Abstract
Our understanding of the role that host genetic factors play in the initiation and severity of infections caused by gram-negative bacteria is incomplete. To identify novel regulators of the host response to lipopolysaccharide (LPS), 11 inbred murine strains were challenged with LPS systemically. In addition to two strains lacking functional TLR4 (C3H/HeJ and C57BL/6JTLR4−/−), three murine strains with functional TLR4 (C57BL/6J, 129/SvImJ, and NZW/LacJ) were found to be relatively resistant to systemic LPS challenge; the other six strains were classified as sensitive. RNA from lung, liver, and spleen tissue was profiled on oligonucleotide microarrays to determine if unique transcripts differentiate susceptible and resistant strains. Gene expression analysis identified the Hedgehog signaling pathway and a number of transcription factors (TFs) involved in the response to LPS. RNA interference–mediated inhibition of six TFs (C/EBP, Cdx-2, E2F1, Hoxa4, Nhlh1, and Tead2) was found to diminish IL-6 and TNF-α production by murine macrophages. Mouse lines with targeted mutations were used to verify the involvement of two novel genes in innate immunity. Compared with wild-type control mice, mice deficient in the E2F1 transcription factor were found to have a reduced inflammatory response to systemic LPS, and mice heterozygote for Ptch, a gene involved in Hedgehog signaling, were found to be more responsive to systemic LPS. Our analysis of gene expression data identified novel pathways and transcription factors that regulate the host response to systemic LPS. Our results provide potential sepsis biomarkers and therapeutic targets that should be further investigated in human populations.
Keywords: endotoxic shock, gram-negative sepsis, inbred murine strains, gene expression, microarray, transcription factor
CLINICAL RELEVANCE.
We have identified novel genetic factors (genes, pathways and transcription factors) that regulate host response to systemic lipopolysaccharide in mice. Our results provide potential sepsis biomarkers and therapeutic targets that should be further investigated in human populations.
Sepsis is the most common cause of death in intensive care units (750,000 cases in the United States in 1995), with a mortality rate of 28 to 40% (1). The sepsis cascade is initiated by the release of bacterial toxins, which leads to an unregulated inflammatory response, systemic inflammatory response syndrome, and ultimately to multiple organ failure. However, antiinflammatory agents have failed in the treatment of sepsis due to the pathophysiologic complexity of the syndrome, which involves cardiovascular, immunological, and endocrine systems. It is therefore important to identify novel therapeutic targets for sepsis and to identify individuals at highest risk for complications from sepsis.
Endotoxin or lipopolysacharide (LPS) on the surface of gram-negative (GN) bacteria activates biologic mediators of shock even at low concentrations. Intravenous LPS induces all of the clinical features of GN sepsis, including fever, shock, leukopenia followed by leukocytosis, disseminated intravascular coagulation, and death (2). These changes can be elicited with LPS from GN bacteria or the intact organisms. Analogous to the “cytokine storm” in systemic inflammatory response syndrome in humans, high doses of LPS given to mice result in the production of proinflammatory cytokines and lead to endotoxic shock (3).
The ability of the host to respond to endotoxin may play an important role in determining the severity of the physiologic and biologic response to this frequently encountered toxin. We have previously shown that polymorphisms in TLR4, the receptor for LPS, predispose humans to GN sepsis (4). However, our previous findings also demonstrate that sequence variants of TLR4 account for only a portion of the LPS phenotype in mice or humans and that other genes are involved in regulating the response to LPS (5, 6). Other researchers have demonstrated that host genetics play a role in the response to bacterial infections (7–9), but the role of host susceptibility in the initiation and severity of infections caused by GN bacteria is incompletely understood.
Genome-wide transcriptional profiling of host cells stimulated with LPS or live bacteria should identify novel candidate genes and pathways involved in innate immunity. Previous genomic studies have identified differentially regulated transcripts in response to purified LPS in peripheral blood of human subjects with severe sepsis (10) and healthy volunteers in response to in vivo or ex vivo stimulation with LPS (11–14). Additionally, Abraham and colleagues found a wide range of sensitivity to LPS in human subjects when they studied neutrophil activation in whole blood, and these differences correlated with differences in neutrophil recruitment to the lungs in response to endobronchial LPS challenge (15). In aggregate, these studies point to the importance of genetic factors in an individual's response to LPS. Transcriptional profiles of multiple organs from mice or rats after systemic administration of LPS (16), intact bacteria (17), or cecal ligation and puncture (18, 19) have also provided insight into the biology of the host gene expression program in response to bacterial pathogens. However, no study to date has combined genetic susceptibility with the gene expression response to LPS to discover novel regulators of the host response to LPS.
In this study, we demonstrated differential susceptibility to systemic LPS among 11 inbred strains of mice. In addition to two strains lacking functional TLR4 (C57BL/6JTLR4−/− and C3H/HeJ), we identified three murine strains with functional TLR4 (C57BL/6J, 129/SvImJ, and NZW/LacJ) that are relatively resistant to systemic administration of LPS. We hypothesized that gene expression profiling of multiple organs from sensitive and resistant strains of mice would lead to identification of novel genetic factors that contribute to differential susceptibility of these strains to systemic LPS. The transcriptional response to LPS in liver, lung, and spleen of the 11 strains was studied, and genes, pathways, and transcription factors that differentiate sensitive from resistant strains in response to systemic LPS were identified. Roles of novel candidate innate immune genes were examined using RNA interference in cultured macrophages and were further evaluated using mouse lines with targeted mutations.
MATERIALS AND METHODS
Ethics Statement
Animal work was approved by Institutional Animal Care and Use Committees at Duke University Medical Center and NIEHS. Every effort was made to ensure that discomfort, distress, and pained injury to animals were limited to that which is unavoidable in the conduct of scientifically sound research. Animals were monitored and cared for by veterinarians at the two institutions.
Animal Model
Male mice (6–8 wk old) from each strain except for the C57BL/6JTLR4−/− strain, were obtained from Jackson Laboratories (Bar Harbor, ME). TLR4-deficient mice were obtained from S. Akira, Osaka University and backcrossed to C57BL/6J mice for 10 generations.
We used an established model of endotoxic shock in which mice are injected with a high dose of LPS with no D-galactosamine sensitization (3). All mice were injected intraperitoneally with 125,000 EU/g, as assessed by the chromogenic Limulus amebocyte lysate kit (Cambrex, East Rutherford, NJ) (range, 15–20 mg/kg body weight of Escherichia coli 0111:B4 LPS) (Sigma Chemical Co., St. Louis, MO) or sterile saline as control. Experimental groups are described in the online supplement.
Statistical Analysis of Morbidity and Cytokine Production
All basic statistical analyses were performed in GraphPad Prism. P values for the comparison of Kaplan-Meier survival curves were calculated using the Mantel-Cox test. P values for cytokine concentration differences were calculated using the two-tailed Student's t test.
Gene Expression Profiling
From the mice killed at 6 hours, total RNA from livers, lungs, or spleens of three animals in each strain/condition was combined in equal amounts to create a pooled RNA sample. Two independent pools were created for each of the 11 murine strains and two conditions (saline and LPS) for all three organs (liver, lung, and spleen) (132 total specimens). Labeling, hybridization, and array scanning were performed according to protocols supplied by Agilent (Santa Clara, CA) and are briefly described in the online supplement (Gene Expression Profiling section).
Microarray Analysis
All primary data have been deposited to the Gene Expression Omnibus database under accession number GSE14675. Data preprocessing is described in the online supplement. Differentially expressed genes between sensitive and resistant strains in response to systemic LPS were identified using two-factor ANOVA with 100 permutations to assess significance. Reported P values were adjusted for multiple comparisons using the false discovery rate approach.
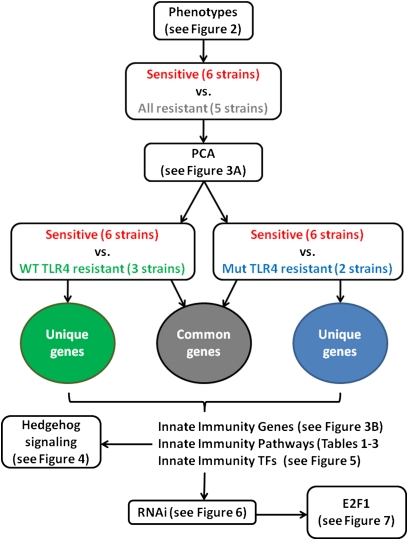
For each organ, two analyses were performed: sensitive strains versus resistant strains lacking functional TLR4 and sensitive strains versus resistant strains with functional TLR4. Genes in common or unique to the two analyses were identified and further explored for significantly represented KEGG pathways using GATHER (20) and for overrepresented transcription factor (TF) binding sites using the PRIMA algorithm (21) implemented in EXPANDER (22). An overview of our analysis strategy is outlined schematically in Figure 1.
Figure 1.
Overview of the approach taken to identify novel innate immune genes in mice. Morbidity and inflammatory (cytokine production in the serum) phenotypes were measured in 11 strains of mice. Strains were grouped based on phenotypes, and two-factor ANOVA analyses were performed to identify significantly differentially expressed genes between defined phenotypic groups (factor 1 in the ANOVA model) in response to lipopolysaccharide (LPS) compared with saline (factor 2 in the ANOVA model). Lists of differentially expressed genes were further analyzed to identify significantly overrepresented pathways or transcription factor binding sites. The involvement of the Hedgehog signaling pathway and the E2F1 transcription factor in innate immunity was validated using siRNA in a macrophage cell line and in mice with targeted deletions of the two genes.
RNA Interference
RNA intereference was performed as described previously (23) and in the online supplement.
Bone Marrow–Derived Macrophage Assays
Bone marrow–derived macrophages (BMDMs) were generated in vitro using standard methodology as described in the online supplement. Differentiated macrophages were stimulated with TLR ligands, and cytokines were measured as described in the online supplement.
RESULTS
Differential Susceptibility of Inbred Murine Strains to Systemic LPS
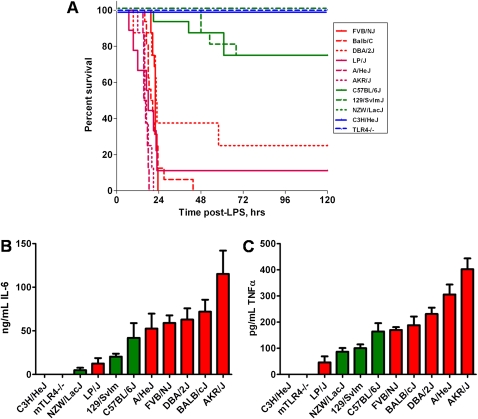
We established differential susceptibility to systemic LPS among 11 strains of mice: 10 commonly used inbred lines and the TLR4 knockout strain on the C57BL/6J genetic background. Six strains were classified as sensitive (> 75% morbidity within 48 h after LPS), and five strains were unresponsive or relatively resistant to LPS (< 25% morbidity at the end of 5 d) (Figure 2A). The two strains that lack functional TLR4 (C3H/HeJ and C57BL/6JTLR4−/−; referred to as mutant or Mut TLR4) were completely resistant to LPS challenge. We also identified three additional strains (129/SvIm, C57BL/6J, and NZW/LacJ; referred to as wild-type or WT TLR4) that were relatively resistant to systemic challenge with LPS. 129/SvIm and C57BL/6J strains have no known polymorphisms in TLR4. The NZW/LacJ strain has been reported to have a polymorphism in TLR4; however, this strain responds normally to inhaled LPS, suggesting that the polymorphic TLR4 is still functional (6). We also measured the production of the proinflammatory cytokines IL-6 and TNF-α, which are typical cytokines produced in animal models of sepsis (3, 24), in the serum 6 hours after administration of LPS and found that sensitivity to systemic LPS correlates with the serum concentrations of IL-6 and TNF-α in almost all strains of mice (Figures 2B and 2C). This is reflected in the correlation coefficient when log (cytokine concentration) is correlated with mean survival for these 11 strains (r2 = 0.64 for IL-6 and r2 = 0.52 for TNF-α). The two exceptions that contribute significantly to these somewhat low correlation coefficients are C57BL/6J mice, which have elevated IL-6 for a resistant strain, and the LP/J strain, which has low concentrations of IL-6 for a sensitive strain. It is worth noting that we did not measure IL-6 concentrations at other times during the course of the experiment and that the relative concentrations of this cytokine in inbred strains may be time dependent. We also measured IL-1β and KC and observed similar trends (data not shown).
Figure 2.
Inbred murine strain phenotypes in response to systemic LPS challenge. (A) Kaplan-Meier survival curves for 11 strains challenged with LPS intraperitoneally (n = 16 for each strains). Two strains lacking functional TLR4 (C3H/HeJ and C57BL/6JTLR4−/−) and one strain with functional TLR4 (NZW/LacJ) are completely unresponsive to systemic administration of LPS (100% survival 5 d after LPS). In addition, 75% of 129/SvIm and C57BL/6J mice survived for 5 days and are also considered resistant to LPS. The six other strains had less than 25% survival and were therefore sensitive to LPS challenge. Sensitive strains as a group (red lines) have significantly lower survival (P < 0.0001 by Mantel-Cox test) than Mut TLR4-resistant strains (blue lines) or WT TLR4-resistant strains (green lines). (B and C) Serum concentrations ± SEM of IL-6 (B) and TNF-α (C) from 11 murine strains challenged with LPS intraperitoneally (n = 8 for each strain). The cytokine concentrations (B and C) correlate with the sensitivity to LPS (A) in the majority of the strains examined (resistant strains depicted in green; sensitive strains in red).
The Hedgehog Pathway Regulates Inflammation
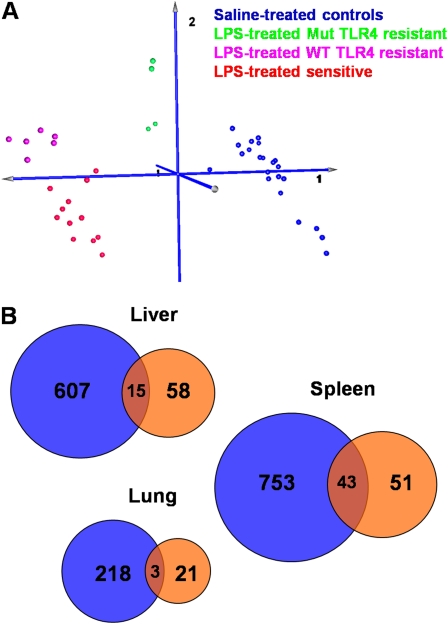
To identify genes that confer sensitivity to systemic LPS, we profiled mRNA from the liver, lung, and spleen of the 11 murine strains 6 hours after exposure to LPS or saline on oligonucleotide arrays that contain probes for approximately 20,000 mouse genes. Many organs are affected in the systemic LPS model; we selected liver because it is a major source of inflammatory mediators in patients with sepsis, lung because of the relevance of the systemic LPS challenge to acute lung injury, and spleen because of its importance in the immune system. We first applied a two-factor ANOVA to identify differentially expressed transcripts in sensitive strains compared with all resistant strains in response to LPS. Principal components analysis of samples using significant genes (P < 0.01 for the ANOVA interaction term) in any of the three organs showed separation of resistant strains based on their TLR4 status (Figure 3A for liver data; lung and spleen data not shown). This grouping of samples suggested that resistance to systemic LPS is mediated by different sets of transcripts in strains with and without functional TLR4.
Figure 3.
Gene expression analysis of lung, liver, and spleen tissue in response to systemic LPS. (A) Principal component analysis of liver samples using interaction-term significant genes (P < 0.01) from the two-factor ANOVA analysis of sensitive compared with all resistant mouse strains (regardless of TLR4 genotype) in response to systemic LPS challenge. Samples are separated into four well defined clusters: a cluster containing saline-treated controls (blue), a cluster of LPS-treated resistant strains that lack functional TLR4 (Mut TLR4, green), a cluster of LPS-treated resistant strains with functional TLR4 (wild-type [WT] TLR4, magenta), and a cluster containing sensitive strains treated with LPS (red). Resistant strains lacking functional TLR4 are closer to unexposed controls, whereas resistant strains with functional TLR4 are closer to but still distinct from sensitive strains, suggesting that different sets of transcripts differentiate the two subgroups of resistant strains from sensitive ones. (B) Venn diagrams showing the number of transcripts that are differentially expressed in response to LPS challenge in sensitive compared with Mut TLR4–resistant (blue circles) or in sensitive compared with WT TLR4–resistant (orange circles) strains in the liver, lung, and spleen tissue. Differentially expressed genes were identified as those having P < 0.01 in the two-factor ANOVA interaction term and 1.5-fold differential expression. Fold change was calculated as the ratio of (exposed/unexposed) for sensitive strains over (exposed/unexposed) for resistant strains. Differentially expressed transcripts are listed in Tables E1, E2, and E3.
To pursue this observation, we identified the genes that were differentially regulated between sensitive mice (A/HeJ, AKR/J, BALB/c, DBA/2J, FVB/NJ, and LP/J) and resistant strains lacking functional TLR4 (Mut TLR4: C3H/HeJ and C57BL/6JTLR4−/−) or those with functional TLR4 (WT TLR4: 129/SvIm, C57BL/6J, and NZW/LacJ). Our analysis revealed a number of differentially expressed transcripts (P < 0.01 and > 1.5-fold change) in all three organs of susceptible strains compared with resistant strains lacking functional TLR4 (Figure 3B; blue circles in the Venn diagrams). More importantly, a smaller but substantial number of genes are up- or down-regulated in sensitive strains compared with resistant mice with functional TLR4 (Figure 3B; orange circles), with relatively little overlap between the two analyses (Figure 3B; red overlap intersections). Complete lists of genes with P values for the ANOVA interaction term and fold changes are presented in Tables E1, E2, and E3 in the online supplement. We chose not to restrict our further analysis to a smaller number of genes that are differentially expressed using more stringent criteria (e.g., higher fold change) because pathway and TF binding site analyses provide a second statistical analysis and associated significance levels (i.e., identify pathways and TFs that are enriched in differentially expressed genes).
To identify molecular pathways activated or repressed in response to systemic LPS, we used the algorithm implemented in the GATHER utility (20) to determine enriched KEGG pathways in the sets of genes shown in Figure 3B. We included genes that are inferred from networks (protein–protein interaction or literature networks implemented in GATHER) in this analysis. The top statistically significant pathways in all analyses are those that have been previously implicated in the host defense against pathogens or purified LPS (Tables 1–3 for liver, lung, and spleen data, respectively; Tables E4–E6 list all genes in each network). Although mostly diverse transcripts differentiate sensitive strains of mice from resistant strains with functional TLR4 or mutant TLR4 (Figure 3B), enriched molecular pathways appear to be similar in all three sets of genes (unique to sensitive versus Mut TLR4 comparison, unique to sensitive versus WT TLR4 resistant comparison, and common to the two comparisons). These pathways include Toll-like receptor signaling (mmu04620), MAPK signaling (mmu04910), cytokine–cytokine receptor interaction (mmu04060), apoptosis (mmu04210), adherens junctions (mmu04520), focal adhesion kinase (mmu04510), coagulation cascade (mmu04610), insulin signaling pathway (mmu04910), and oxidative phosphorylation pathway (mmu00190). One signaling pathway that was not previously known to be involved in LPS-induced inflammation is the Hedgehog signaling pathway, which we observed in the spleen of sensitive mice compared with WT TLR4-resistant mice. This led us to hypothesize that the Hedgehog pathway may uniquely contribute to the hyporesponsive state of the TLR4-sufficient mice that were resistant to systemic challenge with LPS.
TABLE 2.
SIGNIFICANT KEGG PATHWAYS IN THE LUNG IDENTIFIED BY GATHER (P < 005). DIFFERENTIALLY EXPRESSED GENES IDENTIFIED BY TWO-WAY ANOVA WERE USED IN THE ANALYSIS
| Dataset | Kegg Pathway | Number genes | P value | Bayes factor |
|---|---|---|---|---|
| Common | Mmu04080: neuroactive ligand-receptor interaction | 3 | 0.03 | 5 |
| Sensitive vs. functional TLR4 resistant | Mmu04512: ECM-receptor interaction | 7 | 0.05 | 3 |
| Mmu04610: complement and coagulation cascades | 6 | 0.05 | 3 | |
| Sensitive vs. nonfunctional TLR4 resistant | Mmu04060: cytokine-cytokine receptor interaction | 92 | <0.0001 | 59 |
| Mmu04510: focal adhesion | 49 | 0.02 | 8 | |
| Mmu04620: toll-like receptor signaling pathway | 23 | 0.05 | 2 |
Definition of abbreviation: ECM = extracellular matrix.
TABLE 1.
SIGNIFICANT KEGG PATHWAYS IN THE LIVER IDENTIFIED BY GATHER (P < 005). DIFFERENTIALLY EXPRESSED GENES IDENTIFIED BY TWO-WAY ANOVA WERE USED IN THE ANALYSIS
| Dataset | Kegg Pathway | Number of genes | P value | Bayes factor |
|---|---|---|---|---|
| Common | Mmu04060: cytokine-cytokine receptor interaction | 60 | <0.0001 | 59 |
| Mmu04620: Toll-like receptor signaling pathway | 20 | 0.01 | 12 | |
| Mmu04510: focal adhesion | 24 | 0.04 | 4 | |
| Sensitive vs. functional TLR4 resistant | Mmu04510: focal adhesion | 45 | 0.005 | 19 |
| Mmu04910: insulin signaling pathway | 31 | 0.009 | 14 | |
| Mmu04210: apoptosis | 20 | 0.02 | 7 | |
| Mmu04010: MAPK signaling pathway | 36 | 0.03 | 7 | |
| Mmu04520: adherens junction | 15 | 0.05 | 3 | |
| Sensitive vs. nonfunctional TLR4 resistant | Mmu04060: cytokine–cytokine receptor interaction | 90 | 0.003 | 22 |
| Mmu04010: MAPK signaling pathway | 86 | 0.004 | 20 | |
| Mmu04210: apoptosis | 40 | 0.01 | 13 | |
| Mmu04620: Toll-like receptor signaling pathway | 37 | 0.02 | 7 | |
| Mmu04510: focal adhesion | 64 | 0.03 | 5 | |
| Mmu04520: adherens junction | 29 | 0.04 | 5 | |
| Mmu00190: oxidative phosphorylation | 7 | 0.04 | 4 |
TABLE 3.
SIGNIFICANT KEGG PATHWAYS IN THE SPLEEN IDENTIFIED BY GATHER (P < 005). DIFFERENTIALLY EXPRESSED GENES IDENTIFIED BY TWO-WAY ANOVA WERE USED IN THE ANALYSIS
| Dataset | Kegg Pathway | Number of genes | P Value | Bayes Factor |
|---|---|---|---|---|
| Common | Mmu04060: cytokine-cytokine receptor interaction | 46 | 0.0008 | 34 |
| Mmu04610: complement and coagulation cascades | 15 | 0.02 | 10 | |
| Mmu04620: Toll-like receptor signaling pathway | 16 | 0.03 | 7 | |
| Mmu04010: MAPK signaling pathway | 24 | 0.04 | 4 | |
| Mmu04520: adherens junction | 11 | 0.05 | 2 | |
| Sensitive vs. functional TLR4 resistant | Mmu04060: cytokine-cytokine receptor interaction | 45 | 0.009 | 14 |
| Mmu04340: Hedgehog signaling pathway | 17 | 0.01 | 11 | |
| Sensitive vs. nonfunctional TLR4 resistant | Mmu04060: cytokine-cytokine receptor interaction | 108 | 0.001 | 32 |
| Mmu04010: MAPK signaling pathway | 98 | 0.003 | 23 | |
| Mmu04620: Toll-like receptor signaling pathway | 43 | 0.02 | 10 | |
| Mmu04510: focal adhesion | 73 | 0.03 | 6 | |
| Mmu00190: oxidative phosphorylation | 9 | 0.04 | 5 | |
| Mmu04210: apoptosis | 35 | 0.04 | 3 | |
| Mmu04910: insulin signaling pathway | 47 | 0.05 | 3 |
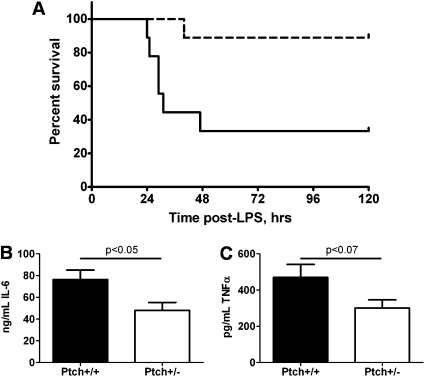
To directly test whether Hedgehog signaling is involved in the innate immune response, we challenged B6;129Ptch1+/− heterozygote mice and the B6;129Ptch1+/+ wild-type littermates with LPS systemically. Patched1 (Ptch1), one of the two receptors for Hedgehog ligands (25), is one of several genes in the Hedgehog pathway that are differentially expressed in our murine data (Ptch1 is up-regulated 1.3-fold in the spleen; Table E6). We monitored morbidity and serum cytokine production in these mice after LPS challenge and determined that B6;129Ptch1+/− heterozygotes have increased survival compared with wild-type littermates (Figure 4A) and produce lower concentrations of TNF-α and IL-6 in the serum (Figure 4B).
Figure 4.
Ptch1 is required for the murine response to systemic LPS. (A) B6;129Ptch+/− mice show decreased morbidity (dashed line) compared with B6;129Ptch1+/+ mice (solid line) followed by systemic LPS challenge (n = 9 in each group). Curve comparison was performed using the Mantel-Cox log-rank test implemented in GraphPad Prism (P < 0.0001). (B) B6;129Ptch1+/− mice produce less serum TNF-α and IL-6 than wild-type control mice after systemic LPS treatment (n = 9 in each group). Two-tailed t test P < 0.07 (trend toward significance) for TNF-α and P < 0.05 (significant) for IL-6.
The E2F1 Transcription Factor Regulates Inflammation
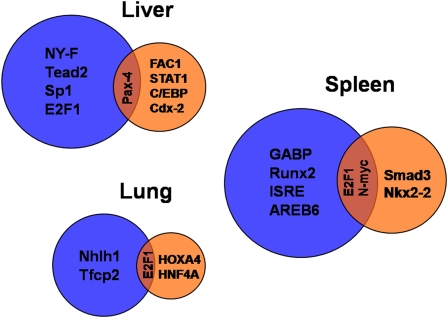
As an additional approach to identify novel regulators of innate immunity, we analyzed the gene expression data to identify transcription factors that regulate the host response to systemic LPS in the three organs. To accomplish this, we searched for overrepresented transcription factor binding sites in the promoters of differentially expressed genes using the PRIMA algorithm (21). Results of this analysis are summarized in the Venn diagram in Figure 5 (details are provided in Table E7). Different transcription factors regulate the response to LPS in sensitive compared with resistant strains with functional or nonfunctional TLR4. This analysis identified several transcription factors already known to affect the host response to LPS or bacterial infections, including C/EBP (26–29), transcription factors that bind to the interferon stimulated response element (28, 29), Smad3 (30–32), Sp1 (26, 33, 34), and Stat1 (35, 36). Additionally, we identified several potentially novel regulators of the response to systemic LPS in the lung, liver, and spleen, including E2F1, Tead2, Pax4, Cdx-2, Nhlh1, Hoxa4, and HNF4A, among others.
Figure 5.
Venn diagrams depicting transcription factor binding sites (TFBS) that are significantly (P < 0.005) overrepresented in the promoters of transcripts (from Figure 2B) that differentiate sensitive compared with Mut TLR4–resistant (blue circles) or sensitive compared with WT TLR4–resistant (orange circles) strains in the liver, lung, and spleen tissue. TRANSFAC accession numbers and P values for each TFBS are given in Table E4.
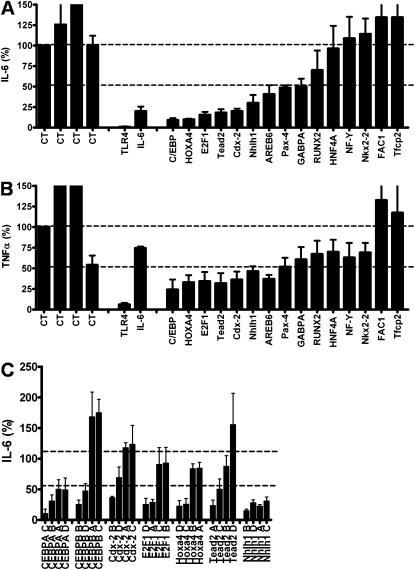
To test these transcription factors for a role in the regulation of the innate immune response, we inhibited 15 of these transcription factor genes using RNA interference in a mouse macrophage cell line and monitored inflammatory cytokine production after LPS treatment. The mouse macrophage cell line J774A.1 was transfected with a pool of four siRNA duplexes for each gene and stimulated with LPS, and cytokine production was measured. As a control, we tested several negative control siRNAs that do not target any gene; these controls had little effect on cytokine production (Figures 6A and 6B). Inhibition of TLR4 strongly diminished the production of IL-6 and TNF-α, whereas inhibition of IL-6 blocked the production of IL-6 but not TNF-α, serving as positive controls for the assay (Figures 6A and 6B). We inhibited 15 transcription factors identified by our analysis, including one known regulator of inflammation as an additional positive control, C/EBP. RNAi-mediated inhibition of six of these transcription factors (CEBPA, Cdx-2, E2F1, Hoxa4, Nhlh1, and Tead2) inhibited the production of IL-6 and TNF-α by at least 50% (Figures 6A and 6B). We performed several additional siRNA experiments with these six genes to confirm the RNAi results. First, we titrated the siRNA concentration down (2, 1, 0.5, and 0.25 μM siRNA) and monitored IL-6 and TNF-α production and RNA knockdown using quantitative PCR. Inhibition of these six transcription factors consistently inhibited production of IL-6 and TNF-α, with the effect lost at lower siRNA concentrations (see Figure E1A in the online supplement). The ability of the siRNAs to inhibit IL-6 production also correlated with the extent of gene expression knockdown (Figure E1B). As an additional confirmation of the RNAi results, we transfected each of the four siRNA duplexes in each siRNA pool individually and demonstrated that at least two of the four individual siRNAs in each pool significantly inhibit production of IL-6 (Figure 6C).
Figure 6.
The effect of RNAi-mediated inhibition of transcription factor genes in J77A4.1 macrophages on IL-6 (A) and TNF-α (B) production in response to LPS stimulation. Pools of four siRNA duplexes per gene (2 μM concentration) were transfected into J77A4.1 cells, LPS was added, and cytokine production was monitored as described in Materials and Methods. Shown are the results for four negative controls (nontargeting siRNAs from Dharmacon; all data are normalized to the first negative control), two positive controls (TLR4 and IL-6), and 15 transcription factors identified by our analysis of gene expression data. To verify that the phenotypes observed were caused by knockdown of that gene, the four siRNA duplexes were transfected individually for the seven genes that had the strongest effect, and IL-6 production was monitored (C). At least two independent siRNAs induced a phenotype for each gene. In all three panels, plotted are means of three independent measurements with error bars representing SD.
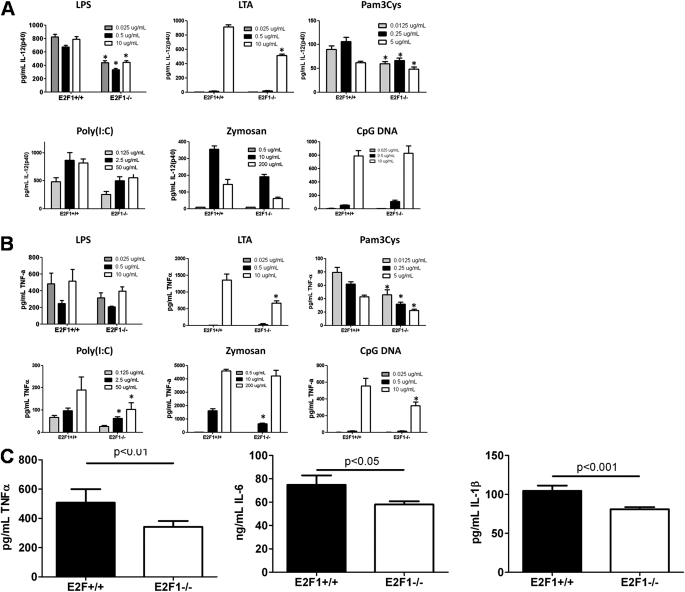
E2F1 is the only transcription factor that affected cytokine production in our siRNA assay whose binding sites are overrepresented in the lung, liver, and spleen gene expression data after stimulation with LPS. We therefore further investigated the role of E2F1 in the murine response to systemic LPS. Based on a recent publication that showed recruitment of E2F1 by NF-kB upon stimulation of a monocytic cell line with LPS (37), we hypothesized that stimulation with any TLR ligand should result in a diminished cytokine production by BMDMs. We observed diminished IL-12(p40) and TNF-α production by BMDMs of B6;129E2F1−/− mice as compared with BMDMs of B6 × 129 control F2 mice when stimulated with three concentrations of six different TLR ligands: LPS (ligand for the TLR4/TLR4 homodimer), Pam 3 Cys-Ser-(Lys) 4 (ligand for the TLR1/TLR2 heterodimer), poly (I:C) (TLR3/TLR3), CpG DNA (TLR9/TLR9), LTA and zymosan (TLR2/6) (Figures 7A and 7B). In addition to these ex vivo studies, we challenged B6;129E2F1−/− knockout mice and B6 × 129 F2 control mice with systemic LPS and found that B6;129E2F1−/− mice had reduced levels of TNF-α, IL-1β, and IL-6 in the serum 3 hours after LPS challenge (Figure 7C).
Figure 7.
E2F1 regulates inflammation. Bone marrow–derived macrophages from B6;129E2F1−/− mice produce less IL-12 (p40) (A) and TNF-α (B) than B6 × 129 F2 control mice (three pools of two mice each) 5 hours after stimulation with LPS, lipotechoic acid (LTA), Pam 3 Cys-Ser-(Lys) 4 (Pam3Cys), poly (I:C), zymosan, and CpG DNA. *P < 0.05 by two-tailed t test. (C) B6;129E2F1−/− mice produce less serum TNF-α, IL-6, and IL-1β than B6 × 129 F2 control mice 3 hours after systemic LPS (n = 12 in each group). Two-tailed t test: P < 0.01 for TNF-α, P < 0.05 for IL-6, and P < 0.005 for IL-1β.
Overlap in the Lung, Liver, and Spleen Gene Expression Profiles
We compared lung, liver, and spleen data to identify overlaps in gene expression in the three organs we studied. We constructed a Venn diagram that compares differentially expressed genes in the lung, liver, and spleen; all genes from Figure 3B organ-specific Venn diagrams were combined into one circle each to represent the three organs (Figure E2A). Ten genes that are differentially expressed in response to LPS in all three organs are listed in the table in Figure E2B. Genes known to be involved in inflammation (chemokine Cxcl5 and colony-stimulating factor 3), hypoxia (HIF-1α), and the response to viruses (Rsad2 or viperin) are among these genes. Novel candidates include Pitpnc1, a gene involved in phosphatidylinositol transfer; a Ras homolog (Rhou); β subunit of inhibitin B, a gene that regulates gonadal stromal cell proliferation has tumor-suppressor activity; and three genes with unknown function.
DISCUSSION
Our analysis of gene expression in the liver, lung, and spleen of inbred strains of mice identified a number of pathways and transcription factors that are differentially regulated in sensitive and resistant strains after systemic LPS challenge. One pathway identified by this approach that was not previously known to be involved in innate immunity is the Hedgehog signaling (mmu04340) pathway. We validated the importance of the Hedgehog pathway by demonstrating that Ptch1+/− mice are more resistant to LPS than wild-type control mice. The in vitro importance of transcription factors was evaluated by RNAi, and E2F1 was further tested in a deficient strain of mice, again demonstrating the importance of this gene in innate immune responsiveness. In aggregate, our results indicate that gene expression from inbred strains of mice can be used to identify novel regulators of innate immune responsiveness that may prove important in humans with GN sepsis.
We have also observed activation of Hedgehog signaling at the transcriptional level in RAW264.7 macrophages upon LPS stimulation (unpublished observation); this more recent analysis was performed using Ingenuity Pathway Analysis (IPA) software, suggesting that our findings hold in a different biological system (macrophage cell line) and using a different statistical analysis approach (IPA instead of GATHER). Published studies have demonstrated that endotoxin impurities in recombinant Sonic Hedgehog (Shh) and purified E. coli LPS activate components of the Hedghehog pathway in human peripheral blood monocytes (38). Moreover, a recent study showed that NF-κB directly regulates Shh in vitro and in vivo and promotes cancer cell proliferation and apoptosis resistance via Shh (39). Another potential explanation for the Hedgehog pathway being involved in innate immune responsiveness is the interaction of the Patched receptor with Tid1 (Dnaja3); Tid1 affects apoptosis and senescence in several ways, including modifying NF-κB and γ-IFN signaling (40, 41). We have also identified two Ptch1 polymorphisms in 129/SvIm and NZW/LacJ mice that may contribute to the hyporesponsivness of these two strains to systemic LPS (unpublished observation), but further investigation is necessary to prove the role these SNPs may play in response to LPS.
In addition to Hedgehog signaling, many pathways known to be involved in innate immune response to LPS—Toll like receptor signaling (mmu04620), MAPK signaling (mmu04910), cytokine–cytokine receptor interaction (mmu04060), and apoptosis (mmu04210) (42)—were identified. Other pathways identified in our analysis have also been implicated in innate immunity and host defense. Infiltration of polymorphonuclear cells into the tissue is a hallmark of the host response to infection and involves signaling between endothelial cells and migrating polymorphonuclear cells, which leads to alterations in the organization of adherens junctions (mmu04520) (43). Focal adhesion kinase is a nonreceptor protein kinase that signals downstream of integrins and was recently shown to interact with Myd88 (44), explaining the function of the Focal adhesion (mmu04510) pathway in response to systemic LPS. Recognizing the link between coagulation activation and inflammation (45), it is also not surprising that the Complement and coagulation cascades pathway (mmu04610) appears in some of the analyses that differentiate LPS-sensitive from LPS-resistant strains. Hyperglycemia is a common feature of the critically ill and has been associated with increased mortality (46), which supports the role the insulin signaling pathway (mmu04910) plays in the host response to systemic LPS. Recent studies at the molecular level have also linked insulin signaling to innate immune pathways (47). Finally, overrepresentation of the oxidative phosphorylation pathway (mmu00190) in the liver and spleen gene expression data can be explained by recent evidence that links the pathogenesis of multiple organ failure in sepsis to mitochondrial damage (48).
Our analysis also identified the E2F1 transcription factor as a novel regulator of the transcriptional response to systemic LPS. E2F1 target genes are overrepresented in the liver, lung, and spleen expression data. E2F1 is a member of the E2F family of transcription factors that plays a crucial role in the control of cell cycle and action of tumor suppressor proteins. E2F1 binds preferentially to the retinoblastoma protein in a cell-cycle–dependent manner and is capable of mediating cell proliferation and apoptosis. E2F1 was recently identified as a transcriptional activator recruited by NF-κB upon TLR4 activation in an LPS-stimulated human monocytic cell line (37). Consistent with published data and our RNAi data in macrophage cell lines, B6;129E2F1−/− mice have a reduced inflammatory response in bone marrow–derived macrophages treated with six different TLR ligands compared with B6 × 129 F2 control mice. Furthermore, our in vivo murine knockout data verify the important role that E2F1 plays in regulating innate immunity.
In addition to C/EBP (a known regulator of innate immunity) and E2F1, four transcription factors were identified as novel potential regulators of the murine response to systemic LPS: Cdx-2 and Tead2 in the liver and HOXA4 and Nhlh1 in the lung gene expression data. Cdx-2 is a homeobox gene that has been implicated as a target of PTEN/phosphatidylinositol 3-kinase signaling and TNF-α signaling via NF-κB activation in the intestine (49). There are no published studies on the function of another homeobox gene HOXA4 in innate immunity, but a recent study showed the mutations in the Caenorhabditis elegans homeobox gene EGL-5 results in a defective response and hypersensitivity to Staphylococcus aureus infection (50). Moreover, HOXA9 and HOXC10 homologs of EGL-5 were shown to regulate NF-κB signaling in human epithelial cell lines in the same study. Very little is known about the transcription factors Tead2 or ETF, but it is known that another member of the gene family, Tead1, is regulated by p38α MAP kinase in proliferating cardiomyocytes (51). Finally, the helix-loop-helix protein Nhlh1 (HEN1) has also never been studied in the context of innate immunity.
By using RNAi-mediated suppression of gene expression in cultured macrophages, we confirmed the significance of C/EBP in controlling proinflammatory cytokine production in response to LPS stimulation. More importantly, five additional transcription factors with less well defined roles in innate immunity (Cdx-2, E2F1, Hoxa4, Nhlh1, and Tead2) were shown to affect cytokine production in macrophages. These five genes represent high-priority candidates, and their function in the regulation of innate immune response to LPS and bacterial infections should be studied in more detail. Two of the five genes belong to the homeobox family of transcription factors. Based on our murine data and recently published findings in C. elegans and human cell lines (50), the role of the other members of the homeobox gene family in innate immunity should be further examined.
Our studies in mouse lines with Ptch1- and E2F1-targeted deletions provide further evidence for the importance of the genes identified in our gene expression analysis in the murine response to LPS. Future studies in human populations are necessary to determine which of the candidates identified in our murine studies could be developed into therapeutic targets and biomarkers for predicting the risk of gram-negative sepsis.
Acknowledgments
The authors thank Jessica Ramsberger, Tatiana Vinogradova, and Sara Weinke for help with animal studies and RNA extractions and Ed Lobenhofer (Cogenics) for assistance with microarray hybridization assays.
This study was supported by the Department of Veterans' Affairs (Merit Review), the National Institute of Environmental Health Sciences grants ES11375 and ES011961 and by the Intramural Research Program of the NIH, National Institute of the Environmental Health Sciences and National Heart Lung and Blood Institute.
Originally Published in Press as DOI: 10.1165/rcmb.2010-0342OC on December 3, 2010
Author Disclosure: S.A. received sponsored grants from National Institutes of Health (NIH) and American Lung Association. B.L. is employed by NIH. D.A.S. received expert witness fees from Wallace and Graham, Brayton and Purcell, Weitz and Luxemberg, and Waters and Kraus. D.A.S. in employed by NIH and National Jewish Health. L.W. is employed by National Jewish Health. I.Y in employed by NIH and National Jewish Health. L.H.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. H.R. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med 2001;29:1303–1310. [DOI] [PubMed] [Google Scholar]
- 2.Ziegler EJ, McCutchan JA, Fierer J, Glauser MP, Sadoff JC, Douglas H, Braude AI. Treatment of gram-negative bacteremia and shock with human antiserum to a mutant escherichia coli. N Engl J Med 1982;307:1225–1230. [DOI] [PubMed] [Google Scholar]
- 3.Buras JA, Holzmann B, Sitkovsky M. Animal models of sepsis: setting the stage. Nat Rev Drug Discov 2005;4:854–865. [DOI] [PubMed] [Google Scholar]
- 4.Lorenz E, Mira JP, Frees KL, Schwartz DA. Relevance of mutations in the TLR4 receptor in patients with gram-negative septic shock. Arch Intern Med 2002;162:1028–1032. [DOI] [PubMed] [Google Scholar]
- 5.Arbour NC, Lorenz E, Schutte BC, Zabner J, Kline JN, Jones M, Frees K, Watt JL, Schwartz DA. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet 2000;25:187–191. [DOI] [PubMed] [Google Scholar]
- 6.Lorenz E, Jones M, Wohlford-Lenane C, Meyer N, Frees KL, Arbour NC, Schwartz DA. Genes other than TLR4 are involved in the response to inhaled LPS. Am J Physiol Lung Cell Mol Physiol 2001;281:L1106–L1114. [DOI] [PubMed] [Google Scholar]
- 7.Cook DN, Pisetsky DS, Schwartz DA. Toll-like receptors in the pathogenesis of human disease. Nat Immunol 2004;5:975–979. [DOI] [PubMed] [Google Scholar]
- 8.Garantziotis S, Hollingsworth JW, Zaas AK, Schwartz DA. The effect of toll-like receptors and toll-like receptor genetics in human disease. Annu Rev Med 2008;59:343–359. [DOI] [PubMed] [Google Scholar]
- 9.Papathanassoglou ED, Giannakopoulou MD, Bozas E. Genomic variations and susceptibility to sepsis. AACN Adv Crit Care 2006;17:394–422. [DOI] [PubMed] [Google Scholar]
- 10.Prucha M, Ruryk A, Boriss H, Moller E, Zazula R, Herold I, Claus RA, Reinhart KA, Deigner P, Russwurm S. Expression profiling: toward an application in sepsis diagnostics. Shock 2004;22:29–33. [DOI] [PubMed] [Google Scholar]
- 11.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, et al. A network-based analysis of systemic inflammation in humans. Nature 2005;437:1032–1037. [DOI] [PubMed] [Google Scholar]
- 12.Feezor RJ, Oberholzer C, Baker HV, Novick D, Rubinstein M, Moldawer LL, Pribble J, Souza S, Dinarello CA, Ertel W, et al. Molecular characterization of the acute inflammatory response to infections with gram-negative versus gram-positive bacteria. Infect Immun 2003;71:5803–5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prabhakar U, Conway TM, Murdock P, Mooney JL, Clark S, Hedge P, Bond BC, Jazwinska EC, Barnes MR, Tobin F, et al. Correlation of protein and gene expression profiles of inflammatory proteins after endotoxin challenge in human subjects. DNA Cell Biol 2005;24:410–431. [DOI] [PubMed] [Google Scholar]
- 14.Wurfel MM, Park WY, Radella F, Ruzinski J, Sandstrom A, Strout J, Bumgarner RE, Martin TR. Identification of high and low responders to lipopolysaccharide in normal subjects: an unbiased approach to identify modulators of innate immunity. J Immunol 2005;175:2570–2578. [DOI] [PubMed] [Google Scholar]
- 15.Abraham E, Nick JA, Azam T, Kim SH, Mira JP, Svetkauskaite D, He Q, Zamora M, Murphy J, Park JS, et al. Peripheral blood neutrophil activation patterns are associated with pulmonary inflammatory responses to lipopolysaccharide in humans. J Immunol 2006;176:7753–7760. [DOI] [PubMed] [Google Scholar]
- 16.Wong ML, O'Kirwan F, Khan N, Hannestad J, Wu KH, Elashoff D, Lawson G, Gold PW, McCann SM, Licinio J. Identification, characterization, and gene expression profiling of endotoxin-induced myocarditis. Proc Natl Acad Sci USA 2003;100:14241–14246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu SL, Chen HW, Yang PC, Peck K, Tsai MH, Chen JJ, Lin FY. Differential gene expression in gram-negative and gram-positive sepsis. Am J Respir Crit Care Med 2004;169:1135–1143. [DOI] [PubMed] [Google Scholar]
- 18.Chinnaiyan AM, Huber-Lang M, Kumar-Sinha C, Barrette TR, Shankar-Sinha S, Sarma VJ, Padgaonkar VA, Ward PA. Molecular signatures of sepsis: multiorgan gene expression profiles of systemic inflammation. Am J Pathol 2001;159:1199–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cobb JP, O'Keefe GE. Injury research in the genomic era. Lancet 2004;363:2076–2083. [DOI] [PubMed] [Google Scholar]
- 20.Chang JT, Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics 2006;22:2926–2933. [DOI] [PubMed] [Google Scholar]
- 21.Elkon R, Linhart C, Sharan R, Shamir R, Shiloh Y. Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res 2003;13:773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shamir R, Maron-Katz A, Tanay A, Linhart C, Steinfeld I, Sharan R, Shiloh Y, Elkon R. Expander–an integrative program suite for microarray data analysis. BMC Bioinformatics 2005;6:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alper S, Laws R, Lackford B, Boyd WA, Dunlap P, Freedman JH, Schwartz DA. Identification of innate immunity genes and pathways using a comparative genomics approach. Proc Natl Acad Sci USA 2008;105:7016–7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osuchowski MF, Welch K, Siddiqui J, Remick DG. Circulating cytokine/inhibitor profiles reshape the understanding of the SIRS/CARS continuum in sepsis and predict mortality. J Immunol 2006;177:1967–1974. [DOI] [PubMed] [Google Scholar]
- 25.Chari NS, McDonnell TJ. The sonic hedgehog signaling network in development and neoplasia. Adv Anat Pathol 2007;14:344–352. [DOI] [PubMed] [Google Scholar]
- 26.Chiang BT, Liu YW, Chen BK, Wang JM, Chang WC. Direct interaction of C/EBPδ and Sp1 at the GC-enriched promoter region synergizes the IL-10 gene transcription in mouse macrophage. J Biomed Sci 2006;13:621–635. [DOI] [PubMed] [Google Scholar]
- 27.Haaxma CA, Kim PK, Andrejko KM, Raj NR, Deutschman CS. Transcription factors C/EBP-α and HNF-1α are associated with decreased expression of liver-specific genes in sepsis. Shock 2003;19:45–49. [DOI] [PubMed] [Google Scholar]
- 28.Burch LH, Yang IV, Whitehead GS, Chao FG, Berman KG, Schwartz DA. The transcriptional response to lipopolysaccharide reveals a role for interferon-gamma in lung neutrophil recruitment. Am J Physiol Lung Cell Mol Physiol 2006;291:L677–L682. [DOI] [PubMed] [Google Scholar]
- 29.Tebo JM, Hamilton TA. Lipopolysaccharide induces DNA binding activity specific for the IFN-stimulated response element in murine peritoneal macrophages. J Immunol 1992;149:2352–2357. [PubMed] [Google Scholar]
- 30.Kanamaru Y, Sumiyoshi K, Ushio H, Ogawa H, Okumura K, Nakao A. Smad3 deficiency in mast cells provides efficient host protection against acute septic peritonitis. J Immunol 2005;174:4193–4197. [DOI] [PubMed] [Google Scholar]
- 31.Le Y, Iribarren P, Gong W, Cui Y, Zhang X, Wang JM. TGF-β1 disrupts endotoxin signaling in microglial cells through smad3 and mapk pathways. J Immunol 2004;173:962–968. [DOI] [PubMed] [Google Scholar]
- 32.McCartney-Francis N, Jin W, Wahl SM. Aberrant toll receptor expression and endotoxin hypersensitivity in mice lacking a functional TGF-β1 signaling pathway. J Immunol 2004;172:3814–3821. [DOI] [PubMed] [Google Scholar]
- 33.Wasiluk KR, McCulloch KA, Banton KL, Dunn DL. Sp1 elements regulate transcriptional activity within the murine toll-like receptor 4 promoter. Surg Infect (Larchmt) 2006;7:489–499. [DOI] [PubMed] [Google Scholar]
- 34.Ye X, Liu SF. Lipopolysaccharide causes Sp1 protein degradation by inducing a unique trypsin-like serine protease in rat lungs. Biochim Biophys Acta 2007;1773:243–253. [DOI] [PubMed] [Google Scholar]
- 35.Joshi VD, Kalvakolanu DV, Chen W, Zhang L, Kang TJ, Thomas KE, Vogel SN, Cross AS. A role for stat1 in the regulation of lipopolysaccharide-induced interleukin-1beta expression. J Interferon Cytokine Res 2006;26:739–747. [DOI] [PubMed] [Google Scholar]
- 36.Lee HJ, Oh YK, Rhee M, Lim JY, Hwang JY, Park YS, Kwon Y, Choi KH, Jo I, Park SI, et al. The role of STAT1/IRF-1 on synergistic ros production and loss of mitochondrial transmembrane potential during hepatic cell death induced by LPS/d-GaLN. J Mol Biol 2007;369:967–984. [DOI] [PubMed] [Google Scholar]
- 37.Lim CA, Yao F, Wong JJ, George J, Xu H, Chiu KP, Sung WK, Lipovich L, Vega VB, Chen J, et al. Genome-wide mapping of RELA(p65) binding identifies E2F1 as a transcriptional activator recruited by NF-κB upon TLR4 activation. Mol Cell 2007;27:622–635. [DOI] [PubMed] [Google Scholar]
- 38.Wakelin SJ, Forsythe JL, Garden OJ, Howie SE. Commercially available recombinant sonic hedgehog up-regulates PTC and modulates the cytokine and chemokine expression of human macrophages: an effect mediated by endotoxin contamination? Immunobiology 2008;213:25–38. [DOI] [PubMed] [Google Scholar]
- 39.Kasperczyk H, Baumann B, Debatin KM, Fulda S. Characterization of sonic hedgehog as a novel NF-κB target gene that promotes NF-κB-mediated apoptosis resistance and tumor growth in vivo. FASEB J 2009;23:21–33. [DOI] [PubMed] [Google Scholar]
- 40.Sarkar S, Pollack BP, Lin KT, Kotenko SV, Cook JR, Lewis A, Pestka S. Htid-1, a human dnaj protein, modulates the interferon signaling pathway. J Biol Chem 2001;276:49034–49042. [DOI] [PubMed] [Google Scholar]
- 41.Tarunina M, Alger L, Chu G, Munger K, Gudkov A, Jat PS. Functional genetic screen for genes involved in senescence: role of Tid1, a homologue of the drosophila tumor suppressor l(2)tid, in senescence and cell survival. Mol Cell Biol 2004;24:10792–10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Remick DG. Pathophysiology of sepsis. Am J Pathol 2007;170:1435–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orrington-Myers J, Gao X, Kouklis P, Broman M, Rahman A, Vogel SM, Malik AB. Regulation of lung neutrophil recruitment by VE-cadherin. Am J Physiol Lung Cell Mol Physiol 2006;291:L764–L771. [DOI] [PubMed] [Google Scholar]
- 44.Zeisel MB, Druet VA, Sibilia J, Klein JP, Quesniaux V, Wachsmann D. Cross talk between MyD88 and focal adhesion kinase pathways. J Immunol 2005;174:7393–7397. [DOI] [PubMed] [Google Scholar]
- 45.Mayr FB, Jilma B. Coagulation interventions in experimental human endotoxemia. Transl Res 2006;148:263–271. [DOI] [PubMed] [Google Scholar]
- 46.Brierre S, Kumari R, Deboisblanc BP. The endocrine system during sepsis. Am J Med Sci 2004;328:238–247. [DOI] [PubMed] [Google Scholar]
- 47.Cuschieri J, Bulger E, Grinsell R, Garcia I, Maier RV. Insulin regulates macrophage activation through activin A. Shock 2008;29:285–290. [DOI] [PubMed] [Google Scholar]
- 48.Piantadosi CA, Carraway MS, Haden DW, Suliman HB. Protecting the permeability pore and mitochondrial biogenesis. Novartis Found Symp 2007;280:266–276, discussion 276–280. [DOI] [PubMed] [Google Scholar]
- 49.Kim S, Domon-Dell C, Wang Q, Chung DH, Di Cristofano A, Pandolfi PP, Freund JN, Evers BM. PTEN and TNF-alpha regulation of the intestinal-specific Cdx-2 homeobox gene through a PI3K, PKB/Akt, and NF-κB-dependent pathway. Gastroenterology 2002;123:1163–1178. [DOI] [PubMed] [Google Scholar]
- 50.Irazoqui JE, Ng A, Xavier RJ, Ausubel FM. Role for β-catenin and hox transcription factors in caenorhabditis elegans and mammalian host epithelial-pathogen interactions. Proc Natl Acad Sci USA 2008;105:17469–17474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ambrosino C, Iwata T, Scafoglio C, Mallardo M, Klein R, Nebreda AR. TEF-1 and C/EBPbeta are major p38alpha MAPK-regulated transcription factors in proliferating cardiomyocytes. Biochem J 2006;396:163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]