Abstract
Non-viral gene delivery vectors were developed for efficient gene transfer to hard to transfect mouse mammary epithelial cells. Ten modified versions of the same base poly(beta-amino ester), poly(1,4-butanediol diacrylate-co-5-amino-1-pentanol), were tested in both traditional 2-D monolayer and in 3-D organotypic cultures. The polymers self-assembled with plasmid DNA encoding enhanced green fluorescent protein to form nanoparticles (~100 nm) used to transfect the cells. Nanoparticle transfection efficacy was tuned by changes in synthesis and fabrication conditions and the transfection efficacy was analyzed using confocal microscopy and flow cytometry. The best performing polymeric nanoparticles transfected 57±6% of the cells in 2-D culture and 6±1% of the cells in 3-D culture. Small modifications to the polymer end-capping molecules and tuning of polymer molecular weight could either significantly enhance the transfection efficacy up to 6-fold or instead abolish efficacy completely. The efficacy of leading polymers was higher than that of the commercial transfection agent FuGENE® HD by a factor of 13 in 2-D and 2 in 3-D. These non-viral nanoparticles may be useful as delivery reagents or targeted therapeutics for breast cancer. This gene delivery strategy is also a promising approach for studying the normal development of the mammary gland.
1. Introduction
A broad spectrum of human diseases including cancer, cardiac disorders and neurodegenerative diseases result from a genetic defect. Gene therapy offers an attractive option to treat these genetic diseases [1]. The introduction of an exogenous therapeutic gene into diseased cells has the potential to override or replace the malfunctioning gene. Although a large number of genetic targets have been identified, the clinical success of gene therapy approaches has been limited by the lack of availability of safe and effective gene delivery vectors [2]. Two major efforts have emerged in the gene delivery community, one focused on developing viral vectors and the other on non-viral strategies. Viral vectors have evolved to transduce many types of mammalian cells with high efficiency, but there are limitations to cell-specificity, cargo capacity, manufacturing, and other challenges [3]. Clinical gene therapy trials have employed viral vectors that carry therapeutic genes and are altered to impair their replication machinery [4–7]. An initially promising viral gene therapy treatment of infants with severe combined immunodeficiency was reported by Cavazzana-Calvo et al. in April 2000 [5]. However, this trial and others have highlighted critical safety issues concerning the immunogenicity [8] and tumorogenicity [9] resulting from the use of viral agents in humans, making them ill-suited for many clinical applications [10, 11].
This setback further motivated the development of non-viral biomaterial-based vectors as a safer alternative for gene delivery. These vectors offer attractive benefits over viral systems such as ease of synthesis and processing, unlimited cargo carrying capacity, structural versatility, repeated transfection ability and biocompatibility [1]. Commercially available vectors such as FuGENE® HD (Roche) and Lipofectamine 2000 (Invitrogen) are routinely used in cell biology research, yet often exhibit lower efficacy and higher cytotoxicity than desired [12–14]. Cationic polymers condense DNA into nanoparticles via electrostatic interaction with the negatively charged DNA backbone. The key events involved in the mechanism of DNA delivery by these non-viral self-assembled polyplexes are, (1) systemic delivery to target cells, (2) endocytic cellular uptake, (3) trafficking through the cytoplasmic machinery, (4) endosomal escape, (5) DNA unpacking and polymer degradation, (6) nuclear translocation of DNA, and (7) gene expression. Each of these steps can pose a barrier that affects the transfection efficiency of these vectors [15]. Polyethylenimine (PEI) and Polylysine (PLL) are two off-the-shelf cationic polymers widely studied for gene delivery applications. However, PEI lacks biodegradable moieties and can cause high cell toxicity and PLL is unable to escape the endosomal compartment as needed for intracellular delivery [16–18].
Poly(beta-amino ester)s (PBAEs) are a newer class of polymeric vectors first developed by David Lynn and co-workers [19, 20]. The main advantage offered by PBAEs over PEI is their biodegradability via the hydrolytically cleavable ester groups. In human primary cells, the reduction in cytotoxicity is over 100-fold on a polymer mass basis [21]. More recently, in an effort to expedite the development of non-viral polymeric vectors, combinatorial polymer library approaches have been used to facilitate the creation of many potentially interesting polymer structures for gene delivery. Using high-throughput synthesis and parallel screening, a large library of over 2,000 PBAEs was created that helped elucidate the effect of small changes in polymer structure on transfection efficiency [22, 23].
In the present study, we chose to focus on identifying polymers optimized for gene delivery to mammary epithelial cells. Mammary epithelial cells are used as a model epithelial system to study epithelial polarity and migration [24], the normal development of the mammary gland [25, 26] and the events that lead to the development of breast cancer [27]. Breast cancer is a leading cause of death in women in the United States with over 40,000 deaths and 190,000 new cases in 2009 alone [28]. Mammary epithelial cells are used in both traditional 2-D cell culture and more recently in 3-D cultures in which primary cells are grown as intact tissues embedded within extracellular matrix (ECM) [29–31]. These 3-D culture techniques produce more “organotypic” results in which the cell and tissue architecture are more typical of the in vivo organ.
2. Materials and Methods
2.1. Materials
1,4-butanediol diacrylate (B4) (Alfa Aesar), 5-amino-1-pentanol (S5) (Alfa Aesar), 2-methylpentane-1,5-diamine (E4), 1-(3-aminopropyl)-4-methylpiperazine (E7), 1-(3-aminopropyl)pyrrolidine (E8), 4-aminophenyl disulfide (E9), cystamine (E10), dimethyl sulfoxide (Sigma-Aldrich), FuGENE® HD (Roche), and Lipofectamine 2000 (Invitrogen), CMV-Luc and EGFP-N1 DNA (Elim Biopharmaceuticals, Hayward, CA), modified Eagle’s medium/F12 (Invitrogen), FBS, 1% v/v penicillin/streptomycin (Sigma P0781), and Alexa-Fluro-568 conjugated Phalloidin (Invitrogen A12380) were used as received.
2.2. Polymer Synthesis and Characterization
The two-step polymer synthesis scheme is described in Figure 1. Acrylate-terminated poly(1,4-butanediol diacrylate-co-5-amino-1-pentanol) base polymer (B4S5) was first synthesized at different acrylate monomer to amine monomer molar ratios, including 1:1.1, 1:1.05, 1:1, 1.05:1, 1.1:1, and 1.2:1. As an example, to prepare the base polymer with 1.05:1 ratio, 3532 mg of 1,4-butanediol diacrylate (17.8 mmol) was added to 1754 mg of 5-amino-1-pentanol (17.0 mmol). The monomers were reacted in the dark by magnetic stirring in glass scintillation vials without addition of solvent (90 °C) or dissolved in dimethyl sulfoxide (DMSO) at 500 mg/mL (40°C). The base polymers at 1:1.05 and 1.05:1 monomer ratios were synthesized by stirring at 40°C for 48 hours. The base polymers at the other monomer ratios were synthesized by stirring at 90°C for 24 hours. In the second step, the diacrylate terminated base polymers were end-capped with amine-containing small molecules E4, E7, E8, E9 and E10. End-capping reactions were performed in 1.5 mL tubes by adding 320 μL of 0.5 M amine solution in DMSO to 480 μL of the base polymer dissolved in DMSO (167 mg/mL). The final polymers were stored in DMSO at 100 mg/mL with desiccant and at −20 °C. Polymers were analyzed by gel permeation chromatography using a Waters Breeze System and 3 Styragel Columns (7.8 × 300 mm) in series: HR 1, HR 3, and HR 4. The eluent was 95% THF/5% DMSO/0.1 M piperidine and was run at 1 mL/min.
Figure 1.
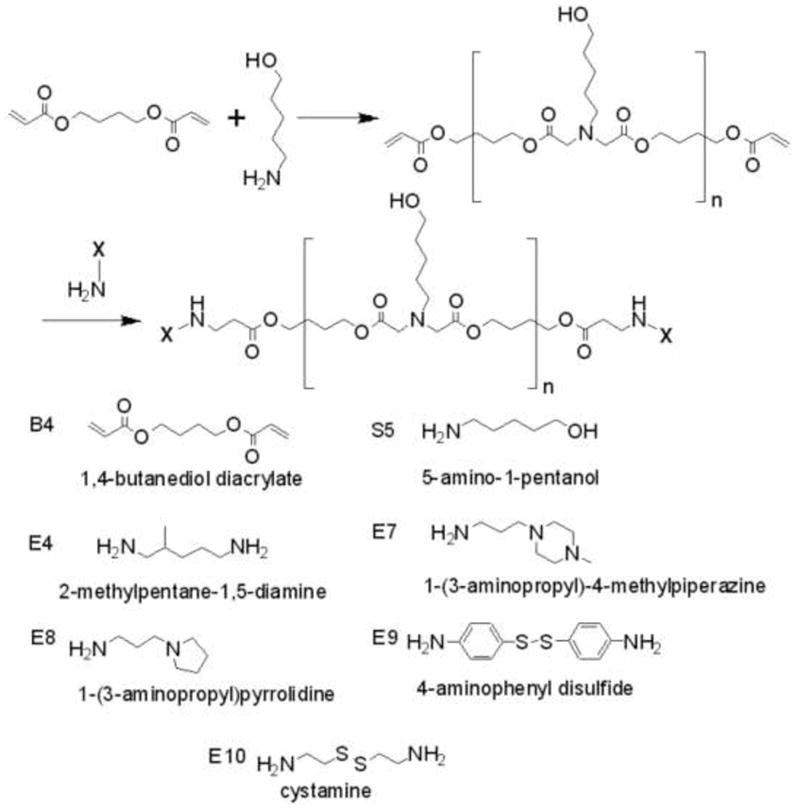
The poly(β-amino ester) (PBAE) synthesis scheme and chemical structures of monomers used for synthesis.
2.3. Particle Synthesis and Characterization
Polymer stock solutions at 100 mg/mL in DMSO and DNA stock solutions at 1 mg/mL in deionized (DI) water were separately diluted in sodium acetate (NaAc) buffer (25 mM, pH = 5) to concentrations required to synthesize particles at a polymer to DNA weight ratio (wt/wt) of 60 wt/wt. Three microliters of polymer stock solution was diluted in 197 μL NaAc buffer and 1.7 μL of DNA stock solution was diluted in 198.3 μL NaAc buffer. Two hundred microliters of diluted polymer solution was mixed vigorously with 200 μL of diluted DNA solution in a 1.5 mL eppendorf tube. The particles were formed by allowing them to self-assemble for 10 minutes and then the particle size was analyzed with two different sizing techniques, dynamic light scattering (DLS) using Malvern Zetasizer NanoZS and nanoparticle tracking analysis (NTA) using Nanosight LM10.
The particle size was first determined by the traditional DLS technique with the Malvern Zetasizer NanoZS. Four hundred microliters of the particle solution was filled into a clean disposable cuvette cell (ZEN0112) and 11 runs were performed per measurement. The Zetasizer NanoZS used a detection angle of 173° and a 633 nm laser to detect the scattering intensity of the particles in Brownian motion. Particle size is reported as the intensity-weighted Z-average of the particle diameter (nm).
The NTA was the second method used for analyzing particle size. A Nanosight LM10-HS (Amesbury, UK) system was used for particle sizing analysis including a high sensitivity Andor camera and a 404 nm laser. The particle solution was diluted 1:20 to 1:120 times in DI water to adjust the sample concentration to 106 – 109 particles/mL such that ~ 30 to 60 light scattering centers are visualized in the visual analysis window. Two hundred microliters of the particle solution was loaded into the sample chamber of the cell using a 1 mL syringe while taking care to avoid introduction of bubbles. For each sample, a 60 second movie containing the Brownian motion tracking of the particles was recorded. The movie was processed using the manufacturer recommended auto settings with manual adjustment of the gain, blur and brightness to enable detection of a least 250 particle tracks per sample. The NTA analysis gives a direct number-averaged distribution of the particle size as well as absolute particle concentration. The mean, standard deviation, and mode of the particles were calculated.
2.4. Cell Culture
EPH4 cells [32] were cultured at 37 °C and 5% CO2 in EPH4 media (Dulbecco’s modified Eagle’s medium/F12 with 10% FBS and 1% v/v penicillin/streptomycin (Sigma P0781)). After polymer transfection, cells were washed twice with 1 mL DMEM and finally incubated in fresh EPH4 media for 48 hours post transfection. Cells were fixed in 4% PFA in PBS for antibody staining. Alexa-Fluro-568 conjugated Phalloidin (Invitrogen A12380) was used to visualize the cell cortex (F-actin) and DAPI was used to stain nuclei.
2.5. Gene Delivery Assays and Flow Cytometry Analysis
2.5.1. 2-D - EPH4 cells
For high throughput transfection screening, EPH4 cells were plated at a density of 150,000 cells/mL in 96-well plates in 100 μL media per well and allowed to adhere overnight. CMV-Luc DNA was diluted in 25 mM sodium acetate (pH = 5) to varying concentrations of 0.06 mg/mL, 0.12 mg/mL and 0.18 mg/mL to respectively vary the DNA dose per well, 0.6 μg/well, 1.2 μg/well and 1.8 μg/well. Polymer stock solutions at 100 mg/mL in DMSO were diluted in the sodium acetate buffer (25 mM, pH = 5) to concentrations required to generate two different polymer to DNA weight ratios (wt/wt), 60 wt/wt and 100 wt/wt. One hundred microliters of diluted polymer solution was mixed vigorously with 100 μL of DNA solution in a 96-well plate using a multichannel pipette. The particles were allowed to self-assemble for 10 minutes, following which 20 μL of each formulation was added to the cells containing 100 μL of complete media per well in quadruplicate. The cells were incubated with the particles for four hours and then the particles were aspirated with a 12-channel aspirating wand. One hundred microliters per well of complete media was added to the cells and they were allowed to grow for two days at 37 °C and 5% CO2. FuGENE® HD and Lipofectamine 2000 were used according to the manufacturer instructions. Two days post-tranfection the CMV-Luc gene expression was measured using Bright-Glo luminescence assay kit (Promega) and a Synergy 2 multilabel plate reader (Biotek) with a one second read time per well. The BCA protein assay kit (Pierce) was used to determine the protein content per well in the Synergy 2 plate reader by measuring the absorbance at 562 nm. The effect of polymer structure, DNA dose and polymer to DNA wt/wt ratio on tranfection efficiency was studied. The results were plotted using GraphPad Prism 5 software and statistical analysis was conducted using one-way Anova and Dunnett’s multiple comparison tests. The graphs show mean ± SEM.
For the scale-up transfections for flow-cytometry studies, EPH4 cells were plated at 150,000 cells/mL in 24-well plates with 500 μL media per well and were allowed to adhere overnight. The EGFP DNA was diluted in 25 mM sodium acetate (pH = 5) to a concentration of 0.12 mg/mL to get a DNA dose per well of 6 μg/well. Polymer stock solutions at 100 mg/mL in DMSO were diluted in the sodium acetate buffer (25 mM, pH = 5) to concentration required to generate polymer to DNA weight ratio of 60 wt/wt. One hundred and ten microliters of diluted polymer solution was mixed vigorously with 110 μL of DNA solution in an eppendorf tube. After allowing the particle to self-assemble for 10 minutes, 100 μL of each formulation was added to the cells in each well of the 24-well plate in duplicates. Again after an incubation period of 4 hours, the particles were aspirated and 500 μL of fresh media was added to each well. The cells were allowed to grow in the incubator at for two days at 37°C and 5% CO2. FuGENE® HD was prepared by following the manufacturer 24-well plate protocol.
After two days, the cells were harvested from the 24-well plate for fluorescence expression quantification using flow cytometry analysis. The media was aspirated from each well, followed by a wash with 500 μL PBS (Gibco) and then 200 μL of 0.05% trypsin/EDTA was added to each well. The cells were trypsinized for 5–10 minutes at 37°C. Five hundred microliters of complete media with serum was added to each well to neutralize the trypsin. The cell suspension was transferred to a 1.5 mL eppendorf tube and was centrifuged in a microcentrifuge (5810 R) at 1000 rpm for 5 minutes. After aspirating the supernatant, the cell pellet was washed twice with PBS and eventually resuspended in 500 μL of flow cytometry buffer containing PBS, 1:50 fetal bovine serum and 1:200 parts propidium iodide (PI). The tube containing the final cell suspension was kept on ice prior to flow cytometry analysis using a BD FACScan flow cytometry scanner (Flow Cytometry Facility, Johns Hopkins School of Medicine). The flow cytometry results were analyzed using FlowJo software (Tree Star, Inc.). Propidium iodide was used to exclude dead cells from the gating. The results were quantified as % live GFP positive cells/live cells. Figure S1 shows a representation of the gating used for analyzing the flow cytometry data.
2.5.2. 3-D – Primary mammary organoid culture
Mammary epithelial cultures were prepared as previously described [26]. Briefly, glands were minced and tissue was shaken for 30 min at 37°C in a 50 mL collagenase/trypsin solution (DMEM/F12 (GIBCO-BRL), 0.1 g trypsin (GIBCO-BRL), 0.1 g collagenase (Sigma C5138), 5 mL fetal calf serum, 25 μL of 10 μg/mL insulin, and 50 μL of 50 μg/mL gentamicin). Following incubation, the collagenase solution was centrifuged at 1500 rpm for 10 min, dispersed through 10 mL DMEM/F12, centrifuged at 1500 rpm for 10 min, and then resuspended in 4 mL DMEM/F12 + 40 μL DNase (2U/μL) (Sigma). The DNase solution was shaken by hand for 2–5 min, then centrifuged at 1500 rpm for 10 min. Organoids were separated from single cells through four differential centrifugations (pulse to 1500 rpm in 10 mL DMEM/F12).
The organoids were plated in 24-well low adherent plates (Nunc Hydrocell #174909) about 30 min prior to transfection. We tested non-treated plates (Falcon #351147) as well as another brand of low adherent plates (Costar #3473) and found that the organoids were often trapped on the surface by the transfection polymer, resulting in major cell losses during handling. We observed minimal to no trapping with Hydrocell plates. Similar to the EPH4 transfection protocol, 110 μL of 0.12 mg/mL EGFP DNA was mixed with 110 μL of diluted polymer in an eppendorf tube to obtain a 60 wt/wt ratio of polymer to DNA. After a 10 min waiting time for particle self-assembly, 100 μL of formulation was added to each organoid well in duplicates per condition. The cells were incubated with the particles for 4 hours, and then the mammary epithelial fragments were collected into eppendorf tubes coated with 2.5% BSA in PBS solution and isolated by centrifugation at 90g for 2 min. Each condition was washed twice with 1 mL DMEM/F12 media followed by centrifugation step to remove residue polymer. Each condition was resuspended in 100 μL Matrigel (~200 organoids/100 mL) in a non-treated 24-well plate (Falcon #351147) on 37°C heat block for 5 min. and followed by 20 min. in a 5% CO2 incubator. Finally, 500 μL branching media (DMEM/F12, 1% v/v insulin, transferrin, selenium (Sigma) and 1% v/v penicillin/streptomycin (100× stock) with 2.5nM FGF2 (Sigma F0291)) was added to each well.
Two days following transfection, the organoids embedded in matrigel were trypsinized (12 wells at a time to avoid over trypsinization) to obtain a single cell suspension for flow cytometry analysis. The media was aspirated from each well, the cells were washed with 500 μL PBS per well and then 200 μL of 0.25% trypsin was added to each well. The Matrigel along with the embedded organoids was trypsinized for ~ 30 minutes at 37°C. The well contents were pipetted up and down several times every 10 minutes to expedite the digestion process. The trypsin in each well was then neutralized with 500 μL of HANKS solution with 4% FBS and the organoid suspension from each well was transferred into 1.5 mL eppendorf tubes. After centrifugation at 1000 rpm for 5 minutes, the pellet was washed with PBS containing 4% FBS and spun again at 1000 rpm for 5 minutes. Finally the cells were resuspended in 500 mL of the flow cytometry buffer. The samples were kept on ice prior to flow cytometry analysis. The flow cytometry analysis was conducted following the same steps as used for the 2-D EPH4 cells.
To optimize subsequent routine use of PBAE polymers for gene manipulation in organoids, we tested the efficiency of transfection in eppendorf tubes and a 24-well plate, using the best polymers identified in this study. Since these explant primary organoid cultures do not attach as a monolayer, it is possible that a tube suspension method would allow the particles to access and get internalized by greater number of cells in the culture. However, both transfection methods were found to be equally effective and subsequently the 24-well plate protocol was followed for screening new polymer conditions.
3. Results & Discussion
In this work we investigated how small molecular changes to a lead poly(beta-amino ester), poly(1,4-butanediol diacrylate-co-5-amino-1-pentanol) (referred to here as B4S5) affect gene delivery in two dimensional mammary cell culture compared to three dimensional primary organotypic cultured mouse mammary tissue. To vary gene delivery efficacy, we tune the polymer molecular weight and polymer terminal group of the polymer by varying the synthesis conditions. Polymer end groups were chosen so that they can facilitate intracellular delivery compared to the parent polymer and the 5-amino-1-pentanol side-chain. End group 2-methylpentane-1,5-diamine (E4) contains a primary amine as a terminal group and can increase polymer-DNA binding [33, 34]. Polymer end-groups 1-(3-aminopropyl)-4-methylpiperazine (E7) and 1-(3-aminopropyl)pyrrolidine (E8) contain tertiary amines and were recently reported to enhance intracellular nucleic acid delivery of DNA [35] and siRNA [36] in 2-D cultures, presumably by buffering the endosome and increasing endosomal escape. Terminal groups 4-amionphenyl disulfide (E9) and cystamine (E10) we introduce here as new end groups that could enable a “smart” triggered release of DNA to the cytoplasm. These new terminal groups can increase DNA binding while the polymers are in an oxidizing space outside the cell or in an endosome, but also decrease DNA binding when the polymers are in the reducing space of the cytoplasm, facilitating intracellular release. The combined effects of changes to base polymer molecular weight and terminal group alterations on the same base PBAE have not been previously investigated and differences between two-dimensional and three-dimensional efficacy are unknown. Here, we show how these changes to molecular structure affect nanoparticle formation and gene delivery in both two-dimensional and three-dimensional epithelial cultures. In both cell systems, the polymers synthesized here are more effective for gene delivery than FuGENE® HD, one of the leading commercially available reagents for non-viral gene delivery. These biomaterials are useful as tools to study the development of mammary epithelial cells and may also be promising as therapeutics for breast cancer.
Although all ten polymers tested were composed of the same base polymer, poly(1,4-butanediol diacrylate-co-5-amino-1-pentanol) (B4S5), small changes to synthesis conditions or to terminal group generated biomaterials with widely different transfection efficacies. These studies highlight how changes to molecular weight (MW) within the same polymer structure (including the same side chain and end-groups such as with B4S5E4s, B4S5E10s, and B4S5s) affect gene delivery. They also show that if the same base polymer with the same molecular weight only has a minor change to its terminal groups (such as B4S5 1:1.1 compared with similar polymers B4S5E4, B4S5E8, and B4S5E10), the result on gene delivery can also be dramatic. Once synthesized, these polymers were used to construct polymeric nanoparticles and the particle sizes were characterized. Subsequently, these nanoparticles were utilized in 2-D culture to find optimal transfection conditions including formulation weight ratio of polymer to DNA and DNA loading per well. Finally, they were used in 3-D culture to transfect organotypic epithelial cells.
3.1. Polymer and Nanoparticle Synthesis and Characterization
Polymers were synthesized according to Figure 1. Through varying the monomer ratios used during synthesis, B4S5-based polymers could be synthesized with molecular weight ranging from less than 10 kDa to greater than 110 kDa as shown in Figure 2A. As expected, monomer ratios closest to unity and higher temperatures produced the largest polymers. However, despite these dramatic changes to polymer size, the nanoparticles formed through self-assembly of these polymers with plasmid DNA were more similar in size and only varied by approximately twofold. Interestingly, the nanoparticle detection method itself was critical in evaluating size of these nanoparticles in aqueous solution. The standard approach to size polymeric gene delivery particles such as these is dynamic light scattering (DLS) [21, 37]. This method gives a Z-average hydrodynamic diameter that is an intensity-weighted mean diameter derived from a single-exponential fit of the intensity autocorrelation function. While this method is very accurate for monodisperse samples, in non-monodisperse samples, the intensity-averaging counts larger particles significantly more than smaller particles. A newer technique for nanoparticle sizing in aqueous conditions is Nanoparticle Tracking Analysis (NTA) [38]. In this method, each individual particle is independently sized so that a direct number-averaged mean can be calculated. As each particle is counted, a mode, or the peak in the number distribution, can also be calculated. Figure 2B shows the size of each of the nanoparticles by DLS (intensity-average) and NTA (number average and mode). While some of the DLS measurements reach into the 250 nm range, it is clear from the NTA data that the majority components of almost all the particle formulations are between 100–150 nm in size. The exceptions are formulations which are smaller than 100 nm and include B4S5 1:1.1 which has a mode of 69 nm and B4S5E9 which has a mode of 77 nm. This analysis also shows that certain distributions are monodisperse (B4S5E4s and B4S5E7), while others are polydisperse to varying degrees (non-end-capped versions of B4S5 formulated near unity as well as B4S5E8 and B4S5E9). However, polymers of a certain type formulated at different monomer ratios and possessing different molecular weights (different versions of B4S5E4s, B4S5E10s, and B4S5s) have equivalent particle uniformity. Thus, the uniformity of particle distribution may be a property of the polymer terminal group itself. All of these formulations are of a size known to facilitate particle uptake by clatharin-mediated endocytosis as well as other internalization mechanism [39]. To our knowledge, this is the first time that NTA has been used for self-assembled polymer/DNA particles, but highlights its utility, especially when combined with traditional DLS analysis.
Figure 2.
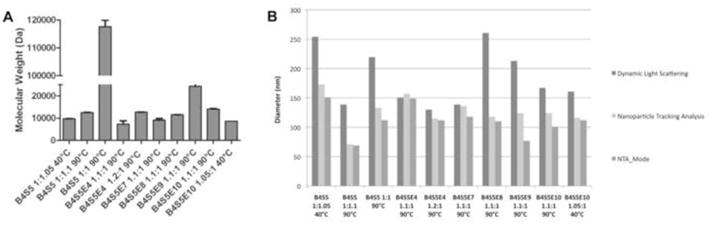
(A) The molecular weight s of the ten PBAE polymers used for transfecting EPH4 cells and organoids. Molecular weight is in Dalton (Da). Ratios are molar monomer ratios (B4:S5) used during polymer synthesis. (B) The particle sizing data of the nanoparticles formed by self-assembly of PBAE with enhanced green fluorescent protein (EGFP) DNA. The particles were sized using two techniques: Dynamic Light Scattering (intensity-weighted mean) and Nanoparticle Tracking Analysis (NTA). NTA was used to determine both the direct number-weighted mean and the mode.
3.2. Gene delivery to 2-D EPH4 cells
The EPH4 cells were initially screened in a 96-well plate by varying polymer to DNA weight ratio (wt/wt) and luciferase DNA dose per well. Cells were plated at a cell density of 150,000 cells/mL and transfected with polymer/DNA complexes formed at two different wt/wt ratios of 60 wt/wt and 100 wt/wt. For 60 wt/wt three different DNA doses per well were tested, 0.6 μg/100 μL, 1.2 μg/100 μL and 1.8 μg/100 μL and for 100 wt/wt two different DNA doses were tested, 0.6 μg/100 μL and 1.2 μg/100 μL. The 60 wt/wt and 1.2 μg/100 μL per well condition gave the highest RLU/g protein (Figure S2). Hence these conditions were chosen for the scale up 24-well plate transfections.
Transfections were next conducted in 24-well plates using GFP DNA so that delivery to individual cells, as well as the cell population overall, could be better ascertained. Figure 3A shows transfection efficacy in 2-D EPH4 cells by confocal microscopy and Figure 4A shows transfection efficacy by flow cytometry. The transfection efficacy is quantified in terms of the percent of live cells positive for GFP expression (% positive). The efficacy of the PBAE polymers is compared to FuGENE® HD, a leading non-viral commercially-available gene delivery vector for cell biology applications. The confocal images show that the EPH4 cells are healthy and agree qualitatively with the quantitative flow cytometry data. The % viability data of EPH4 cells shown in Figure S3 indicates that the PBAE vectors did not generally cause high cytotoxicity, except for B4S5E9 which had a significantly low % viability and low transfection efficacy.
Figure 3.
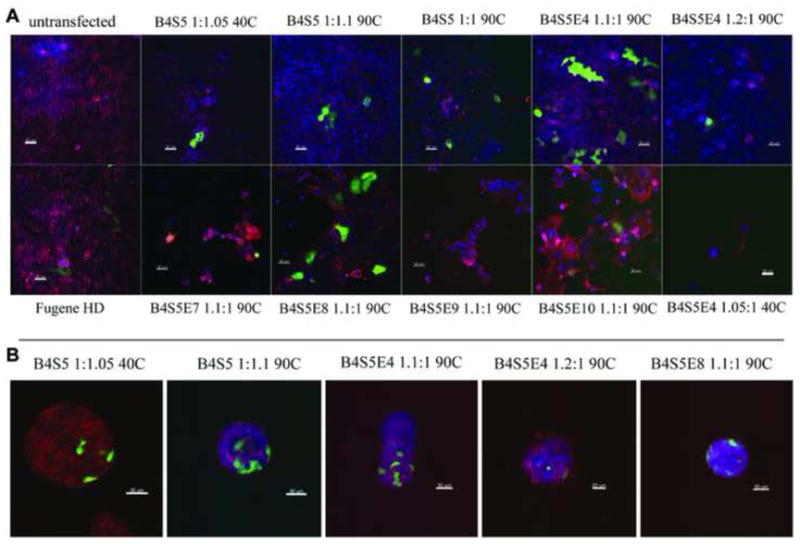
(A) 20X confocal stacks (8 μM) showing transfection efficiency of the ten PBAE polymers in 2-D EPH4 cells 2 days post-transfection. Transfected cells contain EGFP expression plasmid. Cells were fixed with 4% PFA and stained with Phalliodin-Alexa 568 (actin) and DAPI (nuclei). (B) 20X confocal stacks (8 μM) showing transfection of EGFP in mammary epithelial fragments cultured over in a 3-D matrigel. The organoids were fixed with 4% PFA and stained with Phalliodin-Alexa 568 (actin) and DAPI (nuclei).
Figure 4.
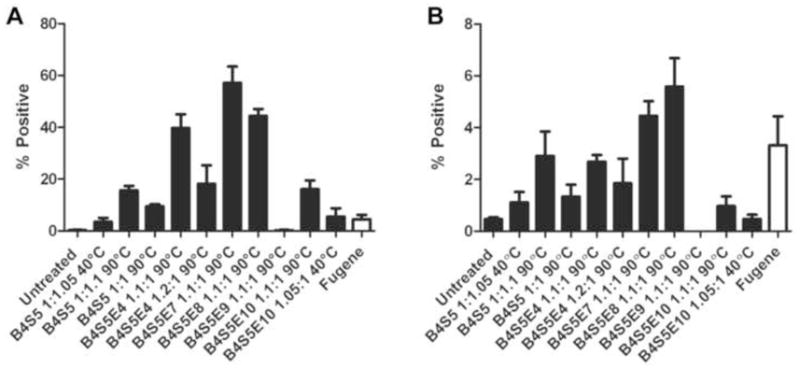
(A) Flow cytometry data showing the transfection efficiency of the ten PBAE polymers in 2-D EPH4 cells. The efficiency is quantified in terms of percentage of live EPH4 cells that are expressing EGFP (mean + SEM, n=4). Fugene HD (white) was used as a positive control. (B) Flow cytometry data showing the transfection efficiency of the ten PBAE polymers in 3-D organoid cultures. The efficiency is quantified in terms of percentage of live cells that are expressing EGFP (mean + SEM, n=4). Fugene HD (white) was used as a positive control.
3.3. Synthesis conditions and polymer molecular weight
We studied the effect of varying polymer synthesis conditions on the transfection efficiency of the PBAE polymers. The synthesis conditions were varied by changing synthesis temperature (40°C and 90°C), diacrylate:amine monomer ratio (1:1.05, 1:1.1 and 1.1:1,) and the type of solvent. The polymer molecular weight (MW) and particle size were determined at different synthesis conditions to analyze how synthesis conditions affect the biophysical properties of the non-viral delivery vectors.
The non-end-capped amino-alcohol terminated B4S5 polymer was synthesized at a lower temperature of 40°C (monomer ratio 1:1.05) and at a higher temperature of 90°C (monomer ratio 1:1.1). While monomer ratio closer to unity also increases molecular weight, synthesis temperature was found to more strongly increase molecular weight in this monomer ratio range (1.05–1.20). The polymer synthesized at a higher temperature showed a four-fold increase in percent transfected cells when compared to the polymer synthesized at a lower temperature. Similarly, for the end-capped cystamine-terminated B4S5E10 polymer, synthesis at a higher temperature of 90°C (at 1.1:1 monomer ratio) resulted in three-fold higher positively transfected cells as compared to synthesis at a lower temperature. Interestingly, the MW data (Figure 2A) shows that polymers synthesized at 90°C have higher MWs than those synthesized at 40°C for both the non-end-capped and end-capped PBAE polymers. This indicates that increasing the synthesis temperature from 40°C to 90°C increases the MW of the polymer and thereby improves the polymer transfection efficiency. This partially mirrors findings with polyethylenimine (PEI), where increased polymer size (molecular weight) was shown to directly increase gene delivery efficacy over a wider range from ~1 kDa – 70 kDa [40]. However, while for a given polymer structure, increasing molecular weight appears to increase transfection efficacy, polymer molecular weight is not the main driver of transfection efficacy overall between polymers with different terminal groups. This is evident as similarly structured polymers with the same MW of ~10 kDa have divergent transfection properties depending on the polymer terminal group. Polymers above ~12 kDa had lower, non-optimal efficacy, possibly due to reduced rates of intracellular DNA release from the polymer as has been demonstrated with higher molecular weight polylysine [41].
3.4. Synthesis procedure
Another variable that can affect the transfection efficiency is the solvent and procedure used during the end-capping step of polymer synthesis. This effect was analyzed with just polymer B4S5E4. In the standard preparation (monomer ratio of 1.1:1), B4S5E4 is end-capped in DMSO (room temperature) and used directly. In the alternative procedure (monomer ratio 1.2:1), B4S5E4 is end-capped in THF (room temperature) and then washed with ethyl ether to remove the THF before subsequently dissolving the polymer in DMSO. While we hypothesized that this extra step would enhance the transfection efficiency of the PBAE polymer in part due to achievement of a higher molecular weight, the THF-synthesized B4S5E4 was found to be lower at delivery (18±5%) compared to the DMSO synthesized B4S5E4 (40±5%) in these cells.
3.5. Terminal group
Six different versions of the PBAE polymer were synthesized at 90°C and monomer ratio of 1.1:1 by varying the small molecule terminal group of the polymers during synthesis. These six PBAE polymers include the amino-alcohol terminated B4S5 version and the five amine terminated end-capped versions B4S5E4, B4S5E7, B4S5E8, B4S5E9 and B4S5E10. These polymers were synthesized at the same monomer ratio 1.1:1 (or 1:1.1 for B4S5) and generally had similar final molecular weights (~10 kDa), although there were differences due to the end-capping groups (most notably B4S5E9 which had a larger 24 kDa size). In terms of particle size, B4S5E7, B4S5E8, and B4S5E10 were very similarly sized at 100–120 nm. B4S5E4 was slightly larger (~150 nm) and B4S5 and B4S5E9 were smaller (~69–77 nm) (Figure 2B). Yet despite these similarities in polymer size and particle size, the small changes to terminal group show dramatic effect on the transfection efficiencies of these polymers (Figure 4A). The B4S5E7 end-capped version had the highest percent GFP positive cells (57±6%), followed by B4S5E8 (44±3%) and B4S5E4 (40±5%). These three best performing polymers showed about 10-fold higher transfection efficiency as compared to that of FuGENE® HD (4±2%). Interestingly, both the E7 and E8 end-capping monomers contain tertiary amine groups, indicating that the presence of tertiary amines could potentially aid in buffering of the endosomal pH and thereby enhance the transfection efficiency. The other amine-terminated versions, B4S5 1:1.1, B4S5E9 1.1:1 and B4S5E10 1.1:1 showed relatively lower transfection efficiencies of 16±2%, 0% and 16±3%, respectively. The B4S5E9 1.1:1 polymer showed high cytotoxicity (Figure S3), which likely limited its efficacy for gene delivery.
The cytoplasmic environment inside the cell is highly reductive. The presence of disulfide-linked end-groups susceptible to degradation by the reductive intracellular environment would enable these polymers to have an enhanced DNA release capacity. The B4S5E9 and B4S5E10 polymers are end-capped with small molecules containing disulfide linkages, 4-aminophenyl disulfide (E9) and cystamine (E10), respectively. The addition of these degradable moieties could allow fine tuning the degradation of the PBAEs and possibly avoid some of the DNA release problems associated with higher molecular weight polymers [41]. While the transfection efficiency of B4S5E10 was four-fold higher than FuGENE® HD for EPH4 cells, additional work is needed to further tune polymeric vectors with multimodal degradation capacities for enhanced intracellular nucleic acid delivery.
3.6. Gene delivery to 3-D organoids
Gene therapy requires delivery to cells within intact tissues. Accordingly we also tested the ability of PBAE based polymers to transfect mammary epithelial cells within intact epithelial fragments (“organoids”). We hypothesized that we might see differences in the magnitude of transfection efficiency and/or in the optimal transfection polymer in the 3-D organoids culture when compared to 2-D EPH4 culture.
Qualitative pre-screening trials performed to optimize the transfection conditions indicated that the most efficient transfection results were obtained for the conditions with high MW versions of polymer, 60 wt/wt formulations, and 6 μg of DNA dose per well in a 24-well plate. These conditions were then used to conduct transfection experiments for quantitative analysis using flow cytometry. Figure 4B shows the flow cytometry data for the transfection efficiency of the ten PBAE polymers used to transfect the organoids culture with GFP plasmid. The transfection efficiency is quantified in terms of percent of live cells positive for GFP expression (% positive). The efficacy of the PBAE polymers is compared to FuGENE® HD as a positive control. Confocal images (Figure 3B) agree with flow cytometry data, highlight multiple transfected cells per organoid, and show good morphology.
3.7. Synthesis conditions and comparison of 3-D to 2-D
In general, the polymers synthesized at a higher temperature had a higher molecular weight (MW) than those synthesized at a lower temperature. The values of positively transfected live cells seen for the organoid cultures are low compared to the 2-D cultures (< 10%) (Figure 4B). This can be explained in part by the fact that the nanoparticles are primarily taken up by the cells in the periphery of the 3-D structure, while most of the cells in the interior remain untransfected. In isolating the organoids into single cells, viable cell recovery is low and many of the transfected cells (as well as the untransfected cells) are lost (data not shown). The non-end-capped B4S5 40°C polymer (MW = 9.6±0.1 kDa) transfected at 1±0.4% transfection efficiency while the B4S5 90°C version (MW = 12.5±0.2 kDa) transfected at 3±1% efficiency. Similarly, the B4S5E10 90°C version (MW = 14.0 kDa±0.4 kDa) gave a slightly higher % GFP positive cells than the 40°C version (MW = 8.63±0.01 kDa). These conditions indicate that for the same polymer, the transfection efficiency increases with increasing molecular weight in 3-D as well as 2-D.
3.8. Terminal groups and comparison of 3-D to 2-D
Changes in the small molecule used for end-capping the base (diacrylate terminated) polymer can have a significant effect on the transfection efficiency of the polymers. To investigate the effect of such small molecular changes on 3-D organoid transfection, six versions of the PBAE polymer were screened. These included the amino-alcohol terminated B4S5 version and the five amine terminated end-capped versions B4S5E4, B4S5E7, B4S5E8, B4S5E9 and B4S5E10 synthesized at 90°C and 1.1:1 monomer ratio. The flow cytometry data (Figure 4B) of the transfected organoid cultures show that the B4S5E8 end-capped version had the highest percent GFP positive cells (6±1%), followed by B4S5E7 (4.0±0.6%) and B4S5 (3.0±0.3%). The transfection efficiency of the leading commercially-available agent, FuGENE® HD, was 3±1%. While these polymers are promising for gene delivery to organoids compared to other reagents, there is a significant need for improved design to allow for increased nanoparticle penetration into the organoids as well as and into tissues in vivo. One possibility is to form nanoparticles dramatically smaller in size and/or employ coating strategies [42] to improve uptake through cell specificity [43] or tissue specificity [44].
Importantly, this work shows that mosaic GFP expression is maintained in cultures transfected with these polymer even after 7 days culture, while GFP expression is absent in the FuGENE® HD condition (Figure 5). Morphology and viability are both good. Continued expression of mosaically labeled cells undergoing morphogenesis is very desirable to contrast individual cell behaviors in the tissue as a whole while studying epithelial polarity and migration. While the absolute level of transfection in 3D organoids is modest, this technology can be used to deliver Cre recombinase mosaically to mammary epithelium from transgenic mice carrying targeted loxp flanked alleles.
Figure 5.
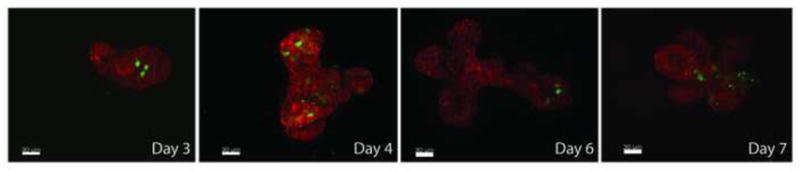
Confocal stacks of B4S5E8 1.1:1 90°C (polymer) based EGFP transfection of 3D organoid mammary epithelium culture. Mosaic EGFP expression is maintained in cultures transfected with PBAE even after 7 days culture. Epithelial fragment was fixed with 4% PFA and stained with Phalliodin-Alexa 568 (actin). Continued expression of mosaically labeled cells undergoing morphgenesis is very desirable to contrast individual cell behaviors in the tissue as a whole. In addition, using floxed gene mammary tissue one can establish mosaic loss of function assays.
In comparing transfection efficacy of 2-D to 3-D cultures, generally data follows a positive linear relationship where transfection in three-dimensional organotypic culture is approximately 10% of transfection efficacy in two dimensional cell culture (Figure 6). Exceptions to this trend include FuGENE® HD (pink downward triangle) which has low overall efficacy, but similar efficacy between 2-D and 3-D cultures. Outliers on the other end are B4S5E4 1.1:1 90°C (blue triangle) and B4S5E7 1.1:1 90°C (red triangle) that although have high 2-D efficacy, have lower 3-D efficacy than predicted. B4S5E8 1.1:1 90°C (green circle) was the most effective for 3-D transfection overall. Interestingly, this polymer was three times as effective for 3-D transfection as B4S5E4 1.1:1 90°C (blue triangle) even though their polymer structures and 2-D efficacies were almost identical.
Figure 6.
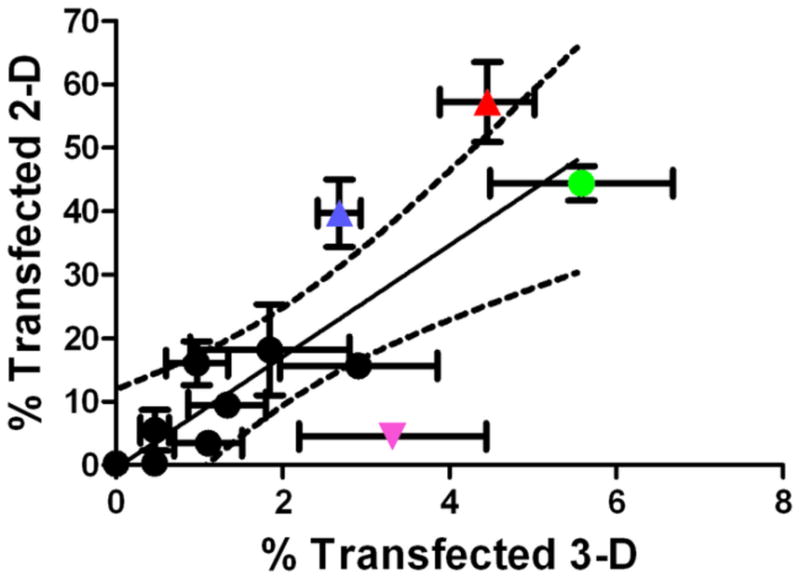
Scatter-plot comparing the transfection efficacy (% positive) of polymeric nanoparticles in 2-D culture and 3-D culture. The colored data points include FuGENE® HD (pink downward triangle), B4S5E4 1.1:1 90°C (blue triangle), B4S5E7 1.1:1 90°C (red triangle) and B4S5E8 1.1:1 90°C (green circle).
The low transfection efficiency in 3-D organotypic cultures achieved with the PBAE vectors is consistent with the results reported by other studies employing 3-D tissue culture models to develop gene delivery strategies for in vivo applications. In one study, Mellor et al. used PEI polyplexes to transfect plasmid DNA into multicellular tumor spheroids (MCTS), an in vitro model of solid tumor. They found spatially limited transfection to cells at the MCTS periphery, as the complexes failed to reach the deeper quiescent regions, even with the use of electroporation [45]. Such spatial limitation of transgene expression is not seen in 2-D monolayer cultures. In another study with the MCTS model, the use of 3-D spheroids revealed the difference in cytotoxicity of PEGylated and non-PEGylated polyplexes that was not detected with the 2-D monolayer cultures, indicating that vital cytotoxicity data can be excluded if vectors are screened solely in 2-D culture systems [46]. Important differences in gene expression have also been noted between 2-D and 3-D systems [47]. In some studies, hydrogels have been used as in vitro scaffolds to support tissue formation by enhanced presentation of specific biological cues through locally controlled gene delivery of tissue inductive factors. Rieux et al. reported the use of fibrin based hydrogel, with cells encapsulated within or seeded onto the hydrogel, to investigate gene delivery efficacy using lipoplexes. They found that the transgene expression increased with time over a period of 11 days in culture and was significantly higher in 2-D as compared to 3-D cultures. Their study suggested that the hydrogel component fibrin interacts with and sequesters the lipoplexes thus limiting cellular internalization and causing a delay in expression [48].
The comparison of transfection efficacy between 2-D and 3-D mammary epithelial cultures indicates that (a) transfection of the 3-D organotypic cultures is more difficult than transfection of 2-D cultures, but likely models some of the key challenges for in vivo gene therapy and (b) many of the same variables that affect transfection efficiency in 2-D (e.g. MW, solvent, polymer structure) also effect transfection efficiency similarly in 3-D, but the magnitude of transfection is lower in 3-D. This study identified highly effective reagents for 2-D transfection and useful reagents and important parameters for 3-D transfection. As a part of future studies, developing improved 3-D assays would be useful for high throughput screening applications [24] to identify next-generation materials. Use of these agents for partial transfection of mammary epithelial cells in vivo would be interesting to study mosaic delivery of Cre recombinase [49]. Partial transfection of a similar cell population may be sufficient for other in vivo applications as well. One such application is suicide gene therapy for tumor regression. In this approach, a gene is delivered that encodes an enzyme that converts a prodrug to its active form. Thus, there is catalytic generation of a cytotoxic anti-cancer agent at the tumor site itself and this agent can diffuse to adjacent cells and deeper into the tumor. This approach has shown promise for breast cancer in early clinical trials [50]. A related strategy is to introduce the suicide genes to cells of a preneoplastic tissue at risk, and these drug susceptible cells will then sensitize all the cells produced in the clonal population [51]. In either case, there is an amplified anti-cancer response following initial cell transfection.
4. Conclusions
Small polymer structural changes were found to tune gene delivery efficacy in both 2-D and 3-D mammary epithelial cell culture. Although all ten polymers tested were composed of the same base polymer, poly(1,4-butanediol diacrylate-co-5-amino-1-pentanol) (B4S5), small changes to synthesis conditions or to terminal group generated biomaterials with widely different transfection efficacies. In general, 2-D transfection positively correlated with 3-D transfection, but there were some exceptions. The degradable polymers synthesized here formed ~100 nm nanoparticles through self-assembly and were more effective than FuGENE® HD for gene delivery in both 2-D culture and 3-D organotypic culture. These polymeric nanoparticles may be useful as reagents and therapeutics for breast cancer.
Supplementary Material
Acknowledgments
Bryan Welm is acknowledged for mammary epithelial cells isolation protocols for FACS analysis. Sunetra Biswas and Erica Dresselhaus are thanked for assistance with preliminary studies. The authors thank the TEDCO MSCRF (2009-MSCRFE-0098-00) for support.
References
- 1.Putnam D. Polymers for gene delivery across length scales. Nat Mater. 2006;5(6):439–51. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- 2.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discovery. 2005;4(7):581–93. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 3.Verma IM, Somia N. Gene therapy -- promises, problems and prospects. Nature. 1997;389(6648):239–42. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 4.Blaese RM, Culver KW, Miller AD, Carter CS, Fleisher T, Clerici M, et al. T lymphocyte-directed gene therapy for ADA- SCID: initial trial results after 4 years. Science. 1995;270(5235):475–80. doi: 10.1126/science.270.5235.475. [DOI] [PubMed] [Google Scholar]
- 5.Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288(5466):669–72. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- 6.Kay MA, Manno CS, Ragni MV, Larson PJ, Couto LB, McClelland A, et al. Evidence for gene transfer and expression of factor IX in haemophilia B patients treated with an AAV vector. Nat Genet. 2000;24(3):257–61. doi: 10.1038/73464. [DOI] [PubMed] [Google Scholar]
- 7.Khuri FR, Nemunaitis J, Ganly I, Arseneau J, Tannock IF, Romel L, et al. a controlled trial of intratumoral ONYX-015, a selectively-replicating adenovirus, in combination with cisplatin and 5-fluorouracil in patients with recurrent head and neck cancer. Nat Med. 2000;6(8):879–85. doi: 10.1038/78638. [DOI] [PubMed] [Google Scholar]
- 8.Hollon T. Researchers and regulators reflect on first gene therapy death. Nat Med. 2000;6(1):6. doi: 10.1038/71545. [DOI] [PubMed] [Google Scholar]
- 9.Check E. Gene therapy put on hold as third child develops cancer. Nature. 2005;433(7026):561. doi: 10.1038/433561a. [DOI] [PubMed] [Google Scholar]
- 10.Cavazzana-Calvo M, Thrasher A, Mavilio F. The future of gene therapy. Nature. 2004;427(6977):779–81. doi: 10.1038/427779a. [DOI] [PubMed] [Google Scholar]
- 11.Check E. Gene therapy put on hold as third child develops cancer. Nature. 2005;433(7026):561. doi: 10.1038/433561a. [DOI] [PubMed] [Google Scholar]
- 12.Gheisari Y, Soleimani M, Azadmanesh K, Zeinali S. Multipotent mesenchymal stromal cells: optimization and comparison of five cationic polymer-based gene delivery methods. Cytotherapy. 2008;10(8):815–23. doi: 10.1080/14653240802474307. [DOI] [PubMed] [Google Scholar]
- 13.Yalvac ME, Ramazanoglu M, Gumru OZ, Sahin F, Palotas A, Rizvanov AA. Comparison and optimisation of transfection of human dental follicle cells, a novel source of stem cells, with different chemical methods and electro-poration. Neurochem Res. 2009;34(7):1272–7. doi: 10.1007/s11064-008-9905-4. [DOI] [PubMed] [Google Scholar]
- 14.Srinivasan C, Burgess DJ. Optimization and characterization of anionic lipoplexes for gene delivery. J Control Release. 2009;136(1):62–70. doi: 10.1016/j.jconrel.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 15.Schaffert D, Wagner E. Gene therapy progress and prospects: synthetic polymer-based systems. Gene Ther. 2008;15(16):1131–8. doi: 10.1038/gt.2008.105. [DOI] [PubMed] [Google Scholar]
- 16.Sonawane ND, Szoka FC, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J Biol Chem. 2003;278(45):44826–31. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 17.Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med. 2005;7(5):657–63. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 18.Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol Ther. 2005;11(6):990–5. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Lynn DM, Langer R. Degradable poly(beta-amino esters): Synthesis, characterization, and self-assembly with plasmid DNA. J Am Chem Soc. 2000;122:10761–8. [Google Scholar]
- 20.Lynn DM, Anderson DG, Putnam D, Langer R. Accelerated discovery of synthetic transfection vectors: Parallel synthesis and screening of a degradable polymer library. J Am Chem Soc. 2001;123:8155–6. doi: 10.1021/ja016288p. [DOI] [PubMed] [Google Scholar]
- 21.Green JJ, Shi J, Chiu E, Leshchiner ES, Langer R, Anderson DG. Biodegradable polymeric vectors for gene delivery to human endothelial cells. Bioconjug Chem. 2006;17:1162–9. doi: 10.1021/bc0600968. [DOI] [PubMed] [Google Scholar]
- 22.Anderson DG, Lynn DM, Langer R. Semi-automated synthesis and screening of a large library of degradable cationic polymers for gene delivery. Angew Chem Int Ed Engl. 2003;42:3153–8. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- 23.Green JJ, Langer R, Anderson DG. A combinatorial polymer library approach yields insight into nonviral gene delivery. Acc Chem Res. 2008;41(6):749–59. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simpson KJ, Selfors LM, Bui J, Reynolds A, Leake D, Khvorova A, et al. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nat Cell Biol. 2008;10(9):1027–38. doi: 10.1038/ncb1762. [DOI] [PubMed] [Google Scholar]
- 25.Fata JE, Mori H, Ewald AJ, Zhang H, Yao E, Werb Z, et al. The MAPK(ERK-1,2) pathway integrates distinct and antagonistic signals from TGFalpha and FGF7 in morphogenesis of mouse mammary epithelium. Dev Biol. 2007;306(1):193–207. doi: 10.1016/j.ydbio.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev Cell. 2008;14(4):570–81. doi: 10.1016/j.devcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waerner T, Alacakaptan M, Tamir I, Oberauer R, Gal A, Brabletz T, et al. ILEI: a cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell. 2006;10(3):227–39. doi: 10.1016/j.ccr.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 28.American Cancer Society. Cancer facts and figures 2009. 2009 Online. Available from URL: http://www.cancer.org.
- 29.Griffith LG, Swartz MA. Capturing complex 3D tissue physiology in vitro. Nat Rev Mol Cell Biol. 2006;7(3):211–24. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 30.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5(9):675–88. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 31.Nelson CM, Bissell MJ. Modeling dynamic reciprocity: engineering three-dimensional culture models of breast architecture, function, and neoplastic transformation. Semin Cancer Biol. 2005;15(5):342–52. doi: 10.1016/j.semcancer.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fialka I, Schwarz H, Reichmann E, Oft M, Busslinger M, Beug H. The estrogen-dependent c-JunER protein causes a reversible loss of mammary epithelial cell polarity involving a destabilization of adherens junctions. J Cell Biol. 1996;132(6):1115–32. doi: 10.1083/jcb.132.6.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Green JJ, Zugates GT, Tedford NC, Huang Y, Griffith LG, Lauffenburger DA, et al. Combinatorial modification of degradable polymers enables transfection of human cells comparable to adenovirus. Adv Mater. 2007;19(19):2836–42. [Google Scholar]
- 34.Zugates GT, Peng W, Zumbuehl A, Jhunjhunwala S, Huang YH, Langer R, et al. Rapid Optimization of Gene Delivery by Parallel End-modification of Poly(beta-amino ester)s. Mol Ther. 2007;15(7):1306–12. doi: 10.1038/mt.sj.6300132. [DOI] [PubMed] [Google Scholar]
- 35.Sunshine J, Green JJ, Mahon K, Yang F, Eltoukhy A, Nguyen DN, et al. Small molecule end group of linear polymer determine cell-type gene delivery efficacy. Adv Mater. 2009;21(48):4947–51. doi: 10.1002/adma.200901718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JS, Green JJ, Love KT, Sunshine J, Langer R, Anderson DG. Gold, poly(beta-amino ester) nanoparticles for small interfering RNA delivery. Nano Lett. 2009;9(6):2402–6. doi: 10.1021/nl9009793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang C, Li H, Goh SH, Li J. Cationic star polymers consisting of [alpha]-cyclodextrin core and oligoethylenimine arms as nonviral gene delivery vectors. Biomaterials. 2007;28(21):3245–54. doi: 10.1016/j.biomaterials.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 38.Filipe V, Hawe A, Jiskoot W. Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm Res. 27(5):796–810. doi: 10.1007/s11095-010-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hillaireau H, Couvreur P. Nanocarriers’ entry into the cell: relevance to drug delivery. Cell Mol Life Sci. 2009;66(17):2873–96. doi: 10.1007/s00018-009-0053-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Godbey WT, Wu KK, Mikos AG. Size matters: Molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle. J Biomed Mater Res. 1999;45(3):268–75. doi: 10.1002/(sici)1097-4636(19990605)45:3<268::aid-jbm15>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 41.Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67(5):598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 42.Shmueli RB, Anderson DG, Green JJ. Electrostatic surface modifications to improve gene delivery. Expert Opin Drug Deliv. 7(4):535–50. doi: 10.1517/17425241003603653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Green JJ, Chiu E, Leshchiner ES, Shi J, Langer R, Anderson DG. Electrostatic ligand coatings of nanoparticles enable ligand-specific gene delivery to human primary cells. Nano Lett. 2007;7(4):874–9. doi: 10.1021/nl062395b. [DOI] [PubMed] [Google Scholar]
- 44.Harris TJ, Green JJ, Fung PW, Langer R, Anderson DG, Bhatia SN. Tissue-specific gene delivery via nanoparticle coating. Biomaterials. 2010;31(5):998–1006. doi: 10.1016/j.biomaterials.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mellor HR, Davies LA, Caspar H, Pringle CR, Hyde SC, Gill DR, et al. Optimising non-viral gene delivery in a tumour spheroid model. J Gene Med. 2006;8(9):1160–70. doi: 10.1002/jgm.947. [DOI] [PubMed] [Google Scholar]
- 46.Oishi M, Nagasaki Y, Nishiyama N, Itaka K, Takagi M, Shimamoto A, et al. Enhanced growth inhibition of hepatic multicellular tumor spheroids by lactosylated poly(ethylene glycol)-siRNA conjugate formulated in PEGylated polyplexes. Chem Med Chem. 2007;2(9):1290–7. doi: 10.1002/cmdc.200700076. [DOI] [PubMed] [Google Scholar]
- 47.Dash A, Inman W, Hoffmaster K, Sevidal S, Kelly J, Obach RS, et al. Liver tissue engineering in the evaluation of drug safety. Expert Opin Drug Metab Toxicol. 2009;5(10):1159–74. doi: 10.1517/17425250903160664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.des Rieux A, Shikanov A, Shea LD. Fibrin hydrogels for non-viral vector delivery in vitro. J Control Release. 2009;136(2):148–54. doi: 10.1016/j.jconrel.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi S, Ito Y, Hatake K, Sugimoto Y. Gene therapy for breast cancer--Review of clinical gene therapy trials for breast cancer and MDR1 gene therapy trial in Cancer Institute Hospital. Breast Cancer. 2006;13(1):8–15. doi: 10.2325/jbcs.13.8. [DOI] [PubMed] [Google Scholar]
- 51.Moolten FL, Vonderhaar BK, Mroz PJ. Transduction of the herpes thymidine kinase gene into premalignant murine mammary epithelial cells renders subsequent breast cancers responsive to ganciclovir therapy. Hum Gene Ther. 1996;7(10):1197–204. doi: 10.1089/hum.1996.7.10-1197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


