Abstract
Recent advances in cellular, molecular, and developmental biology have revolutionized our concepts regarding the process of organogenesis that have important implications for our understanding of both lung formation and pulmonary disease pathogenesis. Pulmonary investigators have long debated whether developmental processes are recapitulated during normal repair of the lung or in the setting of chronic pulmonary diseases. Although the cellular events involved in lung morphogenesis and those causing pulmonary disease are likely to include processes that are distinct, there is increasing evidence that the pathogenesis of many lung disorders involves the same genetic machinery that regulates cell growth, specification, and differentiation during normal lung development.
Keywords: lung, morphogenesis, transcription, respiratory
The mammalian lung represents the evolution of a singular structural solution to the problem of providing constant gas exchange after birth. Perhaps it is not surprising that the lung is a remarkably complex organ requiring multiple cell types for its function (1–5). The precise numbers, positions, and functions of respiratory epithelial cells vary along the dorsal-ventral and rostral-caudal axes of the lung and during development. There has been considerable progress in the elucidation of transcriptional and signaling processes involved in lung morphogenesis, progress being synergized by findings from studies in model organisms. Findings from worms, flies, fish, amphibians, and mammals demonstrate that the signaling systems and transcription factors regulating cell proliferation, migration, specification, and differentiation are highly conserved across diverse phyla, many of which are directly involved in pulmonary organogenesis and disease.
Genetic Networks Regulating Formation of the Respiratory Epithelium
Respiratory epithelial cells are derived from subsets of precursors originating in the anterior foregut endoderm of the early embryo. Formation of definitive endoderm represents a critical step in embryogenesis during which the anterior/posterior (A/P) axis of the embryo is formed, setting the stage for the specification of the many endodermally derived organs that originate along the gut tube. During gastrulation, Sox17 and Foxa2 are critical transcription factors that direct specification of the definitive endoderm (Figure 1) (6). Thereafter, organ-specific cell fates are directed by spatially restricted signaling via multiple signaling pathways, (e.g., sonic hedgehog, wingless [Wnt] family members, growth factors [fibroblast growth factor (FGF), bone morphogenetic protein, transforming growth factors, epidermal growth factors], Notch, and others) that regulate activation of transcription factors that, in turn, direct tissue specific gene expression. Formation of the A/P axis of the endoderm is strongly influenced by Wnt and bone morphogenetic protein signaling. Specification of the posterior endoderm, from which the midgut and hindgut is formed, requires high levels of Wnt signaling, whereas the inhibition of Wnt signaling induces anterior foregut endoderm. Once the A/P axis is established, lung buds arise from the ventral region of the anterior foregut in a process dependent on the precise temporal–spatial regulation of FGF signaling from the cardiac mesenchyme (7, 8) that selectively induces commitment to respiratory lineages that are distinct from those of other organs that form along the foregut (e.g., thyroid, esophagus, parathyroid, thymic epithelium, pancreas, liver, and others) (Figure 1). Cells destined to form the conducting airways and peripheral alveolar regions of the lung are already committed to respiratory cell fates well before the formation of the tracheal/lung buds (9). Respiratory cell lineages are first recognized by the expression of the transcription factor Nkx homeobox (NKX) 2.1 (also known as thyroid transcription factor-1 or TTF-1) (10). TTF-1 regulates gene expression in the nucleus and, via its interactions with other factors, sets the stage for generating the diverse epithelial cell types characteristic of the normal airways. Subsequent differentiation of the respiratory epithelium is dependent on cooperation among many transcription factors, including β-catenin and NKX, forkhead orthologue box (FOX), Sry-related HMG box (SOX), and ETS (E26 Transformation Specific) family members that determine conducting as compared with alveolar regions of the lung (11, 12). These transcription factors are reused to regulate gene expression at various times of development and during repair, where they regulate the expression of epithelial cell-specific genes on which organ function depends. The activation of the transcription factors is determined by binding to regulatory regions of gene targets, which depends on the DNA sequences bound and chromatin structure that makes DNA accessible or inaccessible to transcriptional complexes. DNA and histone methylation, acetylation, and deacetylation strongly regulate transcriptional activity and provide transcriptional marks or “memory” in the reading of the genetic code. Small regulatory RNAs, including microRNAs, provide another layer of control over gene expression regulating mRNA stability controlling groups of genes. The recognition of the importance of this “epigenetic” control of gene expression in development and disease is expanding at a rapid pace.
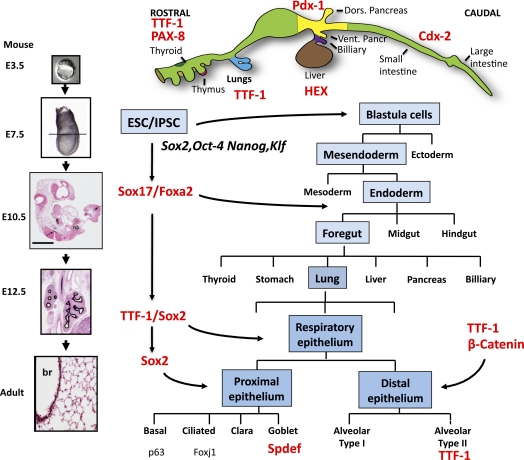
Figure 1.
A schematic of the regions of the mouse embryo from which distinct organs form along the rostral-caudal axis of the gut tube is shown. Lung progenitor cells are derived in the anterior foregut region of the embryo. Mouse lung buds form on embryonic day 10 from the ventral region (blue) marked by expression of thyroid transcription factor 1 (TTF-1), which serves as a critical regulator of lung morphogenesis and differentiation. Developmental stages of the mouse lung morphogenesis are shown at the left; by embryonic day E12.5, the branching structure of the lung is well established as indicated by staining for forkhead ortholog box (Fox)a2, a transcription factor that persists in airways and alveolar type II cells in the adult. Expression of Sry-related HMG box (Sox)2, octamer binding transcription factor 4, homeobox protein Nanog, and Krüpple-like factor can reprogram somatic cells (e.g., fibroblasts) to induce changes in chromatin and gene expression that activate stem cell activity. Embryonic stem cells (ESC) and induced pluripotent stem cells (iPSC) can be programmed or reprogrammed, respectively, and induced to differentiate in vitro or in vivo to multiple, distinct organ lineages, including the lung. Expression of Sox2, Oct4, Nanog, and Klf can reprogram somatic cells (e.g., fibroblasts) to induce changes in chromatin and gene expression that activate stem cell activity. Developmental stages required for the normal lung formation are shown in blue. The sequential expression of transcription factors mediates the process of lung differentiation, shown in red, that lead to the expression of distinct transcription factors (e.g., p63, Foxj1, Spdef) marking the differentiation of airway epithelial cell types.
Critical Role of TTF-1 in Lung Formation and Respiratory Epithelial Cell Differentiation
TTF-1 is a member of the family of NKX homeodomain, containing proteins that serve as transcription factors and that play important roles in organogenesis and cell differentiation in multiple organs and tissues. TTF-1 (or NKX2.1) is selectively expressed in thyroid, the forebrain, and in the lung, where it is the first gene indicating differentiation of the anterior foregut into respiratory epithelial cell progenitors. TTF-1 is expressed throughout the human lung morphogenesis. In conducting airways, TTF-1 is most highly expressed in basal and secretory cells, except goblet cells, and in type II epithelial cells in the alveoli. TTF-1 is required for the formation of the lung parenchyma and for the subsequent differentiation of respiratory epithelial cells, where it plays a critical role in the regulation of gene expression during development and perinatal adaptation to air breathing. In type II alveolar and nonciliated bronchiolar cells, TTF-1 regulates genes mediating pulmonary surfactant and/or innate defense of proteins and fluid homeostasis (13, 14). Germline deletion of TTF-1 blocks the formation of the lung parenchyma, causing tracheal-esophageal fistula and the formation of cyst-like lung tubes lined by undifferentiated epithelial cells (15). TTF-1 interacts with multiple transcription factors and coactivators that activate genes associated with respiratory epithelial differentiation (9). As such, perhaps TTF-1 acts as a “pioneer” lung selective transcription factor and sets the stage for the specification and differentiation of respiratory epithelial cell lineages, the diversity of which is modified by the timing, levels, and duration of transcriptional activity. Gene expression is further modified by interactions of TTF-1 with other transcription factors to influence epithelial cell differentiation. Clinically, mutations in the human TTF-1 gene cause pulmonary, thyroid, and central nervous system disorders (16–18). Pulmonary phenotypes generally are associated with single copy mutations of TTF-1 that cause lung disease of variable severity, ranging from alveolar dysgenesis at birth to postnatal interstitial lung disease.
Role of SOX2 and Notch Signaling in Conducting Airway Epithelial Cell Differentiation
Diverse epithelial cell types (e.g., basal, ciliated, mucus, serous, brush, neuroendocrine, alveolar type I and alveolar type II cells, and others) arise from the lung epithelial committed endodermal progenitors (11, 12, 19). The numbers, patterns, distribution, and behaviors of the multiple cell types lining the respiratory tract are highly stereotypic, varying in precise ways along the airways. The trachea separates from the esophagus early in the embryonic period, esophageal cells adapting a squamous cell phenotype compared with the predominantly columnar epithelial lining airways. The proximal to peripheral (rostral-caudal axis) of the lung is well marked by the differential expression of the transcription factor SOX2 that is selectively expressed in all conducting airways but not in alveolar epithelial cells (Figure 2) (20). SOX2 is required for the normal patterning of conducting airways during early lung morphogenesis, and is used later in development as epithelial progenitors differentiate into diverse cell types (20, 21). The decisions to generate a diversity of epithelial cells from airway progenitors are made in part by cell–cell interactions mediated by Notch signaling (22–24) and the expression of other transcription factors. The sequential activities and interactions among these transcription factors alter chromatin and gene expression to influence subsequent gene expression and epithelial cell differentiation. In the airways, nonciliated progenitors require SOX2, which regulates proliferation and the differentiation of ciliated, Clara (secretory cells), basal, and goblet cells (20, 21). The transcription factors p63, SOX2, and TTF-1 are selectively expressed in basal cells that serve as important progenitors in large conducting airways (25), where they regulate formation of the normal pseudostratified respiratory epithelium characteristic of cartilaginous airways. Deletion of SOX2 in basal or Clara cells during perinatal and postnatal development produces airways devoid of normal differentiated epithelial cell types. Increased expression of SOX2 induced proliferation and respecification of peripheral/alveolar respiratory epithelial cells into those with proximal airway characteristics, indicating the remarkable “plasticity” of respiratory epithelial cell types (26, 27). In the airways, excess Notch signaling induces goblet cell metaplasia, whereas the loss of Notch activity induces ciliated cell differentiation from airway progenitors, supporting the concept that the timing, extent, and specificity of Notch activity plays a critical role in establishing the pattern of the specific cell types lining conducting airways (22–24). Differentiation of the ciliated cells is marked by the expression of the transcription factor FOXJ1, a forkhead transcription factor family member, which is required for the assembly of the ciliary apparatus in airway epithelial cells (28, 29). Disruption of FOXJ1 causes situs inversus and ciliary abnormalities in the mouse, features similar to that of Kartagener syndrome and other ciliopathies that are often associated with chronic pulmonary infection and bronchiectasis (30, 31). Differentiation of airway goblet cells from basal and nonciliated columnar epithelial cells requires the expression of Sam pointed domain Ets-like factor (SPDEF), which is both necessary and sufficient for their differentiation (32, 33). SPDEF regulates the expression of a network of genes associated with mucin biosynthesis and packaging. In the mouse, deletion of SPDEF blocks normal goblet cell differentiation that occurs in the airways in the postnatal period and in submucosal glands (32, 33).
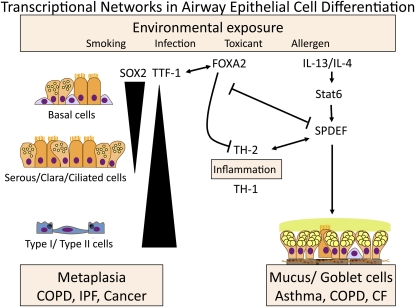
Figure 2.
Transcription factors, regulating lung formation, play important roles in the pathogenesis of common lung diseases. Sry-related HMG box (SOX)2, thyroid transcription factor (TTF)-1, forkhead ortholog box (FOX)A2, and Sam pointed domain Ets-like factor (SPDEF) interact in a transcriptional network that regulates respiratory epithelial cell differentiation during development and in disease. SOX2 is selectively expressed in conducting airway epithelial cells, where it regulates basal, ciliated, and secretory cell differentiation. TTF-1 and FOXA2 are expressed in conducting and alveolar epithelial cells, where they are required for normal cell differentiation. FOXA2 is needed to suppress spontaneous Th2-mediated pulmonary inflammation after birth. Loss of FOXA2 induces SPDEF, a transcription factor regulating goblet cell differentiation and mucus production. SPDEF is expressed during aeroallergen and Th2-mediated lung inflammation and is induced in goblet cells associated with chronic obstructive pulmonary disease (COPD) and cystic fibrosis (CF). SPDEF is expressed in a mutually exclusive manner with TTF-1 and FOXA2. SOX2 is selectively expressed in squamous cell carcinomas, whereas TTF-1 marks the majority of non–small cell pulmonary adenocarcinomas. This transcriptional network both responds to and influences Th1- and Th2-mediated inflammation in the lung. Thus, the respiratory epithelium plays a critical role in instruction of innate immunity involved in the pathogenesis of common lung disease associated with metaplasia of the respiratory epithelium.
Interactions, both direct and indirect, among the various transcription factors: SOX2, NKX2.1, p63, FOXJ1, SPDEF, and FOX family members (including FOXJ1, FOXA1, FOXA2, and FOXA3), provide the basic molecular “highway” critical for formation and differentiation of conducting airway epithelial cells in health and disease (Figure 2). Location and abundance of distinct epithelial cell types is further influenced by diverse signaling pathways, including growth factors and cytokines, that further modify cell fate decisions and the functions of airway epithelial cells.
Developmental Programs Regulating Lung Morphogenesis Instruct the Differentiation of Embryonic Stem Cells and the Reprogramming of Induced Pleuripotent Cells IN VITRO
Identification of the molecular pathways regulating normal lung morphogenesis are providing the scientific insights needed to program embryonic stem (ES) and induced pluripotent stem (iPS) cells to lung lineages. ES cells, and more recently, the ability to reprogram fibroblasts and other cell types into iPS cells with potential for differentiation into cells with characteristics of multiple and diverse organs (34), offer tremendous opportunities to advance our understanding of the molecular and cellular processes involved in lung organogenesis (Figure 1). Fibroblasts and other somatic cells can be reprogrammed by expression of the “Yamanaka” factors, Sox2, Klf4, Nonog, and Oct4. Study of ES and iPS cells also provides the scientific framework for cell-based therapies, organ regeneration, and the study of disease-causing genes in tissue directly relevant to the pathogenesis of inherited and acquired lung diseases. Lessons from the study of organogenesis in model organisms are providing the critical insights needed to induce differentiation of ES and iPS cells in vitro into definitive endoderm. Specification and differentiation of stem cells can be controlled in vitro by provision of developmental signals (for example, the activation and inhibition of critical signaling pathways known to regulate normal morphogenesis) at precise times and concentrations. Mapping the signaling and transcriptional events involved in normal embryonic development has enabled the differentiation of ES and iPS cells into multiple tissue types in vitro. Breakthroughs in ES and iPS cell biology have provided strong support for the concept that cell identity, even of highly differentiated cells, is not rigidly constrained and can be readily redirected to produce a diversity of cell types.
Endodermal differentiation of ES cells can be induced in vitro by the precise regulation of Wnt, FGF, and activin signaling in vitro, which serves to recapitulate the normal developmental signals required for formation of the endoderm. Formation of “definitive endoderm” is marked by the expression of the transcription factors SOX17 and FOXA2. Recent studies demonstrated that this definitive endoderm can be programmed anterior (foregut) or posterior (hindgut) endoderm from which epithelial cells of diverse organ-specific phenotypes can be generated from ES or iPS cells in vitro (35). Highly differentiated intestinal organoids lined by diverse intestinal epithelial cell types have been produced from human ES cells in vitro (36). Although previous in vitro studies supported the ability of ES cells to differentiate into cells with lung cell characteristics, the efficient induction of foregut endoderm, including differentiation of respiratory epithelial cells expressing TTF-1, FOXJ1, mucins, and the surfactant proteins, was recently described (37). Thus, lessons from the study of developmental biology have provided insights and molecular framework that was needed to direct the differentiation of ES/iPS cells into organ-specific cell types that can now be used for the study of gene function and regulation of respiratory epithelial cell differentiation in health and disease. iPS cells derived from patients with pulmonary diseases, for example those bearing mutations or causing lung disorders, will facilitate the study of disease-associated proteins in the context of the cell types causing or contributing to disease pathogenesis. Programming of iPS and ES cells to respiratory epithelial phenotypes will provide in vitro models for testing, perhaps with high-throughput screening, that will uncover novel processes and compounds useful for future lung cell–specific therapies.
Are Transcriptional Networks Reused During Lung Repair and Disease?
After birth, the respiratory tract is constantly exposed to particles, pathogens, and toxicants that cause cell injury, activate inflammatory processes that influence cell proliferation and redifferentiation required for lung repair and homeostasis (Figure 2). Chronic injury and inflammation cause tissue remodeling that underlies the pathogenesis of chronic lung disease in genetically susceptible individuals. The cellularity of the respiratory epithelium is profoundly altered in common chronic lung diseases in which airway cell numbers, morphology, and functions are perturbed. Because innate host defense of the lung depends on mucociliary clearance, the precise regulation of airway fluid homeostasis and the production of innate and acquired defense molecules, abnormalities, and their epithelial cell functions are likely to play critical roles in the pathogenesis of many common lung diseases. Abnormalities in airway epithelial differentiation are pathognomonic features of chronic lung disease (e.g., mucus and squamous metaplasia and hyperplasia) (Figure 2). It is presently unclear whether changes in respiratory epithelial cell differentiation associated with recurrent infection, inflammation, and tissue remodeling represent causal or adaptive responses in chronic lung diseases.
Relevance of the Regulation of Respiratory Epithelial Transcription to Disease Pathogenesis
In spite of years of investigation and progress in the identification of genes associated with acute and chronic lung diseases (e.g., surfactant disorders, idiopathic pulmonary fibrosis, asthma, cystic fibrosis [CF], chronic obstructive pulmonary disease [COPD], and lung cancer), the genetic cellular processes underlying their pathogenesis remain enigmatic; likewise, effective treatments for these common pulmonary diseases remain elusive. Changes in cell differentiation (e.g., squamous and mucus metaplasia), type II cell hyperplasia, and the heterogeneous cell types seen in lung cancer indicate widespread disruption of the process regulating normal epithelial differentiation associated with lung disease.
Distinct Roles for SOX2 and TTF-1 in Lung Disease
The important roles of each of these transcription factors in the development of conducting and peripheral airways in development are replayed in chronic lung diseases. For example, Sox2 is highly expressed in the cells undergoing squamous metaplasia and “bronchiolarized” lesions in idiopathic pulmonary fibrosis and squamous cell carcinoma, but not non–small cell pulmonary adenocarcinomas. The Sox2 gene is commonly amplified in squamous cell lung cancers (38). The finding that Sox2 is activated after airway injury induces differentiation and redifferentiation of respiratory epithelial cells indicates its likely important role in the regulation of airway epithelial cell repair and carcinogenesis. In sharp contrast to Sox2, TTF-1 is commonly expressed in nonsquamous pulmonary adenocarcinomas (∼ 70%). TTF-1 is amplified in approximately 14% of non–small cell adenocarcinomas; whether it plays a role as a tumor initiator or suppressor or both, depending on context, remains unclear at present (39–44). Both Sox2 and TTF-1 can induce proliferation in lung cells. Likewise, TTF-1, but not Sox2, is lost in association with the atypical mucus metaplasia seen in some patients with idiopathic pulmonary fibrosis, wherein Muc5B, but not Muc5AC, is prominently expressed (45) (Figure 2).
SOX2, TTF-1, FOXA2, and SPDEF Interact in a Transcriptional Network Regulating Mucus Metaplasia and Inflammation
The transcription factors FOXA2, TTF-1, and SPDEF, which direct respiratory epithelial cell fate during lung development, are also expressed in a regional and cell-selective manner in the mature lung where they function together in a complex transcriptional network that regulates airway epithelial cell differentiation and function (Figure 2). SPDEF is selectively expressed in both submucosal glands and in airway goblet cells, where it regulates the expression of a group of genes involved in the synthesis, glycosylation, and packaging of mucus (Figure 2). SPDEF mRNA and protein are markedly increased in goblet cells associated with asthma, COPD, and CF (33, 45). SPDEF is regulated by Th2-mediated inflammatory signaling during allergic airway sensitization in the mouse and by IL-13, where it regulates gene expression in a STAT6-dependent manner (Figure 2) (33). SPDEF regulates Muc5AC, Muc16, and a number of enzymes regulating packaging, synthesis of mucin, and its glycosylation (34). SPDEF induces goblet cell metaplasia inhibiting both TTF-1 and FOXA2 in airway epithelial cells. Consistent with the counter-regulatory roles of SPDEF and TTF-1/FOXA2, expression of Foxa2 alone inhibits aeroallergen-induced mucus metaplasia in mouse models (33, 46). Previous findings, that the loss of Foxa2 in airway epithelial cells in the developing mouse lung is sufficient to induce severe Th2-mediated inflammation and goblet cell metaplasia, indicate that the transcription network active in the regulation of airway epithelial cell differentiation and mucus metaplasia also plays an unexpected and critical role in the regulation of innate immunity and Th2-mediated inflammation (47). This intersection between regulation of epithelial-specific gene expression by the SPDEF/TTF-1/FOXA2 network and innate immunity is likely to play an important role in establishing inflammatory responses that will influence the pathogenesis of lung diseases, including asthma, CF, and COPD (Figure 2).
Conclusions
Transcription factors regulating lung formation during development provide the basic molecular framework that determines cell differentiation and gene expression critical for pulmonary homeostasis after birth. Susceptibility to disease is, in part, directly inherited via our genome, which is further influenced by chromatin structure that makes accessible the transcription factors that control cell differentiation and gene expression. Elucidation of the molecular processes that mediate the formation of the lung during development will surely provide scientific insights needed to understand the pathogenesis of common chronic lung diseases and provide the basis for discovery of new treatments of presently intractable disorders in the future.
Footnotes
Supported by the National Institutes of Health grants HL095580 (J.A.W.) and HL100315 (J.A.W.), the University of Cincinnati Postdoctoral Fellow Research Program (Y.M.), and the Medical Research Council, London, United Kingdom (H.M.H.). H.M.H. is an MRC Clinician Scientist.
Originally Published in Press as DOI: 10.1164/rccm.201103-0495PP on June 3, 2011
Author Disclosure: J.A.W. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. H.M.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. Y.M. received an institutional postdoctoral grant.
References
- 1.Warburton D, Tefft D, Mailleux A, Bellusci S, Thiery JP, Zhao J, Buckley S, Shi W, Driscoll B. Do lung remodeling, repair, and regeneration recapitulate respiratory ontogeny? Am J Respir Crit Care Med 2001;164:S59–S62 [DOI] [PubMed] [Google Scholar]
- 2.Shi W, Bellusci S, Warburton D. Lung development and adult lung diseases. Chest 2007;132:651–666 [DOI] [PubMed] [Google Scholar]
- 3.Evans MJ, Van Winkle LS, Fanucchi MV, Plopper CG. The attenuated fibroblast sheath of the respiratory tract epithelial-mesenchymal trophic unit. Am J Respir Cell Mol Biol 1999;21:655–657 [DOI] [PubMed] [Google Scholar]
- 4.Holgate ST, Davies DE, Lackie PM, Wilson SJ, Puddicombe SM, Lordan JL. Epithelial-mesenchymal interactions in the pathogenesis of asthma. J Allergy Clin Immunol 2000;105:193–204 [DOI] [PubMed] [Google Scholar]
- 5.Sorokin SP. The Cells of the Lungs. : Nettesheim P, Hanna MG, Jr, Deatherage JW., Jr Morphology of Experimental Respiratory Carcinogenesis. Pages 3 through 43 with 30 electron micrographs. Proceedings of a Biology Division, Oak Ridge National Laboratory conference; May 13–16, 1970, Gatlinburg, TN: pp. 3–43 [Google Scholar]
- 6.Zorn AM, Wells JM. Vertebrate endoderm development and organ formation. Annu Rev Cell Dev Biol 2009;25:221–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serls AE, Doherty S, Parvatiyar P, Wells JM, Deutsch GH. Different thresholds of fibroblast growth factors pattern the ventral foregut into liver and lung. Development 2005;132:35–47 [DOI] [PubMed] [Google Scholar]
- 8.Cardoso WV, Lü J. Regulation of early lung morphogenesis: questions, facts and controversies. Development 2006;133:1611–1624 [DOI] [PubMed] [Google Scholar]
- 9.Perl AK, Wert SE, Nagy A, Lobe CG, Whitsett JA. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proc Natl Acad Sci USA 2002;99:10482–10487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maeda Y, Davé V, Whitsett JA. Transcriptional control of lung morphogenesis. Physiol Rev 2007;87:219–244 [DOI] [PubMed] [Google Scholar]
- 11.Morrisey EE, Hogan BL. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev Cell 2010;18:8–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warburton D, El-Hashash A, Carraro G, Tiozzo C, Sala F, Rogers O, De Langhe S, Kemp PJ, Riccardi D, Torday J, et al. Lung organogenesis. Curr Top Dev Biol 2010;90:73–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bohinski RJ, Di Lauro R, Whitsett JA. The lung-specific surfactant protein B gene promoter is a target for thyroid transcription factor 1 and hepatocyte nuclear factor 3, indicating common factors for organ-specific gene expression along the foregut axis. Mol Cell Biol 1994;14:5671–5681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeFelice M, Silberschmidt D, DiLauro R, Xu Y, Wert SE, Weaver TE, Bachurski CJ, Clark JC, Whitsett JA. TTF-1 phosphorylation is required for peripheral lung morphogenesis, perinatal survival, and tissue-specific gene expression. J Biol Chem 2003;278:35574–35583 [DOI] [PubMed] [Google Scholar]
- 15.Minoo P, Su G, Drum H, Bringas P, Kimura S. Defects in tracheoesophageal and lung morphogenesis in Nkx2.1(−/−) mouse embryos. Dev Biol 1999;209:60–71 [DOI] [PubMed] [Google Scholar]
- 16.Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox CH, Ward JM, Gonzalez FJ. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev 1996;10:60–69 [DOI] [PubMed] [Google Scholar]
- 17.Galambos C, Levy H, Cannon CL, Vargas SO, Reid LM, Cleveland R, Lindeman R, deMello DE, Wert SE, Whitsett JA, et al. Pulmonary pathology in thyroid transcription factor-1 deficiency syndrome. Am J Respir Crit Care Med 2010;182:549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carré A, Szinnai G, Castanet M, Sura-Trueba S, Tron E, Broutin-L'Hermite I, Barat P, Goizet C, Lacombe D, Moutard ML, et al. Five new TTF1/NKX2.1 mutations in brain-lung-thyroid syndrome: rescue by PAX8 synergism in one case. Hum Mol Genet 2009;18:2266–2276 [DOI] [PubMed] [Google Scholar]
- 19.Shi W, Chen F, Cardoso WV. Mechanisms of lung development: contribution to adult lung disease and relevance to chronic obstructive pulmonary disease. Proc Am Thorac Soc 2009;6:558–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Que J, Luo X, Schwartz RJ, Hogan BL. Multiple roles for Sox2 in the developing and adult mouse trachea. Development 2009;136:1899–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tompkins DH, Besnard V, Lange AW, Wert SE, Keiser AR, Smith AN, Lang R, Whitsett JA. Sox2 is required for maintenance and differentiation of bronchiolar Clara, ciliated, and goblet cells. PLoS ONE 2009;4:e8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guseh JS, Bores SA, Stanger BZ, Zhou Q, Anderson WJ, Melton DA, Rajagopal J. Notch signaling promotes airway mucous metaplasia and inhibits alveolar development. Development 2009;136:1751–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsao PN, Vasconcelos M, Izvolsky KI, Qian J, Lu J, Cardoso WV. Notch signaling controls the balance of ciliated and secretory cell fates in developing airways. Development 2009;136:2297–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morimoto M, Liu Z, Cheng HT, Winters N, Bader D, Kopan R. Canonical Notch signaling in the developing lung is required for determination of arterial smooth muscle cells and selection of Clara versus ciliated cell fate. J Cell Sci 2010;123:213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rock JR, Randell SH, Hogan BL. Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis Model Mech 2010;3:545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Y, Futtner C, Rock JR, Xu X, Whitworth W, Hogan BL, Onaitis MW. Evidence that SOX2 overexpression is oncogenic in the lung. PLoS ONE 2010;5:e11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tompkins DH, Besnard V, Lange AW, Keiser AR, Wert SE, Bruno MD, Whitsett JA. Sox2 activates cell proliferation and differentiation in the respiratory epithelium. Am J Respir Cell Mol Biol (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Knowles HJ, Hebert JL, Hackett BP. Mutation of the mouse hepatocyte nuclear factor/forkhead homologue 4 gene results in an absence of cilia and random left-right asymmetry. J Clin Invest 1998;102:1077–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brody SL, Yan XH, Wuerffel MK, Song SK, Shapiro SD. Ciliogenesis and left-right axis defects in forkhead factor HFH-4-null mice. Am J Respir Cell Mol Biol 2000;23:45–51 [DOI] [PubMed] [Google Scholar]
- 30.Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell 2009;139:663–678 [DOI] [PubMed] [Google Scholar]
- 31.Roy S. The motile cilium in development and disease: emerging new insights. Bioessays 2009;31:694–699 [DOI] [PubMed] [Google Scholar]
- 32.Park KS, Korfhagen TR, Bruno MD, Kitzmiller JA, Wan H, Wert SE, Khurana Hershey GK, Chen G, Whitsett JA. SPDEF regulates goblet cell hyperplasia in the airway epithelium. J Clin Invest 2007;117:978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen G, Korfhagen TR, Xu Y, Kitzmiller J, Wert SE, Maeda Y, Gregorieff A, Clevers H, Whitsett JA. SPDEF is required for mouse pulmonary goblet cell differentiation and regulates a network of genes associated with mucus production. J Clin Invest 2009;119:2914–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663–676 [DOI] [PubMed] [Google Scholar]
- 35.D'Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol 2005;23:1534–1541 [DOI] [PubMed] [Google Scholar]
- 36.Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011;470:105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Green MD, Chen A, Nostro MC, d'Souza SL, Schaniel C, Lemischka IR, Gouon-Evans V, Keller G, Snoeck HW. Generation of anterior foregut endoderm from human embryonic and induced pluripotent stem cells. Nat Biotechnol 2011;29:267–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 2009;41:1238–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka H, Yanagisawa K, Shinjo K, Taguchi A, Maeno K, Tomida S, Shimada Y, Osada H, Kosaka T, Matsubara H, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res 2007;67:6007–6011 [DOI] [PubMed] [Google Scholar]
- 40.Kendall J, Liu Q, Bakleh A, Krasnitz A, Nguyen KC, Lakshmi B, Gerald WL, Powers S, Mu D. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci USA 2007;104:16663–16668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature 2007;450:893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwei KA, Kim YH, Girard L, Kao J, Pacyna-Gengelbach M, Salari K, Lee J, Choi YL, Sato M, Wang P, et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene 2008;27:3635–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris T, Pan Q, Sironi J, Lutz D, Tian J, Sapkar J, Perez-Soler R, Keller S, Locker J. Both gene amplification and allelic loss occur at 14q13.3 in lung cancer. Clin Cancer Res 2011;17:690–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang X, Kadara H, Behrens C, Liu D, Xiao Y, Rice DC, Gazdar AF, Fujimoto J, Moran C, Varella-Garcia M, et al. Abnormalities of the TITF-1 lineage-specific oncogene in NSCLC: Implications in lung cancer pathogenesis and prognosis. Clin Cancer Res 2011;17:2434–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plantier L, Crestani B, Wert SE, Dehoux M, Zweytick B, Guenther A, Whitsett JA. Ectopic respiratory epithelial cell differentiation in bronchiolized distal airspaces in idiopathic pulmonary fibrosis. Thorax (In press) [DOI] [PubMed] [Google Scholar]
- 46.Chen G, Wan H, Luo F, Zhang L, Xu Y, Lewkowich I, Wills-Karp M, Whitsett JA. Foxa2 programs Th2-cell mediated innate immunity in the developing lung. J Immunol 2010;184:6133–6141 [DOI] [PubMed] [Google Scholar]
- 47.Park SW, Verhaeghe C, Nguyenvu LT, Barbeau R, Eisley CJ, Nakagami Y, Huang X, Woodruff PG, Fahy JV, Erle DJ. Distinct roles of FOXA2 and FOXA3 in allergic airway disease and asthma. Am J Respir Crit Care Med 2009;180:603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]