Abstract
Background
Ralstonia pickettii is a nosocomial infectious agent and a significant industrial contaminant. It has been found in many different environments including clinical situations, soil and industrial High Purity Water. This study compares the phenotypic and genotypic diversity of a selection of strains of Ralstonia collected from a variety of sources.
Results
Ralstonia isolates (fifty-nine) from clinical, industrial and environmental origins were compared genotypically using i) Species-specific-PCR, ii) PCR and sequencing of the 16S-23S rRNA Interspatial region (ISR) iii) the fliC gene genes, iv) RAPD and BOX-PCR and v) phenotypically using biochemical testing. The species specific-PCR identified fifteen out of fifty-nine designated R. pickettii isolates as actually being the closely related species R. insidiosa. PCR-ribotyping of the 16S-23S rRNA ISR indicated few major differences between the isolates. Analysis of all isolates demonstrated different banding patterns for both the RAPD and BOX primers however these were found not to vary significantly.
Conclusions
R. pickettii species isolated from wide geographic and environmental sources appear to be reasonably homogenous based on genotypic and phenotypic characteristics. R. insidiosa can at present only be distinguished from R. pickettii using species specific PCR. R. pickettii and R. insidiosa isolates do not differ significantly phenotypically or genotypically based on environmental or geographical origin.
Keywords: Ralstonia pickettii, random amplified polymorphic DNA, PCR, genotyping, High-Purity Water
Background
Ralstonia pickettii, previously called Pseudomonas pickettii and Burkholderia pickettii [1], is ubiquitous in the environment. It has been recovered from a number of water sources and from a wide range of clinical environments [2-5]. R. pickettii has also become recognised in the last decade as a nosocomial pathogen associated particularly with individuals who are debilitated or immunosuppressed [6-8]. These outbreaks have been reported mainly in association with contamination of hospital supplies [9-14] and with contaminated chlorhexidine skin cleansing solutions [15,16]. The emergence of new opportunistic pathogenic microorganisms has been linked to a multiresistance phenotype that makes them refractory to the antibiotics commonly used in clinical practice [17]. The majority of clinical isolates of R. pickettii are characterized by their multiresistance to common antibiotics [17].
The emergence of R. pickettii in High-Purity Water (HPW) systems used in the biopharmaceutical industry necessitates revisiting this organism. R. pickettii has been identified in biofilm formation in industrial plastic water piping [18] and has been isolated from industrial high-purity water [2,19]; laboratory based high-purity water systems [3]; in the Space Shuttle water system [20] and from the Mars Odyssey probe encapsulation facility [21]. It has been shown to produce homoserine lactones [2], the putative cell-cell signalling molecules in biofilm development [22] and has the ability to survive in low nutrient (oligotrophic) conditions [23]. In addition, R. pickettii has been shown to have a wide range of biodegradative abilities that could potentially be used for commercial applications and that may assist in survival and adaption to low nutrient environments [8]. Integrating Conjugative Elements-like elements have been discovered in several isolates of this bacterium [24] indicating a degree of plasticity in their genomes.
Molecular typing methods such as restriction fragment length polymorphism by pulsed-field gel electrophoresis (PFGE) [14,25], random amplified polymorphic DNA (RAPD) analysis by arbitrarily primed PCR [16] and ribotyping [26] have been developed for Ralstonia sp. and have been used to detect relationships between clinical isolates in epidemiological studies. Despite the acknowledged importance of R. pickettii as a nosocomial pathogen, little is known regarding its epidemiology. Studies carried out with limited numbers of bacterial isolates indicated the bacterium appears to have limited diversity [25-27].
Evidence suggests that R. pickettii finds its way into clinical environments through contaminated water supplies [5]. To test this and to determine the level of relatedness between isolates of this bacteria from different environments a comprehensive study of the relatedness of fifty-nine isolates of R. pickettii and R. insidiosa (including soil, water and clinical isolates) using various phenotypic (metabolic activity) and genotypic (flagellin and Interspatial regions typing, BOX-PCR, and RAPD) fingerprinting methods was carried out.
Methods
Bacterial isolates and growth conditions
The fifty-nine isolates used in this study are presented in Table 1. All the isolates were stored at -20°C in Nutrient Broth (Difco) with 50% glycerol. Isolates were grown aerobically on Nutrient Agar (Difco) and incubated overnight at 30°C.
Table 1.
Ralstonia Isolates used in this work
| Strain | Source |
|---|---|
|
R. pickettii JCM5969, NCTC11149, DSM6297, CIP73.23 CCUG3318, CCM2846, CCUG18841 |
Culture Collection |
|
R. pickettii ULC193, ULC194, ULC244, ULC277, ULC297, ULC298, ULC421 |
Microbiology laboratory of Limerick Regional Hospital (Cystic Fibrosis Patients) |
|
R. pickettii ULI788, ULI790, ULI791, ULI796, ULI800, ULI801, ULI804, ULI806, ULI807, ULI818, ULI159, ULI162, ULI165, ULI167, ULI169, ULI171, ULI174, ULI181, ULI187, ULI188, ULI193 |
Isolated from various Industrial Purified water systems (Ireland) |
|
R. pickettii ULM001, ULM002, ULM003, ULM004, ULM005, ULM006 |
Isolated from various Millipore Purified water systems (France) |
|
R. pickettii ULM007, ULM010, ULM011 |
Isolated from various Millipore Laboratory Purified water systems (Ireland) |
|
R. insidiosa ATCC4199, LMG21421 |
Culture Collection |
|
R. insidiosa ULI821, ULI797, ULI785, ULI181, ULI794, ULI185, ULI166, ULI819, ULI784, ULI163, ULI795 |
Isolated from various Industrial Purified water systems (Ireland) |
|
R. insidiosa ULM008, ULM009 |
Isolated from various Millipore Laboratory Purified water systems (Ireland) |
Phenotypic analysis
Oxidase and catalase tests were performed with Oxidase sticks (Oxoid, Basingstoke, UK) and 3% hydrogen peroxide, respectively. A number of classical phenotypic tests were performed that included BioMérieux API 20NE system (BioMérieux UK Limited, Hampshire, UK) and the Remel RapID NF Plus commercial system (Remel, Kansas, USA). A Vitek card; the Non-Fermenter Identification Card (NFC) (BioMérieux), was also used. All of the above tests were carried out as per manufacturer's instructions. Phenotypic relatedness among different isolates of R. pickettii was determined using the API 20NE profiles. Phenotypic characters were scored as discrete variables [0 or 1]; 0, when the character was negative or missing; 1, when character was positive or present). Isolates with the same pattern were grouped into Biotypes numbering 1 to 35. The unweighted pair group method [28] was used for cluster analysis using the MultiVariate Statistical Package (MVSP) software program ver. 3.13 by means of the Jaccard coefficient [29]. The discriminatory power of the biotyping for typing R. pickettii isolates was evaluated by using the discrimination index as described by Hunter and Gaston, as given by the equation: D = 1 - [1/N (N - 1)] ∑nj (nj - 1), where D is the numerical index of discrimination, N is the total number of isolates, and nj is the number of isolates pertaining to the jth type [30].
Genotypic analysis
DNA for all PCR experiments was prepared as described previously [31].
Species-specific PCR and amplification 16S-23S rRNA ISR and fliC gene
The species-specific PCR primers (Rp-F1, Rp-R1 and R38R1) used in this study were designed by Coenye et al., detailed in Table 2[32,33]. The 16SF and 23SR primers were used to amplify the Interspacial Region (ISR) [34] and the Ral_fliC primers (Ral_fliCF and Ral_fliCR) were used to amplify the fliC gene (Table 2), [35]. The PCR assays were performed in 25 μL reaction mixtures, containing 100 ng of template genomic DNA, 1U Taq polymerase, 250 mM (each) deoxynucleotide triphosphate, 1.5 mM MgCl2, 10x PCR buffer (Bioline), and 20 pmol of oligonucleotide primer (MWG Biotech, Ebersberg, Germany) Rp-F1 and 10 pmol of oligonucleotide primers Rp-R1 and R38R1 for the species-specific PCR and 20 pmol each of the primers for the ISR and fliC regions (Table 2). After initial denaturation for 2 min at 94°C, 30 amplification cycles were completed, each consisting of 1 min at 94°C, 1 min at 55°C, and 1 min 30 secs at 72°C. A final extension of 10 min at 72°C was then applied. The PCR products were analysed by electrophoresis in a 1.5% agarose gel (Agarose MP, Roche Diagnostics) for 1 hour (100 V) with ethidium bromide staining in the TBE buffer and photographed under the UV light (UV Products Gel Documentation System Imagestore, Ultra Violet Products, Cambridge). A 200-10000bp DNA ladder (Bioline) was included on all gels to allow standardization and sizing. Following amplification of the ISR and fliC region from test isolates PCR product was purified using the NucleoSpin Extract II kit (Macherey-Nagel, Düren, Germany) according to the manufacturer's instructions and the amplicons sequenced (MWG Comfort Read service).
Table 2.
Oligonucleotides used in this study
| Primer | Oligonucleotide Sequence 5'-3' | Targeta | Product size | Reference |
|---|---|---|---|---|
| Rp-F1 | ATGATCTAGCTTGCTAGATTGAT | 16S rRNA gene | 210 bp | [32] |
| Rp-R1 | ACTGATCGTCGCCTTGGTG | 16S rRNA gene | [32] | |
| R38R1 | CACACCTAATATTAGTAAGTGCG | 16S rRNA gene | 403 bp | [33] |
| 16SF | TTGTACACACCGCCCGTCA | 16S-23S Spacer Region | 860 bp | [34] |
| 23SR | GGTACCTTAGATGTTTCAGTTC | 16S-23S Spacer Region | [34] | |
| Ral_fliCF | CCTCAGCCTCAATACCAACATC | fliC gene | 725 bp | [35] |
| Ral_fliCR | CATGTTCGACGTTTCMGAWGC | fliC gene | [35] | |
| M13 | TTATGTAAAACGACGGCCAGT | RAPD Primer | N\A | [36] |
| P3 | AGACGTCCAC | RAPD Primer | N\A | [37] |
| P15 | AATGGCGCAG | RAPD Primer | N\A | [37] |
| OPA03U | AGTCAGCCAC | RAPD Primer | N\A | [38] |
| BOX-A1R | CTACGGCAAGGCGACGCTGACG | BOX Primer | N\A | [39] |
RAPD and BOX-PCR analysis
For RAPD-PCR each 25 μl PCR sample contained 2.5 μL of 10× buffer, 2 mM of MgCl2, 40 pmol of primer, 200 mM of each of four dNTPs, 200 ng of template genomic DNA and 1 U of Taq polymerase. The PCR reactions were carried out as follows: an initial denaturation at 94°C for 5 min followed by 40 cycles of denaturation at 94°C for 1 min, annealing at 36°C for 1 min and extension at 72°C for 1 min 30 secs. Four different primers were used: M13, P3, P15 and OPA03U are listed in Table 2[36-38].
BOX-PCR typing was carried out with the BOX-A1R primer (Table 2) [39]. 200 ng of template genomic was mixed with 2 U of Taq polymerase, 200 mM of each of four dNTPs, 2.5 μl of dimethyl sulfoxide (DMSO), 0.8 μl of bovine serum albumin (10 mg ml-1) (Promega), 5 μl of 5× Gitschier buffer and 10 pmol of primer in a final volume of 25 μl. After initial denaturation for 2 min at 95°C, 35 amplification cycles were completed, each consisting of 40 secs at 94°C, 1 min at 50°C, and 8 mins at 65°C. A final extension of 8 mins at 65°C was applied.
Amplified products for both procedures were analysed by electrophoresis in a 2% agarose gel containing ethidium bromide at 60 V for 4 hrs and were visualised by UV transillumination. The repeatability of the RAPD and BOX-PCR protocols were tested by studying the isolates in three independent runs.
DNA analysis
The ISR and fliC gene sequences obtained were compared with sequences in the GenBank database using the Basic Local Alignment Search Tool (BLAST) [40] and aligned using the ClustalW program [41]. Phylogenetic and molecular evolutionary analyses were conducted using genetic distance based neighbour joining algorithms [42] within MEGA version 3.1 http://www.megasoftware.net, [43]. The analysis of the RAPD and BOX gels was performed using BioNumerics software (version 5.1 Applied Maths, Kortrijk, Belgium), based on the Pearson correlation coefficient, and clustering by the unweighted pair group method with arithmetic means (UPGMA method) [44]. The isolates that clustered at a cut-off level of more than 80% similarity were grouped together; these were considered clonally related and classified into the same group. The discriminatory power of the BOX and RAPD-PCR for typing R. pickettii isolates was evaluated by using the discrimination index as described by Hunter and Gaston [30].
Accession numbers
DNA sequences were deposited in the EMBL database with accession numbers for sequences from the 16S-23S spacer region are as follows: AM501933-AM501952 and for the FliC genes: FN869041-FN869057.
Results
Species-specific PCR
To confirm that the isolates were in fact R. pickettii a species-specific PCR reaction was carried out using the primers described (Table 2). The results of the experimental analysis of fifty-nine isolates from our study, which include industrial, clinical, laboratory purified water and seven purchased strains are presented in Table 3. Eleven of the industrial high purity water isolates (ULI821, ULI797, ULI785, ULI181, ULI794, ULI185, ULI166, ULI819, ULI784, ULI163, ULI795), two laboratory Millipore water isolates (ULM008, ULM009) and one purchased strain (ATCC42129) were identified as R. insidiosa through use of the species-specific primers (Table 2). The multiplex PCR gave a 403 bp band and a 210 bp band for R. insidiosa and only the 210 bp band for R. pickettii (data not shown).
Table 3.
Characterization of Isolates of Ralstonia sp. using phenotypic assays and whole genome typing
| Strain | API 20 NE | RapID NF Plus | Vitek (NFC) | RAPD | BOX | ||||
|---|---|---|---|---|---|---|---|---|---|
| Biotype | % IDA | % IDA | % IDA | M13 | OPA3OU | P3 | P15 | BOX-A1R | |
| Ralstonia pickettii | |||||||||
| JCM5969 | B1 | 99.00 | 99.94 | 99.00 | A | e | VIII | 13 | F |
| NCTC11149 | B4 | 95.10 | 99.94 | 99.00 | D | a | IX | 13 | F |
| DSM 6297 | B4 | 95.10 | 99.94 | 99.00 | D | e | XX | 13 | F |
| CCUG3318 | B7 | 91.10 | 99.94 | 99.00 | D | a | XIX | 13 | F |
| CIP73.23 | B7 | 91.10 | 99.94 | 99.00 | D | n | XX | 13 | F |
| CCUG18841 | B30 | 00.00 | 99.71 | 99.00 | L | k | VI | 13 | L |
| CCM2846 | B30 | 00.00 | 99.71 | 97.00 | L | k | VI | 13 | L |
| ULI 187 | B3 | 97.70 | 98.34 | 99.00 | I | e | VII | 13 | G |
| ULI 188 | B4 | 95.10 | 99.99 | 99.00 | M | k | VII | 13 | G |
| ULI 798 | B5 | 95.10 | 99.99 | 99.00 | K | k | VII | 13 | H |
| ULI 807 | B10 | 84.10 | 99.99 | 99.00 | K | k | XIX | 13 | F |
| ULI 171 | B10 | 84.10 | 99.99 | 99.00 | I | c | VI | 13 | G |
| ULI 788 | B11 | 80.40 | 99.94 | 99.00 | J | f | XIV | 13 | J |
| ULI 800 | B11 | 80.40 | 99.99 | 99.00 | I | e | XXIII | 13 | A |
| ULI 169 | B11 | 80.40 | 99.99 | 99.00 | K | k | VI | 13 | A |
| ULI 165 | B14 | 67.90 | 99.99 | 99.00 | N | e | XXIV | 13 | D |
| ULI 174 | B14 | 67.90 | 98.34 | 99.00 | A | e | XIX | 13 | A |
| ULI 193 | B15 | 61.70 | 98.38 | 99.00 | A | e | X | 6 | A |
| ULI 796 | B16 | 60.00 | 98.34 | 99.00 | H | e | X | 6 | A |
| ULI 801 | B17 | 56.90 | 99.99 | 99.00 | A | a | X | 6 | A |
| ULI 791 | B17 | 56.90 | 99.99 | 99.00 | B | j | XI | 19 | A |
| ULI 790 | B20 | 44.80 | 98.34 | 99.00 | H | m | X | 10 | B |
| ULI 818 | B21 | 39.50 | 99.94 | 99.00 | H | k | X | 9 | B |
| ULI 804 | B23 | 24.50 | 98.34 | 99.00 | B | a | XI | 19 | B |
| ULI 159 | B29 | 00.00 | 99.94 | 99.00 | F | c | X | 8 | B |
| ULI 806 | B34 | 00.00 | 99.99 | 99.00 | A | a | X | 7 | A |
| ULI 167 | B33 | 00.00 | 99.94 | 99.00 | H | k | X | 9 | A |
| ULI 162 | B30 | 00.00 | 99.99 | 99.00 | A | e | X | 6 | C |
| ULC 298 | B8 | 90.10 | 99.99 | 99.00 | A | b | X | 5 | K |
| ULC 297 | B13 | 70.03 | 99.94 | 99.00 | A | e | X | 2 | K |
| ULC 277 | B15 | 61.70 | 99.99 | 99.00 | A | b | X | 1 | K |
| ULC 244 | B18 | 56.70 | 99.94 | 99.00 | A | e | X | 3 | L |
| ULC 193 | B18 | 56.70 | 98.34 | 99.00 | A | a | X | 4 | K |
| ULC 194 | B18 | 56.70 | 99.99 | 99.00 | A | a | X | 3 | L |
| ULC 421 | B21 | 28.50 | 99.99 | 99.00 | A | a | XVI | 15 | P |
| ULM 001 | B4 | 95.10 | 99.99 | 99.00 | P | h | III | 14 | R |
| ULM 002 | B4 | 95.10 | 99.99 | 99.00 | T | h | XVI | 13 | Q |
| ULM 003 | B9 | 88.60 | 99.28 | 99.00 | R | h | XVI | 13 | |
| ULM 004 | B7 | 91.10 | 99.99 | 99.00 | S | h | XVIII | 13 | Q |
| ULM 005 | B4 | 95.10 | 00.00 | 99.00 | A | e | XVII | 13 | O |
| ULM 006 | B4 | 95.10 | 99.28 | 99.00 | Q | h | XVII | 13 | M |
| ULM 007 | B4 | 95.10 | 99.99 | 99.00 | R | h | XVI | 13 | M |
| ULM 010 | B2 | 99.40 | 99.99 | 99.00 | A | g | XVI | 13 | M |
| ULM 011 | B2 | 99.40 | 99.99 | 99.00 | A | g | XXII | 13 | M |
| Ralstonia insidiosa | |||||||||
| LMG21421 | B15 | 61.70 | 99.94 | 99.00 | E | d | XVII | 13 | H |
| ATCC49129 | B6 | 92.40 | 99.99 | 99.00 | B | b | III | 14 | H |
| ULI 821 | B10 | 84.10 | 99.94 | 99.00 | E | d | XV | 18 | E |
| ULI 797 | B10 | 84.10 | 98.34 | 99.00 | O | e | XXV | 13 | E |
| ULI 785 | B19 | 53.10 | 99.99 | 99.00 | H | l | XXI | 13 | B |
| ULI 181 | B21 | 39.50 | 99.99 | 99.00 | B | f | II | 14 | B |
| ULI 794 | B24 | 06.40 | 34.18 | 99.00 | G | f | II | 14 | B |
| ULI 185 | B25 | 05.70 | 98.34 | 99.00 | U | o | IV | 12 | B |
| ULI 166 | B32 | 00.00 | 99.94 | 99.00 | B | f | I | 17 | B |
| ULI 819 | B26 | 00.00 | 99.99 | 99.00 | C | i | V | 21 | B |
| ULI 784 | B27 | 00.00 | 99.99 | 99.00 | H | e | V | 17 | A |
| ULI 163 | B28 | 00.00 | 98.34 | 99.00 | B | j | VI | 11 | D |
| ULI 795 | B35 | 00.00 | 98.34 | 99.00 | B | f | I | 20 | A |
| ULM 008 | B12 | 80.20 | 99.99 | 99.00 | E | e | XII | 16 | M |
| ULM 009 | B12 | 80.20 | 99.99 | 99.00 | E | d | XII | 16 | M |
A % ID Ralstonia pickettii
Phenotypic characterisation and identification
All isolates were Gram-negative non-fermentative rods and both oxidase and catalase positive. Fifty-nine isolates (eight from culture collections, seven clinical, eleven laboratory purified water and thirty-two industrial isolates and the R. insidiosa type strain LMG21421) were identified initially as R. pickettii (Table 3). These results were confirmed using the Vitek NFC with all isolates being identified as R. pickettii. The Vitek NFC identification rate ranged from 97.0 to 99.0 with two patterns being detected (Table 3). The API 20NE identification rate ranged from 0.00 to 99.4%, with thirty-five different patterns being detected. Most of the purchased culture collection isolates were identified as R. pickettii (except the soil isolates CCUG18841 and CCM2846) with cut-off points higher then 60%, six of the clinical isolates were identified as R. pickettii with cut-off points higher then 50%, while one was identified as Pseudomonas aeruginosa (Table 3). All 11 laboratory purified water isolates were identified as R. pickettii with cut-off points higher then 80%, and seventeen of the thirty-two industrial isolates were identified as R. pickettii species with cut-off points higher then 50%, the rest of the industrial isolates were all identified as non-R. pickettii species. The RapID NF Plus identification rate ranged from 0.00 to 99.9%, with five different patterns being detected. Fifty-seven isolates were identified as R. pickettii, with results of over 98%. The other two were identified as Moraxella sp (Table 3).
The R. insidiosa Type strain LMG21421 was identified as R. pickettii 61.70% ('Low Discrimination' 0050577) with the API 20NE, as R. pickettii 99.94% ('Implicit' 400414) with the RapID NF Plus and as R. pickettii 99% on the Vitek Junior system with the NFC (Table 3).
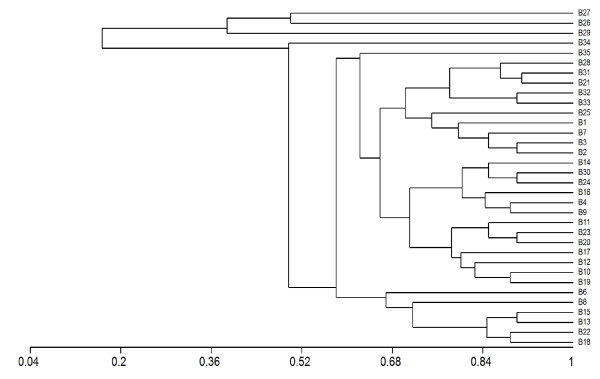
A cluster analysis was carried out using the API 20 NE results and can be seen in Figure 1. The results indicated that the isolates studied are phenotypically very different (The list of tests in the API 20NE can be seen in Additional File 1 Table S1). The 35 biotypes identified are very different with similarity between some of the biotypes being as low as 0.2. The 35 biotypes did not break down based on environment of isolation. These results contradict the results of both the Remel RapID NF Plus and the Vitek NFC, which indicated that R. pickettii was a phenotypically homogenous species with the same phenotypic pattern being found in most isolates. All results are presented in Table 3 and Additional File 1, Table S1. A Simpson's diversity of 0.9813 was calculated for this study using the API 20NE results [30].
Figure 1.
Cluster analysis of API 20NE results. B: Biotype 1 to 35- numbers assigned to API 20NE profile, isolates belonging to each biotype can be seen in Table 1. Scale is a measure of the phenotypic relatedness of isolates.
Genotypic characterisation
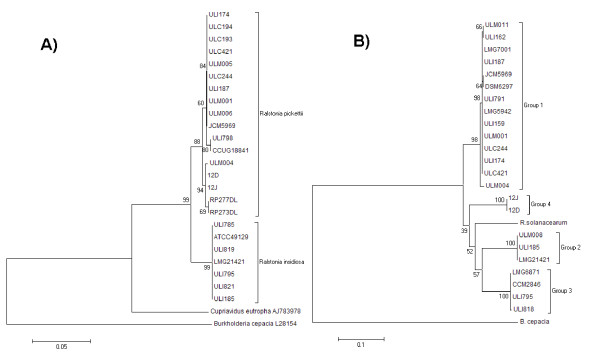
Four different DNA-based typing methods (ISR and fliC gene sequencing, RAPD-PCR and BOX-PCR) were used to compare the isolates at a molecular level. With the analysis of the 16S-23S rDNA ISR a PCR product of approximately 860 bp was obtained for all isolates indicating that the spacer region is highly similar in length in all isolates (data not shown). Sequencing of the ISR of 19 isolates identified phenotypically as R. pickettii, and the type strain of R. insidiosa was carried out. The sequence of several isolates indicated that these were more closely related to R. insidiosa than to R. pickettii sharing greater homology with the R. insidiosa type strain confirming the results obtained from the species-specific PCR reaction (Figure 2a). The ISR comprised a length of 513bp for R. pickettii and 515bp for R. insidiosa. The sequence similarity of the R. pickettii isolates compared to the R. pickettii type strain LMG5942 ranged from 98-100% (Figure 2a) and for all R. insidiosa isolates it was 95% (Figure 2a). All ISR sequences had a GC content of ~52.5%. The Ralstonia ISR spacer region contains two tRNA genes: tRNAIle and tRNAAla comprising 77 and 78 bp respectively. This is a common feature of the ISR in rrn operons in Gram-negative bacteria [45] including R. pickettii [46]. The order observed for sequences generated from our Ralstonia isolates was 16S rRNA - tRNAIle - tRNAAla -23S rRNA. The nucleotide sequences of tRNAIle were identical in all isolates and the tRNAAla gene differed by one nucleotide between R. pickettii and R. insidiosa in the isolates studied. The phylogenetic tree analysis in Figure 2a, supports the positioning of R. pickettii and R. insidiosa as two separate groups (bootstrap values of 91%), with B. cepacia as an out-group. The isolates identified as R. pickettii themselves divide into two different groups (bootstrap value of 99%). However the division into groups did not correlate to clinical or environmental association or indeed on their isolation location.
Figure 2.
Phylogenetic trees. A) Phylogenetic tree of R. pickettii and R. insidiosa 16S-23S ISR of nineteen sequenced isolates and sequence data available on the Genbank database. The tree was rooted with the ISR of Ralstonia solanacearum (Genbank Accession No AJ277280), Cupriavidus necator (AJ783978) and Burkholderia cepacia (L28154). B) Phylogenetic tree of R. pickettii and R. insidiosa fliC genes of nineteen sequenced isolates and sequence data available on the Genbank database. The tree was rooted with the fliC of Burkholderia cepacia (L28154). Cluster analysis was based upon the neighbour-joining method. Numbers at branch-points are percentages of 1000 bootstrap resamplings that support the topology of the tree.
Sequencing was carried out on the fliC gene of sixteen randomly selected isolates of R. pickettii, and the type strain of R. insidiosa. The phylogenetic analysis of the fliC gene can be seen in Figure 2b, with the isolates divided into two branches with B. cepacia as an out-group. The isolates identified as R. insidiosa in-group two grouped together with groups three and four. These however were not supported by high bootstrapping values. Group 1 is made up of R. pickettii isolates from clinical and environmental sources with 97-100% similarity to the R. pickettii type strain. Group 2 is made up of R. insidiosa with 85% similarity to the R. pickettii type strain; Group 3 is made up of both R. insidiosa and R. pickettii with 86-87% similarity to the R. pickettii type strain and Group 4 is made up of the available sequenced R. pickettii strains with 87% similarity to the R. pickettii type strain. The division of the groups did not correlate to clinical or environmental association or on their isolation location. These results indicate that there is variation in the flagellin gene of R. pickettii.
RAPD PCR results and analysis
RAPD analysis was carried out using four different primers, three of which (P3, P15 and M13) have been shown to discriminate between closely related strains of Ralstonia spp. including R. mannitolilytica and Cupriavidus pauculus [Ralstonia paucula] [47,48]. The reproducibility of the RAPD method was tested by repeating the RAPD assays at least three times for each primer used (data not shown). The results revealed that apart from some variations in the band intensity, no significant differences were observed between the profiles obtained, confirming the reproducibility of the method.
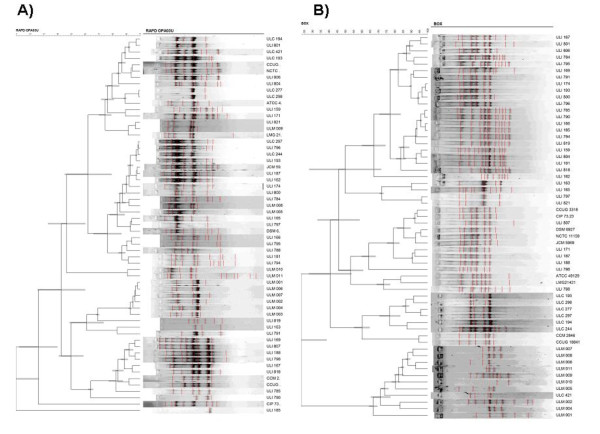
Fifty-nine isolates of R. pickettii and R. insidiosa were characterised by RAPD analysis using all four primers and all isolates were placed into genotypes (Table 3). Percent similarities based on the Pearson correlation coefficients and clustering by the UPGMA method for these isolates using the OPA03U primer is presented in Figure 3a. Dendograms for the other primers (P3, P15 and M13) are presented in Additional File 2, Figure S1, S2 and S3. Fragments ranged from approximately 100 to 1800 bp for all primers. Clusters were distinguished at a similarity cut-off level of 80%. No major differentiation between the clinical, industrial, laboratory purified water and type strains could be observed, as these all fell into separate groups (Table 3) with each primer. For each of the primers there were a number of groups, with M13 there were twenty-one groups, OPA3OU there were 15 groups, P3 there were twenty-five groups and with primer P15 there were twenty-one groups. The clinical isolates grouped together with two of the primers, with M13 they clustered together in Genotype A with the type strain JCM5969, three laboratory water isolates and three industrial water isolates, with P3 they clustered together in Genotype × with nine of the industrial water isolates (including the industrial isolates that grouped together with the clinical isolates in Genotype A with the M13 primer), with primers P15 and OPA30U they fell into several clusters six with P15. The industrial purified water isolates also fell into different groups with all four primers. There were nine groups with primer P15, thirteen groups with primer M13, fifteen groups with primer P3 and eleven groups with primer OPA3OU. The laboratory purified water isolates fell into two different groups with primer P15, six groups with primer M13, five groups with primer P3 and three groups with primer OPA3OU. The isolates identified as R. insidiosa failed to group together with any of the RAPD primers. With the P15 primer there is one large group that contained all the type strains, the soil strains, ten of laboratory water purified isolates and the industrial water isolates, no other primer produced such a large group. The diversity of the bacterial populations studied was calculated using Simpson's Index of Diversity (Di) [30] and the results of each individual primer were M13-0.897, OPA3OU-0.899, P3-0.918 and P15-0.771. The average diversity for the four primers was 0.869. An index (D) of 0.90 or greater is a desirable property of a typing scheme [30]. As can be seen from the results only primer P3 with a D of 0.918 produced a significant D index. The D value indicates that primer P3 would be the best primer to carry out further studies into the diversity of R. pickettii in the future as it is the most discriminatory primer of the four tested.
Figure 3.
RAPD analysis with primer OPA03U and BOX analysis. A) RAPD analysis with primer OPA03U B) BOX analysis. Dendrogram of fifty-nine isolates of R. pickettii and R. insidiosa by the Pearson correlation using the UPGMA linkage method.
BOX-PCR results and analysis
The fifty-nine isolates of R. pickettii and Ralstonia insidiosa were characterised by the BOX-PCR analysis using the BOX-A1R primer [39]. Repeatability of the BOX-PCR was considered good as the isolates showed identical profiles in three independent experiments (data not shown). The results revealed that while there were some variations in the band intensities, no significant differences were observed between the profiles obtained. Percent similarities based on the Pearson correlation coefficients and clustering by the UPGMA method for these isolates are presented in Figure 3b. Clusters were distinguished at a similarity cut-off level of 80%. With the BOX primer eighteen groups were found at this cut-off level. Fragments ranged from approximately 300 to 3000 bp for all primers. The number of groups can be seen in Table 4. The groups, in contrast to the RAPD primers, mostly contained bacteria isolated from the same environments e.g. Group F clustered all the type strains together, Group M the soil strains and groups K, L and P the clinical isolates. The industrial isolates grouped together in-group A, B, C, D, E, G and J. The laboratory water isolates grouped together in groups N, O, Q and R. As with all four RAPD primers the isolates identified as R. insidiosa failed to group together. The Di using BOX-A1R was 0.915. These various primers and techniques demonstrated the limited diversity of the R. pickettii.
Table 4.
No.of Groupings with Four Different RAPD Primers and Box Primer
| Primer | No. of Groupings | Discrimination index |
|---|---|---|
| M13 | 21 | 0.897 |
| OPA3OU | 15 | 0.899 |
| P3 | 25 | 0.918 |
| P15 | 21 | 0.771 |
| BOX | 18 | 0.915 |
Discussion
In the course of this study a number of bacteria previously identified phenotypically as R. pickettii were subsequently identified as R. insidiosa using species-specific PCR. These bacteria are hard to distinguish from each other phenotypically [49]. R. insidiosa, the closest related bacteria to R. pickettii [33], has been isolated from the respiratory tracts of cystic fibrosis patients [33], river and pond water, soil, activated sludge [33] and has also been detected in water distribution systems [50] and laboratory purified water systems [3]. It has also been the causative agent of two cases of serious hospital infection in two immunocompromised individuals [51].
Each of the four DNA-based fingerprinting and sequencing methods were suitable for distinguishing and grouping the isolates, although the sensitivity of the methods varied. Of the three phenotypic methods examined, the API 20NE system was more discriminatory than the Remel RapID NF Plus system or the Vitek NFC. However, the Remel RapID NF Plus system and the Vitek NFC did prove more useful for the accurate identification of R. pickettii isolates, as previously reported [52]. The API 20NE gave thirty-five different biotypes for fifty-nine isolates (Table 3, Figure 1), which grouped together isolates from different environments. These results broadly agree with those of Dimech et al who found homogeneity in physiological parameters [25].
Genotypic studies carried out by both Dimech et al. and Chetoui et al. hinted that R. pickettii also had genotypic homogeneity [25,26]. This was investigated in this study using the methods described above. Our data based on the sequence of 16S-23S spacer regions of nineteen isolates indicated that Ralstonia pickettii is a homogenous species with little difference between isolates from different environmental niches. Clearly using these methods we can however determine differences between R. pickettii and R. insidiosa.
The fliC gene has been used for bacterial strain differentiation in multiple studies such as for Ralstonia solanacearum [35] and Burkholderia cepacia complex [53]. Four different types of flagellin gene have been found in R. pickettii isolates analysed in this study (Groups 1, 2, 3 and 4). This is similar to data from P. aeruginosa where two different types of fliC gene have been found [54] and from the B. cepacia complex where again two different types of fliC gene have also been found [55,56]. The fliC gene appears however not to be useful for distinguishing between R. pickettii and R. insidiosa based on our findings. The division of the groups did not correlate to clinical or environmental association or to their location of isolation. The reasons for the variation between the 16S-23S spacer region and the fliC gene could be potentially due to the structure of the fliC gene. This is demonstrated by Burkholderia flagellin sequences, which exhibit high levels of homology in the conserved terminal regions but differ considerably in the central region [57]. Variation is a common feature of flagellin proteins, which are believed to fold into a hairpin-like conformation, with the terminal domains being responsible for defining the basic filament structure lying on the inner surface and the central, variable region being surface exposed [58].
In a previous epidemiological study involving sixteen isolates of R. pickettii, eight different RAPD profiles were observed for isolates coming from blood culture, distilled water and an aqueous chlorhexidine solution [16]. In another study, involving fourteen isolates of R. pickettii from various biological samples the same RAPD pattern was found in all instances [59], while Pasticci et al., carried out a study involving fifteen isolates of R. pickettii that gave three patterns [27]. The results of our study with a larger number of isolates indicated that there is some diversity in the studied populations but that this is limited and isolates from different environments grouped together.
The results obtained with BOX-PCR showed nineteen different profiles among the fifty-nine isolates examined again demonstrating limited diversity (Figure 3b). To the best of our knowledge this is the first reported study of the diversity of R. pickettii and R. insidiosa carried out with BOX-PCR. A similar study carried by Coenye et al., on ninety-seven B. cepacia Genomovar III isolates found 20 different patterns with a DI value of 0.821 [60].
The molecular fingerprinting methods used here yielded rapid and reproducible fingerprints for clinical and environmental isolates of R. pickettii and R. insidiosa. Presently, little is known regarding the source of R. pickettii isolates occurring in hospital environments. Investigations by other authors have reported no evidence of patient-to-patient transmission, and they suggest that multiple independent acquisitions from environmental sources could be an important mode of transmission of R. pickettii [5]. The most common sites of contamination were blood-sampling tubes, dialysis machines, nebulizers and other items frequently in contact with water [5].
Conclusions
BOX-PCR and RAPD typing was found to be more discriminatory than the typing of genes in R. pickettii such as the fliC gene or the ISR. The majority of isolates were shown to possess similar genotypes by both BOX and RAPD-PCR (Figure 3a, b). The limited diversity of R. pickettii and R. insidiosa isolates observed in this study for fifty-nine isolates is consistent with previous findings and indicates that R. pickettii appears to be a genotypically and phenotypically homogeneous species.
Authors' contributions
MPR conceived the study and its design, carried out the experimental work, performed the analysis and interpretation of the data and wrote the manuscript. JTP participated in conceiving the study and in its design and participated in writing the manuscript. CAA participated in conceiving the study, its design, and participated in writing the manuscript. All authors read and approved the final manuscript. The authors declare no conflict of interest.
Supplementary Material
Table S1. API 20NE and Remel Rapid NF Plus Codes for isolates used in this study and identifiers for biochemical tests.
Figure S1, S2, S3. Dendograms for primers M13, P3 and P15 that were not included in the paper.
Contributor Information
Michael P Ryan, Email: Michael.P.Ryan@ul.ie.
J Tony Pembroke, Email: Tony.Pembroke@ul.ie.
Catherine C Adley, Email: Catherine.Adley@ul.ie.
Acknowledgements
MPR funding was provided by a Postgraduate bursary from the Chemical and Environmental Science Department, Faculty of Science and Engineering, University of Limerick.
References
- Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. Nov.: Proposal of Ralstonia pickettii (Ralston, Palleroni and Doudoroff 1973) comb. Nov., Ralstonia solanacearum (Smith 1896) comb. Nov. and Ralstonia eutropha (Davis 1969) comb. Nov. Microbiol Immunol. 1995;39(11):897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]
- Adley CC, Saieb FM. Biofilm formation in high purity water: Ralstonia pickettii a special case for analysis. Ultrapure Water. 2005;22:14–17. [Google Scholar]
- Adley CC, Ryan MP, Pembroke JT, Saieb FM. In: Biofilms: Persistence and Ubiquity. Mc Bain A, Alison D, Pratten J, Spratt D, Upton M, Verran J, editor. Cardiff: Biofilm Club; 2005. Ralstonia pickettii in high purity water; pp. 261–272. [Google Scholar]
- Gilligan PH, Lum G, Vandamme PAR, Whittier S. In: Manual of Clinical Microbiology. 8. Murray PR, Baron EJ, Pfaller MA, Jorgensen JH, Yolken RH, editor. ASM Press Washington, D.C.; 2003. Burkholderia, Stenotrophomonas, Ralstonia, Brevundimonas, Comamonas, Delftia, Pandoraea and Acidovorax; pp. 729–748. [Google Scholar]
- Ryan MP, Pembroke JT, Adley CC. Ralstonia pickettii: a persistent gram-negative nosocomial infectious organism. J Hosp Infect. 2006;62(3):278–284. doi: 10.1016/j.jhin.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Lacey S, Want SV. Pseudomonas pickettii infections in a paediatric oncology unit. J Hosp Infect. 1991;17(1):45–51. doi: 10.1016/0195-6701(91)90076-K. [DOI] [PubMed] [Google Scholar]
- Wertheim WA, Markovitz DM. Osteomyelitis and intervertebral discitis caused by Pseudomonas pickettii. J Clin Microbiol. 1992;30(9):2506–2508. doi: 10.1128/jcm.30.9.2506-2508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MP, Pembroke JT, Adley CC. Ralstonia pickettii in environmental biotechnology: potential and applications. J Appl Microbiol. 2007;103(4):754–764. doi: 10.1111/j.1365-2672.2007.03361.x. [DOI] [PubMed] [Google Scholar]
- Gardner S, Shulman ST. A nosocomial common source outbreak caused by Pseudomonas pickettii. Pediatr Infect Dis. 1984;3(5):420–422. doi: 10.1097/00006454-198409000-00006. [DOI] [PubMed] [Google Scholar]
- McNeil MM, Solomon SL, Anderson RL, Davis BJ, Spengler RF, Reisberg BE, Thornsberry C, Martone WJ. Nosocomial Pseudomonas pickettii colonization associated with a contaminated respiratory therapy solution in a special care nursery. J Clin Microbiol. 1985;22(6):903–907. doi: 10.1128/jcm.22.6.903-907.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts LA, Collignon PJ, Cramp VB, Alexander S, McFarlane AE, Graham E, Fuller A, Sinickas V, Hellyar A. An Australia-wide epidemic of Pseudomonas pickettii bacteraemia due to contaminated "sterile" water for injection. Med J Aust. 1990;152(12):652–655. doi: 10.5694/j.1326-5377.1990.tb125422.x. [DOI] [PubMed] [Google Scholar]
- Maki DG, Klein BS, McCormick RD, Alvarado CJ, Zilz MA, Stolz SM, Hassemer CA, Gould J, Liegel AR. Nosocomial Pseudomonas pickettii bacteremias traced to narcotic tampering. A case for selective drug screening of health care personnel. JAMA. 1991;265(8):981–986. doi: 10.1001/jama.265.8.981. [DOI] [PubMed] [Google Scholar]
- Raveh D, Simhon A, Gimmon Z, Sacks T, Shapiro M. Infections caused by Pseudomonas pickettii in association with permanent indwelling intravenous devices: four cases and a review. Clin Infect Dis. 1993;17(5):877–880. doi: 10.1093/clinids/17.5.877. [DOI] [PubMed] [Google Scholar]
- Labarca JA, Trick WE, Peterson CL, Carson LA, Holt SC, Arduino MJ, Meylan M, Mascola L, Jarvis WR. A multistate nosocomial outbreak of Ralstonia pickettii colonization associated with an intrinsically contaminated respiratory care solution. Clin Infect Dis. 1999;29(5):1281–1286. doi: 10.1086/313458. [DOI] [PubMed] [Google Scholar]
- Kahan A, Philippon A, Paul G, Weber S, Richard C, Hazebroucq G, Degeorges M. Nosocomial infections by chlorhexidine solution contaminated with Pseudomonas pickettii (Biovar VA-I) J Infect. 1983;7(3):256–263. doi: 10.1016/S0163-4453(83)97196-7. [DOI] [PubMed] [Google Scholar]
- Maroye P, Doermann HP, Rogues AM, Gachie JP, Megraud F. Investigation of an outbreak of Ralstonia pickettii in a paediatric hospital by RAPD. J Hosp Infect. 2000;44(4):267–272. doi: 10.1053/jhin.1999.0691. [DOI] [PubMed] [Google Scholar]
- Zellweger C, Bodmer T, Tauber MG, Muhlemann K. Failure of ceftriaxone in an intravenous drug user with invasive infection due to Ralstonia pickettii. Infection. 2004;32(4):246–248. doi: 10.1007/s15010-004-3033-0. [DOI] [PubMed] [Google Scholar]
- Anderson RL, Holland BW, Carr JK, Bond WW, Favero MS. Effect of disinfectants on pseudomonads colonized on the interior surface of PVC pipes. Am J Public Health. 1990;80(1):17–21. doi: 10.2105/ajph.80.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakov LA, McAlister MB, Ogden KL, Larkin MJ, O'Hanlon JF. Analysis of bacteria contaminating ultrapure water in industrial systems. Appl Environ Microbiol. 2002;68(4):1548–1555. doi: 10.1128/AEM.68.4.1548-1555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig DW, Pierson DL. Microbiology of the Space Shuttle water system. Water Sci Technol. 1997;35(11-12):59–64. doi: 10.1016/S0273-1223(97)00235-7. [DOI] [PubMed] [Google Scholar]
- La Duc MT, Kern R, Venkateswaran K. Microbial monitoring of spacecraft and associated environments. Microb Ecol. 2004;47(2):150–158. doi: 10.1007/s00248-003-1012-0. [DOI] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280(5361):295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- McAlister MB, Kulakov LA, O'Hanlon JF, Larkin MJ, Ogden KL. Survival and nutritional requirements of three bacteria isolated from ultrapure water. J Ind Microbiol Biotechnol. 2002;29(2):75–82. doi: 10.1038/sj.jim.7000273. [DOI] [PubMed] [Google Scholar]
- Ryan MP, Pembroke JT, Adley CC. Novel Tn4371-ICE like element in Ralstonia pickettii and genome mining for comparative elements. BMC Microbiol. 2009;9:242. doi: 10.1186/1471-2180-9-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimech WJ, Hellyar AG, Kotiw M, Marcon D, Ellis S, Carson M. Typing of strains from a single-source outbreak of Pseudomonas pickettii. J Clin Microbiol. 1993;31(11):3001–3006. doi: 10.1128/jcm.31.11.3001-3006.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetoui H, Melin P, Struelens MJ, Delhalle E, Nigo MM, De Ryck R, De Mol P. Comparison of biotyping, ribotyping, and pulsed-field gel electrophoresis for investigation of a common-source outbreak of Burkholderia pickettii bacteremia. J Clin Microbiol. 1997;35(6):1398–1403. doi: 10.1128/jcm.35.6.1398-1403.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasticci MB, Baldelli F, Camilli R, Cardinali G, Colozza A, Marroni M, Morosi S, Pantosti A, Pitzurra L, Repettos A. et al. Pulsed field gel electrophoresis and random amplified polymorphic DNA molecular characterization of Ralstonia pickettii isolates from patients with nosocomial central venous catheter related bacteremia. New Microbiol. 2005;28(2):145–149. [PubMed] [Google Scholar]
- Sneath PHA, Sokal RR. Numerical taxonomy. The principles and practice of numerical classification. WH Freeman & Co: San Francisco, Calif; 1973. [Google Scholar]
- Jaccard P. Étude comparative de la distribution florale dans une portion des Alpes et des Jura. Bull Soc Vaudoise Sci Nat. 1901;37:547–579. [Google Scholar]
- Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26(11):2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenye T, Liu L, Vandamme P, LiPuma JJ. Identification of Pandoraea species by 16S ribosomal DNA-based PCR assays. J Clin Microbiol. 2001;39(12):4452–4455. doi: 10.1128/JCM.39.12.4452-4455.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenye T, Vandamme P, LiPuma JJ. Infection by Ralstonia species in cystic fibrosis patients: identification of R. pickettii and R. mannitolilytica by polymerase chain reaction. Emerg Infect Dis. 2002;8(7):692–696. doi: 10.3201/eid0807.010472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenye T, Goris J, De Vos P, Vandamme P, LiPuma JJ. Classification of Ralstonia pickettii-like isolates from the environment and clinical samples as Ralstonia insidiosa sp. nov. Int J Syst Evol Microbiol. 2003;53(Pt 4):1075–1080. doi: 10.1099/ijs.0.02555-0. [DOI] [PubMed] [Google Scholar]
- Kostman JR, Edlind TD, LiPuma JJ, Stull TL. Molecular epidemiology of Pseudomonas cepacia determined by polymerase chain reaction ribotyping. J Clin Microbiol. 1992;30(8):2084–2087. doi: 10.1128/jcm.30.8.2084-2087.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonfeld J, Heuer H, Van Elsas JD, Smalla K. Specific and sensitive detection of Ralstonia solanacearum in soil on the basis of PCR amplification of fliC fragments. Appl Environ Microbiol. 2003;69(12):7248–7256. doi: 10.1128/AEM.69.12.7248-7256.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torriani S, Zapparoli G, Dellaglio F. Use of PCR-based methods for rapid differentiation of Lactobacillus delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis. Appl Environ Microbiol. 1999;65(10):4351–4356. doi: 10.1128/aem.65.10.4351-4356.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroye P, Doermann HP, Rogues AM, Gachie JP, Mégraud F. Investigation of an outbreak of Ralstonia pickettii in a paediatric hospital by RAPD. J Hosp Infect. 2000;44(4):267–272. doi: 10.1053/jhin.1999.0691. [DOI] [PubMed] [Google Scholar]
- Castle A, Speranzini D, Rghei N, Alm G, Rinker D, Bissett J. Morphological and molecular identification of Trichoderma isolates on North American mushroom farms. Appl Environ Microbiol. 1998;64(1):133–137. doi: 10.1128/aem.64.1.133-137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louws FJ, Fulbright DW, Stephens CT, de Bruijn FJ. Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl Environ Microbiol. 1994;60(7):2286–2295. doi: 10.1128/aem.60.7.2286-2295.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5(2):150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Li WH. Simple method for constructing phylogenetic trees from distance matrices. Proc Natl Acad Sci USA. 1981;78(2):1085–1089. doi: 10.1073/pnas.78.2.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtler V, Stanisich VA. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology. 1996;142(Pt 1):3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- Tyler SD, Strathdee CA, Rozee KR, Johnson WM. Oligonucleotide primers designed to differentiate pathogenic pseudomonads on the basis of the sequencing of genes coding for 16S-23S rRNA internal transcribed spacers. Clin Diagn Lab Immunol. 1995;2(4):448–453. doi: 10.1128/cdli.2.4.448-453.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxboeck F, Stadler M, Assadian O, Marko E, Hirschl AM, Koller W. Characterization of clinically isolated Ralstonia mannitolilytica strains using random amplification of polymorphic DNA (RAPD) typing and antimicrobial sensitivity, and comparison of the classification efficacy of phenotypic and genotypic assays. J Med Microbiol. 2005;54(Pt 1):55–61. doi: 10.1099/jmm.0.45656-0. [DOI] [PubMed] [Google Scholar]
- Moissenet D, Goujon CP, Garbarg-Chenon A, Vu-Thien H. CDC group IV c-2: a new Ralstonia species close to Ralstonia eutropha. J Clin Microbiol. 1999;37(6):1777–1781. doi: 10.1128/jcm.37.6.1777-1781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan MP, Pembroke JT, Adley CC. Differentiating the growing nosocomial infectious threats Ralstonia pickettii and Ralstonia insidiosa. Eur J Clin Microbiol Infect Dis. 2011. in press . [DOI] [PubMed]
- Hoefel D, Monis PT, Grooby WL, Andrews S, Saint CP. Profiling bacterial survival through a water treatment process and subsequent distribution system. J Appl Microbiol. 2005;99(1):175–186. doi: 10.1111/j.1365-2672.2005.02573.x. [DOI] [PubMed] [Google Scholar]
- Van der Beek D, Magerman K, Bries G, Mewis A, Declercq P, Peeters V, Rummens JL, Raymaekers M, Cartuyvels R. Infection with Ralstonia insidiosa in two patients. Clin Microbiol Newsl. 2005;27(20):159–160. doi: 10.1016/j.clinmicnews.2005.09.007. [DOI] [Google Scholar]
- Adley CC, Saieb FM. Comparison of bioMerieux API 20NE and Remel RapID NF Plus, identification systems of type strains of Ralstonia pickettii. Lett Appl Microbiol. 2005;41(2):136–140. doi: 10.1111/j.1472-765X.2005.01737.x. [DOI] [PubMed] [Google Scholar]
- Winstanley C. Improved flagellin genotyping in the Burkholderia cepacia complex. FEMS Microbiol Lett. 2003;229(1):9–14. doi: 10.1016/S0378-1097(03)00721-3. [DOI] [PubMed] [Google Scholar]
- Spangenberg C, Heuer T, Burger C, Tummler B. Genetic diversity of flagellins of Pseudomonas aeruginosa. FEBS Lett. 1996;396(2-3):213–217. doi: 10.1016/0014-5793(96)01099-X. [DOI] [PubMed] [Google Scholar]
- Hales BA, Morgan JA, Hart CA, Winstanley C. Variation in flagellin genes and proteins of Burkholderia cepacia. J Bacteriol. 1998;180(5):1110–1118. doi: 10.1128/jb.180.5.1110-1118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo ST, Tsuchiya K. Genotypic characterization of Burkholderia cenocepacia strains by rep-PCR and PCR-RFLP of the fliC gene. FEMS Microbiol Lett. 2005;245(1):19–24. doi: 10.1016/j.femsle.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Wilson DR, Beveridge TJ. Bacterial flagellar filaments and their component flagellins. Can J Microbiol. 1993;39(5):451–472. doi: 10.1139/m93-066. [DOI] [PubMed] [Google Scholar]
- Hales BA, Morgan JA, Hart CA, Winstanley C. Variation in flagellin genes and proteins of Burkholderia cepacia. J Bacteriol. 1998;180(5):1110–1118. doi: 10.1128/jb.180.5.1110-1118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros N, Gonullu N, Casetta A, Guibert M, Ingrand D, Lebrun L. Ralstonia pickettii traced in blood culture bottles. J Clin Microbiol. 2002;40(7):2666–2667. doi: 10.1128/JCM.40.7.2666-2667.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenye T, Spilker T, Martin A, LiPuma JJ. Comparative assessment of genotyping methods for epidemiologic study of Burkholderia cepacia genomovar III. J Clin Microbiol. 2002;40(9):3300–3307. doi: 10.1128/JCM.40.9.3300-3307.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. API 20NE and Remel Rapid NF Plus Codes for isolates used in this study and identifiers for biochemical tests.
Figure S1, S2, S3. Dendograms for primers M13, P3 and P15 that were not included in the paper.