Abstract
Leukemia/lymphoma related factor/POK erythroid myeloid ontogenic factor (LRF/Pokemon) is a member of the POK family of proteins that promotes oncogenesis in several forms of cancer. Recently, we found higher LRF expression in human breast and prostate carcinomas compared to the corresponding normal tissues. The aim of this study was to examine the regulation of LRF expression in human prostate cells. Epidermal growth factor (EGF) and its receptors mediate several tumorigenic cascades that regulate cell differentiation, proliferation, migration and survival of prostate cancer cells. There was significantly higher level of LRF expression in the nucleus of LNCaP and PC-3 cells than RWPE-1 cells. A significant increase in LRF expression was observed with increasing doses of EGF in more aggressive and androgen-sensitive prostate cancer cells suggesting that EGF signaling pathway is critical in upregulating the expression of LRF/Pokemon to promote oncogenesis.
Keywords: prostate cancer, Pokemon, EGF, LRF, ZBTB7
INTRODUCTION
Despites advances in early detection and diagnosis, the underlying mechanisms leading to the metastasis of prostate cancer have yet to be identified. The secretory epithelial cells of the prostate gland are exposed to many external stimuli, including oxidative stress, mediated by numerous hormones, growth factors and neuropeptides that control their proliferation, differentiation and survival. Leukemia/lymphoma related factor (LRF), also known as POK erythroid myeloid ontogenic factor (POKEMON), is implicated in the pathogenesis of several cancers including gliomas, T-cell a n d B-cell lymphomas, prostate, breast, non-small cell lung, and ovarian carcinomas (Aggarwal et al., 2010; Aggarwal et al., 2011; Apostolopoulou et al., 2007; Jiang et al., 2010; Maeda et al., 2007; Qu et al., 2010; Rovin and Winn, 2005). Indeed, LRF has numerous functions including repression of the transcription of ADH5/FDH, activation of membrane type-1 matrix metalloproteinase in ovarian cancer, p14ARF in lymphomas, cyclin A in adipogenesis, and E-cadherin and NF-κB regulation (Jiang et al., 2010; Maeda et al., 2005a). Little is known, however, as to how the expression of LRF is regulated. One study revealed that specificity protein (Sp1) enhances LRF expression in HepG2 hepatocellular carcinoma cells (Zu et al., 2009). The secretory epithelial cells of the prostate gland are exposed to many external stimuli by numerous hormones and growth factors that control their proliferation, differentiation and survival (Kim et al., 1999; Kumar et al., 1998; Mimeault and Batra, 2006). Thus, it is reasonable to speculate that these hormones or growth factors could be responsible for increased expression of LRF in prostate cells.
Amongst the growth factors, epidermal growth factor (EGF) plays a major role during prostate carcinogenesis because of its autocrine and paracrine secretion and overexpression of its receptor tyrosine kinase (EGFR) on several prostatic cell types (De Miguel et al., 1999; MacDonald and Habib, 1992; Sherwood et al., 1998) which include normal prostate cells RWPE-1, androgen sensitive prostate cancer cells LNCaP and androgen insensitive cancer cells PC-3 (Bello et al., 1997; Webber et al., 2001). In an earlier report, we had shown higher expression of LRF in prostate cancer tissues as compared to benign prostate hyperplasic tissues (Aggarwal et al., 2011). LRF can also act as a co-repressor for the androgen receptor in prostate cancer cells (Cui et al., 2010). The aim of this study was to examine the effect of growth factor, EGF, on the expression of LRF in prostate cell lines.
MATERIALS AND METHODS
Cell Lines and Antibodies
The normal human prostate cells, RWPE-1, and prostate cancer cells, PC-3 and LNCaP, were purchased from the American Type Culture Collection (ATCC; Manassas, VA). RWPE-1 cells were maintained in Keratinocyte Serum Free Medium (K-SFM; Gibco #17005-042) supplemented with 0.05mg/mL bovine pituitary extract (BPE). PC-3 cells were maintained in ATCC-formulated F-12K Medium and LNCaP cells were grown in ATCC-formulated RPMI-1640 medium. Cells were harvested using 0.25% (w/v) trypsin containing 0.53mM EDTA. All cell lines were maintained with 10% FBS, 1% penicillin, and 1% streptomycin at 37°C in a humidified atmosphere containing 5% CO2.
Anti-LRF rabbit polyclonal antibody raised against a peptide derived from the C-terminal region of LRF/Pokemon was prepared by Pacific Immunology (Ramona, CA) and used in our earlier studies (Aggarwal et al., 2010).
EGF treatment
RWPE-1, LNCaP, and PC-3 cells were dispersed by 2–3mL of 0.25% (w/v) trypsin-EDTA solution, re-suspended in respective supplemented media and seeded into 6-well culture plates at a density of 5×103 cells/cm2. After 24 hours, the cell monolayer was washed thoroughly with Dulbecco’s phosphate buffered saline (PBS) and treated with serum free medium for another 24 hours. After that, serum-free with and without EGF (10–100ng/mL) was added to the wells for 24 hours, following which cells were collected for RNA and protein studies. All experiments were done in triplicates in three separate cell samples.
RNA isolation and semi-quantitative reverse-polymerase chain reaction (RT-PCR)
This was performed as previously reported ((Aggarwal et al., 2010; Aggarwal et al., 2011). Briefly, RNA was isolated using Trizol reagent (Sigma) then reverse transcribed using the PCR Master Mix (Promega, Madison, WI). Primers and conditions for amplification of LRF and GAPDH were previously described (Aggarwal et al, 2010). Signals were visualized using the UVP Bioimaging System (Upland, CA).
Western blotting
Cells were lysed in Mammalian Protein Extraction Reagent (Promega) and protein concentrations were determined using the Bio-Rad protein assay (Bio-Rad, Hercules, CA). Equal protein was assayed by immunoblot using rabbit anti-sera against LRF (1:500 dilution) and mouse anti-sera against GAPDH (1:2000 dilution), followed by horse radish peroxidase conjugated goat anti-rabbit IgG (1:2000). Anti-LRF and anti-GAPDH were previously described (Aggarwal et al., 2010). Chemiluminescence was performed using SuperSignal kit (Thermo Fisher Scientific, Rockford, IL).
Immunocytochemistry
Cells were suspended to 5×105 cells/mL PBS and cytospun at 800 rpm for 10 minutes on superfrost/plus slides (Fisher Scientific, St. Louis, MO) using a Shannon Cytopsin. Cells were fixed in acetone for 10 minutes and then washed with PBS 3 times for 5 minutes. Endogenous peroxidase was blocked in the cells by 0.3% hydrogen peroxide for 15 minutes and the cells were immunostained with primary rabbit polyclonal antibody against LRF (1:1600) for 1 hour at room temperature. Negative controls were established by using rabbit pre-immune serum PAC-767 (Pacific Immunology, Ramona, CA) or omission of primary antibody. Cells were washed with PBS and then exposed to biotin-labeled goat anti-rabbit IgG for 30 minutes according to the supplier’s instructions and the immune conjugates were treated with an avidin-biotin complex (PK-6101, Vectastain ABC elite kit, Vector Laboratories, CA). Staining was completed by using diaminobenzidine chromagen (K4010, DAKO) followed by counterstaining with hematoxylin (S3309, DAKO, Carpinteria, CA). Immunostained sections were photographed by an Olympus Dp71 camera at 200x or 400x magnification.
Immunofluorescence
RWPE-1 and PC-3 cells were seeded, in respective serum-supplemented media at a density of 2×103 cells into 6-well plates containing 20×20mm2 cover slips. After 72 hours of culture, the monolayer was treated with serum-free medium for 24 hours. Media with and without EGF (10–100ng/mL) was added to the wells for 24 hours. The coverslip with cultured cells was exposed to 4% paraformaldehyde in PBS for 30 minutes. Excess paraformaldehyde was removed with PBS. Cells were prepared for immunolabeling as described above in the section on Immunocytochemistry.
Statistical Analysis
Densitometric analysis was performed using the UVP Bioimaging System (Upland, CA). Values of all measurements are expressed as the mean ± SE. Regression analysis and single factor ANOVA was used as the statistical method to compare different treatment groups. Single factor ANOVA was used to compare LRF expression levels amongst the 3 cell lines. Microsoft Excel and GraphPad Prism 4.0 programs were used to draw graphs and SPSS was used for statistical analysis. Values of p < 0.05 were considered statistically significant.
RESULTS
LRF is expressed in prostate cell lines
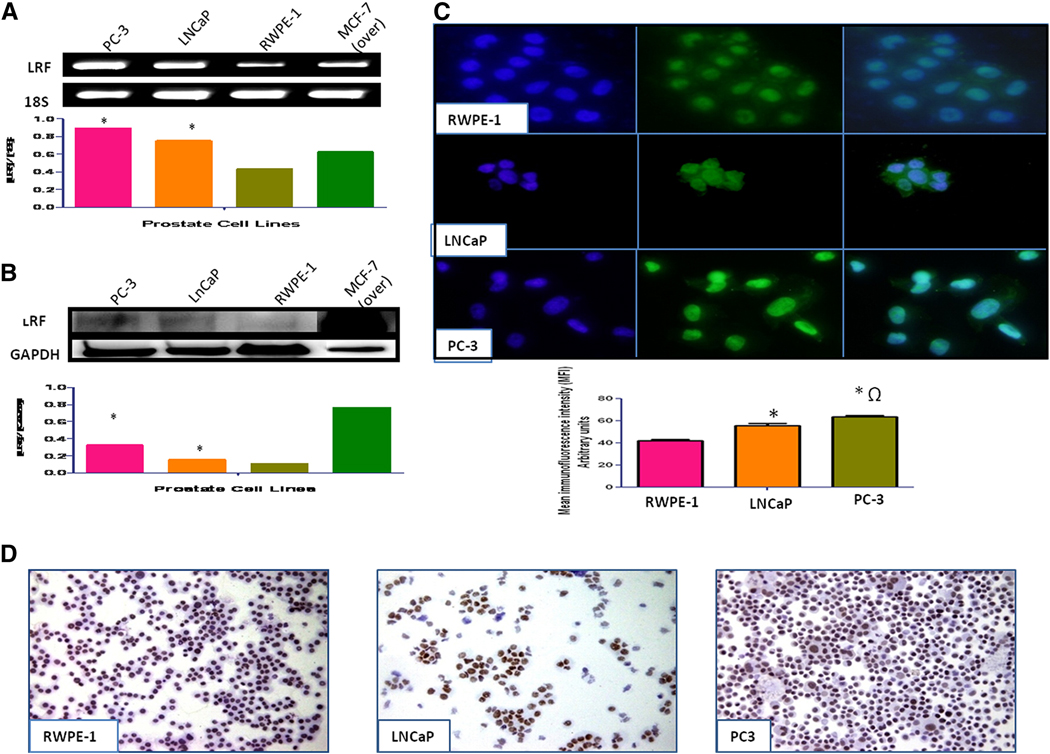
Levels of LRF mRNA transcripts were higher in LNCaP and PC-3 than the RWPE-1 cells (Figure 1a). Quantification of mean fluorescence intensity and Western blot analysis of Pokemon revealed significantly higher levels of LRF in LNCaP and PC-3 cells than RWPE-1 cells (p < 0.01; Figure 1b). The expression level of LRF was higher in PC-3 than LNCaP cells amongst the cancer cells (p < 0.05). Nuclear expression of LRF was observed in LNCaP and PC-3 cancer cells as well as the RWPE-1 normal prostate cells as observed by immunocytochemistry and immunofluorescence (Figures 1c and 1d).
Figure 1. Expression of LRF/Pokemon in human prostate cell lines.
LRF mRNA transcripts (A) and protein expression (B) in prostate cell lines with densitometric analysis indicate that cancer cell lines have significantly higher expression than normal prostate cell lines. MCF-7 cells overexpressing LRF were used as a positive control. (C) Immunohistochemical expression of LRF in prostate cell lines. DAPI (blue) was used as nuclear counterstain (200x–400x). (D) Immunohistochemical expression of LRF in prostate cell lines. DAB was used as a chromogen and hematoxylin as counterstain (200x–400x).
EGF increases expression of LRF in prostate cells
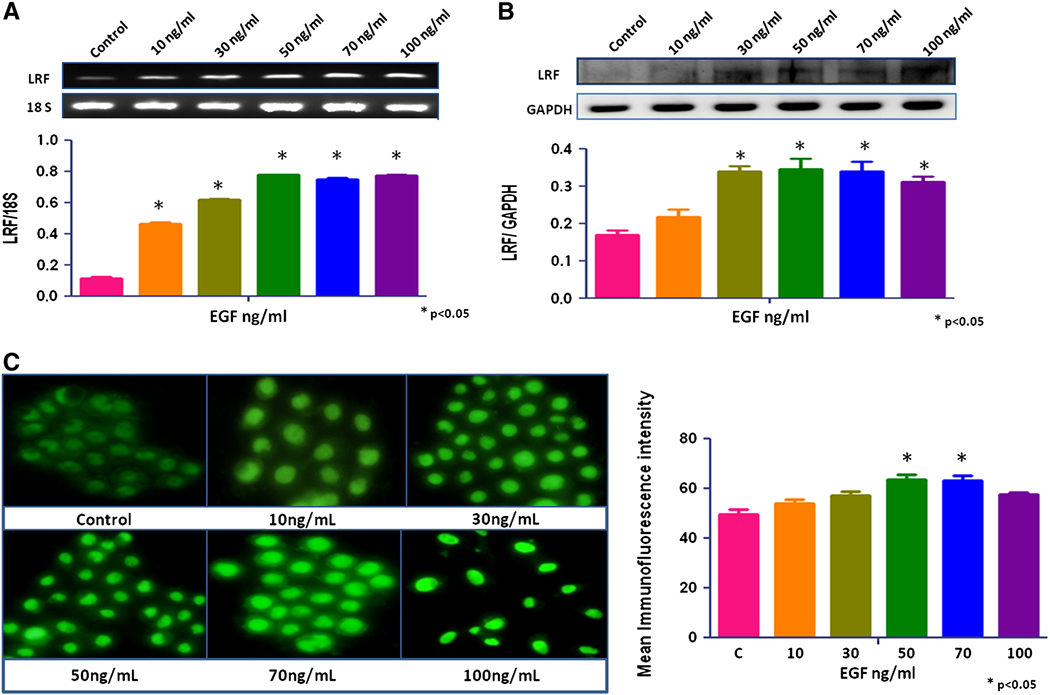
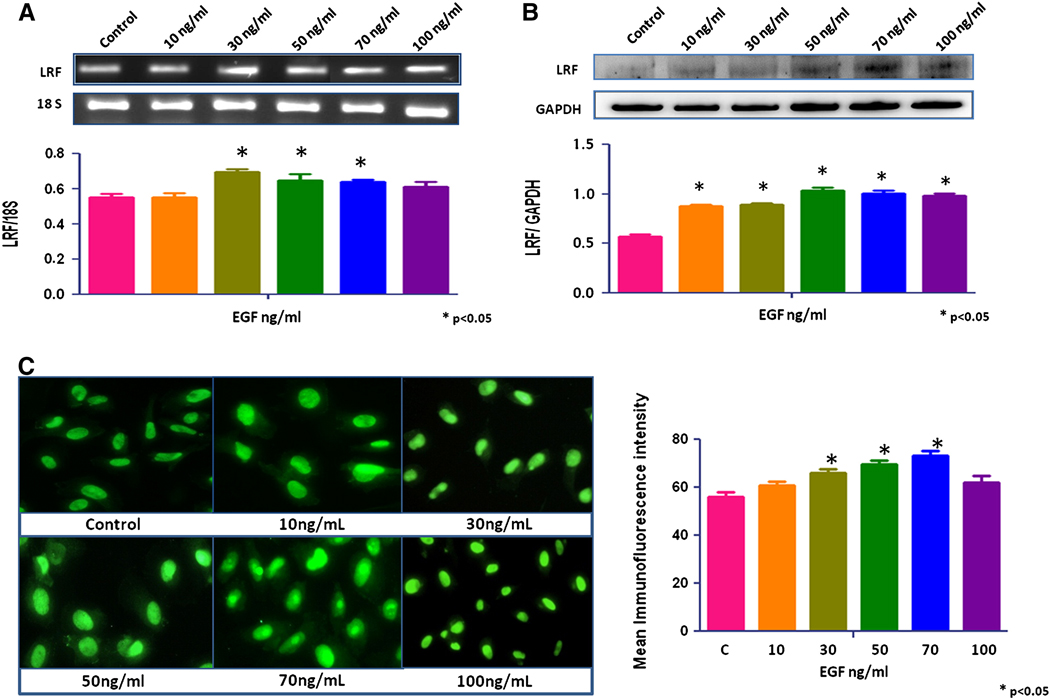
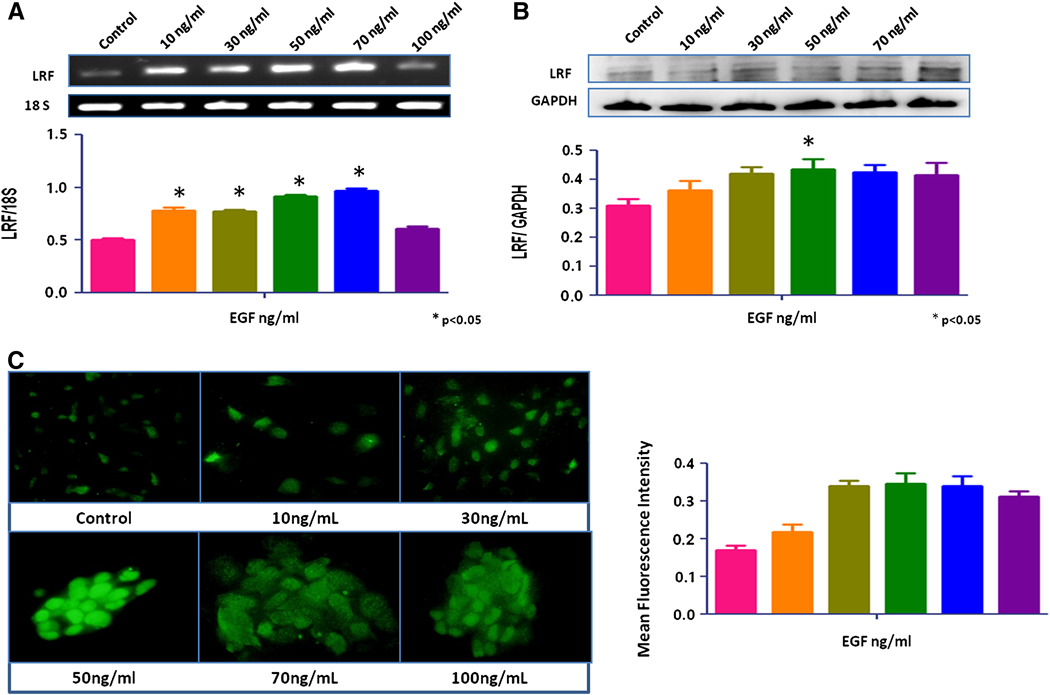
No significant change in LRF expression was seen after treatment with growth factors, IGF-1, TGF-β1, and TGF-β2 (data not shown). EGF increased the expression of LRF mRNA transcripts and protein in RWPE-1 cells in a dose-dependent manner (p < 0.05; Figures 2a, b). The mRNA transcripts and protein expression of LRF also increased significantly in EGF-treated LNCaP and PC-3 cells cells compared to the control group (p < 0.05; Figures 3a,b and 4a,b). Quantitative immunofluorescence also showed an increase in LRF expression in these cells (Figures 2c, 3c, 4c).
Figure 2. Effect of EGF on LRF expression in RWPE-1 cells.
LRF mRNA transcripts (A) and protein expression (B) in RWPE-1 cells with densitometric analysis. The cells were treated with EGF (0–100ng/mL) for 24 hours. (C) Immunohistochemical expression of LRF in RWPE-1 cells after treatment with EGF. DAPI (blue) was used as a nuclear counterstain (400x).
Figure 3. Effect of EGF on LRF expression in LNCaP cells.
LRF mRNA transcripts (A) and protein expression (B) in LNCaP cells with densitometric analysis after treatment with EGF (0–100ng/mL) for 24 hours. (C) Immunohistochemical expression of LRF in LNCaP cells after treatment with EGF. DAPI (blue) was used as a nuclear counterstain (400x).
Figure 4. Effect of EGF on LRF expression in PC-3 cells.
LRF mRNA transcripts (A) and protein expression (B) in PC-3 cells with densitometric analysis after treatment with EGF (0–100ng/mL) for 24 hours. (C) Immunohistochemical expression of LRF in PC-3 cells after treatment with EGF. DAPI (blue) was used as a nuclear counterstain (400x).
DISCUSSION
Increased expression of LRF in several forms of cancers is well documented. However, information regarding its functional expression in solid tumors is limited. Previously, we found an increase in LRF expression in both benign hyperplastic and cancerous lesions of the prostate (Aggarwal et al., 2011). In this study, quantification of LRF expression revealed higher level of LRF expression in cancer than in the normal prostate cells. Another study also noted a positive correlation between LRF expression and increased proliferation and size of the cancerous lesions of the lung by immunohistochemical analysis (Zhao et al., 2008a; Zhao et al., 2008b). Overexpression of LRF induces migration and invasion in ovarian cancer cells (Jiang et al., 2010). This information implies that LRF may be directly involved the proliferation of benign and malignant prostate epithelial cells. A more specific functional role of LRF in the development and progression of prostate cancer is warranted.
LRF expression was seen in normal prostate cells (RWPE-1), androgen sensitive prostate cancer cells (LNCaP) and androgen-insensitive prostate cancer cells (PC-3). A higher expression of LRF was seen in the prostate cancer cells than in the normal cells. Furthermore, the expression of LRF was highest in the PC3 cells which supports the findings of Cui and associates (2010). There are a number of potential explanations for the higher expression of LRF in cancer cells than in the normal cells. First, LRF acts a specific negative regulator of the nodal-tumor suppressor gene, p14ARF (Maeda et al., 2005a; Maeda et al., 2005b). Real-time PCR analysis of a cohort of diffuse large B-cell lymphoma cases revealed that high LRF expression generally correlates with low expression of the p14ARF gene (Kelly and Daniel, 2006). This implies that increased expression of LRF could inactivate the p53 pathway by increasing Mdm2 levels leading to prostate carcinogenesis similar to the report on the p53 mutant in non-small cell lung cancers (Agrawal et al., 2006; Zhao et al., 2008a; Zhao et al., 2008b). The reported frequency of mutation of the p53 tumor suppressor gene in human prostate carcinoma and benign prostate hyperplasia is only up to 26% (Schlechte et al., 1998). Second, the overexpression of LRF has been reported in p53 wild-type cases of lung and ovarian carcinomas (Jiang et al., 2010; Zhao et al., 2008a; Zhao et al., 2008b). This suggests that there must be some additional mechanisms by which LRF exerts its role in the pathogenesis of benign prostate hyperplasia and prostate cancer independent of the p14ARF-Mdm2-p53 axis.
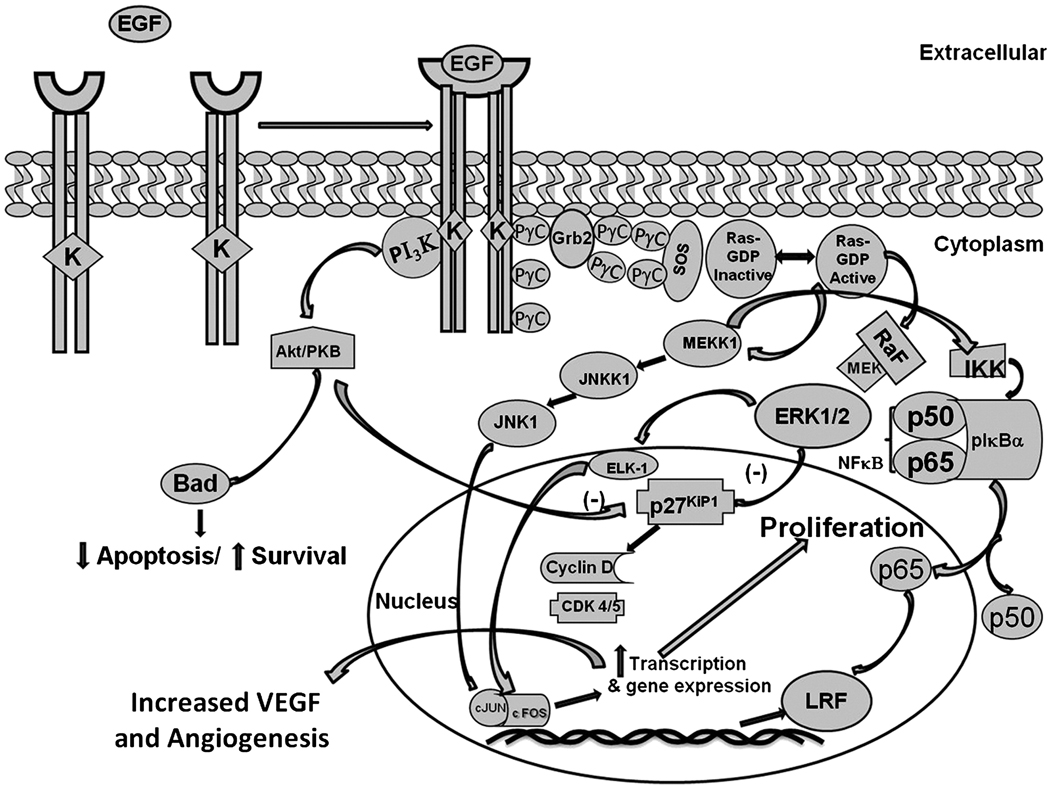
The growth promoting effects of androgens modulate the formation of epidermal growth factor (EGF), keratinocytes growth factor (KGF), and insulin-like growth factors I and II (IGF-1 and IGF-II) in the prostate (Story, 1991). Studies show that there is cross-talk between the androgen receptor and PI3K/Akt pathway and since EGF also regulates the same pathway and also increases expression of LRF (Figure 5), there must be some mechanism leading to prostate oncogenesis (Hu et al., 2009; Wang et al., 2007). Further studies using proliferation, migration, and invasion that regulates the function of LRF in prostate cancer cells are warranted.
Figure 5. Schematic representation of potential mechanism by which EGF could increase the expression of LRF in prostate cells.
EGF binds to its receptor and activates a signaling cascade to transcribe LRF. LRF binds to the p65 subunit of NF-κ which activates other signaling mechanisms leading to the aberrant proliferation of the prostate cells.
REFERENCES
- Aggarwal A, Hunter WJ, III, Aggarwal H, Silva ED, Davey MS, Murphy RF, Agrawal DK. Expression of leukemia/lymphoma-related factor (LRF/POKEMON) in human breast carcinoma and other cancers. Exp Mol Pathol. 2010;89:140–148. doi: 10.1016/j.yexmp.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal H, Aggarwal A, Hunter WJ, III, Yohannes P, Khan AU, Agrawal DK. Expression of leukemia/lymphoma related factor (LRF/Pokemon) in human benign prostate hyperplasia and prostate cancer. Exp Mol Pathol. 2011;90:226–230. doi: 10.1016/j.yexmp.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Yang J, Murphy RF, Agrawal DK. Regulation of the p14ARF-Mdm2-p53 pathway: an overview in breast cancer. Exp Mol Pathol. 2006;81:115–122. doi: 10.1016/j.yexmp.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Apostolopoulou K, Pateras IS, Evangelou K, Tsantoulis PK, Liontos M, Kittas C, Tiniakos DG, Kotsinas A, Cordon-Cardo C, Gorgoulis VG. Gene amplification is a relatively frequent event leading to ZBTB7A (Pokemon) overexpression in non-small cell lung cancer. J Pathol. 2007;213:294–302. doi: 10.1002/path.2222. [DOI] [PubMed] [Google Scholar]
- Bello D, Webber MM, Kleinman HK, Wartinger DD, Rhim JS. Androgen responsive adult human prostatic epithelial cell lines immortalized by human papillomavirus 18. Carcinogenesis. 1997;18:1215–1223. doi: 10.1093/carcin/18.6.1215. [DOI] [PubMed] [Google Scholar]
- Cui J, Yang Y, Zhang C, Hu P, Kan W, Bai X, Liu X, Song H. FBI-1 functions as a novel AR co-repressor in prostate cancer cells. Cell Mol Life Sci. 2010;68:1091–1103. doi: 10.1007/s00018-010-0511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Miguel P, Royuela, Bethencourt R, Ruiz A, Fraile B, Paniagua R. Immunohistochemical comparative analysis of transforming growth factor alpha, epidermal growth factor, and epidermal growth factor receptor in normal, hyperplastic and neoplastic human prostates. Cytokine. 1999;11:722–727. doi: 10.1006/cyto.1998.0443. [DOI] [PubMed] [Google Scholar]
- Hu Y, Sun H, Owens RT, Wu J, Chen YQ, Berquin IM, Perry D, O'Flaherty JT, Edwards IJ. Decorin suppresses prostate tumor growth through inhibition of epidermal growth factor and androgen receptor pathways. Neoplasia. 2009;11:1042–1053. doi: 10.1593/neo.09760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Siu MK, Wong OG, Tam KF, Lam EW, Ngan HY, Le XF, Wong ES, Chan HY, Cheung AN. Overexpression of proto-oncogene FBI-1 activates membrane type 1-matrix metalloproteinase in association with adverse outcome in ovarian cancers. Mol Cancer. 2010;9:318. doi: 10.1186/1476-4598-9-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly KF, Daniel JM. POZ for effect--POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 2006;16:578–587. doi: 10.1016/j.tcb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Kim HG, Kassis J, Souto JC, Turner T, Wells A. EGF receptor signaling in prostate morphogenesis and tumorigenesis. Histol Histopathol. 1999;14:1175–1182. doi: 10.14670/HH-14.1175. [DOI] [PubMed] [Google Scholar]
- Kumar VL, Majumder PK, Gujral S, Kumar V. Comparative analysis of epidermal growth factor receptor mRNA levels in normal, benign hyperplastic and carcinomatous prostate. Cancer Lett. 1998;134:177–180. doi: 10.1016/s0304-3835(98)00256-0. [DOI] [PubMed] [Google Scholar]
- Lee DK, Kang JE, Park HJ, Kim MH, Yim TH, Kim JM, Heo MK, Kim KY, Kwon HJ, Hur MW. FBI-1 enhances transcription of the nuclear factor-kappaB (NF-kappaB)-responsive E-selectin gene by nuclear localization of the p65 subunit of NF-kappaB. J Biol Chem. 2005;280:27783–27791. doi: 10.1074/jbc.M504909200. [DOI] [PubMed] [Google Scholar]
- MacDonald A, Habib FK. Divergent responses to epidermal growth factor in hormone sensitive and insensitive human prostate cancer cell lines. Br J Cancer. 1992;65:177–182. doi: 10.1038/bjc.1992.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Hobbs RM, Merghoub T, Guernah I, Zelent A, Cordon-Cardo C, Teruya-Feldstein J, Pandolfi PP. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005a;433:278–285. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
- Maeda T, Hobbs RM, Pandolfi PP. The transcription factor Pokemon: a new key player in cancer pathogenesis. Cancer Res. 2005b;65:8575–8578. doi: 10.1158/0008-5472.CAN-05-1055. [DOI] [PubMed] [Google Scholar]
- Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, van den Brink MR, Zelent A, Shigematsu H, Akashi K, et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimeault M, Batra SK. Recent advances on multiple tumorigenic cascades involved in prostatic cancer progression and targeting therapies. Carcinogenesis. 2006;27:1–22. doi: 10.1093/carcin/bgi229. [DOI] [PubMed] [Google Scholar]
- Qu H, Qu D, Chen F, Zhang Z, Liu B, Liu H. ZBTB7 overexpression contributes to malignancy in breast cancer. Cancer Invest. 2010;28:672–678. doi: 10.3109/07357901003631007. [DOI] [PubMed] [Google Scholar]
- Rovin RA, Winn R. Pokemon expression in malignant glioma: an application of bioinformatics methods. Neurosurg Focus. 2005;19:E8. doi: 10.3171/foc.2005.19.4.9. [DOI] [PubMed] [Google Scholar]
- Schlechte H, Lenk SV, Loning T, Schnorr D, Rudolph BD, Ditscherlein G, Loening SA. p53 tumour suppressor gene mutations in benign prostatic hyperplasia and prostate cancer. Eur Urol. 1998;34:433–440. doi: 10.1159/000019778. [DOI] [PubMed] [Google Scholar]
- Sherwood ER, Van Dongen JL, Wood CG, Liao S, Kozlowski JM, Lee C. Epidermal growth factor receptor activation in androgen-independent but not androgen-stimulated growth of human prostatic carcinoma cells. Br J Cancer. 1998;77:855–861. doi: 10.1038/bjc.1998.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Story MT. Polypeptide modulators of prostatic growth and development. Cancer Surv. 1991;11:123–146. [PubMed] [Google Scholar]
- Wang Y, Kreisberg JI, Ghosh PM. Cross-talk between the androgen receptor and the phosphatidylinositol 3-kinase/Akt pathway in prostate cancer. Curr Cancer Drug Targets. 2007;7:591–604. doi: 10.2174/156800907781662248. [DOI] [PubMed] [Google Scholar]
- Webber MM, Quader ST, Kleinman HK, Bello-DeOcampo D, Storto PD, Bice G, DeMendonca-Calaca W, Williams DE. Human cell lines as an in vitro/in vivo model for prostate carcinogenesis and progression. Prostate. 2001;47:1–13. doi: 10.1002/pros.1041. [DOI] [PubMed] [Google Scholar]
- Zhao ZH, Wang SF, Yu L, Wang J, Chang H, Yan WL, Fu K, Zhang J. Expression of transcription factor Pokemon in non-small cell lung cancer and its clinical significance. Chin Med J (Engl) 2008a;121:445–449. [PubMed] [Google Scholar]
- Zhao ZH, Wang SF, Yu L, Wang J, Chang H, Yan WL, Zhang J, Fu K. Overexpression of Pokemon in non-small cell lung cancer and foreshowing tumor biological behavior as well as clinical results. Lung Cancer. 2008b;62:113–119. doi: 10.1016/j.lungcan.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Zu X, Yu L, Sun Q, Liu F, Wang J, Xie Z, Wang Y, Xu W, Jiang Y. SP1 enhances Zbtb7A gene expression via direct binding to GC box in HePG2 cells. BMC Res Notes. 2009;2:175. doi: 10.1186/1756-0500-2-175. [DOI] [PMC free article] [PubMed] [Google Scholar]