Abstract
The most recently discovered gel-forming mucin, MUC19, is expressed in both salivary glands and tracheal submucosal glands. We previously cloned the 3′−end partial sequence (AY236870), and here report the complete sequencing of the entire MUC19 cDNA. One highly variable region (HVR) was discovered in the 5′ end of MUC19. A total of 20 different splicing variants were detected in HVR, and 18 variants are able to translate into proteins along with the rest of the MUC19 sequence. The longest variant of MUC19 consists of 182 exons, with a transcript of approximately 25 kb. A central exon of approximately 12 kb contains highly repetitive sequences and has no intron interruption. The deduced MUC19 protein has the bona fide gel-forming mucin structure, VWD-VWD-VWD-“threonine/serine-rich repeats”-VWC-CT. An unusual structural feature of MUC19, which is lacking in other gel-forming mucins, is its long amino terminus upstream of the first VWD domain. The long amino terminus is mostly translated from the sequences in HVR, and contains serine-rich repetitive sequences. To validate the integrity of the MUC19 sequence, primers from both the 3′ and 5′ end were used to demonstrate a similar tissue expression pattern of MUC19 in trachea and salivary glands. In addition, antibodies were developed against either the amino (N) or carboxy (C) terminus of MUC19, and similar antibody staining patterns were observed in both salivary and tracheal submucosal glands. In conclusion, we have cloned and elucidated the entire MUC19 gene, which will facilitate understanding of the function and regulation of this important, yet understudied, mucin gene in airway diseases.
Keywords: mucin, MUC19, airway, epithelium, gland
CLINICAL RELEVANCE.
The present study elucidates the entire gene structure and the gene expression pattern of an important human gel-forming mucin gene-MUC19. It will advance our understanding of the pathogenesis of mucus overproduction in chronic airway diseases.
Mucus, a viscoelastic, gel-like substance, covers the epithelial surface of various mammalian tissues, including the respiratory, digestive, and reproductive tracts. Other than acting as a passive barrier, mucus has many important functions in regulating epithelial homeostasis and innate defense (1). The viscous and elastic properties of the mucus gel have been suggested to be largely caused by the physical properties and structural features of mucin glycoproteins (1, 2). To date, 24 genes have taken the name “MUC” or “Muc”: MUC1, -2, -3A, -3B, -4, -5AC, -5B, -6 through -21, and -24 (http://ncbi.nlm.nih.gov). MUC2, -5AC, -5B, -6, and -19 define a gel-forming mucin subfamily (3, 4). They are all large in size (15–40 kb cDNA), and share a similar structure and sequence homology in the conserved regions, which include multiple “cysteine-rich” Von Willebrand (VW) factor D– or VWC-like domains, a long central repeat region containing threonine/serine–rich repeats, and a C-terminal cystine knot (CT) domain (3, 5). These domains appear to play essential roles in forming disulfide-linked dimers (6, 7) and multimers (3, 8, 9). Alteration of their productions and/or physiological properties can directly affect the composition of mucus and airway homeostasis, which has been implicated in various chronic airway diseases, cancer, and so forth (1, 2, 10).
Previously, we developed a novel “hidden Markov model”–based searching algorithm to screen for the additional gel-forming mucin genes, which led to the discovery of both human and mouse MUC19 (4). This finding was further confirmed by conventional cloning and gene sequencing. Because this search is entirely determined by the coverage of existing databases, another main conclusion of this bioinformatic screening is that MUC19 is the last gel-forming mucin family member in both human and mouse. During the same time, a salivary apomucin-like protein was independently reported through the characterization of a recessive mutation (sublingual gland differentiation arrest) that affects mucous cell development in mouse sublingual glands (11). The sequence of this protein perfectly matches mouse Muc19. Soon after, mouse Muc19 was completely sequenced and shown to have a cDNA length of 22,795 bp encoded by a total of 43 exons and spanning 106 kb of genomic DNA (12). It has a gel-forming mucin structure signal peptide, a large central exon with tandem repeats, VWC, VWD, and C-terminal CT domains. Interestingly, the mouse Muc19 locus contains an additional transcript, submandibular gland protein C (Smgc) (12). Smgc is a major secretory product, and a marker of the type I (terminal tubule) cells of the neonatal rat and mouse submandibular gland, but its expression in the adult is present only in some intercalated duct cells (13). It contains 18 exons. The first exon overlaps with Muc19, and the rest of the sequences are located in intron 1 of Muc19 (12, 13).
Similar to MUC5B/Muc5b, MUC19/Muc19 is expressed by mucous cells of tracheal submucosal glands and salivary glands (4). However, differences between these two gel-forming mucin genes were recently reported in mouse salivary glands. Although both Muc5b and Muc19 were expressed by the minor salivary glands, the major glands (i.e., sublingual and submandibular glands) appear to only express Muc19, but not Muc5b (14). Beyond these two organs, Muc19 was detected in bulbourethral glands (Cowper's glands) in the male reproductive system (14), and MUC19 was detected in lacrimal glands of the ocular system (15). Thus, normal MUC19/Muc19 expression appears to be restricted to the glands of various organ systems. However, under certain disease conditions, it is expressed in the epithelium. Two recently reported examples are increased MUC19 expression in middle ear epithelium from patients having either recurrent otitis media or chronic otitis media with effusion (16), and elevated expression of MUC19 in nasal epithelial cells of patients with allergic rhinitis (17).
Relative to other gel-forming mucins, MUC19 is understudied, particularly in the airway. To date, regulation and potential functional implications of MUC19/Muc19 have only been reported in patients with Sjogren syndrome (15), in cytokine-challenged middle ear epithelium (18), in an allergic mouse model (19), and in a mouse model of mucous cell deficiency in salivary glands (11). One main obstacle is the lack of complete human sequence. The short 3′ end (2.1 kb) that we have reported contains mostly repetitive sequences. The unique sequence, which is suitable for primer design, is very short. Although complete mouse Muc19 has been reported, it has very little use in respiratory research, because MUC19/Muc19 is mainly expressed in the glandular mucous cells of the airway, and the mouse has a very limited submucosal gland structure. To advance the study on this relatively new mucin, we determined to complete the sequence of human MUC19.
MATERIALS AND METHODS
Tissues, RNA, Chemicals, Antibodies, and Kits
Human trachea and salivary gland tissue samples were obtained from the National Disease Research Interchange under an approved protocol. Tissue RNA panel and premium-quality tissue RNA (pooled) from salivary gland and trachea were purchased from Clontech (Mountain view, CA) and used for rapid amplification of cDNA end (RACE) and RT-PCR. The RACE kit was purchased from Roche (Roche Diagnostics Corp., Indianapolis, IN). PCR primers were synthesized by Sigma (St. Louis, MO). Chicken anti-human MUC19 antibodies (hMUC19Ab_C1) was generated by using a C-terminus antigen (CREENYELRDIVLD), and hMUC19Ab_N1 was generated by the N-terminus antigen, CGSYNNKAEDDFMSSQNILEKTSQ. These were made, affinity purified, and ELISA tested by Aves Labs Inc. (Tigard, OR). Horseradish peroxidase–conjugated goat anti-chicken IgG was purchased from Aves Labs Inc. Tyramide signaling amplification (TSA) plus fluorescein kit was purchased from PerkinElmer (Waltham, MA).
5′-RACE
The RACE kit was used to obtain the cDNA ends (4, 20). Briefly, Oligo-dT anchor primer or antisense gene-specific primers corresponding to different regions of the MUC19 message were used to initiate first-strand cDNA synthesis. Then, 5′ tailing with oligo d(G) (or dA, dT, dC) with terminal deoxynucleotidyl transferase was performed on the first-strand cDNA. A PCR was performed using the nested gene-specific primer and the 5′ oligo d(T) anchor primer. The PCR products were subcloned into aTA vector (Invitrogen, Carlsbad, CA) for cloning and DNA sequencing. All primer sequences used in this study are listed in Table 1.
TABLE 1.
RAPID AMPLIFICATION OF CDNA END PRIMERS
| Name | Antisense Sequence |
|---|---|
| M19RC1 | TGGAATCACTGTTTGACTGCTG |
| M19RC2 | GGCCAGCCCTAGTTATTCCACT |
| M19RC3 | TTTCAGCTCCTGTTTTTCCAC |
| M19RC4 | TCTCTGAAGGTGCTGTCTCTGAGG |
| M19RC5 | GTGACTGTTCCATCACTGTTGAAT |
| M19RC6 | TCCTCTACGGCTATTCAGAACAT |
| M19RC7 | TCCTTTGCTCCAGGTAGATA |
RT-PCR Amplification
cDNA was synthesized from total RNA (3 μg) by RT with oligo d(T) primer. The resulting single-strand cDNA was used as a template for PCR amplification by MUC19 gene-specific primers (Table 2). PCR products were TA cloned and sequenced.
TABLE 2.
RT-PCR PRIMERS
| Name | Sense or Antisense | Sequence |
|---|---|---|
| M19RT1 | S | GATTCAAAACTGGCACCTCAGA |
| AS | TGGAATCACTGTTTGACTGCTG | |
| M19RT2 | S | TCTGAAAATTCCACCACAGCA |
| AS | GGCCAGCCCTAGTTATTCCACT | |
| M19RT3 | S | AGGGATCACTGGACCATTTG |
| AS | TTTCAGCTCCTGTTTTTCCAC | |
| M19RT4 | S | AAACGACGAGGTCATGCAAC |
| AS | GGGAAAGTCTGAGCCACTGTATC | |
| M19RT5 | S | AAGTGGTCAGACAGGAACGTG |
| AS | GTCCACCAAAAGAGCATGGAC | |
| M19RT6 | S | GAAATATCTACCTGGAGCAAAGGA |
| AS | GTGACTGTTCCATCACTGTTGAAT | |
| M19RT7 | S | TATCTACCTGGAGCAAAGGAGC |
| AS | CCTTCCACCTTACATCTTCCAG | |
| M19RT8 | S | ACCTGGAGCAAAGGAGCATA |
| AS | TCCTCTACGGCTATTCAGAACAT | |
| M19RT9 | S | GTCCATGCTCTTTTGGTGGAC |
| AS | CCAAAACTGGTGTTTCCAATGT | |
| M19RT10 | S | GATACAGTGGCTCAGACTTTCCC |
| AS | TCCAGTTGTCCTTAACCCTGAA | |
| M19RT11 | S | ACATTGGAAACACCAGTTTTGG |
| AS | CTCTTGATGATGATGGCCTTGT | |
| M19RT12 | S | CAATGGGGCAATCAGATACAAC |
| AS | TGTCCCTAAGCCATCAACATTT | |
| M19RT13 | S | GACTAAATGTTGATGGCTTAGGG |
| AS | ATTGTCCTTTTCACCCCATATG | |
| M19RT14 | S | CAGAAGCTACCAGTGGCACAT |
| AS | AGTAGGTTGGCTTCTCCCTGA | |
| M9RT15 | S | GTCAAACCATCTGCCACATCT |
| AS | CACACTTTGCCATTCCAGTTT | |
| Actin | S | CTCACCCTGAAGTACCCCATC |
| AS | CCTTAATGTCACGCACGATTT |
Definition of abbreviations: AS, antisense; S, sense.
Genomic Walking and Sequencing
Human tracheal DNA was extracted using a genomic DNA kit from Qiagen (Valencia, CA). PCR was performed using the primers (Table 2) designed for cDNA cloning. PCR products were cloned into a TA vector (Invitrogen; for cloning and DNA sequencing) and subject to sequencing. To count for the variation between subjects, the same genomic fragment from the second individual was also cloned and sequenced.
Genomic Structure and Localization
The genomic structure of MUC19 was determined by Blat search of the most up-to-date human genome assembly, GRCh37/hg19 (University of California, Santa Cruz, Santa Cruz, CA).
Phylogenetic Analysis
All nonrepetitive 5′ end peptide sequences from gel-forming mucins of different species were aligned using the ClustalW program (www.ebi.ac.uk/clustalw/). The alignment was edited and the tree was built with the Jalview program (http://www.jalview.org).
Immunofluorescence
For immunofluorescence, human trachea or salivary gland tissues were fixed in 4% paraformaldehyde and then embedded in paraffin. Sections were prepared in the histology facility at Southwest Environmental Health Center (University of Arizona, Tucson, AZ). The tissue sections were incubated with 1:100 diluted anti-MUC19 antibody overnight at 4°C. Anti-chicken IgG secondary antibody (Aves Labs Inc.) and TSA plus fluorescein kit (PerkinElmer) were used to obtain the fluorescence images. The images were acquired by confocal microscopy (LSM 510 meta; Carl Zeiss, Thornwood, NY).
RESULTS
Cloning and Sequencing the Entire MUC19 cDNA
Gel-forming mucins all have evolutionarily conserved cDNA structures: 5′ end unique sequence–large undisrupted central exon 3′ end unique sequence. The reported cDNAs of mouse (12) and pig (21) MUC19 have confirmed this notion. Thus, our general strategy was to start cloning and sequencing from both the 5′ and 3′ ends until reaching the central exon. Because it contains only repetitive sequences and has no intron disruption (12, 22), the central exon is easy to identify by referencing the published genomic sequences in the MUC19 locus.
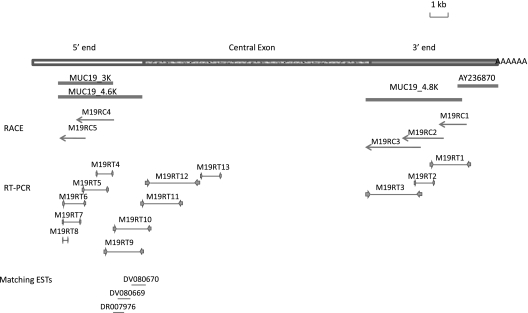
Using our published 3′ end MUC19 sequence (AY236870), we were able to design RACE primers (Table 1) to extend further upstream. Three rounds of RACE (M19RC1, M19RC2, and M19RC3) were performed, and approximately 4.8-kb sequences were identified. At the third round, the sequences were filled with repetitive sequences, suggesting that the central exon had been reached. These results were further verified by RT-PCR (M19RT1–3). We designated those additional sequences starting from M19RC1 (including some overlapped sequences of AY236870) as MUC19_4.8K (Figure 1).
Figure 1.
Cloning strategy. The top rectangular box represents human MUC19 cDNA: the empty portion is the 5′ end, the patterned potion is central exon, and the filled portion is the 3′ end. Four thick lines directly beneath cDNA represent three major cDNA fragments obtained during the cloning process, and AY236870 is previously reported human MUC19 3′-end partial cDNA sequence. The 5′-end rapid amplification of cDNA end (RACE) products are represented by arrows. RT-PCR products are represented by thin lines flanked by two inward arrows indicating a pair of primers. Three thin lines at the bottom represent three matching EST (expressed sequence tag) clones.
We then went on to determine the 5′ end of MUC19. There are usually considerable similarities among the gel-forming mucin orthologs across different species (4). In our previous report, we demonstrated that pig MUC19 (AF005273, also called porcine submaxillary gland mucin) is the closest ortholog of human MUC19 (4). Thus, we used the 5′ end cDNA sequence of pig MUC19 to search for the similar sequences in the genomic sequence of chromosome 12 (http://genome.ucsc.edu/), where the MUC19 locus resides. Indeed, a large piece of the pig MUC19 5′ end sequence matches the sequence from human MUC19 locus, and these matched human sequences were also approximately 18 kb upstream of our published 3′ end of MUC19 (AY236870). Thus, those sequences were very possibly our 5′-end human MUC19. We then designed primers to perform 5′-end RACE to uncover further upstream sequences. Two rounds of RACE (M19RC4 and M19RC5) were performed, and approximately 3 kb additional sequences were identified (designated MUC19_3K) (Figure 1). Using MUC19_3K, we searched dbEST, the Expressed Sequence Tags database (http://www.ncbi.nlm.nih.gov/projects/dbEST/) and uncovered three EST clones (DR007976, DV080669, and DV080670) that overlap with each other and are downstream of MUC19_3K. Those clones were confirmed by resequencing. In addition, we designed various RT-PCR reactions to verify the sequences from the RACE and from the EST clones. A total of 6 RT-PCR products (M19RT4–9) were obtained and sequenced, and the results confirmed a single transcript of approximately 4.6 kb (designated MUC19_4.6K) (Figure 1).
Because the genomic sequences were completed in the MUC19 locus (http://genome.ucsc.edu/), we evaluated the sequences between the MUC19_4.6K (5′ end) and the MUC19_4.8K (3′ end). Indeed, the entire region has only repetitive sequences, and consists of a single giant open reading frame (ORF), suggesting that it should be the central exon. The transitional sequences between the central exon and its 5′/3′ end were confirmed using the primer pairs across the junctions (M19RT3 and M19RT10 primer pairs; see Table 2) (Figure 1). To confirm that it is indeed a single exon, we further designed several pairs of primers inside the central exon. The RT-PCR products (M19RT11–13) indicated the predicted sequence without any intron disruption. However, the sequences downstream of M19RT13 became extremely repetitive and degenerated. Thus, we were unable to test any additional regions in the central exon. Nonetheless, the highly repetitive sequence, the entire sequence being a single ORF, and the pilot PCR verifications all suggested that this was the central exon.
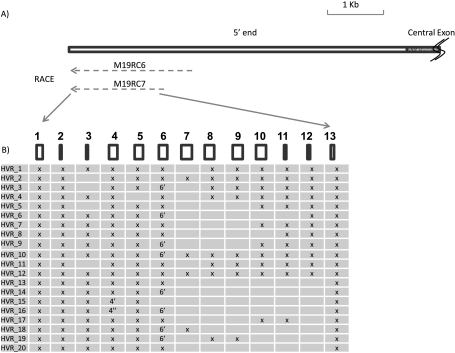
Discovery and Characterization of a 5′-End Highly Variable Region
Although MUC19_4.6K appears to be a single transcript, the products from the additional 5′ RACE (M19RC6) had very complicated compositions, suggesting that they may not be derived from a single transcript. As shown in Figure 2A, We then designed a different reverse primer (M19RC7), which was 301 bp upstream of M19RC6. The RACE result from M19RC7 appeared to be similar to the M19RC6 (i.e., multiple products with overlapping but different compositions), suggesting that the multiple RACE products were not caused by the nonspecific priming. To identify all those transcripts, we cloned and sequenced as many RACE products as possible. A total of 100 transcripts was cloned and sequenced, and 20 nonredundant sequences were identified (Figure 2B). Alignments with the reported human genome sequences (GRCh37/hg19) indicate that they mostly consist of different combinations of 13 exons (Figure 2B). Exon 4 has two other forms with the same 5′ end, but a different 3′ end: 4′ is 20-bp shorter and 4″ is 195-bp longer. In contrast, two forms of exon 6 exist with a different 5′ end but the same 3′ end, and exon 6 is 81-bp longer than exon 6′. The longest transcript has all 13 exons, and the shortest only has 8. Considering the existence of so many alternative splicing transcripts, we named this region the highly variable region (HVR). The sequences of those 20 splicing forms (HVR_1–20) have been deposited in GenBank with the accession numbers HM801843–801862.
Figure 2.
Characterization of highly variable region (HVR). (A) The top rectangular box represents the partial human MUC19 cDNA; the empty portion is the 5′ end, and the patterned potion is the central exon. Both M19RC6 and M19RC7 are 5′ RACE products. Dashed lines indicate the existence of multiple products. (B) Different HVR transcripts. The 13 rectangular boxes represent different exons. The existence of any of those exons is represented by an “X.” Two altered forms of exon 4 are represented by 4′ and 4″, respectively. And one altered form of exon 6 was represented by 6′. The details of those exons are discussed in the Results section and in Table 3.
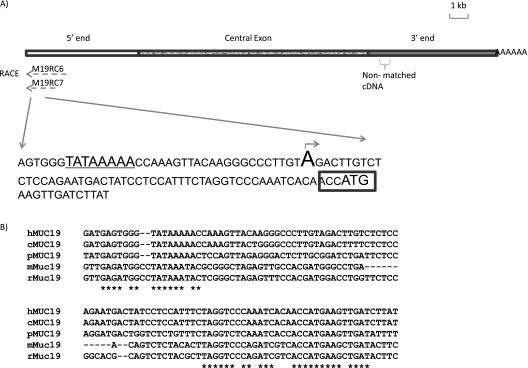
Determination of the Transcription Start Site
Despite the nature of multiple transcripts, both M19RC6 and M19RC7 RACE products stopped at the same nucleotide (Figure 3A), and the additional RACE reactions using the reverse primers designed accordingly to the sequences in both the common and variable regions of HVR produced no further 5′-end sequence (data not shown). In addition, because the RACE product had 5′-end artificial tailing of the nucleotides (dG was added in our original tailing reactions), we tried another three different tailing nucleotides (dA, dT, dC), and confirmed that the “A” was the transcription start site (TSS; Figure 3A). A putative TATA box (TATAAAA) was identified 30-bp upstream of TSS, and a translation start codon (ATG) with Kozak consensus sequence (23) was identified 54-bp downstream of TSS (Figure 3A). Sequence alignment indicates high similarities around the TATA box and the translational start site (ATG) among MUC19 gene of human, chimpanzee, mouse, rat, and pig (Figure 3B).
Figure 3.
Characterization of transcription start site (TSS). (A) TATA box is marked by underline. An arrow and capitalized letter indicates the TSS. The Kozak sequence is marked by a rectangular box. Nonmatched cDNA: cDNA sequences that don't match with GRCh37/hg19, leading to discovery of the additional genomic sequence, HM801863. (B) The alignment of MUC19 sequences near TSS. c, chimpanzee; h, human; m, mouse; p, pig; r, rat. *Identical nucleotides.
Overall MUC19 Genomic Organization and Identification of a Missing Genomic Fragment Downstream of the Central Exon
To obtain the genomic structure, we compared our cDNA sequence with the published human genomic sequences of MUC19 locus (GRCh37/hg19) (Table 3). We found that a fragment of 540bp cDNA (764-bp downstream of the central exon) had no match (showed as “nonmatched cDNA” in Figure 3A). Therefore, we used corresponding RT-PCR primers (Table 2) to perform genomic walking in this region, and identified 7,538-bp additional genomic sequences (deposited in GenBank with accession no. HM801863) that perfectly matched our cDNA sequence, but was missing from the current genome assembly (GRCh37/hg19). To account for human-to-human variation, we further tested the tracheal DNA samples from another individual, and the similar missing genomic sequences were also present in this genome. Thus, the current genome assembly (GRCh37/hg19) appears not to be complete at this location.
TABLE 3.
EXON/INTRON STRUCTURES OF THE LONGEST MUC19 VARIANT (HM801842)
| Exon No. | Intron 3′ Sequence | Exon 5′ Sequence | Exon 3′ Sequence | Intron 5′ Sequence | Exon Size (bp) | Intron Size (bp) | Protein Domains |
|---|---|---|---|---|---|---|---|
| 1 | — | AGACTT | TCAAAG | gtaaga | 106 | 4,380 | — |
| 2 | aaatag | ATGTGG | AATCAG | gtcagt | 33 | 2,130 | — |
| 3 | cttcag | ATGGTA | GATCAG | gtaaat | 27 | 5,957 | — |
| 4* | ctgtag | GTGGCT | CAGCAG | gtactg | 150 | 2,053 | — |
| 5 | ctgtag | GTGGCT | CACCAG | gtaccg | 150 | 3,658 | — |
| 6* | ctgtag | GTGGCT | CAGCAG | gtactg | 150 | 2,286 | — |
| 7 | cagcag | ATAGCA | GCTCAG | gtattc | 186 | 142 | — |
| 8 | cattag | TTGGAA | ATCTAG | gtatta | 165 | 87 | — |
| 9 | ctgcag | GTGATA | GCTCAG | gtattc | 183 | 820 | — |
| 10 | caacag | GTGATT | ACTCAG | gtatcc | 183 | 88 | — |
| 11 | caacag | GTGATT | ACTCAG | gtatcc | 42 | 1,771 | — |
| 12 | cattag | GAGGAA | GAGGAG | gtaaga | 51 | 994 | — |
| 13 | gaatag | AAAAAG | TTATTG | gtaagt | 60 | 974 | VWD |
| 14 | ttatag | GTGAAA | AGAAAG | gtaaat | 229 | 771 | VWD |
| 15 | tttcac | TGTACA | CTCTCG | gtaagt | 118 | 517 | VWD |
| 16 | ttaaag | CTCACT | CTCCAG | gtaaca | 61 | 204 | VWD |
| 17 | tttaag | GGCAAG | ATACAG | gtaaga | 98 | 4,206 | — |
| 18 | ttgtag | CACTGT | TTTGTG | gtaggt | 211 | 757 | — |
| 19 | tccagg | AACACA | CAGAAG | gtagta | 123 | 474 | — |
| 20 | ttctag | GTTATC | TCAGTG | gtatgt | 124 | 1,449 | VWD |
| 21 | taatag | CACATG | GTTCAC | gtatgt | 136 | 1,949 | VWD |
| 22 | ttacag | AATGAA | AATTCT | gtaagt | 93 | 660 | VWD |
| 23 | tattag | TCTGTT | ATTCAG | gtaagt | 85 | 165 | VWD |
| 24 | taacag | ATAAAA | CTGTGG | gtaagt | 138 | 1,000 | VWD |
| 25 | acacag | GACTCT | AAAAAG | gtaata | 150 | 2,184 | — |
| 26 | ttccag | AAAATT | CATGAG | gtatgt | 95 | 426 | — |
| 27 | ttcaag | GAATGC | TATGTG | gtaagc | 127 | 1,385 | — |
| 28 | caacag | AACATT | TAAATG | gtatgg | 247 | 2,199 | — |
| 29 | ttccag | CGTCTG | CCCTAC | gtaagt | 54 | 98 | — |
| 30 | ctgcag | AAAATT | TTTGAT | gtaagt | 101 | 237 | — |
| 31 | ttgtag | GAGAAC | TGAATG | gtgctt | 152 | 660 | VWD |
| 32 | ttttag | TACCTG | CTTGAG | gtaatt | 139 | 1,066 | VWD |
| 33 | ttaaag | GATTAT | TTTCAG | gtacag | 105 | 86 | VWD |
| 34 | ttacag | GATCAG | TGGAAT | gtatgt | 195 | 617 | VWD |
| 35 | atacag | GGCAAA | AACAAG | gtagca | 240 | 73 | VWD |
| 36 | gcctag | GTTGAC | TGTGTC | gtaagt | 157 | 974 | — |
| 37 | ttacag | CTGTCT | TAGAAG | gtaaga | 126 | 844 | — |
| 38 | acacag | GTTGTT | AACATG | gtatat | 142 | 696 | — |
| 39 | tttcag | CTACTG | GCAGTG | gtaagt | 32 | 810 | — |
| 40 | tttcag | AAACTG | TTGCAG | gtaaaa | 30 | 1,494 | — |
| 41 | aaacag | TTTCCA | GAGCAG | gtaaat | 42 | 531 | — |
| 42 | gcatag | CAATTA | GCACAG | gtaagc | 33 | 379 | — |
| 43 | attcag | CAGCAT | CAACAG | gtaggt | 45 | 8,015 | — |
| 44 | ttttag | GAACAA | CTACAG | gtcagt | 39 | 1,466 | — |
| 45 | tctcag | GCATTA | TGGCAG | gtaaat | 99 | 3,107 | — |
| 46 | aagtag | GAACAA | GAGCAG | gtactg | 66 | 615 | — |
| 47 | aattag | GCACAC | TTTCTG | gtaaga | 84 | 218 | — |
| 48 | tttcag | GAAGTA | AATCAG | gtaagt | 78 | 641 | — |
| 49 | tgaaag | GAACTA | TGTCAG | gtgagt | 75 | 242 | — |
| 50 | tgtcag | GTGTAA | GTGGTG | gtgagt | 135 | 384 | — |
| 51 | aattag | GAACAA | CATTGG | gtatgg | 66 | 7,103 | — |
| 52 | gaccag | GTTCAA | CAGCAG | gtaaga | 99 | 1,521 | — |
| 53 | tgaaag | TCTCAG | AATCAG | gcaagt | 102 | 235 | — |
| 54 | aaccag | GGACCC | AACCAG | gtaggt | 102 | 373 | — |
| 55 | ccacag | GTACAC | CACCAG | gtgaga | 96 | 3,388 | — |
| 56 | tgacag | GGGCAA | TAACAG | gtacaa | 12253 | 2,642 | Central exon |
| 57 | attcag | CTACAA | CGTCAG | gtactg | 66 | 107 | — |
| 58 | tctcag | TTGCCA | CTACTG | gtaagt | 54 | 4,072 | — |
| 59 | tttcag | GTACCA | ACATAG | gtgaga | 54 | 656 | — |
| 60 | tttcag | GAACTT | TCTCAG | gtaagt | 54 | 398 | — |
| 61 | tctcag | AGGCTA | CAGGAG | gtgagt | 45 | 249 | — |
| 62 | tcacag | GCACCA | ACACAG | gtaaaa | 54 | 425 | — |
| 63 | ttttag | GTATCA | ACACAG | gtaaag | 54 | 925 | — |
| 64 | tttcag | CTACGA | GTACAG | gtgagt | 54 | 152 | — |
| 65 | tctcag | AGGCCA | CCACAG | gtgagc | 54 | 632 | — |
| 66 | tctcag | AAGCCA | CAGAAG | gtaagc | 54 | 1,382 | — |
| 67 | ttttag | GTACTT | ACACAG | gtagtt | 54 | 135 | — |
| 68 | tctcag | AAGCCA | GAGCAG | gtgagg | 54 | 673 | — |
| 69 | tctcag | AGTCCA | TGACAG | gtgagc | 54 | 669 | — |
| 70 | tctcag | AGGCCA | CAGGAG | gtaagg | 54 | 954 | — |
| 71 | ttttag | GTACCT | ACACAG | gtagtt | 54 | 139 | — |
| 72 | cctcag | AGGCCA | GAACAG | gtgagg | 54 | 2,472 | — |
| 73 | tttcag | CCACCA | AGACAG | gtaagt | 54 | 149 | — |
| 74 | ccttag | AGGCCA | CCACGG | gtgagt | 54 | 599 | — |
| 75 | tctcag | GGGCCA | CAGGAG | gtgagc | 54 | 990 | — |
| 76 | ttttag | GTACTT | ACACAG | gtagtt | 54 | 125 | — |
| 77 | ccacag | AGGCCA | CAGTAG | gtgagg | 54 | 895 | — |
| 78 | tctcag | AGACCA | CCACGG | gtgagc | 54 | 814 | — |
| 79 | taatag | CTACCA | ACACAG | gcaagt | 54 | 121 | — |
| 80 | tctcag | AGGCCA | CAGGAG | gtaagg | 54 | 938 | — |
| 81 | ttgtag | GTACCT | ACACAG | gtagtt | 54 | 132 | — |
| 82 | tctcag | AGGCCA | AGACAG | gtgagg | 54 | 686 | — |
| 83 | tttcag | CCACCA | ATACAG | gtgagt | 54 | 157 | — |
| 84 | tctcag | GGGCCA | CCACCG | gtgagc | 54 | 575 | — |
| 85 | taatag | CTACCA | ACACAG | gcaagt | 54 | 121 | — |
| 86 | tctcag | AGGCCA | CAGGAG | gtaagc | 54 | 966 | — |
| 87 | ttgtag | GTACTT | ACACAG | gtagtt | 54 | 872 | — |
| 88 | tttcag | CCACCA | ACACGG | gtgagt | 54 | 157 | — |
| 89 | tctcag | GGGCCA | CCACCG | gtgagc | 54 | 575 | — |
| 90 | taatag | CTACCA | ACACAG | gcaagt | 54 | 121 | — |
| 91 | tctcag | AGGCCA | CAGGAG | gtaagc | 54 | 1,144 | — |
| 92 | cctcag | AGGCCA | GAACAG | gtgagg | 54 | 1,308 | — |
| 93 | tttcag | CCACCA | AGACAG | gtaagt | 54 | 149 | — |
| 94 | ccttag | AGGCCA | CCACGG | gtgagt | 54 | 606 | — |
| 95 | tctcag | GTGCCA | CAGGAG | gtgagc | 54 | 532 | — |
| 96 | taatag | GTACAA | GGCCAG | gtagga | 54 | 146 | — |
| 97 | ctgaag | GCACCT | AAACAG | gtgagg | 45 | 224 | — |
| 98 | tttag | TTACTA | ACACCG | gtaggt | 54 | 606 | — |
| 99 | tgacag | GCACCC | TGACAG | gtgaag | 45 | 269 | — |
| 100 | tttcag | CCACCA | ACACAG | gtgagt | 54 | 147 | — |
| 101 | tctcag | AGGCCA | TCACAG | gtgagc | 54 | 1,186 | — |
| 102 | ttttag | GTGACA | CCACAG | gtagtt | 54 | 605 | — |
| 103 | tcacag | CAGGCA | AAGCAG | gtgagc | 33 | 263 | — |
| 104 | ttccag | GCACCT | GCACAG | gtgagt | 54 | 144 | — |
| 105 | ttccag | CAGCCA | CTACAA | gtaaga | 54 | 863 | — |
| 106 | ttcaag | AGACCA | AATCAG | gtaaag | 54 | 455 | — |
| 107 | gtggag | CCACCA | GGACAG | gtaaac | 30 | 268 | — |
| 108 | acctag | GCACAG | ACTCAG | gtaaag | 54 | 138 | — |
| 109 | ttgcag | AGGCCA | TCACAG | gttagc | 54 | 1,675 | — |
| 110 | catcag | AGGCCA | AAACTG | gtgaga | 54 | 696 | — |
| 111 | tttcag | GCACCT | GCACAG | gtgagt | 54 | 144 | — |
| 112 | ttccag | CAGCCA | CTACAA | gtaaga | 54 | 874 | — |
| 113 | ttttag | AGACCA | AATTAG | gtaaag | 54 | 503 | — |
| 114 | tttcag | TAGGAA | GGACAG | gtaaac | 36 | 276 | — |
| 115 | gcacag | CTGGAG | ACTCAG | gtaaag | 48 | 138 | — |
| 116 | ttgcag | AGGCCA | TCACAG | gtgagt | 54 | 652 | — |
| 117 | tctcag | AGGCCA | CCAGAG | gtgagc | 54 | 770 | — |
| 118 | ttttag | GTACCA | ACACAG | gtagct | 54 | 142 | — |
| 119 | catcag | AGGCCA | AAATTG | gtgaga | 54 | 411 | — |
| 120 | tcacag | CAGGCA | AAGCAG | gtgagc | 33 | 270 | — |
| 121 | tttcag | GCACCT | GCACAG | gtgagt | 54 | 147 | — |
| 122 | tcgcag | CCACAA | ACTCAG | gtaagc | 51 | 870 | — |
| 123 | tttaag | AGACCA | AATCAG | gtgaag | 54 | 1,043 | — |
| 124 | gcacag | CTGCAG | ACTCAG | gtaaag | 48 | 138 | — |
| 125 | ttgcag | AAGCCA | TCACAG | gtgagc | 54 | 546 | — |
| 126 | tttcag | ATACTA | ACACAG | gtcagg | 54 | 99 | — |
| 127 | tctcag | AGGCCA | CAGGAG | gtgagc | 54 | 767 | — |
| 128 | ttttag | GTACCA | ACACAG | gtagct | 54 | 147 | — |
| 129 | cctcag | AGGCCA | AAACTG | gtgaga | 54 | 743 | — |
| 130 | tttcag | GCACCT | GCACAG | gtgagt | 54 | 155 | — |
| 131 | caatag | CCACAA | CCGCAG | gtaagc | 51 | 873 | — |
| 132 | tttaag | AAACCA | AATCAG | gtaaag | 54 | 502 | — |
| 133 | cattag | GAGCCA | AGACAG | gtaaac | 33 | 270 | — |
| 134 | atctag | GCACAG | ACTCAG | gtaaaa | 54 | 138 | — |
| 135 | ttacag | AGGCCA | TCACAG | gtgagc | 54 | 508 | — |
| 136 | tttcag | GTACTA | ACACAG | gtcagg | 45 | 97 | — |
| 137 | tgtcag | AGGCCA | TGGGTG | gtgagc | 54 | 481 | — |
| 138 | ctgtag | GCGCTT | AAACAG | gtcagc | 45 | 439 | — |
| 139 | cctcag | AGCCCA | AAACTG | gtgaga | 54 | 420 | — |
| 140 | tcacag | TAGGCA | AAGCAG | gtgagc | 33 | 234 | — |
| 141 | tttcag | GCACCT | GCACAG | gtgaat | 54 | 139 | — |
| 142 | tgccag | CAGCCA | CTTCAA | gtaagc | 54 | 887 | — |
| 143 | tttaag | AGACTA | AATCAG | gtaaat | 54 | 682 | — |
| 144 | tattag | GAGCCA | AGACAG | gtagat | 33 | 2,058 | — |
| 145 | tttcag | ATATCA | ACACAG | gtagcc | 54 | 143 | — |
| 146 | cctcag | AGGCCA | AAACTG | gtgaga | 54 | 406 | — |
| 147 | tcacag | CAGGCA | AAGCAG | gtgagc | 33 | 256 | — |
| 148 | tttcag | GCACCT | GCACAG | gtgagt | 54 | 150 | — |
| 149 | tctcag | CAGCTA | CTGCAG | gtaagc | 54 | 971 | — |
| 150 | tttaag | AGACCA | AATCAG | gtgaag | 54 | 510 | — |
| 151 | cattag | GAGCCA | AGACAG | gtaaat | 33 | 255 | — |
| 152 | gcctag | GCACAG | ACTCAG | gtaaag | 54 | 138 | — |
| 153 | tctcag | AGGCTA | AAAATG | ggaagt | 54 | 167 | — |
| 154 | ctacag | GATCTA | AAACAG | gtgagc | 48 | 239 | — |
| 155 | tcttag | TGATTC | TTACAG | gtaagt | 63 | 558 | — |
| 156 | cattag | GAACCA | AAACAG | gtaaat | 48 | 393 | — |
| 157 | tttcag | GCACTA | GTACAG | gtaggc | 54 | 154 | — |
| 158 | ttccag | AAGCCA | AGACAG | gtgaga | 54 | 479 | — |
| 159 | ttttag | GAGCTA | CAATAG | gtaaat | 48 | 590 | — |
| 160 | tctaag | AAGCTA | ATACAG | gtgagc | 54 | 753 | — |
| 161 | ttttag | GTACTA | ACACAG | gtgagt | 54 | 966 | — |
| 162 | cattag | AAGCTA | AAATAG | gtaagc | 48 | 252 | — |
| 163 | ctttag | GCACCA | ACACAG | gtaacc | 54 | 156 | — |
| 164 | acttag | AAGCCA | AAATAG | gtgggc | 54 | 324 | — |
| 165 | ttgcag | TTACCA | ACACAA | gtgagt | 54 | 146 | — |
| 166 | cctcag | AGGCTA | CAACTG | gtgagt | 54 | 6,411 | — |
| 167 | ctttag | GGATCA | AAACAG | gtgagc | 51 | 269 | — |
| 168 | tcccag | GCTACA | GAACAG | gtaagt | 54 | 576 | — |
| 169 | tgtcag | GCAATA | AAGAAG | gtgaga | 42 | 1,101 | — |
| 170 | ttgcag | GAACCA | TCTCAG | gtaata | 54 | 1,248 | — |
| 171 | ttttag | GTCCTT | CCACAG | gtaagt | 54 | 415 | — |
| 172 | aagcag | TCACTG | AAACAG | gtgagt | 51 | 285 | — |
| 173 | tgcaag | GCTCCA | AGCCAG | gtaaaa | 54 | 170 | — |
| 174 | tctcag | AAACCA | AAACAG | gtcagt | 48 | 1,922 | VWC |
| 175 | acacag | AATGTC | CTCCAG | gtaata | 30 | 1,953 | VWC |
| 176 | caacgg | TTTGTC | AAATCT | gtaagt | 32 | 89 | VWC |
| 177 | atgatg | CCTGGA | ACTGTG | gtgtgt | 163 | 331 | VWC |
| 178 | tttcag | AACCAA | TATGAG | gtaagg | 38 | 1,435 | — |
| 179 | tcccag | ATTGGT | AAGCAA | gtcatc | 115 | 1,295 | — |
| 180 | aggaag | ACAGAA | ATACAT | gtgagt | 36 | 654 | CT |
| 181 | ttttag | GTAAGA | TGCCAA | gtgagt | 109 | 659 | CT |
| 182 | ttttag | GTACAA | — | — | 295 | — | CT |
| Alternative exons | |||||||
| 4′ | ctgtag | GTGGCT | TCCAAA | gtcaga | 130 | — | — |
| 4″ | ctgtag | GTGGCT | GCCCTG | gtaata | 345 | — | — |
| 6′ | ctacag | GAGAGG | CAGCAG | gtactg | 69 | — | — |
Definition of abbreviation: bp, base pair.
Exon No. indicates the order of exons (e.g. 1 indicates the first exon); Intron 3′ Sequence indicates part of the nucleotide sequences of the 3′ end of the previous intron at the intron and exon junction; Exon 5′ Sequence indicates the part of the nucleotide sequences of the 5′ end of the current exon at the intron and exon junction; Exon 3′ Sequence indicates part of the nucleotide sequences of the 3′ end of the current exon at the intron and exon junction; Intron 5′ Sequence indicates part of the nucleotide sequence of the 5′ end of the next intron at the intron and exon junction; Exon Size indicates the length of the exon; Intron Size indicates the length of the intron at the 3′ end of the current exon; Protein Domains indicates the documented protein domains.
Alternative exons.
Interestingly, there is a large number of exons (total 122) containing very short sequences (from 32–66 bp, and mostly 54 bp) in the regions of MUC19_4.8K and AY236870 (Table 3). Overall, combined with the exons in HVR, the longest MUC19 transcript has 182 exons (deposited in GenBank with accession no. HM801842) (Table 3), in contrast to mouse Muc19, which has 60 exons (12).
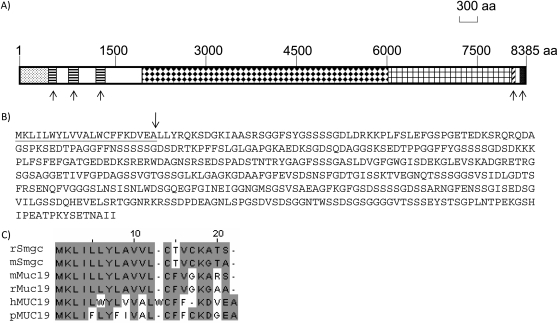
Analysis of MUC19 Protein Structure
The deduced MUC19 protein from the longest transcript has 8,385 amino acids with the classic gel-forming mucin structure: three N-terminal VWD domains, highly repetitive sequences encoded by the central exon (see Table E2.2 in the online supplement), and one VWC domain and one CT domain at the C terminus (Figure 4A). The amino acid sequences encoded by the central exon are serine (S; 13.5%), threonine (T; 23.5%), glycine (G; 21.6%) rich (Table E1.2), and contain numerous potential O-glycosylation sites (www.cbs.dtu.dk). In addition, the repetitive sequences appear to continue downstream of the central exon (Figure 4A), and the amino acid sequences encoded by exons 57–173 are also highly repetitive (Table E2.3) and S (14.5%)/T (24.2%)/G (17%) rich (Table E1.3). In contrast, the amino terminal nonrepetitive sequences of MUC19 contain normal compositions of S, T, or G (Table E1.4).
Figure 4.
Analyses of MUC19 protein. (A) The whole rectangular box indicates the entire MUC19 protein of 8,385 amino acids (aa). The dotted box at the very beginning represents long amino terminus (LAT). The three boxes filled with horizontal lines represent three Von Willebrand (VW) D domains. The box filled with diamonds represents the repetitive sequences encoded by the central exon. The box with the grid represents the repetitive sequences encoded by the exons downstream of the central exon. The box with the diagonal lines represents the VWC domain. The filled box represents the cystine knot (CT) domain. The five upward arrows highlight the five classical mucin domains: three VWD domains, one VWC, and one CT domain. (B) The LAT sequence of MUC19 upstream of the first VWD domain. The underlined sequence is putative signal peptide, and the arrow indicates the potential cleavage site. (C) The alignment of the signal peptide among MUC19 and Smgc proteins. The numbers on top (i.e., 10, 20) indicate the position of the amino acid (e.g., 10 represents the 10th amino acid from the left). Periods represent the gap to facilitate the alignment. Similar sequences are marked by gray boxes. h, human; m, mouse; p, pig; r, rat.
One unusual structure of MUC19 is its long amino terminus (mostly translated from HVR) above the first VWD domain (located in exon 14) (Figures 4A–4B, Table 3), which is missing not only from the other gel-forming mucins, but also from its own orthologs (12, 21). Further analysis of this HVR translated peptide indicates that it also contains several highly repetitive sequences (Table E2.1). However, interestingly, the HRV encodes mostly the serine-rich repeats (Table 1.1), but not the threonine-rich repeats by the central exon (Table 1.2) or the exons downstream of the central exon (Table 1.3). This serine-rich repetitive structure is reminiscent of mouse/rat Smgc (13), a protein encoded by an alternate transcript from the intron 1 of Muc19 (12). The Smgc transcript shares first exon with Muc19, and has an additional 18 exons located in intron 1 of Muc19 (12). Smgc protein contains serine-rich repetitive sequences and resides close to the first VWD domain (in exon 3) of mouse Muc19 (12). The N-terminal signal peptides of Smgc and MUC19 (predicted by SignalIP; www.cbs.dtu.dk [24]) share significant similarities (Figure 4C). Although the relationship could not be established because of the lack of direct sequence similarity in the regions other than signal peptides, the structure and the location related to the VWD domain of MUC19 suggest that the peptide encoded by HVR could be the human counterpart of mouse Smgc.
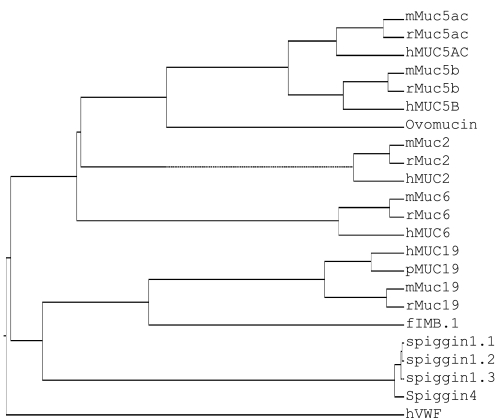
As previously suggested, the gel-forming mucins have significant homology with each other (4, 25). Thus, we compared the protein sequences of family members (MUC2, -5AC, -5B, -6, and -19) from various species. Some related gel-forming mucin-like or mucin-related molecules (chicken ovomucin, frog integumentary mucin [fIBM.1], and VWF and spiggin from fish) were also included to probe potential evolutionary relationships. As shown in Figure 5, Ovomucin, encoding an egg white protein, appears to be the ortholog of both MUC5AC and MUC5B in the chicken, and fIMB.1 and spiggin are orthologs of MUC19. The MUC19 gene family appears to be the first gel-forming mucin to branch out from the common ancestor gene with hVWF. The appearance of four 11p15 gel-forming mucins occurred after the separation of MUC19, and the separation between MUC5AC and MUC5B was the most recent event.
Figure 5.
Phylogenetic analysis of gel-forming mucin family. Protein sequences were obtained from GenBank on the following accession numbers: hMUC2, NP_002448; mMuc2, NP_076055; rMuc2, Q62635; hMUC5AC, P98088; mMuc5ac, NP_034974; rMuc5ac, XP_001063331; hMUC5B, NP_002449; mMuc5b, NP_083077; rMuc5b, XP_238988; hMUC6, NP_005952; mMuc6, EDL18119; rMuc6, XP_215127; hMUC19, HM801842; pMUC19, NP_001106757; mMuc19, NP_997126; rMuc19, XP_002729892; Ovomucin, BAB21488; FIMB.1, CAA69604; Spiggin1_1, BAE92619; Spiggin1_2, BAE92620; Spiggin1_3, BAE92621; Spiggin4, BAE92625; hVWF, AAB59458. h, human; m, mouse; p, pig; r, rat. Phylogenetic analysis methods are described in the Materials and Methods.
Expression of MUC19 Using Both 3′-End and 5′-End Sequence Information
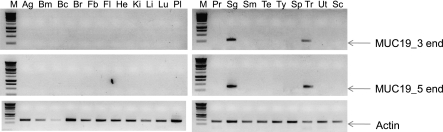
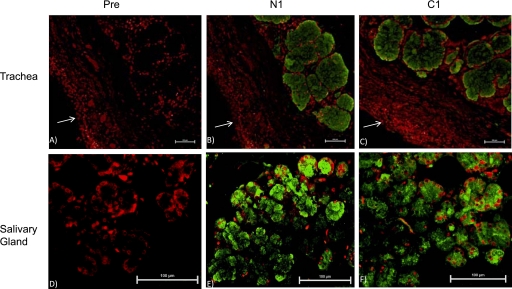
To further confirm that both 3′-end and 5′-end sequences are indeed obtained from a single gene, we first used PCR primers (Table 2) corresponding to either 3′-end (M19RT14) or 5′-end (M19RT15) MUC19 sequences to screen MUC19 expression pattern in a multiple-tissue panel (20 human tissues). The amplification products were confirmed by cloning and sequencing. Both primer sets demonstrated a similar expression pattern in both trachea and salivary glands (Figure 6). This pattern is consistent with the reported Muc19 expression in mouse tissues (14), except that, unlike the mouse panel (14), bulbourethral glands were not present in the human tissue panel. Subsequently, we developed antibodies hMUC19Ab_N1 (against N-terminal region) and hMUC19Ab_C1 (against C-terminal region) to determine protein expression. Both antibodies were affinity purified and verified using ELISA. Immunofluorescent staining indicated similar staining patterns (Figures 7B and 7E were the staining images from hMUC19Ab_N1 and Figures 7C and 7F were the staining images from hMUC19Ab_C1) for both tracheal submucosal gland (Figures 7B–7C) and salivary gland (Figures 7E-7F), whereas the preimmune serum demonstrated no staining (Figures 7A and 7D).
Figure 6.
RT-PCR analyses of MUC19 expression. The pair of primers for MUC19_3 end is M19RT14, and for MUC19_5 end is M19RT15. Actin was used as a control. All primer sequences are listed in Table 2. Ag, adrenal gland; Bc, brain, cerebellum; Bm, bone marrow; Br, brain (whole); Fb, fetal brain; Fl, fetal liver; He, heart; Ki, kidney; Li, liver; Lu, lung (whole); M, molecular marker; Pl, placenta; Pr, prostate; Sc, spinal cord; Sg, salivary gland; Sm, skeletal muscle; Sp, spleen; Te, testis; Tr, trachea; Ty, thymus; Ut, uterus.
Figure 7.
Representative images from immunofluorescence staining. A total of five fields from four sections (prepared from two healthy individuals) was evaluated. (A–C) are trachea sections, and (D–F) are salivary gland section. (A and D) were stained with the preimmune chicken serum (Pre). (B and E) Stained with hMUC19Ab_N1 (N1). (C and F) Stained with hMUC19_C1 (C1). MUC19-positive images were acquired through fluorescein (green) channel. Propidium iodide (red) was used to stain the nuclei. The white arrow (A–C) indicates the epithelial surface. Scale bar, 100 μm.
DISCUSSION
In the gel-forming mucin gene family, MUC19 has an unusual discovery path that appears different from all others. Pig MUC19 (also called porcine submaxillary gland mucin [PSM]) was among the first gel-forming mucins discovered and cloned more than a decade ago (21). It was primarily used as a model to study the biochemical properties of mucus. However, no attempt was made to identify its human or rodent orthologs. Recently, both Culp and colleagues (11, 12) and our group (4) independently discovered the rodent ortholog of Muc19 through different approaches. In addition, using a bioinformatic approach, we further identified and cloned the human MUC19 gene (4). Phylogenetic analysis revealed, for the first time, that PSM is actually the pig MUC19 (4). These findings put this long-time model mucin, PSM, on a par with the other gel-forming mucin family members that have orthologs across mammals.
It has been shown that, among all mucins, gel-forming mucin appeared early in metazoan evolution (25). In addition, epithelial gel-forming mucins have been shown to have a common ancestor with endothelial factor VWF (3). The present study indicates that the MUC19 gene separated from the other gel-forming mucins later than its separation from VWF. Besides, the other four gel-forming mucins (MUC2, -5AC, -5B, and -6) are more related to each other than to MUC19, suggesting their appearances occurred after their separation from MUC19. MUC19 orthologs exist in both fish and amphibian, and no other gel-forming mucins were identified in those species, which further suggests that MUC19 may be more ancient than the other four. Interestingly, MUC19 is located at the same chromosome (chromosome 12) as VWF, whereas the other four mucins are located on chromosome 11 (chromosome 11p15). It is tempting to speculate that it was the first duplication event that created the ancient MUC19 and VWF, and then a translocation event led to the formation of the 11p15 mucin ancestor gene, which further generated MUC2, -5AC, -5B, and -6 through multiple duplication events. Based on our analyses, in chromosome 11p15, it appears that MUC6 was separated first, then MUC2, and the appearance of MUC5AC and MUC5B was the most recent event. The conservation of MUC19 across species, even in primitive species, such as fish and amphibian, suggests that it may have a very important function.
Although the full-length sequences of both pig (21) and mouse (12) MUC19/Muc19 have been reported, the most important ortholog—human MUC19—had only been partially sequenced (4), which is why we determined to completely sequence this mucin in the present study. Interestingly, in contrast to the well held belief that orthologs should have identical structures, human MUC19 has two unique features that are different from its pig or mouse counterparts. First, human MUC19 has an unusually long N terminus, which contains serine-rich repetitive sequences and is encoded by HVR. Multiple alternative splicing forms have been identified from HVR. Because those transcripts were amplified from the mixed RNA samples of multiple adult tracheas or salivary glands, it is still unclear if an individual has all those transcripts, or if they come from different people, which may be an interesting project to pursue in the future. As discussed in the Results section, the peptide encoded by HVR appears to be structurally similar to Smgc (13), a protein encoded by an alternative transcript from the same rodent Muc19 locus (12). In rodent, Smgc is an individual gene and is expressed almost exclusively in the neonatal salivary tissues (12, 13), which are not present in our multiple tissue panel. In adult tissue, even the shortest HVR contains eight additional exons upstream of the first VWD domain. Although most of the HVR transcripts encode peptides in the same ORF with the rest of MUC19, two splicing forms (HVR_14 or HM801856 and HVR_15 or HM801857) can only be translated into truncated peptides, suggesting a potential regulation point of MUC19 expression through alternative splicing at HVR. Another interesting feature of MUC19 is its threonine-rich repeats in the central exon, which differs from the serine-rich repeats in its mouse ortholog (12). In addition, the central exon of human MUC19 encodes mostly the mixed repeats that are similar to human MUC5B (22). In contrast, the central exon of pig MUC19 (21) or mouse Muc19 (12) encodes mostly the identical tandem repeats. Thus, human MUC19 may have a different glycosylation pattern or physical properties than its counterparts in other species.
Through the cloning process, we have identified a missing genomic sequence that covers MUC19 exons 70–81. The repetitive region is usually difficult to assemble when using the popular “shotgun” approach, which assembles DNA fragments based on the similarities at each end (26). Thus, if the DNA ends share considerable similarity (e.g., those located in the repetitive regions of mucin), this approach would have generated an erroneous assembly (26). Thus, genes constraining a large stretch of repetitive sequences should be very prone to assembly errors, and extreme caution should be taken when using the current genome assembly for research on those genes (e.g., mucins). In the present work, because we used published human genome sequences to determine the central exon, it is possible that there are assembly errors in those highly repetitive sequences. Although we have used PCR to confirm some parts of this region, the extreme difficulty in amplifying the repetitive sequences prevented us from examining the entire central exon. In fact, except for the relatively small gel-forming mucins (i.e., MUC2 and MUC6), the central exon sequences of the large mucins are either not completed (e.g., MUC5AC) or conceptually derived from genomic sequences (e.g., MUC5B [22], pig MUC19 [21], mouse Muc19 [12], etc.). Thus, the accuracy of the central exon sequences has actually been determined by the accuracy of human genome assembly. Nonetheless, we have verified MUC19 integrity by using both PCR and antibody staining with primers/antibodies against either 5′ end/N terminus or 3′ end/C terminus. As expected, similar gene expression or staining patterns were observed. Thus, we are confident that we indeed obtained the complete human MUC19 gene sequence.
In addition to verifying the integrity of MUC19 cDNA, the gene expression results (i.e., tracheal submucosal gland and salivary gland) from both mRNA and proteins levels have confirmed the MUC19 tissue expression pattern reported previously (4), which is also identical to the expressions of both mouse Muc19 (14) and pig MUC19 (21). Thus, MUC19 should be one of the major mucin proteins present in both airway mucus and saliva. Interestingly, a recent study (27) found that MUC19 could not be detected in unstimulated human saliva samples, but was present abundantly in stimulated saliva samples from various animals (i.e., horse, pig, cow, rat, and mouse). Because it has been known that the viscosity (also the mucin content) of the stimulated and unstimulated saliva is very different (28) (Dr. David Culp, University of Florida, personal communication), the secretion of MUC19 may well be under neuronal or hormonal control, a concept that will require further study.
In summary, we have cloned and sequenced the human MUC19 gene. Sequence analyses have indicated both the hallmark gel-forming mucin structure (i.e., VWD-VWD-VWD-threonine/serine–rich repeats-VWC-CT) and some distinctive features (i.e., HVR, threonine-dominant repeats). Phylogenetic analysis revealed ancient footage of the MUC19 gene up to fish and amphibian. This information should facilitate future understanding of the function and regulation of MUC19 in health and disease.
Supplementary Material
Acknowledgments
The authors thank Dr. Kenneth B. Adler for editing the early form of the manuscript.
This work was supported in part by National Institutes of Health grant AI061695 and University of Arizona Startup funds.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2010-0312OC on November 12, 2010
Author Disclosure: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Rose MC, Voynow JA. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol Rev 2006;86:245–278. [DOI] [PubMed] [Google Scholar]
- 2.Rubin BK. c. Otolaryngol Clin North Am 2010;43:27–34. (vii–viii.). [DOI] [PubMed] [Google Scholar]
- 3.Desseyn JL, Aubert JP, Porchet N, Laine A. Evolution of the large secreted gel-forming mucins. Mol Biol Evol 2000;17:1175–1184. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y, Zhao YH, Kalaslavadi TB, Hamati E, Nehrke K, Le AD, Ann DK, Wu R. Genome-wide search and identification of a novel gel-forming mucin MUC19/MUC19 in glandular tissues. Am J Respir Cell Mol Biol 2004;30:155–165. [DOI] [PubMed] [Google Scholar]
- 5.Offner GD, Troxler RF. Heterogeneity of high-molecular-weight human salivary mucins. Adv Dent Res 2000;14:69–75. [DOI] [PubMed] [Google Scholar]
- 6.Katsumi A, Tuley EA, Bodó I, Sadler JE. Localization of disulfide bonds in the cystine knot domain of human Von Willebrand factor. J Biol Chem 2000;275:25585–25594. [DOI] [PubMed] [Google Scholar]
- 7.Bell SL, Khatri IA, Xu G, Forstner JF. Evidence that a peptide corresponding to the rat MUC2 C-terminus undergoes disulphide-mediated dimerization. Eur J Biochem 1998;253:123–131. [DOI] [PubMed] [Google Scholar]
- 8.Perez-Vilar J, Eckhardt AE, DeLuca A, Hill RL. Porcine submaxillary mucin forms disulfide-linked multimers through its amino-terminal D-domains. J Biol Chem 1998;273:14442–14449. [DOI] [PubMed] [Google Scholar]
- 9.Perez-Vilar J, Hill RL. Identification of the half-cystine residues in porcine submaxillary mucin critical for multimerization through the D-domains: roles of the CGLCG motif in the D1- and D3-domains. J Biol Chem 1998;273:34527–34534. [DOI] [PubMed] [Google Scholar]
- 10.Evans CM, Koo JS. Airway mucus: the good, the bad, the sticky. Pharmacol Ther 2009;121:332–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fallon MA, Latchney LR, Hand AR, Johar A, Denny PA, Georgel PT, Denny PC, Culp DJ. The sld mutation is specific for sublingual salivary mucous cells and disrupts apomucin gene expression. Physiol Genomics 2003;14:95–106. [DOI] [PubMed] [Google Scholar]
- 12.Culp DJ, Latchney LR, Fallon MA, Denny PA, Denny PC, Couwenhoven RI, Chuang S. The gene encoding mouse MUC19: cDNA, genomic organization and relationship to Smgc. Physiol Genomics 2004;19:303–318. [DOI] [PubMed] [Google Scholar]
- 13.Zinzen KM, Hand AR, Yankova M, Ball WD, Mirels L. Molecular cloning and characterization of the neonatal rat and mouse submandibular gland protein Smgc. Gene 2004;334:23–33. [DOI] [PubMed] [Google Scholar]
- 14.Das B, Cash MN, Hand AR, Shivazad A, Grieshaber SS, Robinson B, Culp DJ. Tissue distibution of murine muc19/Smgc gene products. J Histochem Cytochem 2009. [DOI] [PMC free article] [PubMed]
- 15.Yu DF, Chen Y, Han JM, Zhang H, Chen XP, Zou WJ, Liang LY, Xu CC, Liu ZG. Muc19 expression in human ocular surface and lacrimal gland and its alteration in Sjogren syndrome patients. Exp Eye Res 2008;86:403–411. [DOI] [PubMed] [Google Scholar]
- 16.Kerschner JE. Mucin gene expression in human middle ear epithelium. Laryngoscope 2007;117:1666–1676. [DOI] [PubMed] [Google Scholar]
- 17.Ji C, Guo Y. The expression of mucins gene in the human nasal polyps and allergic rhinitis. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi 2009;23:923–925, 929. [PubMed] [Google Scholar]
- 18.Kerschner JE, Khampang P, Erbe CB, Kolker A, Cioffi JA. Mucin gene 19 (muc19) expression and response to inflammatory cytokines in middle ear epithelium. Glycoconj J 2009;26:1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kouznetsova I, Chwieralski CE, Balder R, Hinz M, Braun A, Krug N, Hoffmann W. Induced trefoil factor family 1 expression by trans-differentiating Clara cells in a murine asthma model. Am J Respir Cell Mol Biol 2007;36:286–295. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Zhao YH, Di YP, Wu R. Characterization of human mucin 5B gene expression in airway epithelium and the genomic clone of the amino-terminal and 5′-flanking region. Am J Respir Cell Mol Biol 2001;25:542–553. [DOI] [PubMed] [Google Scholar]
- 21.Eckhardt AE, Timpte CS, DeLuca AW, Hill RL. The complete cDNA sequence and structural polymorphism of the polypeptide chain of porcine submaxillary mucin. J Biol Chem 1997;272:33204–33210. [DOI] [PubMed] [Google Scholar]
- 22.Desseyn JL, Guyonnet-Duperat V, Porchet N, Aubert JP, Laine A. Human mucin gene muc5B, the 10.7-kb large central exon encodes various alternate subdomains resulting in a super-repeat: structural evidence for a 11p15.5 gene family. J Biol Chem 1997;272:3168–3178. [DOI] [PubMed] [Google Scholar]
- 23.Kozak M. Point mutations define a sequence flanking the aug initiator codon that modulates translation by eukaryotic ribosomes. Cell 1986;44:283–292. [DOI] [PubMed] [Google Scholar]
- 24.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2007;2:953–971. [DOI] [PubMed] [Google Scholar]
- 25.Lang T, Hansson GC, Samuelsson T. Gel-forming mucins appeared early in metazoan evolution. Proc Natl Acad Sci USA 2007;104:16209–16214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng PC, Kirkness EF. Whole genome sequencing. Methods Mol Biol 2010;628:215–226. [DOI] [PubMed] [Google Scholar]
- 27.Rousseau K, Kirkham S, Johnson L, Fitzpatrick B, Howard M, Adams EJ, Rogers DF, Knight D, Clegg P, Thornton DJ. Proteomic analysis of polymeric salivary mucins: no evidence for MUC19 in human saliva. Biochem J 2008;413:545–552. [DOI] [PubMed] [Google Scholar]
- 28.Park MS, Chung JW, Kim YK, Chung SC, Kho HS. Viscosity and wettability of animal mucin solutions and human saliva. Oral Dis 2007;13:181–186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.