Abstract
MicroRNAs (miRNA) are small regulatory RNAs that control gene expression by translational suppression and destabilization of target mRNAs. There is increasing evidence that miRNAs regulate genes associated with fibrosis in organs, such as the heart, kidney, liver, and the lung. In a large-scale screening for miRNAs potentially involved in bleomycin-induced fibrosis, we found expression of miR-29 family members significantly reduced in fibrotic lungs. Analysis of normal lungs showed the presence of miR-29 in subsets of interstitial cells of the alveolar wall, pleura, and at the entrance of the alveolar duct, known sites of pulmonary fibrosis. miR-29 levels inversely correlated with the expression levels of profibrotic target genes and the severity of the fibrosis. To study the impact of miR-29 down-regulation in the lung interstitium, we characterized gene expression profiles of human fetal lung fibroblast IMR-90 cells in which endogenous miR-29 was knocked down. This confirmed the derepression of reported miR-29 targets, including several collagens, but also revealed up-regulation of a large number of previously unrecognized extracellular matrix–associated and remodeling genes. Moreover, we found that miR-29 is suppressed by transforming growth factor (TGF)–β1 in these cells, and that many fibrosis-associated genes up-regulated by TGF-β1 are derepressed by miR-29 knockdown. Interestingly, a comparison of TGF-β1 and miR-29 targets revealed that miR-29 controls an additional subset of fibrosis-related genes, including laminins and integrins, independent of TGF-β1. Together, these strongly suggest a role of miR-29 in the pathogenesis of pulmonary fibrosis. miR-29 may be a potential new therapeutic target for this disease.
Keywords: miR-29, pulmonary fibrosis, basement membrane, profibrotic genes, TGF-β1
Many interstitial lung diseases are associated with pulmonary fibrosis, which results in the thickening of alveolar walls and scarring of the lung. The initiation step has been thought to involve chronic inflammatory responses to a variety of stimuli, including persistent infections, autoimmune reactions, allergic responses, chemical insults, radiation, and tissue injury (1). The disease can be characterized by alveolar epithelial cell injury, loss of basement membrane (BM) integrity, the failure of re-epithelialization leading to epithelial–mesenchymal transition (EMT), inflammatory cell infiltration, fibroblast hyperplasia, deposition of extracellular matrix (ECM), and scar formation (2–4). Fibroblasts and myofibroblasts play essential roles in the pathological process by secreting growth factors, as well as excessive collagens, and other matrix proteins (3).
miRNAs are small regulatory RNAs that control the expression of a large number of genes by translational suppression and destabilization of target mRNAs (5). miRNAs have been implicated in multiple biological processes, including development and disease (6–8). There is increasing evidence that miRNAs are key regulators of genes involved in fibrosis in organs, such as the heart (9, 10), kidney (11), and liver (12). Two recent reports have shown the involvement of let-7 and miR-21 in pulmonary fibrosis by regulating EMT and transforming growth factor (TGF)–β signaling activity, respectively (13).
TGF-β1 plays an important role in the development of pulmonary fibrosis. Increased TGF-β expression and activity have been detected in lung tissue from bleomycin-treated mice (14) and patients with idiopathic pulmonary fibrosis (IPF) (15). Overexpression of active TGF-β1 in the mouse lung in vivo results in severe interstitial and pleural fibrosis (16). Analysis of Smad3-deficient mice indicates that bleomycin-induced pulmonary fibrosis is partially dependent on the integrity of the TGF-β pathway (17). Interestingly, a number of miRNAs the expression of which is influenced by TGF-β have been shown to modulate the TGF-β effects in conditions, such as fibrosis (10, 11, 18, 19).
Here, we performed a large-scale screen for miRNAs potentially involved in pulmonary fibrosis. Using a bleomycin-induced model of pulmonary fibrosis in mice, we identified miR-29 family members are among those most significantly reduced miRNAs in fibrotic lungs. This reduction correlated with increased expression of collagens and ECM-related genes. Moreover, we found that miR-29 is suppressed by TGF-β in IMR-90 cells, and that many fibrosis-associated genes up-regulated by TGF-β are derepressed by miR-29 knockdown. Interestingly, a comparison of TGF-β and miR-29 targets revealed that miR-29 controls an additional subset of profibrotic genes, including laminins and integrins independent of TGF-β. Together, our study supports a role for miR-29 in the pathogenesis of pulmonary fibrosis, and suggests that it may be a candidate therapeutic target in this disease.
MATERIALS AND METHODS
miRNA and mRNA Array Analysis
Affymetrix GeneChip miRNA Arrays and Mouse Gene 1.0 ST Whole Genome Array (Affymetrix, Santa Clara, CA) was used for miRNA and mRNA expression profiling, respectively, according to the manufacturer's protocol. Experiments were performed in triplicate for each condition. A two-sample t test was performed to identify differential gene expression between comparative groups (P < 0.05 considered significant). DAVID (http://david.abcc.ncifcrf.gov/) and Targetscan (http://www.targetscan.org/) databases were used for data analysis. The mRNA array data has been deposited into the GEO database (GSE18651) at the National Center for Biotechnology Information (Bethesda, MD).
Quantitative Real-Time PCR for mRNA and miRNA
Total RNA (1 μg) was subjected to reverse transcription using the SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA). TaqMan Universal PCR Master Mix and Taqman Gene Expression Assays (Applied Biosystems, Carlsbad, CA) were used for quantitative real-time PCR (qRT-PCR). For qRT-PCR of miRNA, 10 ng total RNA was subjected to reverse transcription using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). TaqMan MicroRNA Assays (Applied Biosystems) were used for qRT-PCR. Each experiment was performed in triplicate, and ribosomal protein L23a was used for normalization.
Northern Blot Analysis
The 5′ digoxigenin-labeled locked nucleotide acid (LNA) probes (Exiqon, Vedbaek, Denmark) were used for Northern blot analysis according to manufacturer's protocol. Detailed methods are included in the online supplement.
miRNA In Situ Hybridization on Frozen Sections
We performed in situ hybridization (ISH) on frozen sections using 5′ digoxigenin-labeled LNA probes (Exiqon) according to Obernosterer's protocol (20). Detailed methods are included in the online supplement.
3′ Untranslated Region Luciferase Assay
DNA fragments of the 3′ untranslated regions (UTRs) contain with wild-type or mutated complementary binding sites of miR-29 were cloned downstream of the Renilla luciferase in the psiCHECK2 dual luciferase reporter (Promega, Madison, WI). Reporter plasmids were cotransfected into FG293 cells with either miR-29 Mimic (Dharmacon, Lafayette, CO) or mimic control. Dual-Glo Luciferase Assay System and GloMax Multi-Detection System (Promega) were used for detection. All experiments were performed in triplicate, and firefly luciferase activity expressed from the same psiCHECK-2 vector was used as an internal control. The sequences of PCR primers used are listed in Table E1 in the online supplement.
Native PAGE and Western Analysis
Anti-collagen a1(I) antibody (1:500; Rockland, Gilbertsville, PA) and reagents from Invitrogen were used for native PAGE Western analysis of COL1A1 following the manufacturer's instruction. Detailed methods are included in the online supplement.
Sircol Assay of Soluble Collagen
IMR-90 cells were transfected with miR-29b antisense LNA or scrambled LNA, as mentioned previously here, and maintained in serum-reduced media (0.4%) for 48 hours. Soluble collagen levels in the culture media were measured and compared with a standard curve prepared from rat-tail collagen using the Sircol collagen dye binding assay.
Bleomycin Mouse Fibrosis Model
Male C57Bl/6 mice (8–12 wk old) were intraperitoneally injected with either bleomycin (0.8 U/mouse in 300 μl of sterile PBS; Sigma-Aldrich, St. Louis, MO) or sterile PBS (control) on Days (D) 0, 3, 7, 10, and 14. Mice were killed on D3, 7, 10, 28, 70, and 140. Lungs were collected for RNA isolation, paraffin and frozen embedded, and sectioned.
RESULTS
Expression of miR-29 in Normal and Bleomycin-Treated Lungs
To identify miRNAs potentially involved in pulmonary fibrosis, we first characterized the miRNA profile of bleomycin-treated lungs using Affymetrix miRNA array. The effect of bleomycin in induction of pulmonary fibrosis has been widely reported both in human and animal models (21, 22). Male C57Bl/6 mice (8–12 wk old) were injected intraperitoneally with either bleomycin (0.8 U/mouse in 300 μl of sterile PBS; Sigma-Aldrich) or sterile PBS (control) on D0, 3, 5, 7, 10, and 14. Mice (three for each group) were killed on D3, 7, 10, 28, 70, and 140, and lungs were collected for total RNA isolation and histological analysis.
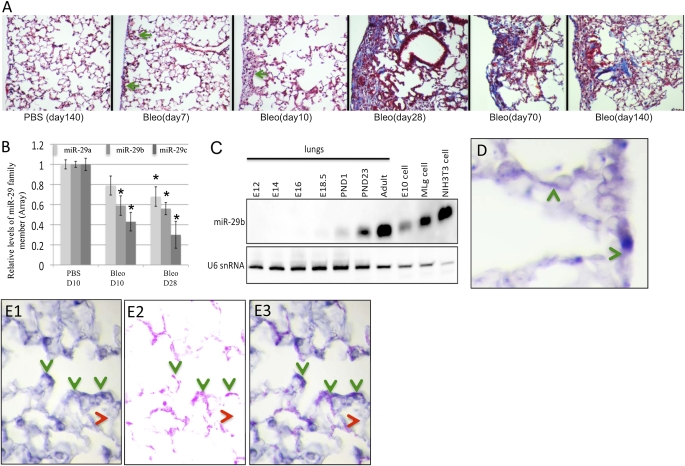
Typical subpleural fibrosis was observed in bleomycin-treated lungs, which showed infiltration of macrophages in alveolar spaces and increased collagen deposition. Trichrome staining revealed localized, small, subpleural fibrotic lesions first observed in lung sections at D7 (Figure 1A). Progressive fibrosis was observed from D10 to D28, with increasing collagen deposition and number of inflammatory cells, mainly monocytes or alveolar macrophages in the distal lung. A large number of foamy alveolar macrophages were found only from D28 onward (Figure 1A). Less obvious lesions were observed in D70 and D140 lungs, suggesting a partial resolution, as previously reported (23) (Figure 1A).
Figure 1.
Expression of miR-29 in normal and bleomycin-treated mouse lungs. (A) Trichrome staining showed subpleural lesions 7 days after bleomycin injection; progressive fibrosis is shown from Day (D) 7 to D28 with increased collagen (Col) staining; a large portion of these lesions were resolved at D70 and D140. (B) miR-29 family members are down-regulated in bleomycin-treated lungs (*P < 0.05). (C). Levels of miR-29 detected by Northern blot increases during lung development, with highest levels in adult lung. Expression of miR-29 is higher in fibroblast cells (Mlg and NIH3T3) than in epithelial cells (E10, type I–like cells). (D) miR-29 is expressed in the alveolar wall and in pleura. (E1–E3) miR-29 is expressed in mesenchymal cells at the entrance of the alveolar duct and costains with α-SMA (B1, miR-29 in situ hybridization [ISH]; B2, α-SMA immunohistochemistry; B3, merged image). Green arrow, subpleural fibrotic lesion; green arrowhead, miR-29–expressing cells; red arrowhead, cells with low or undetectable miR-29 expression.
To identify miRNAs involved in ECM deposition/remodeling in the progressive stages of pulmonary fibrosis, we performed miRNA array analysis of total RNA samples from D10 and D28 bleomycin-treated lungs, and compared them with respective controls. Of 609 miRNAs examined, 49 miRNAs were either up- or down-regulated (P < 0.05; fold changes > 2.0) in bleomycin-treated lungs at at least one time point (Table E2). These include let-7 family members (let-7f and let-7g) that are known to be reduced in human IPF tissues and to play a significant role in EMT in pulmonary fibrosis (13). We also confirmed the reported up-regulation of miR-21 in bleomycin-induced pulmonary fibrosis (24). Expression of several miRNAs, such as miR-184 and miR-192, are specifically altered in D10 and D28 samples, respectively. This suggests stage-specific roles in this process. In addition, we found expression of miR-29 family members significantly reduced in bleomycin-treated lungs (Figure 1B). miR-29 has been implicated in cardiac and skin fibrosis (10, 25), and preferentially targets profibrotic genes. However, little is known about the expression and function of miR-29 in the lung. Thus, first, we examined miR-29 expression in the normal developing lung. We found increasing levels of miR-29 as the lung matures, reaching the highest levels in the adult (Figure 1C). This pattern inversely correlates with the reported decrease in lung collagen expression from developmental stages to adulthood (26). We also detected high levels of miR-29 in two mouse fibroblast cell lines (Mlg and NIH3T3), but low levels in a lung type I–like epithelial cell line (E10) (Figure 1C), suggesting a preferential expression in cells with mesenchymal origin.
Subsequently, we mapped the sites of miR-29 expression in the murine adult lung using double ISH–immunohistochemistry. Our analysis revealed miR-29 expression in distinct domains of the adult lung. First, we found miR-29 expressed in a subset of cells of the alveolar wall and pleura, often sites of pulmonary fibrosis (Figure 1D). Second, miR-29 was found in mesenchymal cells at the entrance of alveolar ducts, colocalizing with α-SMA, strongly suggesting that these are myofibroblasts (Figures 1E1, 2, and 3). Third, miR-29 was expressed in the smooth muscle layer of conducting airways, with near background levels of expression in the epithelium of conducting airways (Figure 3C and data not shown).
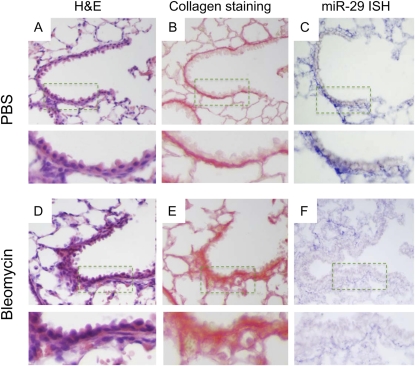
Figure 3.
Reduced expression of miR-29 in subepithelial mesenchymal cells of terminal bronchioles was observed in bleomycin-treated lungs (F) as compared with lungs from mice receiving PBS injection (C). This inversely correlates with collagen staining (B and E) and thickening of subepithelial structures (A and D).
Inverse Correlation of miR-29 and Its Downstream Gene Expression during the Development of Bleomycin-Induced Pulmonary Fibrosis
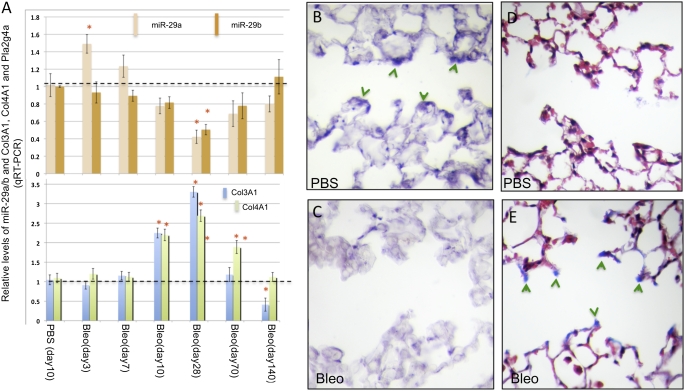
Next we asked whether in the murine lung, miR-29 was expressed in a pattern consistent with a potential role in regulating profibrotic genes during the development of fibrosis. qRT-PCR analysis of miR-29a/b in lungs collected at different time points after bleomycin treatment revealed a gradual reduction in expression, reaching the lowest levels at D28, as shown in our array analysis. miR-29 levels increase subsequently, correlating with remission of the fibrotic lesions in D70 and D140 lungs (Figure 2A and Figure 1A). The down-regulation of miR-29 inversely correlated with increased expression of profibrotic target genes, including ECM collagen (Col3A1), and BM collagen (Col4A1) genes (Figure 2A). ISH confirmed the significant reduction of miR-29 signals in bleomycin-treated lungs at D28. miR-29 down-regulation was observed in mesenchymal cells at the entrance of the alveolar duct and in the subepithelial mesenchyme of terminal bronchioles. Interestingly, these were invariably sites of marked increase in collagen deposition, as shown by Trichrome and Sirius Red staining (Figures 2B–2E and Figures 3A–3F). Thus, our results strongly suggest that derepression of profibrotic gene targets of miR-29 is an important feature of bleomycin-induced fibrosis.
Figure 2.
Levels of miR-29 inversely correlated with its downstream targets in bleomycin-induced pulmonary fibrosis. (A) Levels of miR-29 are gradually reduced after bleomycin injection, reaching lowest levels at D28, and then gradually recover at D70 and D140. Levels of Col3A1 and Col4A1, targets of miR-29, are inversely correlated with miR-29 levels in this process. (B and C) Down-regulation of miR-29 in cells at the entrance of alveolar ducts is found in bleomycin-treated lungs (C) as compared with controls (B), inversely correlating with collagen staining in these cells (D and E). (B and C) ISH of miR-29c; (D and E) Masson's trichrome staining; *P < 0.05.
miR-29 Regulates a Large Number of Genes Associated with Fibrosis
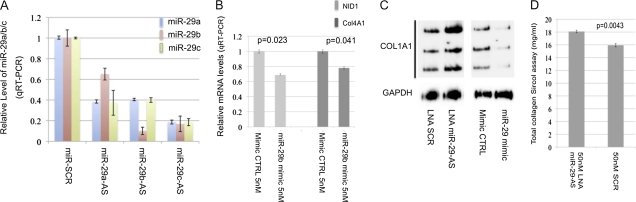
The inverse correlation of miR-29 levels with the severity of bleomycin-induced fibrosis suggests that miR-29 is part of a mechanism that tightly controls expression and activity of profibrotic mediators in the lung mesenchyme. To gain insight into these mechanisms, we investigated the effect of knockdown endogenous miR-29 in IMR-90 cells, a human fetal lung fibroblast cell line widely used to study molecular mechanisms of pulmonary fibrosis. IMR-90 cells were transiently transfected with LNA antisense (Exiqon) oligos designed against miR-29a, miR-29b, or miR-29c (50 nM), as previously described (27). Real-time PCR showed that each of these LNA antisense oligos could knock down the expression of all three family members due to sequence similarity (Figure 4A). Therefore, this approach resulted in an overall reduction in the levels of miR-29.
Figure 4.
miR-29–mediated regulation of target gene expression. (A) Knockdown of miR-29 expression by locked nucleotide acid (LNA) knockdown oligos. Expression of miR-29a, miR-29b, and miR-29c were significantly reduced in IMR-90 cells (human fetal lung fibroblasts) transfected with LNA knockdown oligos either for miR-29a or miR-29b or miR-29c, but not with scrambled LNA oligos. (B) mRNA levels of COL4A1 and Nidgen (NID) 1 are significantly decreased in IMR-90 cells transiently transfected with miR-29 mimic as compared with control oligos. (C) miR-29 knockdown resulted in a significantly increased level of COL1A1 protein, whereas increased miR-29 reduced COL1A1 protein. (D) The level of total soluble collagen in the culture medium is increased in miR-29 knockdown IMR-90 cells.
To investigate the impact of miR-29 knockdown in gene expression, we performed genome-wide array analysis (Affymetrix) of IMR-90 cells 48 hours after transfection of miR-29 antisense or control oligos. We found expression of 830 genes was significantly altered (P < 0.05). Among them, all reported miR-29 targets were significantly up-regulated in miR-29 knockdown cells, including Col1A1, Col1A2, Col3A1, Col4A1, Col4A2, and Col15A1 (Table 1). In addition, a large number of predicted targets were also up-regulated in miR-29 knockdown cells.
TABLE 1.
CATEGORIES OF GENES ENRICHED IN UP-REGULATED GENE LIST IN miR-29 KNOCKDOWN CELLS
| Anti-miR-29/SCR (50 nM) |
TGF-β/CTRL |
||||
|---|---|---|---|---|---|
| Gene Name | Gene Symbol | Fold Changes | P Value | Fold Change | P Value |
| Collagens in ECM formation | |||||
| Collagen, type XV, alpha 1* | COL15A1* | 1.24 | 4.76E-02 | 0.96 | 9.87E-01 |
| Collagen, type I, alpha 1* | COL1A1* | 1.29 | 6.74E-03 | 1.35 | 2.53E-03 |
| Collagen, type I, alpha 2* | COL1A2* | 1.15 | 3.36E-02 | 1.27 | 5.15E-02 |
| Collagen, type III, alpha 1* | COL3A1* | 1.56 | 1.03E-02 | 1.23 | 4.94E-02 |
| Collagen, type V, alpha 1* | COL5A1* | 1.28 | 1.05E-02 | 2.48 | 4.76E-03 |
| Collagen, type V, alpha 2* | COL5A2* | 1.51 | 1.44E-02 | 2.06 | 2.21E-03 |
| Basement membrane–related genes | |||||
| Laminin, alpha 1 | LAMA1 | 1.77 | 8.25E-03 | No data | No data |
| Laminin, alpha 3 | LAMA3 | 1.26 | 4.64E-02 | 1.39 | 1.56E-01 |
| Laminin, beta 1 | LAMB1 | 1.37 | 2.65E-03 | 0.42 | 8.76E-03 |
| Laminin, gamma 1* | LAMC1* | 1.27 | 3.98E-02 | 1.02 | 9.61E-01 |
| Secreted protein, acidic, cysteine-rich* | SPARC* | 1.32 | 8.56E-03 | 1.77 | 4.16E-03 |
| Nidogen 1* | NID1* | 2.05 | 1.20E-02 | 0.86 | 6.83E-01 |
| Collagen, type IV, alpha 1* | COL4A1* | 1.53 | 1.33E-02 | 4.46 | 2.31E-04 |
| Collagen, type IV, alpha 5* | COL4A5* | 1.48 | 1.90E-02 | 0.16 | 7.73E-04 |
| Integrins | |||||
| Integrin, alpha 11* | ITGA11* | 1.38 | 1.03E-02 | No data | No data |
| Integrin, alpha 2 | ITGA2 | 1.47 | 3.62E-02 | 2.10 | 1.76E-01 |
| Integrin, alpha 5 | ITGA5 | 1.35 | 9.61E-03 | 2.47 | 1.20E-03 |
| Integrin, alpha 6* | ITGA6* | 1.24 | 2.56E-02 | 0.83 | 7.02E-01 |
| Integrin, alpha 8 | ITGA8 | 1.80 | 1.59E-02 | 0.58 | 1.94E-01 |
| Integrin, alpha V | ITGAV | 1.13 | 4.04E-02 | 2.11 | 1.18E-03 |
| Integrin, beta 5 | ITGB5 | 1.16 | 1.46E-02 | 0.71 | 5.03E-02 |
| Metallopeptidase, collagenase and other proteinases in ECM remodeling | |||||
| ADAM metallopeptidase domain 12* | ADAM12* | 1.66 | 2.25E-02 | 19.71 | 6.77E-05 |
| ADAM metallopeptidase domain 19* | ADAM19* | 1.39 | 1.28E-02 | 4.86 | 1.23E-03 |
| ADAM metallopeptidase with thrombospondin type 1 motif, 12 | ADAMTS12 | 1.85 | 1.43E-03 | 1.09 | 2.72E-01 |
| ADAM metallopeptidase with thrombospondin type 1 motif, 2 | ADAMTS2 | 1.63 | 1.61E-03 | 1.29 | 1.24E-01 |
| ADAM metallopeptidase with thrombospondin type 1 motif, 9* | ADAMTS9* | 1.43 | 1.03E-02 | 0.91 | 2.81E-01 |
| Bone morphogenetic protein 1 | BMP1 | 1.25 | 4.64E-02 | 2.14 | 5.56E-03 |
| Carboxypeptidase D | CPD | 1.42 | 1.61E-02 | 0.87 | 5.62E-01 |
| Membrane metallo-endopeptidase | MME | 2.08 | 4.20E-03 | 0.29 | 1.68E-02 |
| Matrix metallopeptidase 14 | MMP14 | 1.54 | 4.17E-03 | 0.77 | 4.07E-01 |
| Matrix metallopeptidase 2* | MMP2* | 1.33 | 9.78E-04 | 0.88 | 1.42E-01 |
| Pregnancy-associated plasma protein A, pappalysin 1 | PAPPA | 1.24 | 1.01E-02 | 0.83 | 4.79E-01 |
| IL-1 pathway | |||||
| Interleukin -1 receptor, type I (IL1R1), mRNA | IL1R1 | 1.63 | 3.96E-02 | 0.27 | 3.81E-05 |
| Interleukin 1 receptor, type II (IL1R2) | IL1R2 | 1.22 | 4.72E-02 | 1.00 | 3.74E-01 |
| Interleukin 1 receptor accessory protein (IL1RAP)* | IL1RAP | 1.25 | 8.79E-02 | 2.42 | 8.85E-03 |
| Interleukin 1 receptor accessory protein-like 1 (IL1RAPL1) | IL1RAPL1 | 1.38 | 1.70E-02 | 0.44 | 1.13E-01 |
| Other genes in ECM formation | |||||
| Fibrillin 1* | FBN1* | 1.24 | 2.58E-02 | 1.95 | 3.06E-02 |
| Lysyl oxidase-like 2* | LOXL2* | 1.23 | 5.02E-03 | 1.84 | 1.41E-03 |
| Lumican | LUM | 1.61 | 1.23E-02 | 0.61 | 5.97E-02 |
| Follistatin-like 1* | FSTL1* | 1.22 | 2.93E-02 | 1.80 | 2.92E-02 |
| Platelet-derived growth factor receptor C | PDGFC* | 1.22 | 2.24E-04 | 1.65 | 2.87E-02 |
| TNF receptor-associated factor 4* | TRAF4* | 1.21 | 4.26E-04 | 1.03 | 7.76E-01 |
| Glycoprotein (transmembrane) nmb | GPNMB | 3.38 | 1.50E-02 | 0.29 | 1.64E-01 |
| Amphiregulin | AREG | 2.11 | 1.00E-02 | 0.91 | 4.84E-01 |
| Serpin peptidase inhibitor, clade H, member 1* | SERPINH1* | 1.44 | 5.49E-03 | 2.43 | 5.76E-03 |
Definition of abbreviations: CTRL, control; ECM, extracellular matrix; miR, miRNA; SCR, scrambled control; TGF, transforming growth factor.
Bold text indicates that the levels of mRNAs are up-regulated by TGF-β1 in IMR-90 cells; italicized text indicates that the levels of mRNAs are up-regulated in bleomycin-treated lungs.
Predicted targets of miR-29
To validate our results further, we compared the effect of miR-29 knockdown with that of overexpression on COL4A1 (a known target) and Nidogen 1 (NID1; a predicted target). miR-29 levels were increased in IMR-90 cells by transient transfection of an miR-29b mimic (Dharmacon), as previously described (28). As expected, mRNA levels of COL4A1 and NID1 were significantly reduced by the mimic, the opposite of the effect of knockdown of miR-29 (Figure 4B). Thus, our approach allowed the reliable identification of additional miR-29 targets and other downstream genes that are related to fibrosis in IMR-90 cells.
We used the DAVID (EASE) program to find the biological processes, pathways, and cellular function of the genes significantly altered in miR-29 knockdown cells. We found that genes coding for components of BM (Laminins, NID1, SPARC, Col4A1, and Col4A5), integrins (IGTA2, ITGA8, ITGA11, ITGA6, and ITGB5), and genes involved in proteolysis and ECM remodeling (ADAM12, ADAM19, ADAMTS9, ADAMTS12, MMP2, MMP14, and BMP1) were up-regulated by knockdown of miR-29 (Table 1). Among the 45 genes that fit into these categories, 23 (51%) were predicted targets of miR-29, as determined by Targetscan (Table 1). Altered expression of these genes has been associated fibrosis in different organs (4, 29–31). These data indicate that besides a large number of collagens, miR-29 also controls expression of a broad range of genes involved in fibrosis.
New Direct Targets of miR-29 in Pulmonary Fibrosis
We then wanted to determine whether the genes derepressed in miR-29 knockdown cells (Table 1) were increased in bleomycin-treated lungs. For this, we compared our gene list with that of Affymetrix Array data sets (GSE2640; GEO) of C57/Bl6 mouse lungs treated with bleomycin (32). We identified 19 (42%; Table 1, italicized) of these genes were also up-regulated in bleomycin-treated lungs, and most were predicted targets of miR-29 (Table 1).
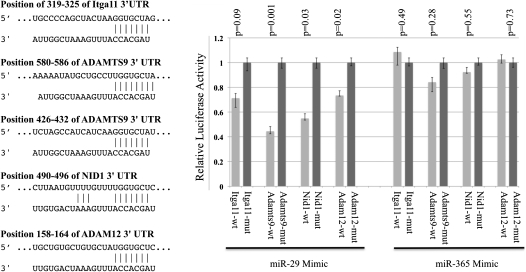
To test whether these genes were direct targets of miR-29, fragments of the 3′ UTRs of several predicted targets (Table 1) containing wild-type or mutated miR-29 complementary sites were cloned into the psiCHECK2 dual luciferase reporter plasmid. Luciferase reporters were cotransfected with miRNA mimic of miR-29 or miR-365, an unrelated miRNA with no complementary binding site in these luciferase reporters. Luciferase analysis revealed that the activity of reporters containing the wild-type 3′ UTRs of ITGA11, ADAM12, ADAMTS9, and NID1 were significantly suppressed by miR-29, but not by miR-365. Suppression by miR-29 was abolished by mutating miR-29 complementary sites in these reporters (Figure 5). This confirmed that ITGA11, ADAM12, ADAMTS9, and NID1 are direct targets of miR-29.
Figure 5.
ITGA11, ADAMTS9, ADAM12, and NID1 are direct targets of miR-29. FG293 cells were cotransfected with individual 3′ untranslated region (UTR) luciferase reporters with wild-type (wt) or mutated (mut) miR-29–binding sites, along with miRNA mimics of miR-29 or miR-365. Firefly luciferase activity from the same construct was used to normalize and to generate relative activity of Renilla luciferase, which was subject to the regulation of cloned 3′ UTR. miR-29 significantly suppressed luciferase activities of reporters harboring wild-type 3′ UTR sequences, but not for reporters with mutated miR-29–binding sites. miR-365, an unrelated miRNA, has no significant effect on the luciferase activities of reporters with either wild-type or mutant 3′ UTRs.
We then asked whether miR-29 knockdown affected the protein levels of its targets, because translational suppression is another mechanism mediated by miRNAs. COL1A1 is a confirmed miR-29 target, and a major component of pulmonary fibrosis. COL1A1 mRNA levels were increased by 30% in miR-29 knockdown cells (Table 1). Native gel Western blot analysis of protein samples from IMR-90 cells revealed that knocking down miR-29 resulted in a threefold increase of COL1A1 protein. In contrast, increased miR-29 by a mimic oligo (Dharmacon) resulted in a fivefold reduction in COL1A1 protein level (Figure 4C). This indicates that miR-29 has a greater impact on COL1A1 protein production than on mRNA level. Analysis of total soluble collagen in the culture medium using Sircol assay revealed an 11% increase in the culture medium of miR-29 knockdown cells as compared with control cells (Figure 4D). We conclude that miR-29 has a profound effect on the protein synthesis of its target genes.
miR-29 Is a Mediator of the TGF-β Effect on Expression of Profibrotic Genes
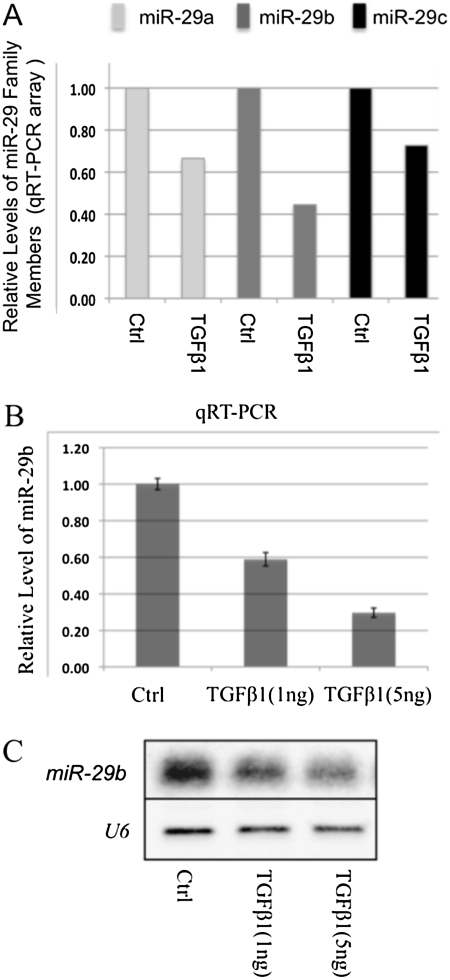
miR-29 expression is suppressed by TGF-β treatment in cardiac fibroblasts (10). To determine whether this was also true in lung fibroblasts, we examined the expression of miR-29 in another miRNA array database in which we characterized downstream miRNA targets of TGF-β1 (1 ng, 48 hrs) in IMR-90 cells. We found that all miR-29 family members were down-regulated by TGF-β1, with more than a twofold reduction of miR-29b (Figure 6A). These results were further confirmed by qRT-PCR and Northern blot analysis (Figures 6B and 6C).
Figure 6.
Expression of miR-29 is down-regulated by transforming growth factor (TGF)–β1 in IMR-90 cells. Down-regulation of miR-29a, -b, and -c were detected by qRT-PCR array (A), and confirmed by quantitative real-time PCR and Northern blot (B and C).
We reasoned that down-regulation of miR-29 may result in derepression of the genes under the control of TGF-β. Expression profiles of the 45 genes up-regulated in miR-29 knockdown cells were examined in the array data of TGF-β1–treated IMR-90 cells at 48 hours (GSE17518). We found that 18 genes (40%) were significantly up-regulated by TGF-β (Table 1, bold text). Interestingly, except for BMP1 and IGTAV, 16 genes (89%) were predicted targets of miR-29 (Table 1). The up-regulation of genes, such as COL1A1, COL3A1, and COL1A2, by TGF-β1 was likely a consequence of reduced miR-29, as miR-29 knockdown alone resulted in a similar degree of up-regulation (Table 1). By contrast, expression of genes, ADAM12 and ADAM19, was far more up-regulated by TGF-β1 treatment as compared with miR-29 knockdown alone, suggesting that down-regulation of miR-29 will facilitate TGF-β–mediated up-regulation. All these factors strongly suggest that down-regulation of miR-29 synergizes with the TGF-β effects. We propose that down-regulation of miR-29 mediates the up-regulation of profibrotic genes by TGF-β in IMR-90 cells.
We also found that more than half of the genes derepressed in miR-29 knockdown cells (Table 1) are not up-regulated by TGF-β. The majority of these genes (68%) are not predicted targets of miR-29. These include four laminins, four integrins, two MMPs, and three ADAMs. Up-regulation of these genes is likely a secondary effect of miR-29 knockdown. Thus, miR-29 controls an additional subset of ECM- and BM-associated genes independent of TGF-β.
DISCUSSION
miR-29 has been implicated in EMT, skeletal muscle cell and osteoblast differentiation, as well as in cardiac fibrosis and systemic sclerosis (10, 25, 27, 33, 34). In nasopharyngeal carcinomas and myocardial infarction–associated cardiac fibrosis, down-regulation of miR-29 inversely correlates with increased expression of several predicted targets, including collagens (10, 28). Here, we report results consistent with a novel and significant role of miR-29 in bleomycin-induced pulmonary fibrosis, and it may represent an attractive therapeutic target for this disease.
We found that TGF-β1 down-regulates miR-29 in IMR-90 cells, consistent with findings in other systems (10, 25). Little is known about how expression of miR-29 is regulated by TGF-β. Although we found increased phosphorylated Smad2 levels, it is unknown whether Smad has a role in TGF-β–mediated down-regulation of miR-29. No Smad binding cis elements could be identified in the conserved regions of genomic DNA surrounding miR-29 alleles.
Other cytokines, such as PDGF-B and IL-4 have been shown to suppress miR-29 expression in human skin fibroblasts (25). In skeletal muscle progenitor cells (C2C12), the NF-κB and YY1 pathways have been implicated in the suppression of the miR-29b/c allele (33). Recent publications suggest a role of TGF-β in the activation of NF-κB in liver and breast cancer, and in osteoclast survival (35–37). PDGF also can activate NF-κB through RAS/PI(3)K and Akt pathways (38). The possible involvement of NF-κB in the down-regulation of miR-29 in TGF-β–treated IMR-90 cells, and the mechanism of miR-29 down-regulation in bleomycin-treated lungs, will be part of our future studies.
By characterizing genome-wide downstream targets of endogenous miR-29, we show that miR-29 not only controls the expression of many collagens, as previously reported in cardiac and skin fibrosis, but also controls genes associated with BM, as well as a significant number of integrins and ADAMs in ECM remodeling. Analysis of TGF-β and miR-29 targets suggests that miR-29 may be an important facilitator of TGF-β–mediated regulation of ECM-related profibrotic genes. However, miR-29 also controls a subset of ECM- and BM-associated genes independent of TGF-β. More importantly, the expression of a large number of these profibrotic targets is also increased in bleomycin-induced fibrotic lungs, strongly suggesting a derepression of miR-29 fibrotic targets in the development of fibrosis. The strength of our study is the characterization of genome-wide targets of miR-29, which strongly suggests an important role in ECM deposition and remodeling. However, it is unknown whether manipulation of miR-29 expression in vivo has a significant impact on the development of pulmonary fibrosis. Therefore, examination of the effect of miR-29 in vivo will be the priority of future studies.
The screening and study of miRNA involvement in pulmonary fibrosis is still in initial stages. Two recently published studies have associated the expression of let-7 and miR-21 with pulmonary fibrosis (13, 24), and both were identified in our miRNA array analysis. let-7 is significantly reduced in human IPF tissues. Inhibition of let-7 in epithelial cell lines in vitro and in vivo resulted in increased expression of mesenchymal-specific genes, such as vimentin and α–smooth muscle actin, strongly suggesting a role in EMT in fibrosis (13). Levels of miR-21 are up-regulated in myofibroblast cells of bleomycin-treated lungs. Knockdown of miR-21 both in vivo and in vitro attenuated the profibrogenic activity of TGF-β1 that plays a central role in fibrotic diseases. This strongly suggests that miR-21 regulates fibrosis by modulating TGF-β signaling activity (24). Here, we showed that miR-29 is downstream of TGF-β, serving as a mediator in up-regulating the expression of profibrotic genes. Together, these studies reveal related yet distinct roles for these individual miRNAs in the development of pulmonary fibrosis.
Supplementary Material
Acknowledgments
The authors are grateful for the comments and suggestions provided by Mary Williams, Jerome Brody, Alan Fine, Xingbin Ai, Arjun Guha, Felicia Chen, and Matthew Layne.
This work was supported by grants from National Heart, Lung, and Blood Institute.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2010-0323OC on October 22, 2010
Author Disclosure: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Maher TM, Wells AU, Laurent GJ. Idiopathic pulmonary fibrosis: multiple causes and multiple mechanisms? Eur Respir J 2007;30:835–839. [DOI] [PubMed] [Google Scholar]
- 2.Moore BB, Hogaboam CM. Murine models of pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol 2008;294:L152–L160. [DOI] [PubMed] [Google Scholar]
- 3.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol 2008;214:199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strieter RM, Mehrad B. New mechanisms of pulmonary fibrosis. Chest 2009;136:1364–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol 2010;220:126–139. [DOI] [PubMed]
- 7.van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest 2007;117:2369–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA–cancer connection: the beginning of a new tale. Cancer Res 2006;66:7390–7394. [DOI] [PubMed] [Google Scholar]
- 9.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, et al. MicroRNA-21 contributes to myocardial disease by stimulating map kinase signalling in fibroblasts. Nature 2008;456:980–984. [DOI] [PubMed] [Google Scholar]
- 10.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA 2008;105:13027–13032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung AC, Huang XR, Meng X, Lan HY. miR-192 mediates TGF-beta/Smad3–driven renal fibrosis. J Am Soc Nephrol 2010;21:1317–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu AS, Friedman JR. A role for microRNA in cystic liver and kidney diseases. J Clin Invest 2008;118:3585–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, Konishi K, Yousem SA, Singh M, Handley D, et al. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2010;182:220–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raghow B, Irish P, Kang AH. Coordinate regulation of transforming growth factor beta gene expression and cell proliferation in hamster lungs undergoing bleomycin-induced pulmonary fibrosis. J Clin Invest 1989;84:1836–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broekelmann TJ, Limper AH, Colby TV, McDonald JA. Transforming growth factor beta 1 is present at sites of extracellular matrix gene expression in human pulmonary fibrosis. Proc Natl Acad Sci USA 1991;88:6642–6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sime PJ, Xing Z, Graham FL, Csaky KG, Gauldie J. Adenovector-mediated gene transfer of active transforming growth factor–beta1 induces prolonged severe fibrosis in rat lung. J Clin Invest 1997;100:768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao J, Shi W, Wang YL, Chen H, Bringas P Jr, Datto MB, Frederick JP, Wang XF, Warburton D. Smad3 deficiency attenuates bleomycin-induced pulmonary fibrosis in mice. Am J Physiol Lung Cell Mol Physiol 2002;282:L585–L593. [DOI] [PubMed] [Google Scholar]
- 18.Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, Cheng JQ. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol 2008;28:6773–6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato M, Putta S, Wang M, Yuan H, Lanting L, Nair I, Gunn A, Nakagawa Y, Shimano H, Todorov I, et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol 2009;11:881–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obernosterer G, Martinez J, Alenius M. Locked nucleic acid–based in situ detection of microRNAs in mouse tissue sections. Nat Protoc 2007;2:1508–1514. [DOI] [PubMed] [Google Scholar]
- 21.Santrach PJ, Askin FB, Wells RJ, Azizkhan RG, Merten DF. Nodular form of bleomycin-related pulmonary injury in patients with osteogenic sarcoma. Cancer 1989;64:806–811. [DOI] [PubMed] [Google Scholar]
- 22.Lazo JS, Hoyt DG, Sebti SM, Pitt BR. Bleomycin: a pharmacologic tool in the study of the pathogenesis of interstitial pulmonary fibrosis. Pharmacol Ther 1990;47:347–358. [DOI] [PubMed] [Google Scholar]
- 23.Borzone G, Moreno R, Urrea R, Meneses M, Oyarzun M, Lisboa C. Bleomycin-induced chronic lung damage does not resemble human idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2001;163:1648–1653. [DOI] [PubMed] [Google Scholar]
- 24.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. Mir-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med 2010;207:1589–1597. [DOI] [PMC free article] [PubMed]
- 25.Maurer B, Stanczyk J, Jungel A, Akhmetshina A, Trenkmann M, Brock M, Kowal-Bielecka O, Gay RE, Michel BA, Distler JH, et al. miR-29 is a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum 2010;62:1733–1743. [DOI] [PubMed]
- 26.Bradley K, McConnell-Breul S, Crystal RG. Collagen in the human lung: quantitation of rates of synthesis and partial characterization of composition. J Clin Invest 1975;55:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gebeshuber CA, Zatloukal K, Martinez J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep 2009;10:400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sengupta S, den Boon JA, Chen IH, Newton MA, Stanhope SA, Cheng YJ, Chen CJ, Hildesheim A, Sugden B, Ahlquist P. MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc Natl Acad Sci USA 2008;105:5874–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pardo A, Selman M. Matrix metalloproteases in aberrant fibrotic tissue remodeling. Proc Am Thorac Soc 2006;3:383–388. [DOI] [PubMed] [Google Scholar]
- 30.Goodwin A, Jenkins G. Role of integrin-mediated TGFbeta activation in the pathogenesis of pulmonary fibrosis. Biochem Soc Trans 2009;37:849–854. [DOI] [PubMed] [Google Scholar]
- 31.Fernandes DJ, Bonacci JV, Stewart AG. Extracellular matrix, integrins, and mesenchymal cell function in the airways. Curr Drug Targets 2006;7:567–577. [DOI] [PubMed] [Google Scholar]
- 32.Haston CK, Tomko TG, Godin N, Kerckhoff L, Hallett MT. Murine candidate bleomycin induced pulmonary fibrosis susceptibility genes identified by gene expression and sequence analysis of linkage regions. J Med Genet 2005;42:464–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, Cheng A, Hall BM, Qualman SJ, Chandler DS, et al. NF-kappaB–YY1–miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 2008;14:369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapinas K, Kessler CB, Delany AM. miR-29 suppression of osteonectin in osteoblasts: regulation during differentiation and by canonical wnt signaling. J Cell Biochem 2009;108:216–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arsura M, Panta GR, Bilyeu JD, Cavin LG, Sovak MA, Oliver AA, Factor V, Heuchel R, Mercurio F, Thorgeirsson SS, et al. Transient activation of NF-kappaB through a TAK1/IKK kinase pathway by TGF-beta1 inhibits AP-1/Smad signaling and apoptosis: implications in liver tumor formation. Oncogene 2003;22:412–425. [DOI] [PubMed] [Google Scholar]
- 36.Neil JR, Schiemann WP. Altered TAB1:I kappaB kinase interaction promotes transforming growth factor beta–mediated nuclear factor–kappaB activation during breast cancer progression. Cancer Res 2008;68:1462–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gingery A, Bradley EW, Pederson L, Ruan M, Horwood NJ, Oursler MJ. TGF-beta coordinately activates TAK1/MEK/AKT/NFKB and Smad pathways to promote osteoclast survival. Exp Cell Res 2008;314:2725–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romashkova JA, Makarov SS. NF-kappaB is a target of Akt in anti-apoptotic PDGF signalling. Nature 1999;401:86–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.