Abstract
Multipotential (MP) differentiation is one characteristic of a tissue-specific stem cell (TSC). Lineage tracing of tracheobronchial basal cells after naphthalene (NA) injury or in the postnatal period demonstrated that basal cells were MP progenitors for Clara-like and ciliated cells. These studies, as well as reports of spatially restricted, label-retaining basal cells, and MP differentiation by human bronchial cells support the hypothesis that a TSC maintained and repaired the tracheobronchial epithelium. However, differences in basal cell phenotype (keratin [K] 5+ versus K14+), age (postnatal versus adult), health status (normal versus injured), and injury type (acid, detergent, NA) limited comparisons among studies and thus diminished the strength of the TSC argument. The finding that K14 was up-regulated after NA injury was a caveat to our previous analysis of reparative (r)K14-expressing cells (EC). Thus, the present study lineage traced steady-state (s)K14EC and evaluated differentiation potential in the normal and repairing epithelium. We showed that sK14EC were unipotential in the normal epithelium and MP after NA, sK14EC-dervied clones were not restricted to putative TSC niches, sK14EC cells were a direct progenitor for Clara-like and ciliated cells, MP-sK14EC clones accumulated over time, and sK14EC-derived Clara-like cells were progenitors for ciliated cells.
Keywords: basal, clara-like, ciliated, differentiation potential, lineage tracing, tissue-specific stem cell
CLINICAL RELEVANCE.
We used lineage tracing methods to demonstrate that progenitor–progeny relationships are altered by epithelial injury in vivo. These data support a model in which multipotential differentiation of the basal cell progenitor is regulated by environmental influences. Development of interventions that promote multipotential differentiation has the potential to enhance epithelial repair that is mediated by endogenous progenitor cells.
A tissue-specific stem cell (TSC) is defined as a long-lived cell that cycles infrequently and has a differentiation potential equivalent to the cellular diversity of its home tissue (1). Differentiation potential is evaluated by lineage tracing, a method that introduces a genetic tag into a cell that is defined by expression of a specific gene. Clones of genetically tagged cells are then evaluated for the presence of differentiated cell types. Clones that contain all of the differentiated cell types that are characteristic of a tissue are termed “multipotential” (MP).
The adult mouse trachea serves as a model for evaluation of the cellular mechanisms that regulate repair of the human tracheobronchial (TB) epithelium. Like the human TB epithelium, the mouse tracheal epithelium is pseudostratified and is composed of basal cells (∼30%), secretory cells (∼35%), and ciliated cells (∼35%). The secretory epithelial cell is termed “Clara-like cell” and has limited structural and functional similarity to the bronchiolar Clara cell (2). These differences may be a consequence of Clara-like cell derivation from a basal cell progenitor (3). Histological markers used to define tracheal epithelial cell types are keratin (K)5 and -14 for basal cells, Clara cell secretory protein (CCSP) for Clara-like cells, and acetylated tubulin (ACT) for ciliated cells.
Several studies supported the argument that a TSC repaired the TB and tracheal epithelium. Engelhardt demonstrated that human bronchial basal cells were a MP progenitor for the submucosal gland (SG) and surface epithelium (4, 5). Lineage tracing analysis after naphthalene (NA) injury demonstrated that the adult mouse K14-expressing cell (K14EC) served as a MP progenitor (6, 7). Hogan and colleagues (8) extended these studies to the developing trachea by lineage-traced K5-expressing cells (K5EC). They demonstrated that K5EC were MP progenitors during the postnatal growth period. Randell and colleagues used the label-retention assay (9, 10) to evaluate the cell cycle frequency of basal cell progenitors that proliferated in response to acid- or detergent injury (11). The label-retaining cells were limited to the SG duct junctions (SGDJ) and intercartilagenous (ICR) regions of the tracheal epithelium. These data suggest that a spatially restricted subset of basal cells served as a TSC. Although compelling, differences in basal cell phenotype (K5+ versus K14+), age (postnatal versus adult), health status (normal versus injured), and injury models (acid, detergent, NA) limited comparisons among studies and thus the strength of the TSC argument. Below, we define the outstanding issues that were addressed in this study.
Basal cells are typically thought of as K5/K14 dual-positive cells (12). However, we demonstrated that only 20% of adult basal cells were K14+ and that 100% of basal cells express K14 after NA injury (13). Thus, differences in basal cell phenotype prevented comparison of studies that lineage traced K14EC (6, 7) and K5EC (8). Additional differences in these studies included age (adult versus postnatal) and injury status (normal versus NA injury). To remedy this situation, we lineage traced sK14EC and determined their differentiation potential in the normal and repairing epithelium.
A hallmark of TSC is sequestration in a protective niche (14). The label retention study (11) demonstrated that cells with the TSC characteristic of infrequent cell division were located in the SGDJ and ICR regions. However, we demonstrated that reparative basal cells were uniformly distributed along the proximal to distal axis of the trachea after NA injury (13). These data, in combination with detection of multiple rK14EC-derived clone types, caused us to question whether certain clone types were spatially restricted while others were uniformly distributed. To evaluate this issue, we determined the location of sK14EC-derived clones in the normal and repairing epithelium.
Multiple mechanisms may lead to the generation of a MP clone. Reevaluation of MP rK14EC-derived clones demonstrated that they contained numerous basal cells but few ciliated and Clara-like cells. The latter two cell types were segregated from each other. These data suggest that an individual basal cell differentiated directly into a ciliated cell while a second basal cell was the progenitor for the Clara-like cell. These cell progenitor/progeny relationships were evaluated by clone analysis over time in vivo. Lineage tracing in air–liquid interface (ALI) cultures (15) allowed analysis of an enriched population of basal cells and temporal separation of basal and Clara-like cell differentiation.
Lineage tracing studies typically focus on a the MP clone. However, rK14EC generated MP clones as well as bipotential clones (containing basal and ciliated cells) and unipotential clones (containing only basal cells) (6, 7). Whether the three K14EC-derived clone types represented subclasses of K14EC or a sequence of differentiation intermediates was not determined. We addressed this issue by quantifying sK14EC-derived clones in the steady state and as a function of time after single or repeated NA injury.
Previous analyses demonstrated that the Clara-like cells and Clara cells were a progenitor for ciliated cells in the normal epithelium and after injuries that targeted the ciliated cell (16). The finding that rK14EC were the progenitor for ciliated cells after NA injury raised the possibility that Clara-like cells were not a ciliated cell progenitor after NA injury. This question was addressed by lineage-tracing nascent Clara-like cells (17) in vitro and in vivo.
MATERIALS AND METHODS
Methods are presented in greater detail in the online supplement.
Animal Care and Treatment
All procedures involving animal use were reviewed and approved by the National Jewish Health Animal Care and Use Committee. Mice were 6 to 8 weeks old.
Lineage Tracing of sK14EC
A ligand-regulated Cre recombinase system was used to evaluate differentiation potential of sK14EC (6, 7).
Experimental Design
Figure 1 illustrates the experimental design. The no-exposure group (Group 1) included untreated monotransgenic mice (Figure 1A). In the single NA exposure group, mono- or bitransgenic mice were treated with intraperitoneally with 4 mg tamoxifen (TAM) in corn oil. Group 2 mice (n = 4) were the unexposed control. Tissue was recovered on Day 20. Groups 3 and 4 were used to evaluate sK14EC differentiation potential after a single NA exposure. Mice were exposed to 275 mg/kg NA on Day 0 (21-d TAM washout period). The extent of NA-mediated injury and repair was assessed as previously reported (13) (see the online supplement). Groups 3 (n = 4 mice) and 4 (n = 4) were killed on recovery Days 20 and 40, respectively. Mice in the double NA exposure group (Figure 1C) were used to evaluate sK14EC differentiation potential after repeated NA exposure. All mice were treated with TAM. Groups 5, 6, and 7 were exposed to 275 mg/kg NA on Day 0. Group 5 (n = 3) was the single exposure control and was killed on recovery Day 58. Groups 6 (n = 4) and 7 (n = 3) were reexposed to 275 mg/kg NA on recovery Day 58. Group 6 was killed on Day 80, 22 days after the second NA exposure. Group 7 was killed on Day 120, 62 days after the second NA exposure.
Figure 1.
Experimental design. (A) No-exposure group. (B) Lineage tracing of steady-state keratin 14–expressing cells (sK14EC) in untreated mice (Group 2) and in naphthalene (NA)-treated mice that were recovered at 20 days (Group 3) or 40 days (Group 4). (C) Lineage tracing of sK14EC after a repeated NA exposure. Single NA exposure control (Group 5), repeated NA–exposure with a 22-day recovery (Group 6), or a 62-day recovery (Group 7). n = sample size; TAM = tamoxifen.
Lineage Tracing of Nascent CCSP-EC
A cell type–specific Cre recombinase system was used to evaluate the differentiation potential of nCCSP-EC (17).
β-Galacosidase Detection
The β-galactosidase (β-gal) reporter was detected using standard X-galactosidase (X-gal) staining methods (6, 7).
Histological Detection of Differentiation Markers
Differentiation markers were detected using standard immunohistochemical and immunofluorescence methods (18).
Quantification of Lineage-Traced Clones
Lineage-traced clones were evaluated by two observers who were blinded to the experimental groups. Clones identified on at least four adjacent serial sections were analyzed. Clones were scored for the presence of at least one cell that coexpressed the β-gal reaction product and CCSP, the ciliated cell marker acetylated tubulin (ACT), or the basal cell markers K14 or K5. Representation of each colony type was expressed as a percent of all clones. The following numbers of clones were analyzed: steady state: n = 24; single NA exposure + 20-day recovery: n = 32; double NA exposure + 22-day recovery: n =40; double NA exposure + 62-d recovery: n = 109.
ALI Cultures
Mouse tracheal cell cultures (15) were generated from wild-type FVB/n, K14-cre:ert/RS, or CC10-cre/RS trachea.
Lineage Tracing In Vitro
Recombination was induced in K14-cre:ert/RS cultures by exposure to 1 μg/ml 4-hydroxy TAM (4-OH-Tam) (CalBiochem, Gibbstown, NJ) in ethanol on culture Days 1 through 5. 4-OH-Tam is the active form of TAM. Control cultures were treated with 95% ethanol.
RESULTS
Differentiation Potential of sK14EC
sK14EC were genetically tagged using a modification of previously described lineage-tracing methods (6, 7). Untreated wild-type mice or bitransgenic mice exposed to corn oil exhibited β-gal activity in peritracheal glands (Figures 2A and 2B, white arrows). This proximally restricted β-gal activity was detected in all mice regardless of transgene or exposure status. This pattern reflects endogenous enzyme activity. β-gal activity was not detected in other regions of wild-type or untreated bitransgenic trachea (Figures 2A and 2B).
Figure 2.
Lineage tracing of sK14EC without and with a single NA exposure. White arrows: endogenous β-galactosidase (β-gal) activity. Black arrows: Single β-gal+ cells. Red arrows: Single-layered group of β-gal+ cells. Yellow arrows: Multilayered regions of β-gal+ cells. Green arrows: Cuboidal cells that are negative for differentiation markers. Blue arrows: Example of a clone that did not span adjacent sections. (A–D) Groups 1 through 4. Genotype is indicated in each panel. β-gal activity (blue) in tracheal tissue whole mounts at 10× magnification. (E–G) Group 4. 30× magnification. Asterisks indicate intercartilaginous (ICR) regions. (H–J) Immunohistochemical analysis (brown stain) of differentiation markers on histological sections at 200× magnification. (H) Keratin 5. (I) Acetylated tubulin. (J) Clara cell secretory protein (CCSP). 1, 2, 3, and 3′ indicate clones referred to in Results. (K) Quantification and distribution of clone types (group 4). Tracheal subregions: ICR (black), midcartilagenous (MCR, gray).
Tamoxifen treatment of steady-state bitransgenic mice resulted in recombination in single cells (Figure 2B, black arrows) or in clusters of three to five cells in the distal trachea (Figure 2B, red arrow). All recombined cells were positive for K5 (data not shown). Histochemical analysis demonstrated that lineage-traced cells were adjacent to the basement membrane and were in single-layered regions and that the cells had basal cell morphology. These data indicate that sK14EC are a self-renewing population that was involved in maintenance of the K5+ basal cell population.
Differentiation Potential of sK14EC after NA Injury
The differentiation potential of sK14EC was evaluated after NA-mediated injury and a 20- or 40-day recovery period (Figures 2C–2J). Comparison of recombination in uninjured and NA-injured tracheal tissue demonstrated that clone size increased in response to treatment (Figure 2B versus 2C).
Because previous analysis indicated that label-retaining basal cells were limited to the SMDJ and the ICR (11), the distribution of tagged clones was determined. sK14EC-derived clones were located throughout the trachea, including the ventral-lateral surface from cartilaginous ring 3 to the carina, the dorsal membranous region, and the bronchi. Clones spanned adjacent ICR and midcartilaginous (MCR) zones and were in single-layered or multilayered regions (Figure 2C, yellow arrow).
Clones persisted in all regions of the trachea for at least 40 days (Figure 2D). However, the size of the tagged clusters decreased between recovery Days 20 and 40. Clones were detected throughout the surface epithelium but were more frequent in MCR (72%) than in ICR (28%). Clones that spanned adjacent ICR and MCR zones were detected (Figure 2E) and were present in single-layered and multilayered structures on the ventral surface (Figure 2E, yellow arrow), on the dorsal surface (Figure 2F), and in the bronchi (Figure 2G). Recombined cells were detected in SG ascini and ducts (data not shown). However, recombined cells were not detected in the surface epithelium adjacent to the glands. These data indicate that SG myoepithelial cells did not contribute to epithelial repair after NA injury.
Four types of lineage-traced clones were detected on recovery Day 40. Nineteen percent of clones contained basal, secretory, and ciliated cells (Figures 2H–2J; clone 1) and were classified as multipotential (Figure 2K). Twenty-five percent of clones contained basal and secretory cells (Figures 2H–2J; clone 2) and were classified as bipotential: basal-secretory (Figure 2K). Six percent of clones contained basal and ciliated cells and were classified as bipotential: basal-ciliated (Figure 2K). Fifty percent of clones contained only basal cells (Figures 2H–2I; clone 3, 3′) and were classified as unipotential-basal (Figure 2K). Some basal-only clones spanned the abluminal–luminal axis and contained cells with cuboidal morphology (Figures 2H–2J, green arrows). In contrast with previous analysis of rK14EC (6, 7), sK14EC did not generate bipotential clones containing only secretory and ciliated cells. None of the four clone types was localized specifically in the ICR or MCR regions of the dorsal trachea (Figure 2K, black versus gray bars).
Direct sK14EC to Ciliated Cell Differentiation
Evaluation of the MP clones generated by rK14EC (6, 7) or sK14EC (Figures 2H–2J; clone 1, brown arrows) demonstrated that ciliated and Clara-like cells were rare constituents of the clone and that these two cell types were not adjacent to each other. These data suggest that mouse rK14EC and sK14EC, like human basal cells (4, 5), differentiated directly into ciliated cells. However, differentiation of ciliated and Clara-like cells overlapped in time and space (13), making analysis of cellular mechanisms difficult. Consequently, the progenitor–progeny relationships leading to ciliated cell replacement were evaluated using tracheal ALI cultures (15).
The kinetics of proliferation and differentiation in ALI cultures were evaluated using dual immunofluorescence staining methods (see Figure E1 in the online supplement). The cultures were composed primarily of K5+ basal cells and a rare subset of K14+ cells on culture Days 2 and 3 (Figures E1A and E1B). Cell density increased over time, and a confluent monolayer of K5+ basal cells was established on culture Day 5 (Figure E1C). Most cells were Ki67+ on culture Day 5, indicating extensive proliferation (Figure E1D). Proliferation diminished greatly by culture Day 7, and mitotic cells were concentrated at the culture edge from this time point through culture Day 17 (data not shown). Rare CCSP+ (Figure E1E) were detected on culture Day 3. Many of these Clara-like cells were abnormally large, suggesting cell death (Figure E1E). No CCSP+ cells were detected on culture Day 10 (Figure E1F), but this cell type was detected on culture Days 14 (Figure E1G) and 17 (Figure E1H). Rare ACT+ ciliated cells were detected on culture Day 3 (Figure E1I). Numerous ACT+ cells were detected on culture Days 10 to 17 (Figures E1J–E1L). These data indicated that ciliated cell and Clara-like cell differentiation could be temporally separated in vitro: Ciliated cells differentiated by culture Day 10, whereas Clara-like cells differentiated on culture Day 14.
To evaluate K14EC differentiation, ALI cultures were generated from K14-cre:ert/RS mice. Because TAM cannot be metabolized to its active hydroxy-form in vitro, cultures were treated with 4-OH-Tam (Figure E2). Recombination was not observed in vehicle-treated cultures (Figure E2B). Range-finding experiments demonstrated that maximal recombination occurred when 4-OH-Tam was supplied throughout the proliferation period (culture Days 0–5) (Figure E3A). This induction period was used for subsequent analyses. Clones were distributed throughout the culture (Figures E2A and E3B). The average number of cells per clone increased slightly during the differentiation period, but the changes were not statistically significant (Figures E2C–E2F and E3C–E3F).
Lineage-traced K14EC were detected on culture Day 5, but these cells did not express ACT or CCSP (Figures 3A and 3B; X-gal reaction product is pseudocolored white [white arrows]). On culture Day 11, lineage-traced K14EC expressed ACT (Figures 3C and 3D, red arrows) but did not express CCSP. These data supported the conclusion that basal cells differentiated directly into ciliated cells. On culture Day 17, lineage-traced cells expressed ACT or CCSP (Figures 3E and 3F). These data indicated that K14EC also differentiated into Clara-like cells.
Figure 3.
-Lineage tracing of K14EC in vitro. (Top) Experimental design. Air–liquid interface cultures were generated from K14-cre:ert/RS trachea. The 20-day culture period is divided into two phases: proliferation (Days 0–5) and differentiation (Days 6–20). Differentiation was initiated on culture Day 5. Ciliated cells were first detected on culture Days 6 and 7. Clara-like cells were detected on culture Days 9 and 10. (A–F) Analysis of K14EC differentiation as a function of time. β-gal (white pseudocolor) and differentiation markers (color code indicated at the top of each column). (A and B) Proliferation stage, Day 5. (C and D) Differentiation stage, Day 11. (E and F) Differentiation stage, Day 17. All images are en face views at 200× original magnification. A', 400× magnification of the region indicated by the arrows in A. C', 400× magnification of the region indicated by the arrows in C. E', 400S magnification of the region indicated by the green arrows in E. White arrows: β-gal+ cells. Red arrows: β-gal/ACT dual-positive cells. Green arrows: β-gal/CCSP dual-positive cells.
Differentiation Potential of sK14EC after Repeated NA Exposure
The in vivo and in vitro lineage-tracing studies suggested that ciliated and Clara-like cells were acquired as a function of time after NA injury. Thus, the representation of multipotential colonies might also increase as repair progresses. To address this issue, sK14EC were lineage traced in the steady state. These mice were then treated with NA, recovered after 58 days, and treated with a second dose of NA (see Figure 1C). These mice lost 25% of their body weight within 2 days of the second NA exposure, indicating effective secondary injury. Lineage-tagged clones similar to those detected in the single injury model were detected on recovery Days 22 (Figure E4) and 62 (Figure E5). β-gal+ cells were not identified between cartilaginous ring 3 and the carina in monotransgenic tracheas (Figures E4 and E5) at either time point.
Three of four bitransgenic tracheas contained X-gal+ clones on recovery Day 22 (Figures E4A–E4D). These clones were located mainly in the ventral/distal (Figure E4A), dorsal/distal (Figure E4B), and lateral surfaces (Figures E4C and E4D). Smaller clones of cells were detected throughout the trachea. On recovery Day 62, all bitransgenic tracheas contained β-gal+ clones in the ventral (Figure E5A), dorsal (Figure E5B), and lateral (Figures E5C and E5D) surfaces. These clones were similar in size to those detected at the 22-day recovery time point (compare Figures E4A–E4D with Figures E5A–E5D).
The differentiation potential of sK14EC-derived cells after single or repeated NA exposure was evaluated by immunohistochemical staining of serial adjacent cross-sections. Examples of the major clone types detected 44 days after the second NA injury are shown in Figure E6. As previously demonstrated for the single-injury model (see Figures 1H–SJ), multipotential clones in twice injured tracheas contained nonadjacent ciliated and Clara-like cells (Figures E6J–E6L). These data indicated that direct basal to ciliated and basal to Clara-like cell differentiation contributed to epithelial repair after a second NA injury.
Location of Clones along the Proximal to Distal Axis
Previous analysis suggested that MP cells were limited to the SGDJ and ICR (11). After single or repeated NA injury, MP, bipotential: basal-ciliated, and bipotential: basal-secretory clones were located throughout the tracheal epithelium (Figure 4A). Unipotential: basal, unipotential: secretory, and unipotential-ciliated cell clones were too infrequent to accurately determine their distribution. Bipotential and MP sK14EC clones were biased to the proximal trachea. This region also contained the majority of all clone types. Consequently, the spatial skewing could be a function of prevalence rather than spatial restriction.
Figure 4.
Differentiation of sK14EC after repeated NA exposure. See Figure 1C for experimental design. (A) Distribution of clone types along the proximal to distal axis of the trachea 62 days after the second NA exposure. Cross-sections through three or four complete trachea were evaluated. (B) Quantification of the clone types detected in the steady state (black), after a single NA exposure and a 40-day recovery (NA-1X: long term, gray), after a second NA exposure and a 22-day recovery (NA-2X: short-term, stippled) or a 62-day recovery (NA-2X: long-term, hatched).
Frequency of Clone Types as a Function of Time
Representation of the various clone types was compared 22 and 62 days after the second NA injury (Figure 4B). The fraction of MP clones increased approximately 2-fold over this interval. Bipotential: basal-ciliated and unipotential: ciliated clone representation was not changed. Bipotential: basal-secretory clones decreased approximately 5-fold, and unipotential: basal clones decreased approximately 2-fold. These data suggest that sK14EC differentiation to ciliated and secretory cells increased over time.
Restoration of the Clara-Like Cell Progenitor Cell Pool after NA Injury
To determine whether the Clara-like/ciliated cell lineage was restored after NA-mediated injury, Clara-like cells were lineage traced in vivo. These experiments used the previously validated CC10-cre/RS model (17). CC10 is synonymous with CCSP and SCGB1A1. Monotransgenic mice were β-gal− (Figure E7). The CCSP-dependent lineage tag was detected in steady-state Clara-like and ciliated cells (Figures 5A and 5B). These data confirmed the previously reported lineage relationship between these two cell types (16, 19). The lineage tag was also detected in less than 2% of luminal K5+ basal cells (Figures 5C and 5D). These data are consistent with the report that rare cell types coexpress K14 and CCSP (6) and with results from Rock and colleagues (8). K14+ cells were infrequent in the normal CC10-cre/RS trachea (Figures 6C and 6D); however, rare regions containing K5+/K14+ cells (Figures 6E and 6F) were detected. These K5+/K14+ cells, as well as adjacent hyperplastic K5+/K14− cells, were β-gal−. The CCSP-dependent lineage tag was not detected in SG cells (data not shown).
Figure 5.
Lineage tracing of CCSP-expressing cells in vivo. Bitransgenic mice harboring a CC10-cre transgene and the RS recombination substrate were evaluated in the steady state and 13 days after NA exposure. Recombined cells and their progeny were analyzed by β-gal staining and immunohistochemical detection of differentiation antigens. (A–F) Paired images showing codetection of β-gal (white, pseudocolored brightfield) and differentiation antigens (fluorescence) in sections from untreated CC10-cre/RS mice. (A, C, E) Merged brightfield and dual immunofluorescence images. (B, D, F) Dual immunofluorescence images. Antigens and the detection color are indicated within each panel. Arrows are color coded according to the antigen and serve as fiduciary points for images A and B, C and D, and E and F. Scale bar, 50 μm. (G–J) Codetection of β-gal (blue) and differentiation antigens (brown) in sections from NA-treated CC10-cre/RS bitransgenic mice. Antigens and detection color are noted in each panel. Black arrows: β-gal− basal cells. Blue arrows: β-gal+ cells that coexpress CCSP or ACT. Scale bar, 50 μm.
Figure 6.
Lineage tracing of nascent CCSP-expressing cells in vitro. ALI cultures were generated from CC10-cre:RS trachea. Differentiation kinetics were similar to those indicated in Figure 3. Cells were stained for β-gal activity and coexpression of differentiation antigens at the indicated culture time points. (A) The percentage of lineage-traced cells that coexpressed CCSP was quantified as a function of time in culture. Mean ± SEM (n = 3). *P = 0.005 with respect to Day 5. (B) Percentage of lineage-traced cells that coexpress ACT. Mean ± SEM (n = 3). *P = 0.005 with respect to Day 4.
To determine whether nascent CCSP+ cells served as a progenitor for ciliated cells after tracheal injury, CC10-cre/RS mice were exposed to 275 mg/kg NA and recovered after13 days. NA injury resulted in 95% depletion of tracheobronchial Clara-like and ciliated cells on recovery Day 3 (data not shown), which was similar to previous analysis (13). On recovery Day 13, nascent Clara-like (Figure 5I) and ciliated cells (Figure 5J) were detected, and both cell types were β-gal+. These data indicated that Clara-like to ciliated differentiation was reestablished after NA injury. Neither K5+ (Figure 5G) nor K14+ (Figure 5H) basal cells were positive for β-gal. These data indicated that lineage-traced CCSP+ and ACT+ cells did not contribute to the basal cell pool after NA injury.
Clara-Like to Ciliated Cell Differentiation In Vitro
To determine whether Clara-like cells served as a progenitor for ciliated cells in vitro, ALI cultures were generated from CC10-cre/RS mice. Preliminary studies demonstrated that cultures were 99% basal cells on culture Day 5 (data not shown) and that rare (< 0.5%) lineage-traced cells were detected on culture Days 4 and 6 (Figures E8A and E8B). Two to ten percent of the lineage-traced cells expressed CCSP or ACT (Figures 6A and 6B), and many of the CCSP+ cells were abnormally large, suggesting cell death. A 3-fold increase in CCSP+/β-gal+ cells was detected between culture Days 6 and 8 (Figures E8B versus E8C; Figure 6A). Because ALI were approximately 95% basal cells on culture Days 4 through 6 (Figure E1), these data supported the previous conclusion (see Figure 3) that CCSP+ cells were derived from a basal cell. On culture Days 8 through 25, approximately 30% of lineage-traced cells were CCSP+ (Figures 6A and E8E–E8G). These data indicated that the majority of basal cell–derived CCSP+ cells accumulated after basal cell–derived ciliated cells.
Differentiation of Clara-like cells into ciliated cells was analyzed on culture Days 4 through 20 (Figure E8H). On Days 4 through 6, rare β-gal+/ACT+ cells (Figure 6B) were detected, and the representation of these cells was similar to that of β-gal+/CCSP+ cells (Figure 6A). A 5-fold increase in the number of lineage-traced ciliated cells was detected between culture Days 8 and 11. From this point through culture Day 20, approximately 50% of ciliated cells were β-gal+. These data indicated that half of the ciliated cells were derived from a CCSP-expressing progenitor.
DISCUSSION
In the present study, sK14EC were lineage traced to overcome a caveat to our previous analysis of rK14EC (i.e., up-regulation of K14 after NA injury). We provide the following conclusions: (1) The differentiation potential of sK14EC expanded from basal-only to MP after NA treatment. The differentiation potential of sK14EC after NA and rK14EC was similar. The only exception was that sK14EC did not generate Clara-like/ciliated only clones. (2) sK14EC-derived clones were not spatially restricted to putative TSC niche. In the normal trachea, sK14EC-dervied clones were more frequent in MCR than in ICR. After NA, the clones were similarly distributed to the MCR and ICR and were not found in SGDJ. A proximal bias was not noted. (3) sK14EC were a direct progenitor for ciliated cells and for Clara-like cells after NA. Thus, novel lineage relationships were established after injury. (4) The MP sK14EC-derived clone type was enriched over time and after a second NA injury. These results suggested that clones accumulated differentiated cell types over time. Therefore, the various clone types were likely to be part of a differentiation series. (5) The Clara-like cell progenitor was reestablished after NA. These results indicated that ciliated cells within the repaired epithelium were derived from two precursor cell types, the basal and the Clara-like cell.
Expansion of sK14EC Differentiation Potential after NA Injury
The increased differentiation potential of sK14EC suggested that sK14EC differentiation was repressed in the steady state. Previous analysis demonstrated that the Clara-like cell was the ciliated cell progenitor after NO2 or O3–mediated injury (20). Because the basal cell did not contribute to the nascent ciliated cell population under these conditions, these data suggest that ciliated cells did not regulate basal cell differentiation. Similarly, generation of ciliated cells in vitro did not block basal to Clara-like cell differentiation. These data suggest that the Clara-like cell is a negative regulator of sK14EC differentiation potential.
Several signaling pathways may limit the basal cell progenitor to self-renewal. Evans and colleagues suggested that O3 injury results in the release of FGF from the extracellular matrix (21). Rock and colleagues identified Notch signaling (8). In humans, the distribution of EGF- and EGFR-positive cells changes after injury (22). Although the present study did not evaluate these factors, it did identify basal cell differentiation potential as an outcome measure that could be used to test functions for these signaling pathways in genetically modified mice or in vitro.
sK14EC-Derived Clones Are Not Localized to the Putative TSC Niche
In contrast with previous analysis of label-retaining basal cells, the present study indicated that MP sK14EC were distributed throughout the tracheal epithelium. Preliminary label retention analysis in the NA model demonstrated that slow-cycling cells were located throughout the proximal–distal axis of the trachea after NA injury (M. Ghosh, unpublished observations). Differences between these analyses may be related to the extent of injury (23).
The broad distribution of MP cells reported herein is similar to that described for the interfollicular epidermis (24, 25) and to reports that gut stem cells maintain only a portion of the crypt–vilus axis (26). These data suggest that a TSC is responsible for the maintenance and repair of a relatively small area of the epithelium. In each of these tissues, the TSC niche is defined by the location of the TSC. However, a precise anatomical location was not defined. These observations are consistent with our preliminary finding that purified TSCs create their own niche in vitro (M. Ghosh, unpublished observations).
Direct Differentiation of Basal Cells to Ciliated or Clara-Like Cells
Direct differentiation of basal cells to ciliated cells, in the absence of a secretory intermediate, was indicated by detection of basal-ciliated clones after a single NA injury (see Figure 2). This interpretation was substantiated by K14 lineage tracing in vitro (see Figure 3). However, these studies could not eliminate participation of a transient Clara-like cell progenitor. The observation that the first cohort of ciliated cells generated in the CC10-cre/RS ALI studies (see Figure 4) were untagged supported a direct basal to ciliated cell differentiation pathway.
Direct basal to Clara-like cell differentiation was supported by the presence of basal-secretory clones in vivo (see Figure 2). The late appearance of tagged CCSP+ cells in the K14-cre:ert ALI model substantiated this finding (see Figure 3). These data indicated that basal cells were a direct progenitor for Clara-like cells. Lineage tracing using the CC10-cre/RS model in vivo (see Figure 5) and in vitro (Figure 6) indicated that the basal cell–derived Clara-like cells were a progenitor for a second set of ciliated cells.
Demonstration that the Clara-like progenitor cell pool was established late in culture suggested that a population of “early” ciliated cells was generated directly from the basal cell progenitor. In contrast, “late” ciliated cells are a product of Clara-like to ciliated cell differentiation as well as continued basal to ciliated cell differentiation. These data suggest that two pools of ciliated cells are present after injury: those derived from the basal cell and those descended from the Clara-like cell.
It is unknown whether the two cohorts of ciliated cells are functionally equivalent. However, it is known that bronchial and bronchiolar ciliated cells beat with distinct frequencies (27). This physiological difference is thought to be a functionally important component of mucociliary clearance. Analysis of human bronchial cells in vitro demonstrated that taste receptors are located on motile cilia and that receptor subtypes were located on distinct cells (28). These studies indicate greater diversity within the ciliated cell population than previously appreciated (29). We suggest that part of this diversity is a consequence of lineage relationships between the ciliated cell and the basal or Clara-like progenitor cell types.
The Multipotential Phenotype Is Acquired over Time
The lineage tracing data provided evidence for MP sK14EC after a single or repeated NA injury (see Figure 4). However, as shown for lineage-traced rK14EC, most derivatives of a sK14EC were basal cells (Figure E6). Only rare cells within the clone expressed ciliated cell or Clara-like cell markers. Furthermore, the frequency of basal to ciliated or basal to Clara-like cell differentiation increased as a function of time after repeated injury (see Figure 4). This increase was due in part to the reestablishment of a Clara-like cell progenitor, which then generated ciliated cell progeny. However, ongoing direct differentiation of basal cells to ciliated cells was also likely, as indicated by maintenance of the basal-ciliated clone type frequency. These data suggest that a sK14EC-derived colony acquired differentiated cell types over time.
The frequency of bipotential and MP sK14EC-derived colonies also increased after a second NA exposure. Because body weight loss and gene expression parameters indicated the two injuries were equivalent, it was unlikely that a population of NA-resistant Clara-like and ciliated cells survived the second NA exposure. It is possible that initial NA injury resulted in selection of a multipotential basal cell progenitor. However, ciliated cells and Clara-like cells remained rare constituents of all colony types even after the second injury. Limited representation of ciliated or Clara-like cells within each clone type indicated that basal cell differentiation was a consequence of cellular context rather than an intrinsic property of a specific subtype of sK14EC. These data suggest that MP differentiation was regulated at the cellular level.
Context-Dependent Regulation of Tracheal Basal Cell Differentiation
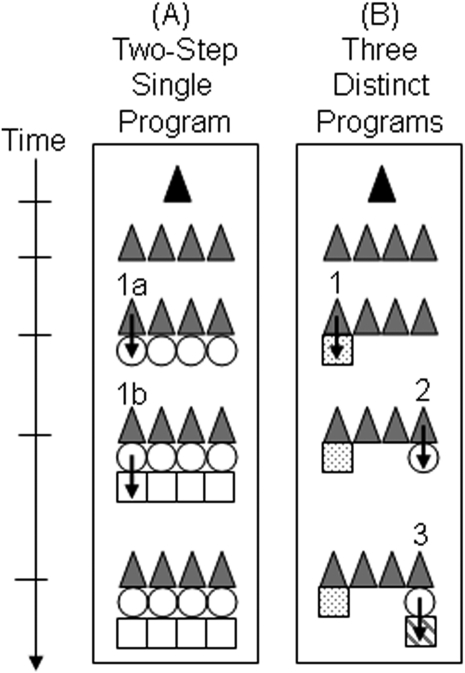
The “epidermal-influenced” model of multipotential differentiation (30) (Figure 7A) predicts that a multipotential clone contains uniformly distributed differentiated cells. This model suggests the existence of a single differentiation program that proceeds via two phases: phase 1a, replacement of an intermediate cell type and phase 1b, replacement of terminally differentiated cells.
Figure 7.
Models of multipotential differentiation. Time after NA injury is represented by the vertical axis. Black triangles: Multipotential basal cell. Gray triangles: Progeny of the multipotential basal cell. Circles: Clara-like cell. Squares: Ciliated cell. Arrows: Progenitor–progeny relationship. (A) A single differentiation program with two steps that are labeled 1a and 1b. This model is refuted by the data presented herein. (B) Three differentiation programs result in replacement of ciliated and Clara-like cells. Program 1: basal–to–ciliated cell differentiation (1). Program 2: basal–to–Clara-like cell differentiation (2). Program 3: Clara-like–to–ciliated cell differentiation (3). This model is supported by the present data set.
The results of this study indicate that the epidermal differentiation model cannot be applied to replacement of ciliated and Clara-like cells after NA exposure. Multipotential clones derived from sK14EC contained numerous basal cells and rare differentiated cell types (Figure 7B). This observation suggested that two distinct programs regulated basal cell differentiation into ciliated (program 1) or Clara-like (program 2) cells. Finally, the observation that Clara-like to ciliated differentiation was reestablished after NA injury indicated that a third differentiation program participated in epithelial repair.
Supplementary Material
This work was supported by funding from the Asthma Foundation of Western Australia and the NHMRC Centre of Research Excellence in Pulmonary and Environmental Medicine and by NIH Award Number R01HL075585 (S.D.R.).
This article contains an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2010-0283OC on December 3, 2010
Author Disclosure: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Morrison SJ. Stem cell potential: can anything make anything? Curr Biol 2001;11:R7–R9. [DOI] [PubMed] [Google Scholar]
- 2.Stripp BR, Reynolds SD. Clara cells. In: Shapiro S, Laurent G, editors. Encyclopedia of respiratory medicine. St. Louis, MO: Elsevier; 2006. pp. 471–477.
- 3.Stripp BR, Reynolds SD. Maintenance and repair of the bronchiolar epithelium. Proc Am Thorac Soc 2008;5:328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engelhardt JF, Schlossberg H, Yankaskas JR, Dudus L. Progenitor cells of the adult human airway involved in submucosal gland development. Development 1995;121:2031–2046. [DOI] [PubMed] [Google Scholar]
- 5.Engelhardt JF, Allen ED, Wilson JM. Reconstitution of tracheal grafts with a genetically modified epithelium. Proc Natl Acad Sci USA 1991;88:11192–11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. Basal cells are a multipotent progenitor capable of renewing the bronchial epithelium. Am J Pathol 2004;164:577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. In vivo differentiation potential of tracheal basal cells: evidence for multipotent and unipotent subpopulations. Am J Physiol Lung Cell Mol Physiol 2004;286:L643–L649. [DOI] [PubMed] [Google Scholar]
- 8.Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BL. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA 2009;106:12771–12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 1990;61:1329–1337. [DOI] [PubMed] [Google Scholar]
- 10.Cotsarelis G, Cheng SZ, Dong G, Sun TT, Lavker RM. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: implications on epithelial stem cells. Cell 1989;57:201–209. [DOI] [PubMed] [Google Scholar]
- 11.Borthwick DW, Shahbazian M, Krantz QT, Dorin JR, Randell SH. Evidence for stem-cell niches in the tracheal epithelium. Am J Respir Cell Mol Biol 2001;24:662–670. [DOI] [PubMed] [Google Scholar]
- 12.Hutton E, Paladini RD, Yu QC, Yen M, Coulombe PA, Fuchs E. Functional differences between keratins of stratified and simple epithelia. J Cell Biol 1998;143:487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole BB, Smith RW, Jenkins KM, Graham BB, Reynolds PR, Reynolds SD. Tracheal basal cells: a facultative progenitor cell pool. Am J Pathol 2010;177:362–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs E, Tumbar T, Guasch G. Socializing with the neighbors: stem cells and their niche. Cell 2004;116:769–778. [DOI] [PubMed] [Google Scholar]
- 15.You Y, Richer EJ, Huang T, Brody SL. Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am J Physiol Lung Cell Mol Physiol 2002;283:L1315–L1321. [DOI] [PubMed] [Google Scholar]
- 16.Evans MJ, Shami SG, Cabral-Anderson LJ, Dekker NP. Role of nonciliated cells in renewal of the bronchial epithelium of rats exposed to no2. Am J Pathol 1986;123:126–133. [PMC free article] [PubMed] [Google Scholar]
- 17.Reynolds SD, Zemke AC, Giangreco A, Brockway BL, Teisanu RM, Drake JA, Mariani T, Di PY, Taketo MM, Stripp BR. Conditional stabilization of beta-catenin expands the pool of lung stem cells. Stem Cells 2008;26:1337–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reynolds SD, Giangreco A, Power JH, Stripp BR. Neuroepithelial bodies of pulmonary airways serve as a reservoir of progenitor cells capable of epithelial regeneration. Am J Pathol 2000;156:269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rawlins EL, Okubo T, Xue Y, Brass DM, Auten RL, Hasegawa H, Wang F, Hogan BL. The role of scgb1a1+ clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell 2009;4:525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans MJ, Cabral-Anderson LJ, Freeman G. Role of the clara cell in renewal of the bronchiolar epithelium. Lab Invest 1978;38:648–653. [PubMed] [Google Scholar]
- 21.Evans MJ, Fanucchi MV, Baker GL, Van Winkle LS, Pantle LM, Nishio SJ, Schelegle ES, Gershwin LJ, Miller LA, Hyde DM, et al. Atypical development of the tracheal basement membrane zone of infant rhesus monkeys exposed to ozone and allergen. Am J Physiol Lung Cell Mol Physiol 2003;285:L931–L939. [DOI] [PubMed] [Google Scholar]
- 22.Voynow JA, Fischer BM, Roberts BC, Proia AD. Basal-like cells constitute the proliferating cell population in cystic fibrosis airways. Am J Respir Crit Care Med 2005;172:1013–1018. [DOI] [PubMed] [Google Scholar]
- 23.Engelhardt JF. Stem cell niches in the mouse airway. Am J Respir Cell Mol Biol 2001;24:649–652. [DOI] [PubMed] [Google Scholar]
- 24.Ghazizadeh S, Taichman LB. Organization of stem cells and their progeny in human epidermis. J Invest Dermatol 2005;124:367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen UB, Lowell S, Watt FM. The spatial relationship between stem cells and their progeny in the basal layer of human epidermis: a new view based on whole-mount labelling and lineage analysis. Development 1999;126:2409–2418. [DOI] [PubMed] [Google Scholar]
- 26.Wong MH, Saam JR, Stappenbeck TS, Rexer CH, Gordon JI. Genetic mosaic analysis based on cre recombinase and navigated laser capture microdissection. Proc Natl Acad Sci USA 2000;97:12601–12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salathe M. Regulation of mammalian ciliary beating. Annu Rev Physiol 2007;69:401–422. [DOI] [PubMed] [Google Scholar]
- 28.Shah AS, Ben-Shahar Y, Moninger TO, Kline JN, Welsh MJ. Motile cilia of human airway epithelia are chemosensory. Science 2009;325:1131–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinnamon SC, Reynolds SD. Cell biology: using taste to clear the air(ways). Science 2009;325:1081–1082. [DOI] [PubMed] [Google Scholar]
- 30.Blanpain C, Fuchs E. P63: revving up epithelial stem-cell potential. Nat Cell Biol 2007;9:731–733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.