated cell sorting (FACS) analyses45 have confirmed that expansion occurs in postmitotic neurons, and the degree of expansion differs substantially among somatic tissues and brain regions44. The expansions in post-mortem brain tissue from patients with HD are so substantial that, in addition to the inherited TNRs, the CAG tract length in somatic brain cells apparently influences the onset of disease46.
Expansions in coding CAGs in spinocerebellar ataxia type 1 (SCA1) and HD share properties (TABLE 1). Expansions in patients with SCA147 and in a mouse model48 occur most often through paternal transmission (up to 28 repeats have been observed), and offspring of affected mothers are more likely to show no change or to show contractions in CAG tract size47. Instability during transmission by female patients with SCA1 or in transgenic animals48 increases with maternal age and instability is not observed in oocytes from young female mice, which suggests that expansion occurs in arrested oocytes48. Somatic changes in TNR tract length occur in patients with SCA1, but there is little increase in heterogeneity with increasing tract length.
Thus, coding TNRs also have consistent patterns: the largest expansions are observed in non-dividing somatic cells, and smaller instabilities are observed under both dividing and non-dividing conditions.
Comparing cell-type dependence in coding and non-coding TNR expansion
Because TNR alleles are subject to genetic selection, it is difficult to define a ‘one size fits all’ explanation for expansion. That is, long coding TNRs are likely to be selected against if the encoded gene product is too dysfunctional for viability, whereas TNR expansions in untranslated or intronic regions are likely to be better tolerated. Nonetheless, the features of human disease suggest that coding and non-coding TNRs share mechanisms.
In terminally differentiated somatic cells, the sizes of the expansions are surprisingly comparable for coding and non-coding TNRs (TABLE 1). For example, somatic cells in patients with DM1 can harbour non-coding TNRs that are hundreds to thousands of units long. Similarly, in post-mortem brain samples from patients with HD, coding CAG expansions of up to thousands of units (increasing with age) have been observed in the striatum, which is the brain region that is most vulnerable to cell death in HD39,40. The cells harbouring the longest expansions are the most likely to die first. Thus, the frequency of large expansions observed in the brains of patients with HD may be higher, but cannot be measured. Also, in at least two mouse models of HD, animals maintain long CAG repeats of greater than 700 units in the coding sequence49. Thus, in terminally differentiated somatic cells, coding and non-coding TNRs are capable of expanding to roughly the same degree, which suggests similar repair-dependent mechanisms.
Non-coding TNRs can expand in oocytes, and the age-dependent changes in these quiescent cells imply a repair-dependent expansion mechanism. The underlying mechanism for TNR instability is more difficult to assess in male germ cells, as larger repeat tracts in either coding or non-coding repeats seem to be selected against26. Either pre-mutation alleles undergo successive, small expansions to the disease range or smaller expansions are the residual products of large deletions that occur when TNR tracts are long. In either case, the changes are detected during proliferation.
Thus, a mechanistic explanation must be found for the following three patterns of human mutation: the occurrence of the largest expansions in non-dividing cells; the occurrence of the longest TNR deletions in dividing cells; and the observation of smaller instabilities with or without cell division. In the following sections I discuss how the currently proposed repair and replication models of expansion fit with these three human mutation patterns. For expansion to occur, single-strand loops must form to provide the ‘extra’ DNA, and then the loops must be incorporated into duplex DNA. Therefore, I first consider how loops might form in the context of human development. I then discuss how loops might be incorporated into DNA, which may occur by distinct mechanisms from loop formation.
Large expansions in non-dividing cells
It is surprising and perhaps anti-intuitive that the largest expansions occur under non-dividing conditions. So, what is happening in meiotically arrested oocytes or in terminally differentiated neurons to generate large looped intermediates? Gains of 100–1,000 units in DM1 and FXS require a mechanism that opens 300–3,000 DNA base pairs (either as a few large loops or several smaller loops) and is energetically untenable under non-dividing conditions without some sort of DNA break.
Double-strand breaks (DSBs) occur in quiescent human oocytes in the pachytene stage of meiosis13, and occur around the time that expansion is observed. However, repair of DSBs by homologous recombination, crossover or non-homologous end joining apparently does not influence expansion in humans50,51 or in mice28. Current evidence suggests that loop formation arises instead from repair of single-strand breaks (SSBs) during excision of damaged DNA bases.
Base excision repair
SSBs are generated as intermediates during the process of removing chemically damaged bases from DNA by base excision repair (BER)52 (FIG. 2). In a mouse model of HD, age-dependent TNR expansion in somatic cells occurs concomitantly with the accumulation of oxidized DNA bases45, and in a mouse model of FXS, treatment with potassium bromide, a powerful oxidant, increases CGG expansion in vivo53. Also, in human HD fibroblasts, which do not normally expand their repeats in culture, exposure to peroxide can induce expansion45. Together these results imply that expansion arises during removal of oxidized DNA bases. In support of that idea, loss of 7,8-dihydro-8-oxoguanine DNA glycosylase (OGG1), the main DNA glycosylase responsible for removal of 7,8-dihydro-8-oxoguanine (8-oxoG) (the most common oxidized base in DNA), suppresses expansion in HD mice45. In fact, the stoichiometry of BER proteins correlates with the differential somatic instability in the striatum and cerebellum in a mouse model of HD54. These findings imply that BER is involved in expansion under non-dividing conditions, but how might it happen?
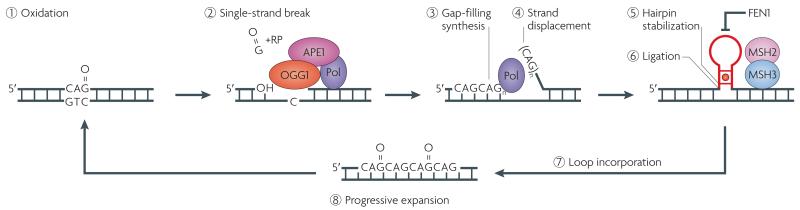
Figure 2. Loops formed during base excision repair by strand displacement: the toxic oxidation cycle.
In base excision repair (BER)52, 7,8-dihydro-8-oxoguanine DNA glycosylase (OGG1) (red oval) recognizes and removes oxidized guanines (O = G) in the DNA template (steps 1 and 2). OGG1 has two enzymatic functions: glycosylase activity and lyase activity. The glycosylase activity cleaves the C1 glycosidic bond between the ribose sugar and the base. Removal of the oxidized guanine creates an apurinic site in which the widowed cytosine (C) has no partner. OGG1 can nick the phosphodiester backbone between the C3 bond of the ribose ring and the phosphate on the 3′ side of the apurinic site. The resulting single-strand break (step 2) leaves behind a residual ribose phosphate (+RP) group. The RP is not recognized by most the polymerases. However, apurinic/apyrimidinic endonuclease 1 (APE1, also known as APEX1) (dark pink oval) processes and removes the RP site and produces a 3′ hydroxyl group (OH) suitable for extension by a DNA polymerase (Pol) (purple oval). The RP site can also be removed by Pol β, which has its own lyase activity. The trinucleotide repeat (TNR) strand is displaced during gap-filling synthesis (step 3) and TNRs from the displaced ‘flap’ (step 4) can fold back into a hairpin (a flap containing CAG is shown as an example). In normal BER, flap endonuclease 1 (FEN1) excises the flap. However, the folded 5′ end of the stable TNR hairpin prevents cleavage by FEN1 (T bar). Binding of the mismatch repair recognition complex MutS homologue 2 (MSH2)–MSH3 (light pink and blue ovals) to the A-A mismatched bases (red circle in hairpin stem) may further stabilize the repetitive hairpin (step 5) and contribute to the prevention of flap removal. The hairpin DNA is ligated (step 6) and expansion occurs after the DNA hairpin loop is incorporated into duplex DNA (step 7). Base oxidation occurs daily and throughout life. Thus, continual rounds of oxidation–excision–expansion form a ‘toxic oxidation cycle’45 and lead to TNR growth and progressive somatic mutation with age (step 8). If two oxidized bases occur on opposite strands, the hairpin on one strand may be copied by the polymerase into duplex DNA during gap-filling synthesis on the strand opposite the hairpin (not shown).
After removal of the oxidized base, BER is completed by DNA synthesis to restore the correct Watson–Crick base and ligation to reconnect the broken end (FIG. 2). In BER, gap-filling synthesis can occur using either a long patch or a short patch mechanism52. However, only the long patch repair pathway generates displaced single-strand flaps that are of sufficient size for loop formation45,52 (FIG. 2). Moreover, CAG tracts facilitate strand displacement45, and the DNA repair polymerase β (Pol β), which can fill in the gap, is stimulated by fork-like flap structures55. Normally, a flap generated by BER is removed by flap endonuclease 1 (FEN1), which has 5′ to 3′ exonuclease activity56-58 (FIG. 2). However, in vitro, a looped intermediate, such as a hairpin, ‘hides’ the 5′ end of the DNA and prevents FEN1-mediated cleavage in vitro56,58 and, presumably, in vivo56,59.
In long-patch BER, Pol β59,60 or one of the replicative polymerases (polymerase δ or polymerase ε)52,60,61 typically incorporates two to 15 nucleotides into the repair patch, which is too small a number to generate a large expansion in a single step. However, oxidation of DNA bases occurs approximately 50,000 times per day52, providing ample opportunity for many SSBs. Repeated rounds of oxidation–repair–expansion at TNRs would promote a toxic oxidation cycle for loop formation and progressive expansion in many sequential steps45,62 (FIG. 2).
However, there are problems with a BER-dependent model for large expansions. OGG1 is the only glycosylase that is known to promote TNR expansion in mice, but it is not the only glycosylase that removes 8-oxoG52,63, nor is 8-oxoG the only oxidized lesion possible within CNG (N = C, A, G) tracts52. Because G and C occur with equal frequency, oxidation is predicted to induce formation of 5-hydroxycytosine as well as 8-oxoG within CNG tracts52. Human endonuclease III-like protein 1 (NTH1)52 has a preference for removal of 5-hydroxycytosine and, theoretically, should be able to induce TNR expansion via the same SSB mechanism as OGG1 (ref. 52). However, loss of NTH1 does not reduce expansion of CAG tracts in an HD mouse model45. Moreover, other glycosylases, such as endonuclease VIII-like 1 (NEIL1), NEIL2 and NEIL3 (ref. 64) can serve as ‘back-ups’ for OGG1 in vivo65 and would also be expected to induce expansion if it occurs by BER. Enhanced oxidation might increase the demand for OGG1, or other proteins might cooperate with OGG1 to increase specificity for the TNR tract. Thus, the specific importance of OGG1 in promoting expansion is not yet fully understood.
Nucleotide excision repair
Nucleotide excision repair (NER) also induces formation of single-strand flaps, and has been implicated in TNR instability. NER consists of two subpathways (recently reviewed in refs 66,67): global genome repair (GGR) is used for lesion correction throughout the genome, whereas transcription-coupled repair (TCR) corrects lesions within actively transcribing genes66,67 (FIG. 3). A specialized TCR-related process called differentiation associated repair (DAR) is less well understood but seems to promote lesion removal from the non-transcribed strand in neurons68. Although these subpathways have distinct lesion recognition steps, the repair steps are identical (FIG. 3).
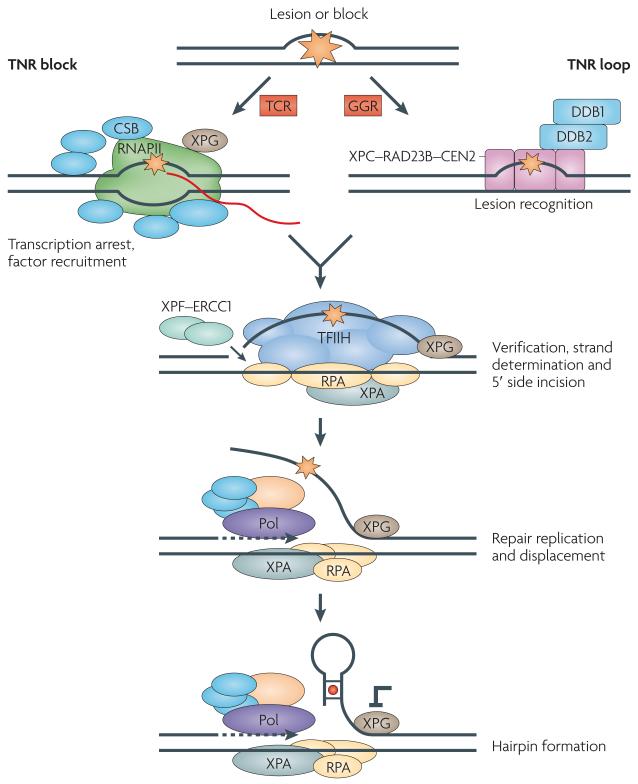
Figure 3. Nucleotide excision repair and trinucleotide repeat loop formation.
Nucleotide excision repair has two branches: transcription-coupled repair (TCR) (left) and global genome repair (GGR) (right). In a model for the involvement of TCR in trinucleotide repeat (TNR) loop formation, the GC-rich TNR sequences block (orange star) progression of RNA polymerase II (RNAPII) (the red line represents mRNA). TCR rescues the stalled polymerase through the combined action of a group of accessory proteins (blue), including Cockayne syndrome protein CSB (also known as ERCC6). See ref. 67 for further details. Xeroderma pigmentosum complementation group A (XPA) stabilizes the transcription bubble. The TNR tract should eventually be removed from DNA by the action of two nucleases: the XPF–ERCC1 complex (composed of XPF and excision cross complementing repair 1 (ERCC1)) and the endonuclease XPG. However, the incision by XPF–ERCC1 is made first. XPG has been implicated in TNR instability; it both binds and stabilizes the open transcription ‘bubble’. XPG is recruited to the opened DNA together with TFIIH (a transcription factor complex associated with RNAPII). Helicase and ATPase activities within TFIIH stimulate further opening of the bubble. Replication protein A (RPA) and XPA protect the ssDNA in the denatured bubble and stabilize the complex. Gap-filling repair replication (involving DNA polymerase (Pol) and associated proteins) and strand displacement from the 5′ side incision begin before the 3′ side incision is made by XPG. Thus, the transient flap created during strand displacement repair can fold back to form a stable hairpin loop at TNRs (mismatched bases in the stem of the hairpin are indicated by a red circle). In principle, GGR could also contribute to expansion, but it is likely to operate only if a looped-out structure is already formed (structural block indicated by an orange star) and constitutes a bulky lesion. However, neither of the two lesion sensing complexes of GGR has been implicated in expansion: XPC (part of the XPC–RAD23B–centrin 2 (CEN2) complex) has no effect on expansion in mice. The second sensor — a complex of DNA damage binding protein 1 (DDB1) and DDB2 — has not yet been tested for its effects on expansion. Figure is modified from ref. 67 © (2008) Macmillan Publishers Ltd. All rights reserved.
A complex of xeroderma pigmentosum complementation group F (XPF) and excision cross complementing repair 1 (ERCC1) makes an incision 5′ to the lesion and the endonuclease XPG makes an incision 3′ to the lesion66,67. This excises a patch consisting of 10–20 nucleotides. In vivo, the 5′ incision by XPF–ERCC1 is made before the 3′ incision by XPG69, so ssDNA containing the TNR has the potential to form a looped intermediate, at least transiently (FIG. 3). In theory, either GGR or TCR can initiate flap formation in this way. However, in mice, loss of XPC — which is specific to GGR (FIG. 3) — has little effect on CAG expansion in HD, suggesting that GGR is unlikely to cause expansion70. By contrast, loss of Cockayne syndrome protein CSB (also known as ERCC6) — which is specific to TCR (FIG. 3) — suppresses contractions in human cells, suggesting that TCR is a more relevant mechanism for TNR instability71.
Indeed, in Drosophila melanogaster, transcriptional induction of a transgene (under the control of regulatory protein GAl4) that contained exon 10 of ataxin 3 (ATXN3, also known as SCA3) resulted in expansions and contractions of the TNR tract within this exon, and loss of XPG in this line reduced instability72. In human cell lines, using a selection system for contractions (which does not detect expansions), contraction of long CAG repeats within an intron of an active hypoxanthine guanine phosphoribosyl transferase (HPRT) minigene71,73 is reduced by small interfering RNA knockdown of ERCC1, XPG and CSB. XPG binding, but not its enzymatic activity, is required at the transcription bubble to stimulate the ATPase activity of CSB74, which could explain why both XPG and CSB are involved in TNR instability. However, XPG and XPF bind together at the opened transcriptional bubble66,67, and loss of XPG may also influence bubble stability, the extent of XPF–ERCC1 incision and, consequently, TNR flap formation.
There also are problems with TCR being the mechanism for expansion. The single-strand flap that forms in TCR66,67 is typically 10–20 nucleotides long and is too small to generate large loops in a single step. For TCR to be a plausible mechanism to generate loops (a few large loops or multiple single-strand loops), TNRs must either block XPG incision (thus allowing abnormal progression of a transcription complex and large strand displacements (FIG. 3)) or TNR tracts must have an unusually high frequency of TCR-derived incisions. The latter case implies that transcriptional pausing occurs frequently within or nearby TNR tracts. In support of this idea, CTG and CGG tracts at the human DM1 and FXS loci75 and GAA repeats at frataxin (FXN)76 alter or inhibit elongation by RNA polymerase.
There also is ‘crosstalk’ between NER and BER77,78. For example, CSB cooperates with components of BER machinery79-81 (including OGG1) to remove 8-oxoG from transcribed genes. Furthermore, the BER glycosylase NEIl1 is more efficient at removal of oxidation damage that is in bubble DNA — a classic substrate for NER — than elsewhere82. Alternative pathways provide additional mechanisms to ensure removal of oxidative DNA damage, but the role of hybrid pathways in flap formation is unknown.
Small expansions in spermatogonia
The observations of small expansions in coding and non-coding TNRs in spermatogonia suggest that there are mechanisms for expansion that either depend on replication or that occur during replication.
Simple slippage
Small increases and decreases of a few bases can occur at any repetitive sequence by classic polymerase slippage8,9,83-86 (FIG. 4). For example, of the HD pre-mutation alleles that change during transmission, 78% are increases of one to three CAG units and 22% are contractions of one CAG unit87. The DNA polymerase pauses as it encounters the TNR sequence84-86 (FIG. 4). ‘Stuttering’ and misalignment on the daughter strand generates an increase in length after the next round of replication83,84. On the template strand, the advancing helicase generates ssDNA behind it, which forms a hairpin as the polymerase passes over it (FIG. 4); deletion occurs after a second round of replication.
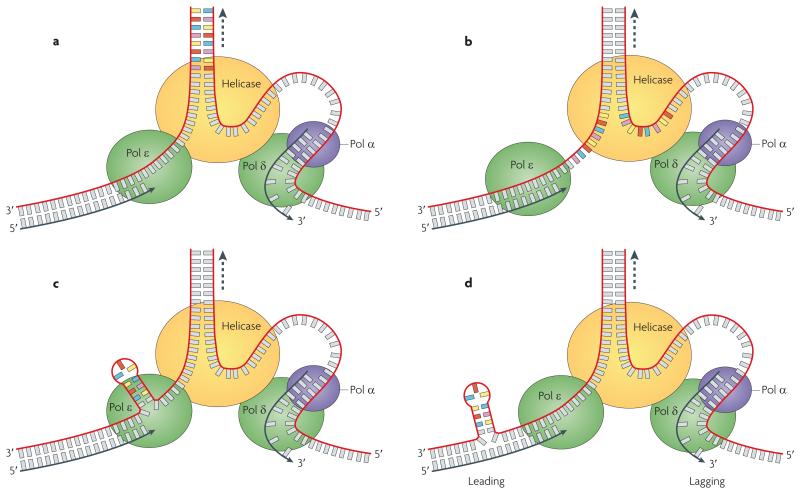
Figure 4. Slippage model for small trinucleotide repeat length changes.
Illustrated is a slippage error on the leading strand template. a | Before reaching the CAG repeat sequence (coloured bars), the replicative DNA polymerase (Pol ε on the leading strand) and DNA helicase are tightly coupled and coordinate with each other at the replication fork. b | When the polymerase encounters the CAG tract, polymerase progression is slowed but the helicase speed is unaffected (space between green and yellow ovals). c | To avoid uncoupling of the polymerase and the helicase, the leading strand polymerase bypasses a segment of unreplicated CAG template on the leading strand. d | Coupling stress is relieved when the fork has moved through the CAG repeat region and resumes copying random DNA sequence. Base pairing and processing of the looped strand results in instability of the CAG tract (not shown). Deletions occur if the loops form on the template strand and insertions occur when loop formation is on the daughter strand (not shown). Slippage can occur in both leading and lagging strand orientation, although instability (most often involving deletions) is highest when the repeats are on the lagging strand template (Pol α is the primase that synthesizes primers on the lagging strand). Figure is modified, with permission, from ref. 86 © (2008) The American Society for Biochemistry and Molecular Biology.
However, simple slippage fails to explain the expansion sizes of coding and non-coding TNRs in the disease range. Polymerase slippage is constrained by the thermodynamics of base-pairing, which limits the slippage size to a few bases8,9,88. Larger expansion could theoretically arise as the sum of many small replication slippage errors. However, in yeast and bacteria (which are good models for dividing cells), deletions of repetitive DNA and TNRs in the disease range are favoured roughly ten- to 100-fold over expansion during proliferation2,88-90, and long TNR tracts are difficult to sustain during active growth2,84,86,88-90. This is in sharp contrast to the sperm of patients with DM1 or HD, in which expansions are favoured roughly ten- to 100-fold over contractions23,30,32,33,35. In DM, for example, 80% of the changes in pre-mutation length alleles are expansions, which can be as large as 300 repeats23. Therefore, the pattern of mutation in humans does not match the profiles of TNR changes that occur in dividing yeast and bacterial cells. Furthermore, there is no correlation between the rate of expansion and the rate of division in mammalian cells91. Therefore, if large or multiple small TNR loops occur during proliferation in spermatogonia, they are more likely to arise from a repair-dependent mechanism that occurs during cell proliferation.
Replication restart
In dividing yeast, there is strong evidence for a replication-dependent repair mechanism92. Two-dimensional gel analysis of replication intermediates provides evidence that DNA polymerase has difficulty traversing and stalls on long tracts of CAG repeats93, CGG repeats94 or GAA repeats95,96. In such cases, the replication fork can ‘back-up’97, and the polymerase uses the daughter on the opposite strand to synthesize enough DNA to ‘bootstrap’ synthesis through a TNR tract (FIG. 5A). TNR loops can form as the polymerase reverses to copy the daughter strand or if TNRs misalign during restart. Electron microscopy provides direct evidence that DNA polymerase spontaneously regresses when moving through CTG repeats, which results in the predicted four-way ‘chicken foot’ intermediate98 (FIG. 5A). Replication restart is consistent with the evidence that DNA polymerase inhibitors suppress expansion94,99 and that expansion does not depend on DSB repair pathways28, as discussed above. If the leading strand polymerase copies the daughter on the lagging strand, the size of the expansion would not be expected to exceed the length of the Okazaki fragment (typically 100–200 nucleotides in eukaryotes). This is consistent with the size of many expansions in the pre-mutation range (55–200 repeats) observed in spermatogonia of males with DM1 (ref. 23). An expansion mechanism reminiscent of replication restart has been reported recently for long GAA tracts in yeast96.
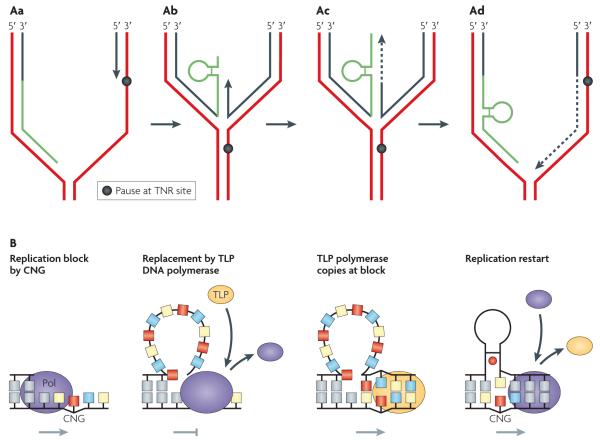
Figure 5. A model for loop formation based on polymerase stalling and restarting within a trinucleotide repeat.
A | Bypass of a lesion. Aa | The trinucleotide repeat (TNR) tract prevents polymerase passage on the leading strand template and it stalls (black circle), but the lagging strand can continue synthesis (green line). Ab | To overcome the block, the fork ‘backs-up’, forming a four-way junction resembling a ‘chicken foot’. Ac | The leading strand polymerase uses the newly synthesized daughter on the lagging strand as a template to synthesize enough DNA to pass the long non-coding TNR tract block (dashed line). Ad | TNR loops can occur during replication fork reversal or restart. In this example, a hairpin within the TNR tract (in green) is shown on the daughter strand of the lagging strand template, which is trapped to form an expansion intermediate. The template strand is in red; the nascent strand is in black. B | Multiple restart attempts by DNA polymerase (Pol) (purple oval) may induce large single-strand loops (second panel from left), which signal an SOS response. The SOS response induces expression of nucleotide excision repair machinery and translesion polymerases (TLPs), which restart replication. A CNG (N = C, A, G) TNR tract or a structural intermediate can be bypassed via a poorly understood ‘switching’ mechanism. A TLP (yellow oval) replaces a stalled replicative polymerase until lesion is bypassed. In a second switch, the TLP is displaced by the replicative polymerase and synthesis can resume. The TNR loops that are left behind form hydrogen bonded intermediates (hairpin with red circle). Grey arrows represent polymerase progress; the grey T bar represents polymerase stalling.
Large deletions in spermatogonia
How then do large deletions in non-coding TNRs occur in dividing spermatogonia? Answering this question is more difficult because deletion cannot be easily distinguished from genetic selection in vivo. However, the TNR deletion bias observed in the spermatogonia of males with DM1 (ref. 23) or FXS19,20 is consistent with the mutation pattern observed in proliferating yeast or bacterial cells (FIG. 4). This observation implies that large deletions could occur by multiple slippage events that arise as the DNA polymerase copies the TNR tracts. If the DNA polymerase fails to traverse a repeat tract, despite repeated attempts, it may leave behind unrepaired single-strand loops that — in both bacteria100 and eukaryotes101 — can induce an SOS response. This response upregulates expression of specialized ‘translesion’ polymerases (TLPs) to bypass the block and complete replication (FIG. 5B). How TLPs bypass blocks is poorly understood. However, there is evidence for a ‘switching’ mechanism100-103: a TLP replaces a stalled replicative polymerase until a block is bypassed, then the replicative polymerase displaces the TLP and synthesis resumes (FIG. 5B). Because the large single-strand loops (generated by DNA polymerase attempting to restart replication) would occur in the template strand, they would be deleted in the next round of replication. The deletion is unlikely to perfectly restore the inherited TNR length. Thus, the residual deletion products may contribute to TNR size heterogeneity in spermatogonia and may even be taken as expansions.
Loop incorporation: completing expansion
The current replication and repair models, discussed above, explain how large and small extrahelical loops might form in distinct cell types but do not address how they are incorporated into DNA. The mechanisms of loop formation and loop incorporation may not be equivalent, and independent pathways for these two parts of expansion should be considered.
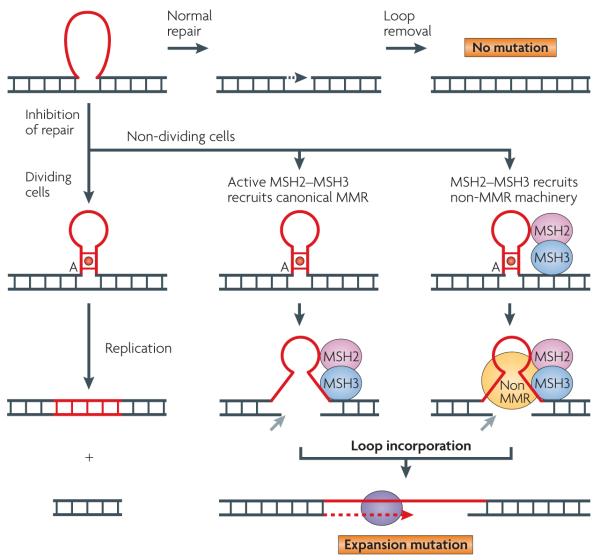
Normally, in dividing cells, small loops are removed by the mismatch repair (MMR) system in which a 5′ to 3′ exonuclease (EXO1) removes the loop, a DNA polymerase fills the gap, and the DNA duplex is faithfully resealed without mutation104 (FIG. 6, ‘normal repair’). In vitro, human cell extracts can catalyse error-free repair of CAG or CTG hairpins, in a manner similar to MMR105,106. However, for expansion to occur, uncorrected loops must be maintained to provide the ‘extra’ DNA that is eventually the expansion. Thus, at its most basic definition, expansion is a failure to remove loops (FIG. 6, ‘inhibition of repair’).
Figure 6. Three models for loop incorporation into duplex DNA.
Normally, mismatch repair (MMR) machinery removes small loops in DNA (red) during replication without mutation (top panels, ‘no mutation’). However, failure to remove the loops (‘inhibition of repair’) can result in expansion. In dividing cells, the uncorrected loop (shown here as a hairpin with an A mismatch (red circle)) is copied into DNA during the next round of replication. Therefore the trinucleotide repeat (TNR) sequence increases by the size of the DNA (red ‘ladder’). In non-dividing cells, loop incorporation is not coordinated with the replication fork. The mechanism of de novo loop removal is unknown. However, two models for loop incorporation have been proposed, both of which involve a single-strand break (nicking) on the strand opposite the hairpin (grey arrow). In the canonical MMR model (centre panels), the hairpin recruits catalytically active MutS homologue 2 (MSH2)–MSH3. The MMR machinery then introduces a nick on the opposite strand from the hairpin and a polymerase (purple oval) incorporates the loop after gap-filling synthesis. In an alternative model (right panels) MSH2–MSH3 binds to the mispaired bases in the stem of the hairpin. The interaction with the hairpin alters the function of MSH2–MSH3 such that it does not successfully carry out canonical MMR. Instead, it acts as adaptor to recruit non-MMR machinery to complete loop incorporation through a crosstalk pathway.
So, how are the uncorrected loops incorporated into duplex DNA? The bulk of the evidence suggests that TNR incorporation also involves MMR104,107. The R6/1 mouse model of HD carries the portion of the human huntingtin (HTT) gene that contains the CAG repeats41, and if these mice are crossed with mice lacking the MMR protein MutS homologue 2 (MSH2), expansion is abolished43,108. Similarly, in a DM1 mouse model, loss of the MSH2–MSH3 complex or its binding partner, PMS2, suppresses expansion109. In mice, expansion of the 5′-CTG-3′ repeat in the 3′ non-coding region of the human DMPK transgene110 and expansion of the 5-′CAG-3′ tract in the coding sequence of a truncated human HTT transgene111 both depend on MSH2–MSH3 but not on the other eukaryotic MMR complex, MSH2–MSH6. Apparently, MSH2–MSH3 has a dominant role in causing rather than correcting expansion mutations. Two current models for how this might occur are hotly debated: either an active MMR pathway aids loop incorporation or MSH2–MSH3 aids loop formation and TNR loop incorporation occurs by a non-canonical pathway.
Loop incorporation by active MMR
The MSH proteins couple DNA binding with ATP hydrolysis to carry out the first steps of MMR104. MSH2–MSH3 and MSH2–MSH6 bind and hydrolyse ATP within two conserved Walker-type nucleotide-binding domains104. Mutations in these domains can inactivate ATP binding or ATP hydrolysis or both104. If ATP hydrolysis in the MSH2 subunit were essential for loop incorporation, then loss of this function would inhibit expansion. Indeed, crossing a DM1 mouse model carrying a highly unstable CTG tract (>300 units) with mice harbouring a G693A mutation in MSH2 (which is predicted to abolish ATPase activity) stops expansion112. In yeast, expression of the same msh2-G693A mutation suppresses large and small GAA deletions113. Downstream signalling by MSH2–MSH3 involves a complex involving the MutL homologue (MLH) proteins and PMS1 (ref. 113). In yeast, a mutation in PMS1 that is predicted to impair complex formation has similar effects to the MSH2 mutation113. Purified MSH2–MSH3 binds ATP114,115 and retains at least some ATP hydrolytic activity in vitro when bound to a CAG hairpin111,114. Collectively, these findings support the notion that the ATPase function of the MSH2 subunit and the MSH2–MSH3–MLH–PMS complex are necessary for TNR instability in yeast.
Loop incorporation by alternative pathways
However, if it causes expansion, active MMR cannot be working properly. Canonical MMR removes small loops without mutation104 (FIG. 6, ‘normal repair’), whereas expansion requires the presence of an unrepaired loop precursor (FIG. 6, ‘inhibition of repair’). Indeed, the patterns of mutation during human development indicate that loops are not removed correctly during mitosis or meiosis. In an MMR-competent yeast strain, hairpins that are stabilized by hydrogen-bonding (for example, CAG, CTG or GAC repeats) escaped repair in vivo, whereas loops formed from random sequences or triplets without the capacity to form stable structures (for example, GTT repeats) were efficiently removed during meiosis without error116. Thus, in these strains, MMR was operational but the hairpin loops that were able to form stable structures were not removed. In human cell extracts, in vitro repair of loops in synthetic templates is similar whether or not MSH2 is present106, supporting the idea that MSH2 is required for loop formation rather than loop incorporation. Indeed, a model in which MSH2–MSH3 binds and stabilizes a CAG hairpin loop has been proposed36,111.
Therefore, a second scenario is that MSH2–MSH3 is essential for loop formation, but a non-canonical MMR pathway is used for loop incorporation. Several models are possible. TNR hairpins harbour mispaired bases in their stems that bind MSH2–MSH3 in vitro, but MSH2–MSH3 may fail to remove them efficiently by canonical MMR36,111. In this scenario, the bound MSH2–MSH3 acts as an ‘adaptor’ to recruit non-canonical machinery to complete repair (FIG. 6). In non-dividing cells, loop incorporation probably requires a nick on the opposite strand to the loop, loop melting and gap-filling synthesis, which leads to expansion. Enzymes that carry out these functions in the absence of a replication fork are unknown. Loops might be incorporated during the gap-filling synthesis step of BER if strand displacement occurs on the opposite strand from the hairpin. No specific enzymes that carry out loop incorporation in non-dividing cells have been definitively identified. However, it could be envisioned that, in an alternative mechanism, MSH2 retains all or part of its ATPase activity (in agreement with the mouse data discussed above), but failure to couple catalytic activity to productive repair results in recruitment of a non-canonical pathway and expansion.
Perspectives and future directions
In summary, coding and non-coding TNRs in humans apparently share mechanisms of expansion. However, the cell type and its status of division impose constraints on which mechanisms are used. Shorter changes of premutation-length alleles can apparently occur in any cell type. Whether expansions, in some cases, are residual deletion products should be considered. However, longer expansions or deletions are observed under particular circumstances. In humans, the largest expansions of coding or non-coding alleles occur in quiescent cells and involve DNA repair-dependent mechanisms. Deletions of the largest alleles are most prominent in dividing cells. Thus, the biology of human disease not only provides insights into where and when repeat instability occurs but also suggests that expansion and deletion, of at least long alleles, occur by distinct mechanisms.
Many questions and puzzling features remain. However, to continue progress, there is a crucial need to draw conclusions from appropriate experimental systems that closely reflect the properties of the cell type in which expansion occurs. It will also be important to integrate the results of different experimental approaches, as they may lead to different conclusions. For example, the role of MMR in expansion is a work in progress that will benefit greatly by considering more than one set of data. The MSH2-G693A112 mouse model provides compelling evidence for the importance of ATPase activity in expansion. However, the biochemistry of the MSH2-G693A mutant protein needs to be explored further: does it bind ATP, does the mutation influence the activity of MSH3 or might the ATPase activity of MSH3 compensate for the mutation in MSH2? In vitro repair assays are powerful systems for evaluating the enzymology of loop repair. However, cell extracts contain the machinery of many active DNA repair pathways, and discerning which ones are operating at the TNR loops is an important and challenging issue. Biochemical measurements of MMR and of other enzymes implicated in expansion need to be integrated with DNA repair assays to link enzymology with function.
In general, we are only beginning to think about the possibility of crosstalk among DNA repair pathways and their relationship with TNR expansion78. However, tantalizing pieces of evidence suggest that hybrid pathways might be important. For example, MSH2–MSH6 and the BER glycosylase, MUTYH, form a physical complex117. It can be speculated, though not yet demonstrated, that MSH2–MSH3 and OGG1 interact, which could explain the involvement of BER and MMR in TNR length changes. Future work that explores the interactions among the components of different repair pathways will be informative.
More attention needs to be paid to TLPs and their role in TNR instability. Interestingly, TLPs are involved in BER and NER — both of which are candidate pathways for expansion — and may also be involved in generating large deletions (in replication restart). In NER, the DNA damage binding proteins DDB1 and DDB2 (also known as XPE)118, which recognize cyclobutane-pyrimidine-dimers (CPDs) and UV-induced (6-4) pyrimidine photoproducts, and Pol η and Q119,120 are TLPs. In BER, Pol λ serves as a back-up for OGG1 and has fivefold greater DNA synthesis fidelity than Pol β121,122. Its use may result in robust flap formation if oxidized bases block Pol β progression at TNRs122. The majority of evidence suggests that oxidized bases in random DNA sequences do not inhibit replicative DNA polymerases, but it will be important to test whether oxidized bases in CG-rich TNR tracts promote polymerase stalling. It should also be tested whether TLPs can promote strand displacement or enhance lengthening of a single-strand flap in either BER or TCR. The use of TLPs may be a common factor in large changes in TNR length and should be explored more thoroughly.
Finally, all of these mechanisms for expansion must operate within the context of chromatin, and there is growing interest in exploring how chromatin structure and epigenetic modifications influence expansion. For example, a study of the human ATXN7 locus in transgenic mice has established a link between binding of CCCTC-binding factor (CTCF, a regulatory protein implicated in DNA conformation and genomic imprinting) and regulation of repeat instability123. Currently, the links among epigenetic changes and expansion remain enigmatic, but the influence of genome locus, post-translational modification of histones and DNA methylation on TNR expansion will be key issues to explore4,5,124,125.
In conclusion, TNR expansions of ten to 10,000 units add a stunning 30 to 30,000 base pairs to DNA during transmission and during somatic growth, and both contribute to disease onset. Thus, blocking expansion at various developmental stages is likely to be beneficial. The inherited TNR tract determines whether an individual develops disease, but progressive somatic mutation may influence, at least in part, when disease occurs. The ability to modulate expansion raises hope that the severity of pathophysiology might be reduced or its onset delayed, thereby widening the therapeutic window for these deadly TNR diseases.
Acknowledgements
I would like to thank J. Majka, V. Platt, W. Lang, E. Xun, C. Canaria and S. Bernstein for critical discussions and comments. This work is supported by US National Institutes of Health grants NS062384, NS40738, GM066359, NS060115, NS069177 and CA092584.
Glossary
- Threshold length
In the context of trinucleotide repeat alleles, the number of trinucleotide repeats at which the tract becomes unstable.
- Pre-mutation
The length of a pre-mutation trinucleotide repeat tract is within the normal range, but the pre-mutation allele has increased susceptibility to mutation in a subsequent transmission. Individuals with a pre-mutation allele will exhibit a normal phenotype but may have offspring who have a higher number of trinucleotide repeats and who might be affected by the disease.
- Anticipation
The propensity of trinucleotide repeats above a certain threshold to increase (expand) during transmission to offspring, often causing increases in disease severity and decreases in the age of onset.
- Fragile X syndrome
(Also known as Martin–Bell syndrome.) A genetic disorder caused by the expansion of a single trinucleotide gene sequence (CGG) on the X chromosome, which leads to loss of expression of the FMR1 gene.
- Full mutation
A repeat tract that is unstable and of a length that is typically associated with disease. The term is often used to distinguish such cases from alleles of a shorter, pre-mutation length that are not associated with disease.
- Normal length
In the context of trinucleotide repeat alleles, the range of repeat lengths in which the repeat tract is stable as the gene is passed to the next generation.
- Myotonic dystrophy type 1
(Also known as Steinert’s disease.) A chronic, slowly progressive, inherited, multi-systemic disease that has a severe congenital form and a milder childhood-onset form.
- Huntington’s disease
A neurodegenerative disease that is caused by an abnormal trinucleotide repeat (CAG) in the huntingtin (HTT) gene. Uncoordinated, involuntary movements and decline in mental cognition characterize the disease.
- Homologous recombination
A type of genetic recombination in which nucleotide sequences are exchanged between two similar or identical molecules of DNA. It is most widely used by cells to accurately repair harmful breaks that occur on both strands of DNA (known as double-strand breaks).
- Crossover
The exchange of material between two chromosomes. It is one of the final phases of genetic recombination and occurs during prophase I of meiosis (diplotene) in a process called synapsis. Crossover usually occurs when matching regions on matching chromosomes break and then reconnect to the other chromosome.
- Non-homologous end joining
A pathway that repairs double-strand breaks in DNA using a non-homologous chromosome. It typically uses short homologous DNA sequences, called microhomologies, to guide repair. Microhomologies are often present in single-strand overhangs on the ends of double-strand breaks.
- Base excision repair
The cellular mechanism that is primarily responsible for removing small, non-helix-distorting base lesions from the genome. It is important for removing damaged bases that could otherwise cause mutations by mispairing or lead to breaks in DNA during replication.
- Long patch
In the context of base excision repair, long patch repair is when the gap-filling polymerase induces strand displacement at the single-strand break and restores normal Watson–Crick pairing by replacing a patch of two to 15 nucleotides.
- Short patch
In the context of base excision repair, short patch repair is when the gap-filling polymerase replaces the damaged base with a single nucleotide to restore normal Watson–Crick pairing.
- Polymerase β
The major DNA repair polymerase that is used in base excision repair.
- Polymerase δ
The major polymerase that copies DNA on the discontinuous or lagging strand template during cell proliferation.
- Polymerase ε
The major polymerase that copies the leading strand template during cell proliferation.
- Apurinic
A site in duplex DNA that has lost guanine or adenine.
- Polymerase slippage
The misalignment of the DNA polymerase during cell proliferation, typically at repetitive DNA sequences. Misalignment on the template strand causes a loss in DNA bases and misalignment on the daughter strand leads to a gain in DNA bases.
Footnotes
Competing interests statement
The author declares no competing financial interests.
References
- 1.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 2.Kovtun IV, McMurray CT. Features of trinucleotide repeat instability in vivo. Cell Res. 2008;18:198–213. doi: 10.1038/cr.2008.5. [DOI] [PubMed] [Google Scholar]
- 3.La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nature Rev. Genet. 2010;11:247–258. doi: 10.1038/nrg2748. A comprehensive Review of recent progress in understanding the pathophysiology of expansion disease.
- 4.Dion V, Wilson JH. Instability and chromatin structure of expanded trinucleotide repeats. Trends Genet. 2009;25:288–297. doi: 10.1016/j.tig.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumari D, Usdin K. Chromatin remodeling in the non-coding repeat expansion diseases. J. Biol. Chem. 2009;284:7413–7417. doi: 10.1074/jbc.R800026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Day LW, Ranum LP. RNA pathogenesis of the myotonic dystrophies. Neuromuscul. Disord. 2005;15:5–16. doi: 10.1016/j.nmd.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 7.Slattery JP, Murphy WJ, O’Brien SJ. Patterns of diversity among SINE elements isolated from three Y-chromosome genes in carnivores. Mol. Biol. Evol. 2000;17:825–829. doi: 10.1093/oxfordjournals.molbev.a026361. [DOI] [PubMed] [Google Scholar]
- 8.Petruska J, Hartenstine MJ, Goodman MF. Analysis of strand slippage in DNA polymerase expansions of CAG/CTG triplet repeats associated with neurodegenerative disease. J. Biol. Chem. 1998;273:5204–5210. doi: 10.1074/jbc.273.9.5204. [DOI] [PubMed] [Google Scholar]
- 9.Hartenstine MJ, Goodman MF, Petruska J. Base stacking and even/odd behavior of hairpin loops in DNA triplet repeat slippage and expansion with DNA polymerase. J. Biol. Chem. 2000;275:18382–18390. doi: 10.1074/jbc.275.24.18382. [DOI] [PubMed] [Google Scholar]
- 10.Fu YH, et al. Variation of the CGG repeat at the Fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- 11.Sullivan AK, Crawford DC, Scott EH, Leslie ML, Sherman SL. Paternally transmitted FMR1 alleles are less stable than maternally transmitted alleles in the common and intermediate size range. Am. J. Hum. Genet. 2002;70:1532–1544. doi: 10.1086/340846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helderman-van den Enden AT, et al. Monozygotic twin brothers with the fragile X syndrome: different CGG repeats and different mental capacities. J. Med. Genet. 1999;36:253–257. [PMC free article] [PubMed] [Google Scholar]
- 13.Tripathi A, Kumar KV, Chaube SK. Meiotic cell cycle arrest in mammalian oocytes. J. Cell. Physiol. 2010;223:592–600. doi: 10.1002/jcp.22108. [DOI] [PubMed] [Google Scholar]
- 14.Rifé M, et al. Analysis of CGG variation through 642 meioses in fragile X families. Mol. Hum. Reprod. 2004;10:773–776. doi: 10.1093/molehr/gah102. This study provided evidence that the earliest detected CGG expansions in female patients with FXS occurred in the primary oocytes, suggesting that expansion occurred in non-dividing cells in humans.
- 15.Sermon K, et al. Preimplantation diagnosis for fragile X syndrome based on the detection of the non-expanded paternal and maternal CGG. Prenat. Diagn. 2000;19:1223–1230. [PubMed] [Google Scholar]
- 16.Ashley-Koch AE, et al. Examination of factors associated with instability of the FMR1 CGG repeat. Am. J. Hum. Genet. 1998;63:776–785. doi: 10.1086/302018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bontekoe CJ, et al. Instability of a (CGG)98 repeat in the Fmr1 promoter. Hum. Mol. Genet. 2001;10:1693–1699. doi: 10.1093/hmg/10.16.1693. [DOI] [PubMed] [Google Scholar]
- 18.Entezam A, et al. Regional FMRP deficits and large repeat expansions into the full mutation range in a new fragile X premutation mouse model. Gene. 2007;15:125–134. doi: 10.1016/j.gene.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reyniers E, et al. The full mutation in the FMR-1 gene of male fragile X patients is absent in their sperm. Nature Genet. 1993;4:143–146. doi: 10.1038/ng0693-143. [DOI] [PubMed] [Google Scholar]
- 20.Malter HE, et al. Characterization of the full fragile X syndrome mutation in fetal gametes. Nature Genet. 1997;15:165–169. doi: 10.1038/ng0297-165. This work provided definitive evidence that the absence of the full mutation in male patients with FXS was due to deletion of CGG tracts in their germ cells at fetal stages of development.
- 21.Dean N, Tan S, Ao A. Instability in the transmission of the myotonic dystrophy CTG repeat in human oocytes and preimplantation embryos. Fertil. Steril. 2006;86:98–105. doi: 10.1016/j.fertnstert.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 22.De Temmerman N, et al. Intergenerational instability of the expanded CTG repeat in the DMPK gene: studies in human gametes and preimplantation embryos. Am. J. Hum. Genet. 2004;75:325–329. doi: 10.1086/422762. This study indicated that the earliest detected CTG expansions in female patients with DM1 also occurred in the primary oocytes, suggesting that large expansion did not require replication.
- 23.Martorell L, et al. Germline mutational dynamics in myotonic dystrophy: allele length and age effects. Neurology. 2004;62:269–274. doi: 10.1212/wnl.62.2.269. The results indicated that the largest length changes in CTG tracts in male patients with DM1 were observed for pre-mutation alleles and the highest frequency of contractions were observed in full mutation alleles.
- 24.Ashizawa T, et al. Characteristics of intergenerational contractions of the CTG repeat in myotonic dystrophy. Am. J. Hum. Genet. 1994;54:414–423. [PMC free article] [PubMed] [Google Scholar]
- 25.Lavedan C, et al. Myotonic dystrophy: size- and sex-dependent dynamics of CTG meiotic instability, and somatic mosaicism. Am. J. Hum. Genet. 1993;52:875–883. [PMC free article] [PubMed] [Google Scholar]
- 26.Jansen G, et al. Gonosomal mosaicism in myotonic dystrophy patients: involvement of mitotic events in (CTG)n repeat variation and selection against extreme expansion in sperm. Am. J. Hum. Genet. 1994;54:575–585. [PMC free article] [PubMed] [Google Scholar]
- 27.Savouret C, et al. MSH2-dependent germinal CTG repeat expansions are produced continuously in spermatogonia from DM1 transgenic mice. Mol. Cell. Biol. 2004;24:629–637. doi: 10.1128/MCB.24.2.629-637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savouret C, et al. CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J. 2003;22:2264–2273. doi: 10.1093/emboj/cdg202. This work provided evidence from knockout mice that dispelled the notion that the homologous recombination and non-homologous end joining pathways were major mechanisms for CTG expansions.
- 29.Fortune MT, Vassilopoulos C, Coolbaugh MI, Sicilliano MJ, Mockton DG. Dramatic, expansion-biased, age-dependent, tissue-specific somatic mosaicism in a transgenic mouse model of triplet instability. Hum. Mol. Genet. 2000;9:439–445. doi: 10.1093/hmg/9.3.439. [DOI] [PubMed] [Google Scholar]
- 30.Kremer B, et al. Sex-dependent mechanisms for expansions and contractions of the CAG repeat on affected Huntington disease chromosomes. Am. J. Hum. Genet. 1995;57:343–350. [PMC free article] [PubMed] [Google Scholar]
- 31.Wheeler VC, et al. Factors associated with HD CAG repeat instability in Huntington disease. J. Med. Genet. 2007;44:695–701. doi: 10.1136/jmg.2007.050930. This analysis of a large cohort of patients with HD is a comprehensive analysis of the factors governing expansion in HD. These factors include length, age-dependence and gender-dependence of expansion.
- 32.Norremolle A, Sorensen SA, Fenger K, Hasholt L. Correlation between magnitude of CAG repeat length alterations and length of the paternal repeat in paternally inherited Huntington’s disease. Clin. Genet. 1995;47:113–119. doi: 10.1111/j.1399-0004.1995.tb03941.x. [DOI] [PubMed] [Google Scholar]
- 33.Telenius H, et al. Molecular analysis of juvenile Huntington disease: the major influence on (CAG)n repeat length is the sex of the affected parent. Hum. Mol. Genet. 1993;2:1535–1540. doi: 10.1093/hmg/2.10.1535. [DOI] [PubMed] [Google Scholar]
- 34.Kovtun IV, Therneau TM, McMurray CT. Gender of the embryo contributes to CAG instability in transgenic mice containing a Huntington’s disease gene. Hum. Mol. Genet. 2000;9:2767–2775. doi: 10.1093/hmg/9.18.2767. [DOI] [PubMed] [Google Scholar]
- 35.Telenius H, et al. Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nature Genet. 1994;6:409–414. doi: 10.1038/ng0494-409. [DOI] [PubMed] [Google Scholar]
- 36.Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nature Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 37.Yoon SR, Dubeau L, de Young M, Wexler NS, Arnheim N. Huntington disease expansion mutations in humans can occur before meiosis is completed. Proc. Natl Acad. Sci. USA. 2003;100:8834–8838. doi: 10.1073/pnas.1331390100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leeflang EP, et al. Analysis of germline mutation spectra at the Huntington’s disease locus supports a mitotic mutation mechanism. Hum. Mol. Genet. 1999;8:173–183. doi: 10.1093/hmg/8.2.173. [DOI] [PubMed] [Google Scholar]
- 39.Kennedy L, et al. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum. Mol. Genet. 2003;12:3359–3367. doi: 10.1093/hmg/ddg352. This work provided the first strong evidence for the importance of age-dependent somatic instability in HD pathogenesis. The results demonstrated that increases of up to 1,000 CAG repeats occurred in human HD striatal cells early in the disease course. These increases might influence the age of disease onset.
- 40.Shelbourne PF, et al. Triplet repeat mutation length gains correlate with cell-type specific vulnerability in Huntington disease brain. Hum. Mol. Genet. 2007;16:1133–1142. doi: 10.1093/hmg/ddm054. [DOI] [PubMed] [Google Scholar]
- 41.Mangiarini L, et al. Instability of highly expanded CAG repeats in mice transgenic for the Huntington’s disease mutation. Nature Genet. 1997;15:197–200. doi: 10.1038/ng0297-197. [DOI] [PubMed] [Google Scholar]
- 42.Wheeler VC, et al. Length-dependent gametic CAG repeat instability in the Huntington’s disease knock-in mouse. Hum. Mol. Genet. 1999;8:115–122. doi: 10.1093/hmg/8.1.115. [DOI] [PubMed] [Google Scholar]
- 43.Kovtun IV, Thornhill AR, McMurray CT. Somatic deletion events occur during early embryonic development and modify the extent of CAG expansion in subsequent generations. Hum. Mol. Genet. 2004;13:3057–3068. doi: 10.1093/hmg/ddh325. [DOI] [PubMed] [Google Scholar]
- 44.Gonitel R, et al. DNA instability in postmitotic neurons. Proc. Natl Acad. Sci. USA. 2008;105:3467–3472. doi: 10.1073/pnas.0800048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kovtun IV, et al. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. This work provided evidence for a direct causative link among oxidative DNA damage, BER and expansion during mouse development. Expansion occurred in the process of removing oxidized bases and depended on OGG1.
- 46.Swami M, et al. Somatic expansion of the Huntington’s disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum. Mol. Genet. 2009;18:3039–3047. doi: 10.1093/hmg/ddp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koefoed P, et al. Mitotic and meiotic instability of the CAG trinucleotide repeat in spinocerebellar ataxia type 1. Hum. Genet. 1998;103:564–569. doi: 10.1007/s004390050870. [DOI] [PubMed] [Google Scholar]
- 48.Kaytor MD, Burright EN, Duvick LA, Zoghbi HY, Orr HT. Increased trinucleotide repeat instability with advanced maternal age. Hum. Mol. Genet. 1997;6:2135–2139. doi: 10.1093/hmg/6.12.2135. [DOI] [PubMed] [Google Scholar]
- 49.Morton AJ, et al. Paradoxical delay in the onset of disease caused by super-long CAG repeat expansions in R6/2 mice. Neurobiol. Dis. 2009;33:331–341. doi: 10.1016/j.nbd.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 50.Richards RI, et al. Fragile X syndrome: genetic localisation by linkage mapping of two microsatellite repeats FRAXAC1 and FRAXAC2 which immediately flank the fragile site. J. Med. Genet. 1991;28:818–823. doi: 10.1136/jmg.28.12.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolff RK, Plaetke R, Jeffreys AJ, White R. Unequal crossing over between homologous chromosomes is not the major mechanism involved in the generation of new alleles at VNTR loci. Genomics. 1989;5:382–384. doi: 10.1016/0888-7543(89)90076-1. [DOI] [PubMed] [Google Scholar]
- 52.Bjelland S, Seeberg E. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat. Res. 2003;531:37–80. doi: 10.1016/j.mrfmmm.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 53.Entezam A, Lokanga AR, Le W, Hoffman G, Usdin K. Potassium bromate, a potent DNA oxidizing agent, exacerbates germline repeat expansion in a fragile X premutation mouse model. Hum. Mutat. 2010;31:611–616. doi: 10.1002/humu.21237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goula AV, et al. Stoichiometry of base excision repair proteins correlates with increased somatic CAG instability in striatum over cerebellum in Huntington’s disease transgenic mice. PLoS Genet. 2009;5:e1000749. doi: 10.1371/journal.pgen.1000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daube SS, Arad G, Livneh A. Translesion replication by DNA polymerase β is modulated by sequence context and stimulated by fork-like flap structures in DNA. Biochemistry. 2000;39:397–405. doi: 10.1021/bi991443m. [DOI] [PubMed] [Google Scholar]
- 56.Spiro C, et al. Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol. Cell. 1999;4:1079–1085. doi: 10.1016/s1097-2765(00)80236-1. [DOI] [PubMed] [Google Scholar]
- 57.Liu Y, et al. Coordination between polymerase β and FEN1 can modulate CAG repeat expansion. J. Biol. Chem. 2009;284:28352–28366. doi: 10.1074/jbc.M109.050286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henricksen LA, Veeraraghavan J, Chafin DR, Bambara RA. DNA ligase I competes with FEN1 to expand repetitive DNA sequences in vitro. J. Biol. Chem. 2002;277:22361–22369. doi: 10.1074/jbc.M201765200. [DOI] [PubMed] [Google Scholar]
- 59.Asagoshi K, et al. DNA polymerase β dependent long patch base excision repair in living cells. DNA Repair. 2010;9:109–119. doi: 10.1016/j.dnarep.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beard WA, Prasad R, Wilson SH. Activities and mechanism of DNA polymerase β. Meth. Enzymol. 2006;408:91–107. doi: 10.1016/S0076-6879(06)08007-4. [DOI] [PubMed] [Google Scholar]
- 61.Kunkel TA. Evolving views of DNA replication (in) fidelity. Cold Spring Harb. Symp. Quant. Biol. 2009;74:91–101. doi: 10.1101/sqb.2009.74.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaplan S, Itzkovitz S, Shapiro E. A universal mechanism ties genotype to phenotype in trinucleotide diseases. PLoS Comput. Biol. 2007;3:e235. doi: 10.1371/journal.pcbi.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rolseth V, et al. Widespread distribution of DNA glycosylases removing oxidative DNA lesions in human and rodent brains. DNA Repair. 2008;7:1578–1588. doi: 10.1016/j.dnarep.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hazra TK, Mitra S. Purification and characterization of NEIL1 and NEIL2, members of a distinct family of mammalian DNA glycosylases for repair of oxidized bases. Meth. Enzymol. 2006;408:33–48. doi: 10.1016/S0076-6879(06)08003-7. [DOI] [PubMed] [Google Scholar]
- 65.Takao M, et al. A back-up glycosylase in Nth1 knock-out mice is a functional Nth (endonuclease III) homologue. J. Biol. Chem. 2002;277:42205–42213. doi: 10.1074/jbc.M206884200. [DOI] [PubMed] [Google Scholar]
- 66.Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nature Rev. Genet. 2009;10:756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- 67.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nature Rev. Mol. Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 68.Nouspikel T. DNA repair in differentiated cells: some new answers to old questions. Neuroscience. 2007;145:1183–1448. doi: 10.1016/j.neuroscience.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 69.Staresincic L, et al. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 2009;28:1111–1120. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dragileva E, et al. Intergenerational and striatal CAG repeat instability in Huntington’s disease knock-in mice involve different DNA repair genes. Neurobiol. Dis. 2009;33:37–47. doi: 10.1016/j.nbd.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol. Cell. Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jung J, Bonini N. CREB-binding protein modulates repeat instability in a Drosophila model for polyQ disease. Science. 2007;315:1857–1859. doi: 10.1126/science.1139517. This work provided evidence that CAG instability could occur by a TCR process.
- 73.Lin Y, Dion V, Wilson JH. Transcription promotes contraction of CAG repeat tracts in human cells. Nature Struct. Mol. Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 74.Sarker AH, et al. Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: insights for transcription-coupled repair and Cockayne syndrome. Mol. Cell. 2005;20:187–198. doi: 10.1016/j.molcel.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 75.Parsons AM, Sinden RR, Izban MG. Transcriptional properties of RNA polymerase II within triplet repeat-containing DNA from the human myotonic dystrophy and fragile X loci. J. Biol. Chem. 1998;273:26998–27008. doi: 10.1074/jbc.273.41.26998. [DOI] [PubMed] [Google Scholar]
- 76.Grabczyka E, Usdin K. The GAA•TTC triplet repeat expanded in Friedreich’s ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 2000;28:2815–2822. doi: 10.1093/nar/28.14.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fousteri M, Mullenders LHF. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 2008;18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- 78.Kovtun IV, McMurray CT. Crosstalk of DNA glycosylases with pathways other than base excision repair. DNA Repair. 2007;6:517–529. doi: 10.1016/j.dnarep.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 79.Wong H-K, et al. Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res. 2007;35:4103–4113. doi: 10.1093/nar/gkm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thorslund T, et al. Cooperation of the Cockayne syndrome group B protein and poly(ADP-ribose) polymerase 1 in the response to oxidative stress. Mol. Cell. Biol. 2005;25:7625–7636. doi: 10.1128/MCB.25.17.7625-7636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Khobta A, Kitseraa N, Speckmanna B, Epe B. 8-Oxoguanine DNA glycosylase (Ogg1) causes a transcriptional inactivation of damaged DNA in the absence of functional Cockayne syndrome B (Csb) protein. DNA Repair. 2009;8:309–317. doi: 10.1016/j.dnarep.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 82.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 83.Kang S, Jaworski A, Ohshima K, Wells RD. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nature Genet. 1995;10:213–218. doi: 10.1038/ng0695-213. This work demonstrated that slippage could generate small increases and decreases in TNR tracts depending on the direction of replication.
- 84.Kang S, Ohshima K, Shimizu M, Amirhaeri S, Wells RD. Pausing of DNA synthesis in vitro at specific loci in CTG and CGG triplet repeats from human hereditary disease genes. J. Biol. Chem. 1995;270:27014–27021. doi: 10.1074/jbc.270.45.27014. [DOI] [PubMed] [Google Scholar]
- 85.Viguera E, Canceill D, Ehrlich DS. Replication slippage involves DNA polymerase pausing and dissociation. EMBO J. 2001;20:2587–2595. doi: 10.1093/emboj/20.10.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Delagoutte E, Goellner GM, Guo J, Baldacci G, McMurray CT. Single-stranded DNA-binding protein in vitro eliminates the orientation-dependent impediment to polymerase passage on CAG/CTG repeats. J. Biol. Chem. 2008;283:13341–13356. doi: 10.1074/jbc.M800153200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Goldberg YP, et al. Molecular analysis of new mutations for Huntington’s disease: intermediate alleles and sex of origin effects. Nature Genet. 1993;5:174–179. doi: 10.1038/ng1093-174. This work demonstrated how transmitted repeat tracts change at the threshold for HD and showed that pre-mutation alleles underwent roughly equal small losses and gains of repeats. However, after the repeat grew past the threshold, expansion became the dominant change.
- 88.Schweitzer JK, Livingston DM. The effect of DNA replication mutations on CAG tract stability in yeast. Genetics. 1999;15:953–963. doi: 10.1093/genetics/152.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kroutil LC, Kunkel TA. Deletion errors generated during replication of CAG repeats. Nucleic Acids Res. 1999;27:3481–3486. doi: 10.1093/nar/27.17.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pinder DJ, Blake CE, Lindsey JC, Leach DR. Replication strand preference for deletions associated with DNA palindromes. Mol. Microbiol. 1998;28:719–727. doi: 10.1046/j.1365-2958.1998.00831.x. [DOI] [PubMed] [Google Scholar]
- 91.Gomes-Pereira M, Fortune MT, Monckton DG. Mouse tissue culture models of unstable triplet repeats: in vitro selection for larger alleles, mutational expansion bias and tissue specificity, but no association with cell division rates. Hum. Mol. Genet. 2001;10:845–854. doi: 10.1093/hmg/10.8.845. [DOI] [PubMed] [Google Scholar]
- 92.Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Samadashwily GM, Raca G, Mirkin SM. Trinucleotide repeats affect DNA replication in vivo. Nature Genet. 1997;17:298–304. doi: 10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- 94.Voineagu I, Surka CF, Shishkin AA, Krasilnikova MM, Mirkin SM. Replisome stalling and stabilization at CGG repeats, which are responsible for chromosomal fragility. Nature Struct. Mol. Biol. 2009;16:226–228. doi: 10.1038/nsmb.1527. This work provided definitive evidence that long repeat tracts blocked polymerase passage in vivo and suggested that expansion can depend on replication-dependent repair mechanisms.
- 95.Krasilnikova MM, Mirkin SM. Replication stalling at Friedreich’s ataxia (GAA)n repeats in vivo. Mol. Cell. Biol. 2004;24:2286–2295. doi: 10.1128/MCB.24.6.2286-2295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shishkin AA, et al. Large-scale expansions of Friedreich’s ataxia GAA repeats in yeast. Mol. Cell. 2009;35:82–92. doi: 10.1016/j.molcel.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pomerantz RT, O’Donnell M. Direct restart of a replication fork stalled by a head-on RNA polymerase. Science. 2010;327:590–592. doi: 10.1126/science.1179595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fouché N, Özgür S, Roy D, Griffith JD. Replication fork regression in repetitive DNAs. Nucleic Acids Res. 2006;34:6044–6050. doi: 10.1093/nar/gkl757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang Z, Lau R, Marcadier JL, Chitayat D, Pearson CE. Replication inhibitors modulate instability of an expanded trinucleotide repeat at the myotonic dystrophy type 1 disease locus in human cells. Am. J. Hum. Genet. 2003;73:1092–1105. doi: 10.1086/379523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Maul RW, Sutton MD. Roles of the Escherichia coli RecA protein and the global SOS response in effecting DNA polymerase selection in vivo. J. Bacteriol. 2005;187:7607–7618. doi: 10.1128/JB.187.22.7607-7618.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Guo C, Kosarek-Stancel JN, Tang TS, Friedberg EC. Y-family DNA polymerases in mammalian cells. Cell. Mol. Life Sci. 2009;66:2363–2381. doi: 10.1007/s00018-009-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Waters LS, et al. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Edmunds CE, Simpson LJ, Sale JE. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol. Cell. 2008;30:4519–4529. doi: 10.1016/j.molcel.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 104.Modrich P. Mechanisms in eukaryotic mismatch repair. J. Biol. Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hou C, Chan NL, Gu L, Li GM. Incision-dependent and error-free repair of (CAG)n/(CTG)n hairpins in human cell extracts. Nature Struct. Mol. Biol. 2009;16:869–675. doi: 10.1038/nsmb.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Panigrahi GB, Lau R, Montgomery SE, Leonard MR, Pearson CE. Slipped (CTG)*(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nature Struct. Mol. Biol. 2005;12:654–662. doi: 10.1038/nsmb959. [DOI] [PubMed] [Google Scholar]
- 107.McMurray CT. Hijacking of the mismatch repair system to cause CAG expansion and cell death in neurodegenerative disease. DNA Repair. 2008;7:1121–1134. doi: 10.1016/j.dnarep.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Manley K, Shirley TL, Flaherty L, Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nature Genet. 1999;23:471–473. doi: 10.1038/70598. This work provided the first evidence that the MMR system causes rather than corrects repeat expansions.
- 109.Gomes-Pereira M, Fortune MT, Ingram L, McAbney JP, Monckton DG. Pms2 is a genetic enhancer of trinucleotide CAG.CTG repeat somatic mosaicism: implications for the mechanism of triplet repeat expansion. Hum. Mol. Genet. 2004;13:1815–1825. doi: 10.1093/hmg/ddh186. [DOI] [PubMed] [Google Scholar]
- 110.van den Broek WJ, et al. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet. 2002;11:191–198. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- 111.Owen BA, et al. [CAG]n-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nature Struct. Mol. Biol. 2005;12:663–670. doi: 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- 112.Tomé S, et al. MSH2 ATPase domain mutation affects CTG•CAG repeat instability in transgenic mice. PLoS Genet. 2009;5:e1000482. doi: 10.1371/journal.pgen.1000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim HM, et al. Chromosome fragility at GAA tracts in yeast depends on repeat orientation and requires mismatch repair. EMBO J. 2008;27:2896–2906. doi: 10.1038/emboj.2008.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tian L, Gu L, Li GM. Distinct nucleotide binding/hydrolysis properties and molar ratio of MutSα and MutSβ determine their differential mismatch binding activities. J. Biol. Chem. 2009;284:11557–11562. doi: 10.1074/jbc.M900908200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Owen BAL, Lang WH, McMurray CT. The nucleotide binding dynamics of human MSH2-MSH3 are lesion dependent. Nature Struct. Mol. Biol. 2009;16:550–557. doi: 10.1038/nsmb.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Moore H, Greenwell PW, Liu CP, Arnheim N, Petes TD. Triplet repeats form secondary structures that escape DNA repair in yeast. Proc. Natl Acad. Sci. USA. 1999;96:1504–1509. doi: 10.1073/pnas.96.4.1504. This work demonstrated that repeats that could form structures were not repaired during meiosis, whereas unstructured repeats were removed. This indicated that the DNA secondary structure prevented the removal of looped expansion intermediates.
- 117.Gu Y, et al. Human MutY homolog, a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins human MutS homolog 2/human MutS homolog 6. J.Biol. Chem. 2002;277:11135–11142. doi: 10.1074/jbc.M108618200. [DOI] [PubMed] [Google Scholar]
- 118.Kulaksiz G, Reardon JT, Sancar A. Xeroderma pigmentosum complementation group E protein (XPE/DDB2): purification of various complexes of XPE and analyses of their damaged DNA binding and putative DNA repair properties. Mol. Cell. Biol. 2005;25:9784–9792. doi: 10.1128/MCB.25.22.9784-9792.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Masutani C, et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 120.McCulloch SD, Kokoska RJ, Garg P, Burgers PM, Kunkel TA. The efficiency and fidelity of 8-oxo-guanine bypass by DNA polymerases δ and η. Nucleic Acids Res. 2009;37:2830–2840. doi: 10.1093/nar/gkp103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yoshimura M, et al. Vertebrate POLQ and POLβ cooperate in base excision repair of oxidative DNA damage. Mol. Cell. 2006;24:115–125. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Braithwaite EK, Kedar PS, Lan L. DNA polymerase λ protects mouse fibroblasts against oxidative DNA damage and is recruited to sites of DNA damage/repair. J. Biol. Chem. 2005;280:31641–31647. doi: 10.1074/jbc.C500256200. [DOI] [PubMed] [Google Scholar]
- 123.Libby RT, et al. CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination. PLoS Genet. 2008;4:e1000257. doi: 10.1371/journal.pgen.1000257. This work was the first to definitively demonstrate that chromosomal position could determine whether a repeat tract was unstable. The evidence suggested that chromatin context was a key factor in expansion.
- 124.Brock GJ, Anderson NH, Monckton DG. Cis-acting modifiers of expanded CAG/CTG triplet repeat expandability: associations with flanking GC content and proximity to CpG islands. Hum. Mol. Genet. 1999;8:1061–1067. doi: 10.1093/hmg/8.6.1061. [DOI] [PubMed] [Google Scholar]
- 125.Engel N, Thorvaldsen JL, Bartolomei MS. CTCF binding sites promote transcription initiation and prevent DNA methylation on the maternal allele at the imprinted H19/Igf2 locus. Hum. Mol. Genet. 2008;17:1306–1317. doi: 10.1093/hmg/ddl237. [DOI] [PubMed] [Google Scholar]