Abstract
Transition Path Sampling is a well known technique that generates reactive paths ensembles. Due to the atomic detail of these reactive paths, information about chemical mechanisms can be obtained. We present here a comparative study of Bacillus stearothermophilus and human heart homologs of lactate dehydrogenase. A comparison of the transition path ensemble of both enzymes revealed that small differences in the active site reverses the order of the particle transfer of the chemical step. Whereas the hydride transfer preceded the proton transfer in the heart heart LDH, the order is reversed in the Bacillus stearothermophilis homolog (in the direction of pyruvate to lactate). In addition, transition state analysis revealed that the dividing region that separates reactants and products, the separatrix, is likely wider for BsLDH as compared to human heart LDH. This would indicate a more variable transition process in the Bacillus enzyme.
1. INTRODUCTION
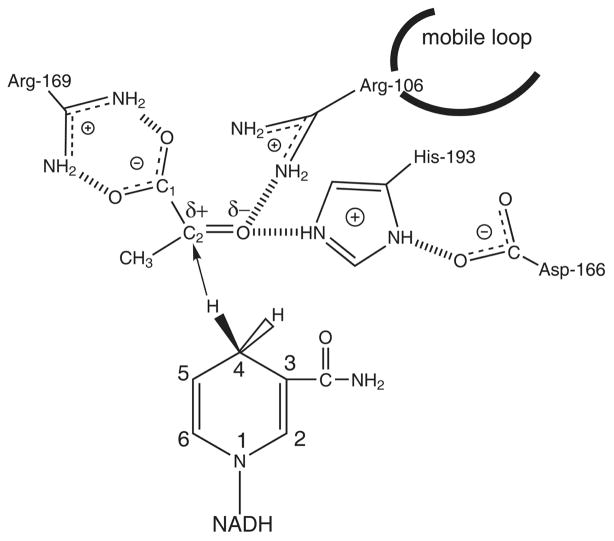
Lactate dehydrogenase, an enzyme that interconverts pyruvate and lactate, does so in a stereospecific manner using the NADH as its cofactor. As shown in Figure 1, the reaction involves a proton transfer from the protonated histidine in the active stie to the carbonyl oxygen atom of pyruvate, as well as a hydride transfer from the nicotinamide ring of the NADH cofactor to the carbonyl oyxgen atom of the substrate. Many studies on this enzyme have established details of the mechanism of this catalytic reaction. The roles of residues surronding the active site have been revealed from site directed mutatgensis experiments[1–3]. The order of binding of the substrate and cofactor has been clarified; the cofactor is followed by the substrate which is then followed by closure of the active site by a mobile loop. This step has been shown to be rate limiting in the wild type enzyme[4, 5]. While many details of the mechanism for this enzyme have been resolved, details regarding the chemical step of the enzyme continue to be eluciated [6–10].
Figure 1.
Diagram of the binding site of LDH with bound NADH and pyruvate showing hydrogen bonds between the substrate and catalytically important residues of the protein. The catalytic event involves the hydride transfer of the C4 hydrogen of NADH from the pro-R side of the reduced nicotinamide ring to the C2 carbon of pyruvate and proton transfer from the imidozole group of His-195 to pyruvate’s keto oxygen substrate
The Transition Path Sampling algorithm developed by Chanlder and coworkers [11–15] offers many advantages in studying mechanisms of enzymatic reactions. Mainly, there is no predefined reaction coordinate that governs the system. The paths generated from TPS are true dynamical trajectories from which reaction mechanisms can be obtained in atomic detail. Using TPS, our group has been able to elucidate details of the reaction mechanism of human heart LDH on an atomic level. The complete transformation of reactant to the product state can be observed through the generation of ensemble of unbiased reactive trajectories without prior knowledge of the reaction coordinate. In addition, through committor analysis[13], transition states can be identified and new details of the reaction coordinate can be revealed. The results of our TPS study from the human heart LDH found that while a concerted mechanism exists, a stepwise mechanism dominates where the hydride always transfered prior to the proton in the transformation from pyruvate to lactate. Also, residue motions along the hydride donor–acceptor axis were intimately connected with the hydride and proton transfers. A compression of residues 31–33 and 65–66 behind the nicotinamide ring of the NADH cofactor and of residues 106 and 195 behind the substrate brought the donor acceptor atoms of the hydride closer together. Further compression of residues behind the NADH combined with a relaxation of residues 106 and 196 behind the substrate completed the transfer reaction[16]. Transition state analysis revealed that even for the reactive paths where the proton transfer lags behind the hydride transfer, the transition state was always near the hydride transfer region. Committor distribution analysis also proved that the residues along the donor-acceptor axis of the hydride were part of the reaction coordinate and that the chemistry of the active site was not enough sufficient for defining the reaction coordinate[17].
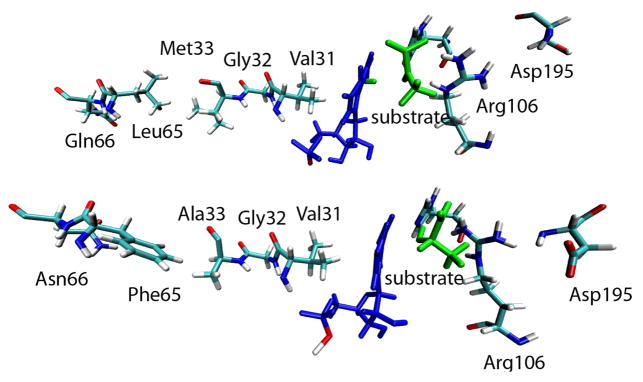
The LDH from Bacillus stearothermophilus is one of the most widely studied lactate dehydrogenases. The 32.73% sequence identity between the human heart and bacterial lactate dehydrogenase homologs shows that the bacterial and human are structurally very similar. Upon alignment of these two enzymes there is only a 1.58 Å RMS difference, clearly demonstrating the strong similarity. These enzymes also share the same residues surrounding the active site. As mentioned above, previous work in our group has elucidated the role of specific residues that extend beyond the active site, along the donor-acceptor axis of the hydride, which are included in the reaction coordinate of this reaction. Figure 2 shows that this residue architecture exists for both the human and the bacterial enzymes. Both have a similar line up of residues along the donor–acceptor axis that could produce similar dynamic motions. While the specific residues vary along the axis, both the human and bacterial enzyme have a valine immediately behind the NADH and an arginine on the acceptor side.
Figure 2.
Comparison of the axis residues along the donor-acceptor axis of the hydride. Top: human heart LDH. Bottom: BsLDH
The purpose of this paper is to determine whether the small difference between the bacterial and human enzymes leads to differences in the mechanism. Using the Transition Path Sampling method we obtained an ensemble of trajectories. Chemistry of the active site was probed as well as residue motions along the donor–acceptor axis. Finally we performed a transition state analysis. Our results demonstrate that small changes within the active site reverse the order of hydride and proton transfer as compared to the human heart LDH. In addition, transition state analysis reveals differences between concerted and stepwise reactions that were not observed the human heart LDH.
2. METHODS
2a. Simulation Details
We used the crystal structure of lactate dehydrogenase from Bacillus stearothermophillus enzyme solved by Wigley et al. (Brookhaven PDB ID: 1LDN)[18]. The structure was complexed with the allosteric activator (FBP), NADH and oxamate as the substrate analog. Each monomer consists of 316 residues, NADH and oxamate and crystallographic waters. For computational reasons we used the monomer and FBP was removed since its role for dimers has been shown to be negligible[19]. To generate reactive potential energy surfaces, Quantum Mechanical/Molecular Mechanical calculations were performed on a Linux Cluster using the CHARMM[20]/MOPAC[21] interface with the CHARMM27, all hydrogen force field. The CHARMM27 force field includes specific parameters for NAD+/NADH. Oxamate (NH2COCOO), an inhibitor of LDH, is an isosteric, isoelectronic mimic of pyruvate with similar binding kinetics. Changes to the PDB file included substitution of the oxamate nitrogen with carbon to create pyruvate and replacement of the active site neutral histidine with a protonated histidine to establish appropriate starting conditions with pyruvate and NADH in the active site. Glu198, a residue that is close to Asp166 which directly interacts with the active site histidine was protonated as well[9]. A total of 39 atoms were treated with the AM1 potential: 17 or 16 atoms of the NADH or NAD+ nicotinamide ring, 13 or 12 atoms of the protonated or neutral histidine imidazole ring, and 9 or 11 atoms of the substrate pyruvate or lactate, respectively. While a higher level of theory would be optimal, due to computational expenses and since we were not comparing with experimental results, the AM1 semiemperical method was used. The Generalized Hybrid Orbital (GHO)[22] method was used to treat the two covalent bonds which divide the Quantum Mechanical and Molecular Mechanical regions. The two GHO boundary atoms are the histidine Cα atom and the NC1 carbon atom of the NAD+/NADH adenine dinucleotide structure which covalently bonds to the nicotinamide ring. This is a gas phase calculation and while solvation effects are important to dynamics, it is likely on the timescale studied, no significant changes would be found.
Minimization, equilibration, and dynamics followed a standard protocol. First the structure was minimized using only the CHARMM27 Molecular Mechanical potential for 5000 steps using steepest descent to remove bad van der Waals contacts created by the addition of hydrogens. Then 20,000 further steps of minimization were performed using an adapted-basis Newton-Raphson algorithm (ABNR), achieving a change in energy < 0.001 kcal/mol and a RMS gradient < 0.001 kcal/mol-Å. The MM-MM nonbonding cutoffs were 14 Å and there were no cutoffs for QM-MM nonbonding interactions. The enzyme was heated to 300 K over 10 ps, equilibrated with velocities assigned from a Gaussian distribution every 100 steps at 300 K for another 10 ps and then dynamically equilibrated for a final 10 ps. The equations of motion were propagated using leapfrog Verlet integration with a 1 fs time step, using SHAKE constraints.
2b. Generation of the Transition Path Ensemble
The reactive trajectory surface was divided into Pyruvate, Lactate, and Transition regions. The pyruvate region included all configurations where the bond length of the reactive proton and the reactive nitrogen of the active site histidine (NE2) was 1.3 Å or shorter and the bond length of the reactive hydride and the reactive carbon (NC4) of the NADH coenzyme was 1.3 Å or shorter. The lactate region included all configurations where the bond length of the reactive proton and the reactive substrate oxygen (O) was 1.3 Å or shorter and the bond length of the reactive hydride and reactive substrate carbon (C2) was 1.3 Å or shorter. The transition region included all configurations where neither of the above combined bond lengths were satisfied. A reactive trajectory was defined as a single dynamics simulation which connected the pyruvate region to the lactate region or vice versa. A nonreactive trajectory connected the same basin, i.e., lactate/lactate or pyruvate/pyruvate[16].
An initial trajectory was generated by restraining the hydride and proton donor-acceptor distances of an equilibrated structure to approximately 3 Å each and repositioning the hydride and proton midway between their respective donors and acceptors. Velocities were taken from a 300 K equilibration run and were used with the above coordinates to initiate propagation forward and backward in time. This produced a 500 fs trajectory that began in the lactate region and ended in the pyruvate region.
As we have previously described [16,17], the TPS algorithm was implemented within CHARMM. A microcanonical ensemble of reactive trajectories was generated. This algorithm is described in detail elsewhere[15] but we will summarize it here. An initial 500 fs trajectory, including both co-ordinates and momenta, was used as a seed for further generation of trajectories. New trajectories were generated through the use of the shooting algorithm on randomly chosen time slices from the 500 fs trajectory. A small random displacement chosen from a zero mean Gaussian distribution, multiplied by a constant factor was added to the momenta. The momenta were rescaled to conserve linear momentum, angular momentum, and energy. Dynamics was run forward and backward in time to complete a 500 fs trajectory. The initial and final conformations were determined to be in the Pyruvate, Lactate, or Transition region through inspection of the order parameter values. If the new trajectory was reactive, it was used as a seed for generation of a new reactive trajectory. If the trajectory was unreactive, a new time slice was randomly chosen from the old trajectory until a new trajectory was generated. Using this method, we achieved a 24% acceptance ratio with trajectories decorrelating in about thirty five successful shooting trials.
2c. Transition State Analysis
The committor probability is a statistical quantity that can be calculated along the reactive path and measures the probability of a structure to land in the reactant or product region. At the beginning of the path, when the structure is in the reactant region, the probability of that structure landing in the product region would be zero. However, as the trajectory evolves to the product region, the probability of that structure landing in the product region will increase to one. The transition state is defined here as a structure that given a random velocity has an equal probability of ending either at the reactant or product region[13].
The transition paths generated from the TPS method are ideal for this kind of analysis because there is a guaranteed transition from reactant to product. To be truly unbiased, each configuration along each 500 fs trajectory from reactants to products should be considered as a potential transition state. However, this would be computationally expensive and configurations far from the transition surface can be ruled out quickly. Our approach has been instead to evaluate the committor values in regions around where either the hydride and proton, or both, transfer i.e. the region of the trajectory where the hydride and proton are equidistant from the donors and acceptors respectively. This significantly narrowed the search region.
The shooting algorithm was used to determine committor values along the trajectory. A random velocity chosen from a Maxwell Boltzmann distribution was assigned to a configuration and trajectories were run. Up to 100 trajectories were run per time slice. The configurations where the probability of forming lactate and pryruvate were both in the 0.4 to 0.6 region were considered to be parts of the transition state ensemble.
3. RESULTS AND DISCUSSION
3a. Transition Path Ensemble
From examination of the ensemble trajectories generated for the BsLDH three different reaction mechanisms were observed. A concerted reaction involving simultaneous transfer of both the hydride and the proton was observed for 54% of the paths generated. 25% of the trajectories showed a stepwise mechanism (stepA) where the proton transfered to the substrate prior to the hydride. 21% of the trajectories showed a third mechanism (stepB) in which the proton displayed a two step process of transferring to the substrate. It first transferred from the active site histidine to the carbonyl oxygen of the carboxamide side chain of the NADH. Following this, the proton then transfered to the substrate. In the stepB mechanism the hydride transfer still followed after the complete proton transfer. These results are significantly different from those observed for the human heart LDH. For human heart LDH, while both concerted and stepwise mechanisms were observed, the order of particle transfer was reversed with the hydride transferring prior to the proton. In addition, the stepB mechanism that was observed for the BsLDH was never observed for the human heart LDH.
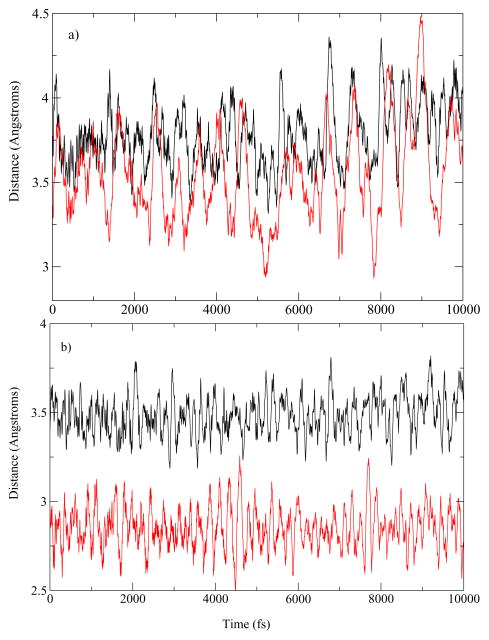
To understand the differences in the mechanisms observed for the Bacillus homolog, we examined small differences in the organization of the active site of both the human heart LDH and the BsLDH when complexed with pyruvate. The donor–acceptor distances of the homologs are plotted in Figure 3. For the human heart LDH, the average hydride donor–acceptor distance was 3.78 Å and the average proton donor–acceptor distance was 3.55 Å. For BsLDH, the average donor–acceptor distance of the hydride was 3.48 Å while the average donor–acceptor distance of the proton is only 2.85 Å. While residue motions along the donor–acceptor axis of the hydride likely brought donor and acceptor atoms closer together in a similar manner as the human heart LDH, the proton donor and acceptor atoms were already less than 3 Å apart and enabled transfer between the two atoms independent of the residue motions along the donor–acceptor axis of the hydride. This difference in donor–acceptor distance is likely a contributing factor to why in the case of BsLDH the proton transfer is observed prior to the hydride transfer.
Figure 3.
a. Human heart LDH b. BsLDH Comparison of the donor acceptor distances of the hydride and the proton in the pyruvate basin. In black is the distance of the donor–acceptor of hydride and in red is the donor–acceptor distance of the proton.
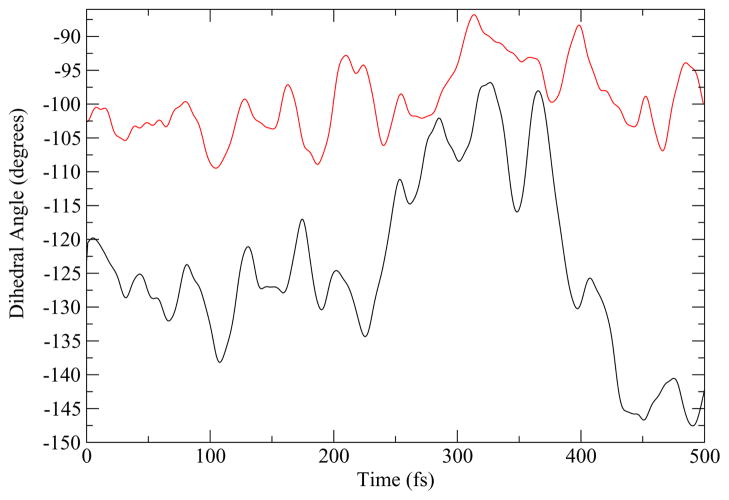
We sought an additional explanation for why in the stepB reaction mechanism we observed that the proton did not transfer directly to the substrate but rather first protonated the carbonyl oxygen of the carboxamide side chain of the NADH. Figure 4 shows a comparison of the range of the dihedral rotation of the active site histidine over the course of a reactive path for the both the human heart and Bacillus homologs. For the human heart LDH, the histidine can rotate about 20°, whereas the bacterial LDH the histidine has a range of motion of about 50°. This larger range of motion permits the active site histidine to orient towards NADH and not only towards the substrate. The stepB mechanism of the BsLDH takes advantage of a range of motion which enables the histidine to transfer its proton to the carbonyl oxygen of the carboxamide side chain of NADH before bonding to the substrate oxygen.
Figure 4.
The rotation of the dihedaral angle along a reaction path from the human heart LDH (red) and BsLDH (black) homologs.
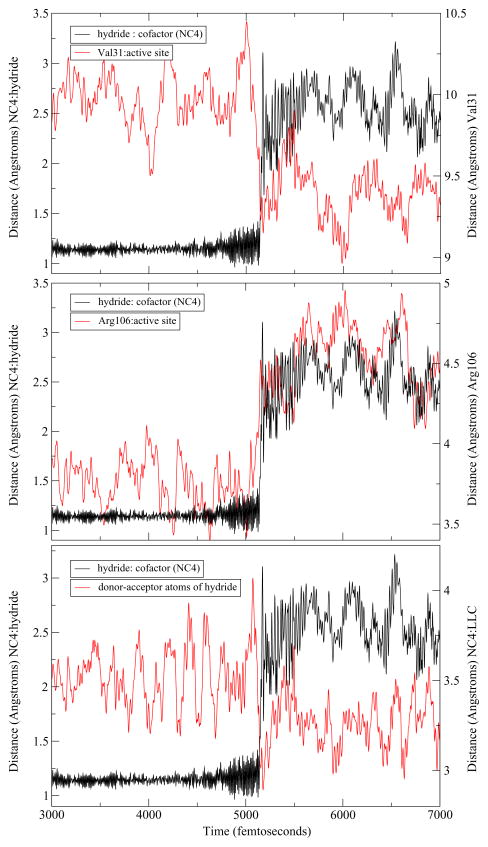
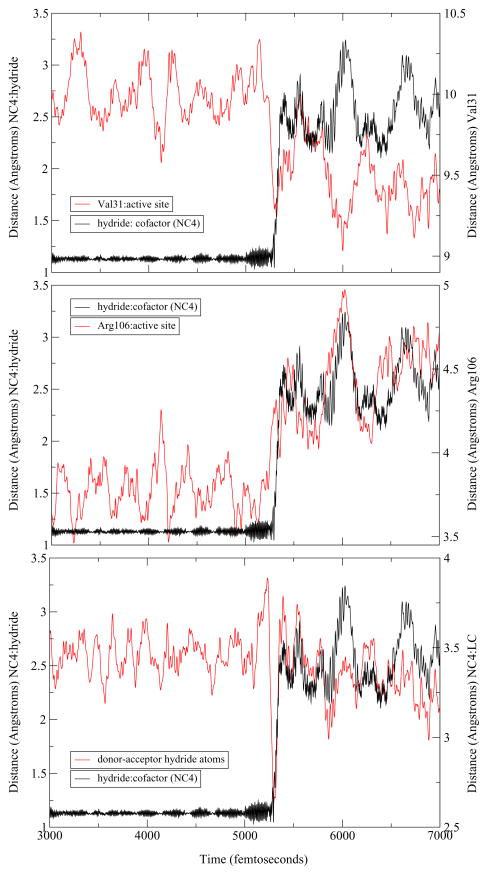
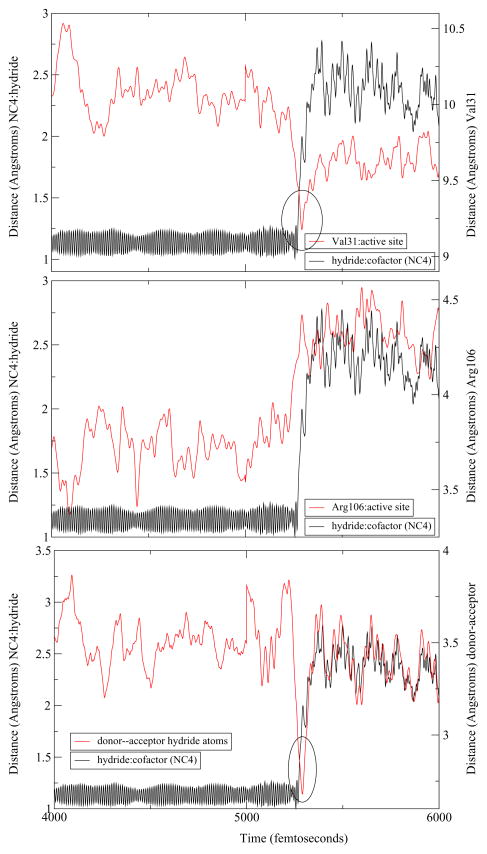
The differences between the human heart and Bacillus homologs that have been described thus far were limited to the active site. To examine enzyme wide changes, we extended a representative trajectory of each of the three mechanisms observed to study the dynamics of the residues along the donor-acceptor axis. As mentioned above, BsLDH shows a similar architecture along the donor–acceptor axis of the hydride as compared to the human heart LDH. Indeed, upon examination of the extended trajectories, a similar compression–relaxation motion was observed. However, its role differed slightly among the three mechanisms observed. For both the concerted and StepA mechanisms, the hydride transfer appears to be directly correlated with the dynamics of the axis residues. Val31 compresses towards the active site while Arg106, once it reaches its minimum, relaxes away ensuring the reaction a complete hydride transfer. However, there is a subtle difference the dynamics of the stepB mechanism. In this case, while compression and relaxation motions exist within the enzyme, they appear not as well correlated to the timing of the hydride transfer. Specifically, the hydride begins to transfer before Val31 has reached its minimum and before the Arg106 has completely relaxed away (Figure 5, Figure 6, Figure 7).
Figure 5.
An example of a trajectory displaying a concerted mechanism. From the top: hydride transfer shown as the distance of the hydride to the C4 of the NADH (NC4) is compared to Val31 compression to the active site, the Arg106 relaxation away from the active site, and the distance of the hydride donor–acceptor atoms (NC4 and substrate carbon (LC).
Figure 6.
An example of a reactive trajectory that displays the stepA mechanism. From the top: the hydride transfer shown as the distance of the hydride to the C4 of the NADH (NC4) is compared to to the Val31 compression, the Arg106 relaxation, and the distance of the hydride donor-acceptor atoms.
Figure 7.
An example of a trajectory with the StepB mechanism. From the top: we compare the hydride transfer shown as the distance of the hydride to the C4 of the NADH (NC4) to the Val31 compression, the Arg106 relaxation, and the distance of the hydride donor-acceptor atoms.
3b. Transition State Analysis
A transition state analysis was performed to determine whether the differences in the dynamics between the concerted, stepA and stepB mechanisms would change the transition state structure. We began our search for transition states for BsLDH using the same procedure as previously employed for the human heart LDH.[17] Among the trajectories that displayed a concerted mechanism, transition states were identified by shooting 100 trajectories that were 250 fs in length from structures in the transition region. One transition state was found whose committor value for both lactate and pyruvate was between 0.4–0.6 for each reactive path.
However, finding the transition states for reactive trajectories where there was a lag between the proton and hydride transfer proved to be more difficult. For the reactive paths that displayed the stepA mechanism, once a configuration left the pyruvate basin (after the proton transfer and before hydride transfer), trajectories of 250 fs that were “shot” from those configurations were an insufficient amount of time to land in a stable basin. Rather, shooting trajectories landed in an region where the proton transferred to the substrate oxygen but the hydride had not yet transferred. In the region of the path where the hydride transfer occurred, committor probabilities were obtained that showed half of the trajectories landed in the lactate region (product basin) and half did not commit to stable basin. However, for a transition state to be identified a structure must have equal probability of landing in the reactant and product basin. In the end, a transition state was only identified using a longer shooting length of 5 ps. Using trajectories of this length we were able to identify a transition state that had a committor probability for both lactate and pyruvate in the 0.4–0.6 region.
We encountered the same difficulties for locating the transition states of reactive paths that displayed the stepB mechanism. Once the trajectory moved out of the pryruvate basin (the proton was transferred from the active site histidine but before the hydride transfer) shooting trajectories of 250 fs could not drive a configuration to either pyruvate or lactate. Similar to the results for the stepA mechanisms, for structures near the hydride transfer region, when 250 fs shooting trajectories were used, half of the configurations landed in the lactate region and half the trajectories landed in an undefined region. Transition states were only identified upon using longer shooting trajectories of 30 ps in length.
The table below highlights three examples of transition states identified, one from each of the three different mechanisms observed in the transition path ensemble. The distances between the important atoms of the active site and axis residues are shown. While there is variation among each of the transition states found, their overall structures are similar. The results obtained for the transition state ensemble (TSE) for BsLDH were consistent with the results obtained from the human heart LDH. There was only one transition state for each reactive trajectory and the transition states identified were near the hydride transfer region independent of the order of particle transfer. The main difference between the two transition state ensembles obtained was in how each TSE was acquired. For the human heart LDH the length of shooting trajectory required was the same for both concerted and stepwise trajectories. However, for BsLDH, the length of the shooting trajectory depended on the mechanism. In addition, each transition state identified from the heart heart LDH was in an advanced state of hydride transfer. For BsLDH among the transition state identified, there was more variability of the degree of hydride transfer. These differences likely reflect the shape of the separatix that divdes the reactant and product regions. While the separatrix for the human heart LDH was shown to be very narrow, the width of this region is larger and therefore could encorporate more variablity in structure and also require different lengths of shooting trajectories depending where the transition state is located on the separatrix.
4. CONCLUSION
While the structure of the human heart and bacterial LDH homologs are very similar, small changes within the active site reversed the order of particle transfer from that observed in the human heart LDH. While this result differs from that which was observed previously, which described the process as concerted where the hydride transfer is prior to the proton transfer[9] it is likely due to the fact that in their work the potential energy surface was generated from a reduced reaction coordinate though it was corrected with high level correction terms. Using TPS the paths generated are true dynamic trajectories that are not biased by any reaction coordinate.
We have also shown that the path of proton transfer to the substrate manifested in two different ways which was due to the increased mobility of the active site histidine. Transition state analysis revealed that in contrast to the narrow separatrix region previously defined from analysis of the TSE from the human heart LDH, for BsLDH, the separatrix region is wider and therefore different shooting trajectory lengths would be required to identify the transition state for each mechanism. While rate constant calculations have not yet been performed, this present work would predict that the different mechanisms would each have a different rate for the chemical step. While it is highly dangerous to speculate on evolutionary pressure on the rapid chemical step of enzymes in which chemistry is not rate limiting, it is clear that the chemical mechanism in the mamalian LDH is more perfectly tuned to efficiently produce product than in the Bacillus enzym enzyme.
Table 1.
TS parameters
| DISTANCE | Concerted | StepA | StepB |
|---|---|---|---|
| cofactor(NC4):hydride | 1.54 | 1.16 | 1.47 |
| hydride:substrate carbon | 1.61 | 1.52 | 1.27 |
| histidine (NE2):proton | 1.43 | 1.78 | 2.10 |
| proton:substrate oxygen | 1.49 | 1.26 | 1.07 |
| Val31:active site | 9.23 | 9.29 | 9.44 |
| Arg106:active site | 4.51 | 4.26 | 4.20 |
| dihedral-His195 (degrees) | −107.26 | −107.05 | −105.69 |
Acknowledgments
This work was supported by National Institutes of Health Grant GM068036. S.L.Q. was supported by National Institutes of Health Grant T32 GM008572-12.
References
- 1.Kedzierski P, Moreton K, Clarke A, Holbrook JJ. Biochemistry. 2001;40:7247–52. doi: 10.1021/bi0026775. [DOI] [PubMed] [Google Scholar]
- 2.Nobbs TJ, Cortes A, Holbrook J, Atkinson T, Scawen M, Nicholls DJ. Biochem J. 1994;300:491–9. doi: 10.1042/bj3000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng H, Zheng J, Clarke A, Holbrook J, Callender R, Burgner J. Biochemistry. 1994;33:2297–305. doi: 10.1021/bi00174a042. [DOI] [PubMed] [Google Scholar]
- 4.Clarke A, Wigley D, Chia W, Barstow D, Atkinson T, Holbrook J. Nature. 1986;324:699–702. doi: 10.1038/324699a0. [DOI] [PubMed] [Google Scholar]
- 5.Dunn C, Wilks H, Halsall D, Atkinson T, Clarke A, Muirhead H, Holbrook J. Phil Trans R Soc Lond B. 1991;332:117–184. doi: 10.1098/rstb.1991.0047. [DOI] [PubMed] [Google Scholar]
- 6.Ranganathan S, Gready JE. J Phys Chem B. 1997;101:5614–5618. [Google Scholar]
- 7.Moliner V, Turner A, Williams I. Chem Commun. 1997:1271–1272. [Google Scholar]
- 8.Turner A, Moliner V, Williams I. Phys Chem Chem Phys. 1999;1:1323. [Google Scholar]
- 9.Ferrer S, Ruiz-Pernia JJ, Tunon I, Moliner V, Garcia-Viloca M, Conzalez-Lafont A, Lluch JM. J Chem Theory Comput. 2005;1:750–761. doi: 10.1021/ct050016l. [DOI] [PubMed] [Google Scholar]
- 10.Ferrer S, Tunon I, Martf S, Moliner V, Garcia-Viloca M, Conzalez-Lafont A, Lluch J. J Am Chem Soc. 2007;128:16851–16863. doi: 10.1021/ja0653977. [DOI] [PubMed] [Google Scholar]
- 11.Dellago C, Bolhuis P, Csajka F, Chandler D. J Chem Phys. 1998;108:1964–1977. [Google Scholar]
- 12.Geissler Phillip L, Chandler CDD. J Phys Chem B. 1999;103:3706–3710. [Google Scholar]
- 13.Bolhuis P, Dellago C, Chandler D. Proc Nat Acad Sci USA. 2000;97:5877–5882. doi: 10.1073/pnas.100127697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolhuis P, Chandler D, Dellago C, Geissler P. Annu Rev Phys Chem. 2002;53:291–318. doi: 10.1146/annurev.physchem.53.082301.113146. [DOI] [PubMed] [Google Scholar]
- 15.Dellago C, Bolhuis P, Geissler P. Adv Chem Phys. 2001;123:1–86. [Google Scholar]
- 16.Basner JE, Schwartz SD. J Am Chem Soc. 2005;127:13822–13831. doi: 10.1021/ja043320h. [DOI] [PubMed] [Google Scholar]
- 17.Quaytman S, Schwartz S. Proc Natl Acad Sci USA. 2007;104:12253–12258. doi: 10.1073/pnas.0704304104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wigley DB, Gamblin S, Turkenburg J, Dodson E, Piontek K, Muirhead H, Holbrook J. J Mol Biol. 1992;223:317–335. doi: 10.1016/0022-2836(92)90733-z. [DOI] [PubMed] [Google Scholar]
- 19.Iwata S, Ohta T. J Mol Biol. 1993;230:21–27. doi: 10.1006/jmbi.1993.1122. [DOI] [PubMed] [Google Scholar]
- 20.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 21.Field M, Bash P, Karplus M. J Comp Chem. 1990;11:700–733. [Google Scholar]
- 22.Gao J, Amara P, Alhambra C, Field M. J Phys Chem A. 1998;102:4714–4721. [Google Scholar]