Abstract
The murine surfactant-associated protein B (Sftpb) gene promoter, spanning nucleotides −653 to +42, is composed of functionally distinct proximal and distal regions. Although both regions contain consensus/putative activator protein 1 (AP-1) sites, the distal, but not the proximal, region mediates the inhibition by jun proto-oncogene (JUN) of Sftpb promoter activity. In transient cotransfection assays, JUN inhibited the luciferase reporter activity of plasmid constructs containing Sftpb promoter fragments that lacked the distal putative AP-1 site, indicating that another regulatory motif mediates JUN-dependent inhibition. Electrophoretic mobility shift assays and in silico analyses identified a DNA target sequence (Sftpb nucleotides −339 to −316) and transcription factors that regulate Sftpb promoter activity. The identified sequence contains a CCAAT/enhancer-binding protein (C/EBP) consensus recognition element. Mutation of the site reduced Sftpb promoter activity and sensitivity to inhibition by JUN. Purified recombinant JUN, which did not recognize the −339 to −316 target sequence when added alone, supershifted the mobility of in vitro translated C/EBP-α and C/EBP-β proteins complexed with the identified cis-regulatory element. These findings support the idea that heterodimerization between JUN and C/EBP-α and/or C/EBP-β targets JUN to the Sftpb promoter, thereby mediating its inhibitory regulatory role.
Keywords: surfactant protein B, acute lung injury, gene regulation, pulmonary surfactant metabolism dysfunction type 1, pulmonary alveolar proteinosis
CLINICAL RELEVANCE.
One of the key events in acute lung injury is the loss of functional surfactant. Thus, surfactant has long been investigated in the treatment of acute lung injury. However, surfactant replacement has yet to be demonstrated as an effective therapy for acute lung injury. An alternative strategy may involve the development of treatments that maintain or restore the production of endogenous surfactant. We analyzed the core surfactant-associated protein B (Sftpb) promoter to identify transcription factors and recognition sites that contribute to the inhibition of SFTPB.
The Sftpb gene product (SFTPB) is one of the protein components of pulmonary surfactant. The other protein components include SFTPA, SFTPC, and SFTPD. These proteins represent a small fraction (∼ 10%) of surfactant, and were initially considered contaminants (1). SFTPB and SFTPC constitute approximately 1% of the total surfactant mass (2). However, the surfactant-associated proteins, and in particular SFTPB, are essential for normal lung function and survival. Because SFTPB is essential for surfactant function, lamellar body formation, and the processing of SFTPC, SFTPB deficiency causes congential alveolar proteinosis and respiratory failure (3–6).
The expression levels of SFTPB may be reduced because of genetic disorders or during acute lung injury after exposure to toxicants. Recently, we reported that the maintenance of SFTPB expression is critical to survival during nickel-induced lung injury in mice (7). Nickel increased jun proto-oncogene (JUN) transcripts in murine lung and alveolar type II epithelial cells (MLE-15), and the induction of JUN inhibited Sftpb gene promoter activity. Studies aimed at understanding the molecular basis of SFTPB inhibition may thus advance our ability to develop strategies that reverse the loss of SFTPB during lung injury.
The 5′-flanking region of the murine Sftpb gene contains functionally important proximal (nucleotides −132 to −1) and distal (nucleotides −382 to −283) promoter regions (8). Both promoter regions contain a number of transcription factor recognition sites, including activator protein 1 (AP-1) recognition sequences. JUN-related proteins bind to and activate AP-1 regulatory elements in the promoter and enhancer regions of several mammalian genes (9). Despite the presence of a consensus AP-1 site in the Sftpb proximal promoter region and a putative AP-1 site in the distal promoter region, we found that the distal, but not the proximal, Sftpb promoter region mediated the JUN-dependent inhibition of promoter activity (7). The mechanism of the JUN-mediated inhibition of Sftpb transcription is unclear.
The transcription factor JUN participates in regulating a variety of biological processes, including cell proliferation, survival, apoptosis, tumorigenesis, tissue remodeling, and development (10–16). JUN controls these diverse processes through its ability to regulate the transcription and activity of numerous target genes and gene products. JUN, which was originally identified as AP-1 (17–19), belongs to a large family of proteins known as bZIP. These bZIP proteins are functionally related proteins with homologous sequences containing a basic DNA-binding domain and a leucine zipper region. JUN interacts with more than 50 related bZIP proteins and with structurally unrelated transcription factors, forming homodimeric and heterodimeric protein complexes (20, 21). The multiplicity of combinatorial JUN–protein interactions and the sequence compositions of DNA recognition sites determine target-gene specificity and regulatory selectivity in a cell type–dependent manner.
Another group within the bZIP transcription factor family is the CCAAT/enhancer-binding protein (C/EBP) subfamily. C/EBP subfamily members (α, β, and δ), in addition to C/EBP-γ and C/EBP-ζ, are expressed and play important roles in lung development, gene regulation, and acute lung injury (22–26). The deletion of C/EBP-α is perinatally lethal, in part because of lung abnormalities attributable to the hyperproliferation of alveolar type II cells (27). C/EBP-α–deficient newborn mice also exhibited increased surfactant associated protein A, B, and C mRNA, indicating a role for C/EBP-α in the control of alveolar cell proliferation/differentiation and the regulation of lung-specific target genes (28). In contrast, a deficiency of C/EBP-β and C/EBP-δ did not lead to lung abnormalities (29). However, the expression levels of C/EBP-β, C/EBP-δ, and C/EBP-regulated inflammatory mediators are increased in LPS-induced, bleomycin-induced, and oxidative stress–induced acute lung injury (26). Here, we investigated the role of C/EBP proteins in the inhibition by JUN of Sftpb promoter activity.
MATERIALS AND METHODS
Promoter Reporter Plasmid Constructs and Transient Transfection Assays
To analyze Sftpb promoter regulation, the region spanning nucleotides −653 to +42 (numbering according to Bruno and colleagues (8)) and its mutant variants were inserted into the luciferase reporter pGL4–10 (catalogue number E6651; Promega, Madison, WI). The plasmids pCMV6-XL4 and pCMV-Jun (pJun; Origene Technologies, Inc., Rockville, MD) and the luciferase reporters were transfected into MLE-15 cells (a gift of Dr. Jeffrey Whitsett) (30). The efficiency of transfection was normalized using pCMV-β–Gal. To determine promoter luciferase reporter activity, cells were lysed using Glolysis buffer, assayed in 96-well plates using the Bright-Glo luciferase or Beta-Glo systems (catalogue numbers E2610 and E4720, respectively; Promega), and luminescence was measured (Fusion α; Packard Bioscience/Perkin Elmer Life and Analytical Science, Waltham, MA) (see the online supplement for additional details).
Electrophoretic Mobility Shift Assays
To investigate protein–promoter binding, biotinylated oligonucleotides were used for electrophoretic mobility shift assays (EMSAs). A probe containing a binding site for AP-1 JUN homodimer and JUN/FOS heterodimeric complexes (5′-CGCTTGATGACTCAGCCGGAA-3′ annealed with 3′-GCGAACTACTGAGTCGGCCTT-5′) served as a positive control for purified recombinant JUN binding. Nuclear protein extracts (2.5 μg) were incubated with biotinylated probes and competitors, and were analyzed (see the online supplement for additional details).
Chromatin Immunoprecipitation Assay
To examine whether the bZIP proteins D site albumin promoter binding protein (DBP), C/EBP-α, and C/EBP-β bind to the endogenous Sftpb promoter, chromatin immunoprecipitation (ChIP) was performed with MLE-15 chromatin (ChIP-IT Express, catalogue number 53008; Active Motif, Inc., Carlsbad, CA) (see the online supplement for additional details) and control rabbit IgG (sc-2027), anti–C/EBP-α (sc-61), anti–C/EBP-β (sc-150), or anti-DBP (sc-98411) antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). The immunoprecipitated chromatin, after repeated washings, was eluted, reverse cross-linked, and treated with proteinase K. The supernatant and eluate fractions were analyzed by PCR amplification for the Sftpb promoter region −159 to −541, using the primer pair forward 5′-CCACAGGGGACACAGAAATC-3′ and reverse 5′-CGATGTCGGTTCCTAGTCCT-3′ (31).
In vitro Transcription/Translation
C/EBP-α and C/EBP-β cDNA expression constructs (plasmids 12550 and 12557, respectively; Addgene, Cambridge, MA) and negative control pSP72 (catalogue number P2191; Promega) were linearized to produce in vitro translated proteins (catalogue number L4610; Promega). In addition, full-length Dbp (catalogue number 4195116), NKX2–1 (catalogue number 3941576), POU2F1 (catalogue number 2966289), and Jund (catalogue number 4456297) cDNAs (Open Biosystems/Thermo Scientific, Huntsville, AL) were subcloned into pSP72 for coupled in vitro transcription/translation. The reactions were incubated for 90 minutes at 30°C. For EMSA analysis, 1 μl of the transcription/translation product that had been diluted 4-fold or 8-fold was used.
Identification of Transcription Factor Binding Sites
The Transcription Element Search System (32) and JASPAR (33) were used to identify putative transcription factor binding sites in the Sftpb promoter region −339 to −316.
Statistical Analysis
Groups were compared by one-way ANOVA with the Holm-Sidak all pairwise multiple comparison procedure (SigmaStat Program; SPSS, Inc., Chicago, IL). Relative luciferase activity units of each Sftpb promoter reporter construct are expressed as folds of the vector pGL4–10-ΔBgl II/Hind III or percentages of control values. Transient reporter assays were performed at least three times each, in triplicate or more. Results are expressed as means ± SE, and P < 0.05 was considered significantly different from the appropriate control.
RESULTS
The Sftpb Distal Promoter Region Functions as an Independently Active Promoter
Previous studies reported that the distal Sftpb promoter region mediated the inhibition by JUN of Sftpb promoter activity (34). We further demonstrated that JUN inhibited the distal, but not the proximal, promoter region (7). Because the Sftpb distal promoter region contains a putative AP-1 site, whereas the proximal region contains a consensus recognition site (nucleotide sequences −370 to −364 and −16 to −10, respectively), the mechanism underlying JUN's regulatory selectivity for the distal region was not known. To investigate the mechanism of JUN's inhibition of Sftpb promoter activity, luciferase reporter constructs under the control of the Sftpb promoter region, encompassing nucleotides −653 to +42 and mutant variants (Figure 1A), were analyzed. The enzyme reporter activity of the distal promoter region −653 to −194 was greater than the activity of the proximal promoter region −198 to +42 (4.6-fold versus 1.4-fold, respectively) compared with the control vector (Figure 1B). Upon subsequent deletion of the −653 to −194 promoter fragment, the reporter activity of the −400 to −275 was enhanced (4.6-fold versus 7.2-fold, respectively), whereas reporter activity of the promoter fragments −653 to −401 and −274 to −194 was reduced compared with the −653 to −194 fragment. These results indicate the presence of inhibitory cis-acting regulatory sequences in the −653 to −401 or −274 to −194 promoter regions. In contrast to the −653 to −194 fragment, deletion of the −198 to +42 promoter region reduced reporter activity. These results demonstrate that both the proximal and distal Sftpb promoter regions are functionally active and distinguishable, enabling further investigation of the molecular basis of the inhibition by JUN of Sftpb promoter activity.
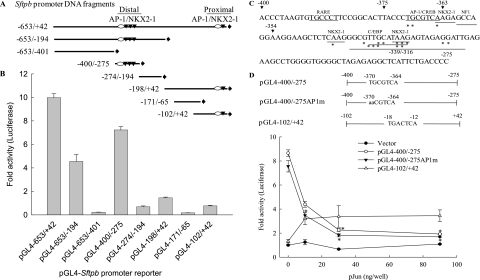
Figure 1.
Transient transfection analysis of surfactant-associated protein B (Sftpb) promoter reporter activity. (A) Schematic representation of the Sftpb promoter DNA fragments analyzed. Reporter constructs were generated by inserting promoter DNA fragments in the luciferase reporter plasmid pGL4–10. The Sftpb 5′ and 3′ nucleotide sequence numbers and the proximal and distal promoter regions containing the putative activator protein–1/Nk2 homeobox–1 (AP-1/NKX2.1) regions (ovals and inverted triangles, respectively) are shown. Arrowhead indicates the orientation of the DNA fragments ligated to the luciferase gene reporter. (B) The Sftpb distal promoter region is active, independent of the proximal promoter. Sftpb promoter reporter activity (fold activity versus vector control) was determined by transient transfection of MLE-15 cells (n = 8–16). (C and D) JUN inhibits Sftpb promoter reporter activity of a construct containing site-directed mutations in the distal putative AP-1 site (nucleotides −370 to −364). (C) Nucleotide sequence of the mouse Sftpb distal promoter region −400 to −275 denotes consensus/putative recognition elements for retinoic acid receptor (RARE), AP-1/cyclic AMP-responsive element binding protein (AP-1/CREB), Nk2 homeobox 1 (NKX2–1), nuclear factor 1 (NF1), and CCAAT/enhancer-binding protein (C/EBP) (underlined). The 5′ ends of deletion mutants (arrows), the region of sequence −339 to −316, and the nucleotides altered by site-directed mutagenesis (asterisks) to analyze promoter activity (Figures 2–5) are also indicated. (D) Schematic representation of the constructs used and the mutations introduced are shown (top). Dose–response relationships of JUN co-expression on Sftpb proximal and distal promoter reporter activities were assayed by transfecting MLE-15 cells with pGL4–10ΔBgl II/Hind III (vector), pGL4−102/+42 (proximal), pGL4−400/−275, and pGL4−400/−275AP-1m (distal putative WT and AP-1 site mutant [AP-1m], respectively) constructs. MLE-15 cells were cotransfected with reporter constructs and increasing concentrations of plasmid pCMV-Jun (pJun). The total amount of DNA was adjusted, using empty plasmid. JUN inhibited the distal putative AP-1 mutant and WT reporters (n = 4). *Decreased compared with control with no pJun (P < 0.05), as determined by one-way ANOVA with Holm-Sidak all pairwise multiple comparisons procedure.
Inhibition by JUN of Sftpb Promoter Reporter Activity Occurs Independent of the Distal Putative AP-1 Site
A previous study suggested that the distal promoter region contained an AP-1 element that is part of a composite binding site wherein AP-1, the cyclic AMP response element binding protein (CREB), thyroid transcription factor–1 (also known as NKX2–1), and nuclear factor I (NF1) (Figure 1C) interact (34). The nucleotide sequence of the Sftpb distal putative AP-1 site (TGCGTCA) differs from that of the proximal AP-1 site (TGACTCA) in the third and fourth nucleotides, raising the possibility that this property may underlie the selective JUN inhibition on the distal compared with the proximal promoter region. To investigate the contribution of the distal putative AP-1 site to JUN's inhibition of Sftpb promoter activity, a reporter construct with point mutations in the distal AP-1 site was generated. Dose-dependently, JUN inhibited both the wild-type and the AP-1 mutant −400 to −275 promoter reporters (pGL4−400/−275, pGL4−400/−275AP-1m), but not the proximal promoter reporter (pGL4−102/+42) (Figure 1D).
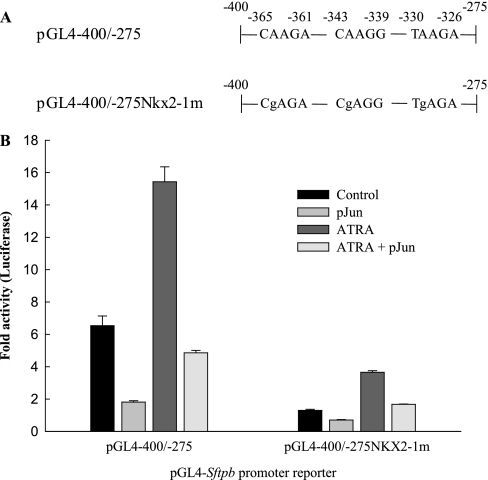
The protein JUN represses glucocorticoid-receptor and retinoic acid receptor gene regulatory activities by direct protein–protein interaction (35, 36). In a recent study (37), interactions of the TGF-β signaling protein SMAD3 with NKX2–1 and FOXA1 reduced binding to cognate DNA sites, leading to Sftpb gene repression by TGF-β. Retinoic acid and NKX2–1 recognition elements play important roles in Sftpb gene regulation (38–40). One of the NKX2–1 consensus core sites in the distal Sftpb promoter region overlaps the putative AP-1 site and retinoic acid receptor sites. To investigate the contribution of NKX2–1 recognition sites to JUN inhibition, a reporter with point mutations in the NKX2–1 sites (pGL4−400/−275 NKX2–1m; Figure 2A) was constructed. The mutant construct exhibited reduced reporter activity. The treatment of transfected MLE-15 cells with all trans-retinoic acid stimulated wild-type and mutant reporter activity, which was inhibited by JUN (Figure 2B).
Figure 2.
JUN inhibits basal and all-trans-retinoic acid (ATRA)–inducible Sftpb promoter reporter activity. Control (pGL4−400/−275) and a mutant Sftpb reporter construct containing three site-directed mutations (pGL4−400/−275NKX2–1m) were cotransfected into MLE-15 cells with either empty plasmid (control) or JUN expression plasmid pJun (30 ng/well). The day after transfection, serum-containing medium was replaced with serum-free medium in the absence or presence of ATRA (1 μg/ml) for 20–24 hours. (A) Schematic representation of promoter fragments shows mutated nucleotides. (B) Sftpb promoter reporter activity was determined by transient transfection of MLE-15 cells (n = 4).
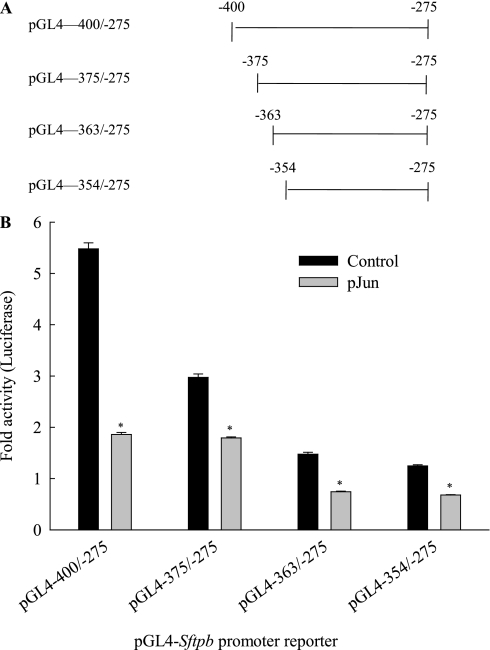
To rule out the possibility that point mutations neither abolished the binding of an inhibitory AP-1 complex nor interfered with protein–protein interactions, rendering mutant reporter activity sensitive to JUN inhibition, we generated deletion mutants lacking the AP-1 and upstream NKX2–1 consensus core sites. Further deletion of the distal reporter pGL4−400/−275 reduced reporter activity. Moreover, despite the lack of a putative AP-1 site, the deletion reporters remained responsive to inhibition by JUN (Figures 3A and 3B). These results suggest that JUN exerts its inhibitory effect either through Sftpb promoter sites independent of the targeted putative AP-1 recognition sequence, or through protein–protein interactions.
Figure 3.
JUN inhibits Sftpb promoter reporter activity of constructs lacking the distal AP-1 site. Deletion mutant reporters (pGL4−375/−275, pGL4−363/−275, and pGL4−354/−275) were constructed after exonuclease III treatment of linearized pGL4–Sftpb−400/−275. MLE-15 cells were cotransfected with reporter constructs plus either empty plasmid (Control) or pJun (30 ng/well). (A) Schematic representation of promoter DNA fragments analyzed. The Sftpb 5′ and 3′ nucleotide sequence numbers are indicated. (B) Sftpb promoter reporter activity was determined by transient transfection of MLE-15 cells (n = 4). *Decreased compared with control with no pJun (P < 0.05), as determined by one-way ANOVA with Holm-Sidak all pairwise multiple comparisons procedure.
Recombinant JUN Enhances the Intensity of DNA–Protein Complexes Formed by Incubating the Sftpb −339/−316 Probe with MLE-15 Nuclear Protein Extract
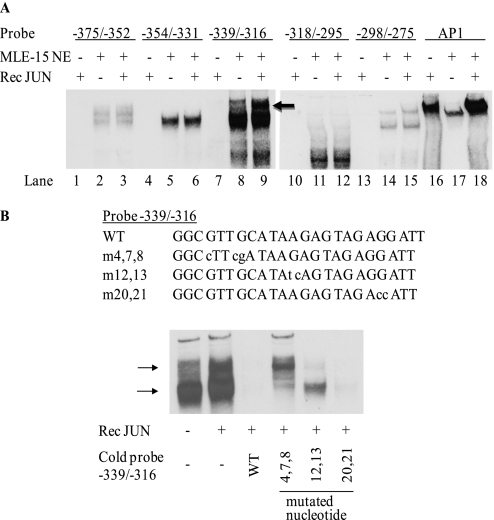
After finding that the Sftpb distal putative AP-1 and NKX2–1 recognition sites may not be critical for JUN inhibition, we sought to determine whether JUN directly or indirectly influenced the formation of DNA–protein complexes. Oligonucleotide probes (24-mer) spanning the Sftpb promoter region −375 to −275 were incubated in the absence or presence of recombinant JUN, with or without MLE-15 cell nuclear extracts, and were analyzed by EMSAs (Figure 4A). All probes formed DNA–protein complexes. The recombinant JUN, when used alone, interacted with an AP-1 consensus oligonucleotide probe included as a positive control (Figure 4A, lane 16), whereas no interaction was evident with Sftpb promoter–derived oligonucleotides (Figure 4A, lanes 1, 4, 7, 10, and 13). However, the presence of recombinant JUN increased the intensity of the probe–protein complexes formed by incubating the Sftpb oligonucleotide probe −339/−316 with MLE-15 nuclear protein extract (in particular, the slow-migrating complex) (Figure 4A, lane 9, arrow).
Figure 4.
MLE-15 cell nuclear proteins form complexes with oligonucleotides corresponding to Sftpb distal promoter region. (A) Identification of a region whose complex-forming property is altered in the presence of purified recombinant JUN. Electrophoretic mobility shift assays (EMSAs) were performed with 24-mer oligonucleotides (spanning Sftpb promoter nucleotide sequence −375 to −275) and an AP-1 consensus probe. The oligonucleotide probes were incubated in the presence or absence of purified recombinant JUN (Promega), with or without MLE-15 cell nuclear extract. Addition of JUN to the probe −339/−316 and MLE-15 nuclear extract mix increased the intensity of DNA–nuclear protein complexes formed, and in particular the slow-migrating complex (arrow). (B) Multiple proteins (arrows) bind to the Sftpb oligonucleotide probe −339/−316. EMSAs were performed with wild-type (WT) probe (−339/−316) without (lane 1) or with (lanes 2–6) recombinant JUN in the absence or presence of 100-fold excess unbiotinylated WT (lane 3) or mutant (lanes 4–6) oligonucleotides. The DNA–protein complexes formed in the absence and presence of JUN (lanes 1 and 2) were competed by excess WT and mutant (m) 20,21 oligonucleotides. Although m4,7,8 preferentially inhibited the formation of the fast-migrating complex, m12,13 inhibited the slow-migrating complex, indicating the presence of more than one protein species.
To identify potential transcription factor binding sites in Sftpb promoter region −339 to −316, in silico analysis was performed using TESS (32) and JASPAR (33). Several transcription factor recognition sites were predicted (ABF1, DBP, POU1F1a, EFII, HOXA5, GAL4, DDIT3∷C/EBP-α, BRCA1, NFIL3, HLF, POU5F1, C/EBP-α, FOXL1, SOX2, GATA2, HLTF, TBP, NKX2–5, FOXC1, EN1, ZNF354C, MZF1_1–4, SPIB, and ETS1). To locate regions within the 24-mer oligonucleotide that may be critical to the formation of DNA–protein complexes, mutations were introduced in the −339/−316 oligonucleotide and designated m4,7,8, m12,13, and m20,21 (Figure 4B). To test the effects of the mutations on the formation of DNA–protein complexes, a 100-fold molar excess of unlabeled wild-type and mutant oligonucleotide competitors was used. The formation of faster and slower migrating complexes was inhibited by wild-type and m20,21 oligonucleotide −339/−316 (Figure 4B, lanes 3 and 6). In contrast, oligonucleotides −339/−316 m4,7,8 and m12,13 inhibited the formation of complexes differentially. Whereas −339/−316 m4,7,8 inhibited the faster migrating complex, −339/−316 m12,13 inhibited the slower migrating complex (Figure 4B, lanes 4 and 5). These results suggested the presence of multiple protein species with partial selectivity to subregions of the −339/−316 region of the Sftpb promoter and the presence of exogenously added recombinant JUN modulated DNA–protein interactions.
The Recognition Site in the Sftpb Distal Promoter Region −339 to −316 Plays a Key Role in the Regulation of Sftpb Promoter Activity
The Sftpb promoter region −339 to −316 contained nucleotide sequences that highly matched the consensus recognition site for the D-site albumin promoter binding protein (DBP, a member of the proline-rich and acid-rich bZIP [PAR-bZIP] protein subfamily) and C/EBP-α transcription factor (a member of the C/EBP subfamily) (Figure 5A) (41, 42). The putative DBP and C/EBP recognition elements were selected for further analysis because both DBP and the C/EBP proteins belong to the bZIP protein family, of which JUN is a member. Members of different bZIP protein subfamilies homodimerize and heterodimerize, expanding the repertoire of target sites recognized by bZIP proteins.
Figure 5.
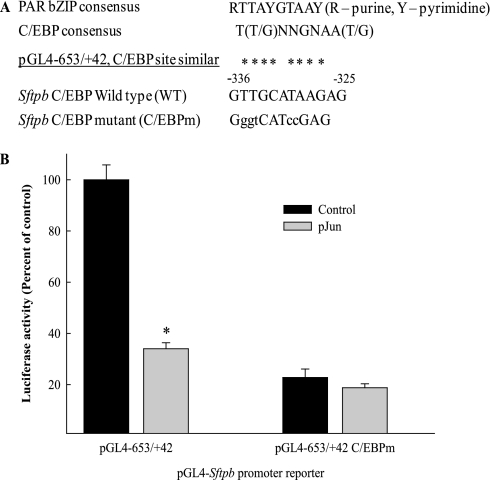
Mutation of the C/EBP recognition site reduces Sftpb promoter reporter activity and sensitivity to JUN-mediated inhibition. (A) The Sftpb C/EBP recognition site and mutated nucleotides (nucleotides −336 to −325) are shown in relation to the PAR-bZIP and C/EBP consensus recognition sequences. (B) Wild-type (pGL4−653/+42) and C/EBP site mutant (pGL4−653/+42 C/EBPm) plasmid reporter constructs were cotransfected with either empty plasmid (Control) or the expression plasmid pJun (30 ng/well) into MLE-15 cells. The pGL4 −653/+42 C/EBPm reporter was less active (4.5-fold) and less sensitive to JUN-mediated inhibition compared with the pGL4−653/+42 reporter (n = 5 and 6, respectively). *Decreased compared with control sample with no pJun (P < 0.05), as determined by one-way ANOVA with Holm-Sidak all pairwise multiple comparisons procedure.
To examine the role of the putative DBP and C/EBP site in the Sftpb promoter, we generated a reporter construct containing point mutations (pGL4−653/+42 C/EBPm; Figure 5A). The mutations introduced were intended to target the two halves of the recognition site, and are different from the mutations used in the competitive EMSA (Figure 4B). The wild-type and mutant reporter constructs were transfected into MLE-15 cells in the presence or absence of the JUN expression plasmid pJun (Figure 5B). Mutagenesis of the putative DBP and C/EBP site reduced Sftpb promoter activity by 4.5-fold, indicating that this region plays a critical role in regulating Sftpb promoter activity. Further, the pGL4−653/+42 C/EBPm reporter was less sensitive to JUN-mediated inhibition (Figure 5B). These results suggested that JUN forms complexes with Sftpb gene transactivators or repressors.
C/EBP-α and C/EBP-β Proteins Bind to the Sftpb Promoter
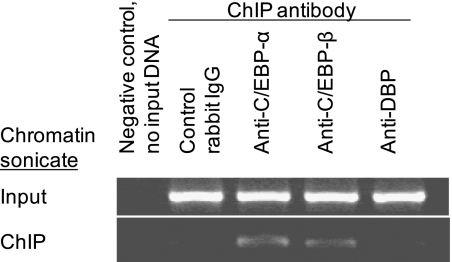
To examine whether the bZIP proteins DBP, C/EBP-α, and C/EBP-β bind to the endogenous Sftpb promoter, ChIP was used. The DNA–protein complex immunoprecipitated using anti–C/EBP-α and anti–C/EBP-β antibodies was enriched compared with either control IgG or anti-DBP antibodies, as determined by PCR analysis (Figure 6). These results suggest that C/EBP-α and C/EBP-β proteins bind to the murine Sftpb promoter.
Figure 6.
C/EBP-α and C/EBP-β proteins bind to the endogenous Sftpb promoter. Cross-linked chromatin isolated from MLE-15 cells was immunoprecipitated with nonimmune rabbit IgG (lane 2), anti–C/EBP-α (lane 3), anti–C/EBP-β (lane 4), or anti–D-site albumin promoter binding protein (anti-DBP) (lane 5) antibodies. Eluates of the immunoprecipitated DNA samples were PCR-amplified with a primer pair specific to the murine Sftpb promoter region, spanning nucleotides −159 to −541. The supernatant fraction of the immunoprecipitated chromatin samples was used to check input DNA. A PCR reaction mix with no chromatin added served as negative control (lane 1). ChIP, chromatin immunoprecipitation.
C/EBP-α, C/EBP-β, and DBP Proteins Bind to the Sftpb Promoter Region −339 to −316
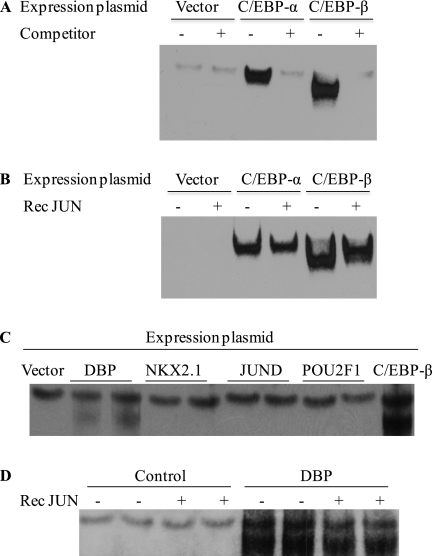
Because of the DNA fragment sizes of the chromatin sonicate used as a template for PCR analysis (100 to 1,000 bp) in the ChIP assay and the presence of multiple putative C/EBP sites in the Sftpb promoter, the exact location of the region mediating C/EBP binding could not be determined using ChIP. EMSA was used to demonstrate direct C/EBP protein binding to the Sftpb −339/−316 region in which C/EBP-α and C/EBP-β proteins were expressed, using a rabbit reticulocyte lysate system, and incubated with biotinylated probe −339/−316. The presence of C/EBP-α or C/EBP-β translation products shifted probe mobility (Figure 7A, lanes 3 and 5), and complex formation was competed by 100-fold excess unbiotinylated −339/−316 oligonucleotide (Figure 7A, lanes 4 and 6). In contrast, the nonspecific complex formation detected in the negative control sample was not competed by the addition of unbiotinylated oligonucleotide (Figure 7A, lane 2).
Figure 7.
In vitro translated C/EBP-α, C/EBP-β, and DBP proteins bind to the Sftpb promoter −339/−316 probe. (A) In vitro transcription/translation products, generated using vector (lanes 1 and 2), C/EBP-α (lanes 3 and 4), or C/EBP-β (lanes 5 and 6) expression plasmid DNA templates, were incubated with the biotinylated probe −339/−316. Samples were preincubated in the absence or presence of 100-fold excess unbiotinylated oligonucleotides (lanes 2, 4, and 6) to determine specificity. The nonspecific band (lane 1) was not competed by the addition of unbiotinylated probe (lane 2). (B) Incubation of the DNA probe −339/−316 and in vitro translation products in the presence of recombinant JUN supershifted the C/EBP–DNA complexes formed (lanes 4 and 6). (C) In vitro transcription/translation products, generated using vector (lane 1), DBP (lanes 2 and 3), NKX2–1 (lanes 4 and 5), jun proto-oncogene related gene d (JUND) (lanes 6 and 7), POU class 2 homeobox 1 (POU2F1) (lanes 8 and 9), or C/EBP-β (lane 10) expression plasmid DNA templates, were incubated with the biotinylated Sftpb −339/−316 probe. DBP formed a complex with the DNA probe (lanes 2 and 3). (D) The migration pattern of the DBP/DNA complex formed remained similar in the absence or presence of recombinant JUN (lanes 5 and 6 versus 7 and 8).
To test whether C/EBP binding to the promoter region −339/−316 mediated JUN binding, the in vitro translation products were preincubated with purified recombinant JUN. The presence of recombinant JUN supershifted the mobility of the C/EBP-DNA complex (Figure 7B, lanes 4 and 6). These results, in combination with the effect of recombinant JUN on EMSAs of nuclear proteins (Figure 4), suggest that C/EBP-α and/or C/EBP-β proteins bind to the identified Sftpb promoter region, and JUN binding is mediated through the formation of heteromeric complexes.
In addition, DBP, NKX2–1, POU2F1, and JUND binding to the Sftpb −339/−316 probe was examined by EMSA, because one of the putative NKX2–1 sites overlaps the C/EBP site, and JUND and POU2F1 (also known as OCT1) regulate Sftpb and Clara cell secretory protein genes, respectively (34, 43). DBP, NKX2–1, POU2F1, and JUND translation products were incubated with the Sftpb −339/−316 region and analyzed by EMSA. DBP, but not NKX2.1, POU2F1, or JUND, bound the −339/−316 oligonucleotide (Figure 7C).
To examine DBP binding further, the formation of DBP–DNA complexes in the absence or presence of JUN was compared. The migration pattern of the DBP-biotinylated oligonucleotide complex remained similar in the absence or presence of JUN (Figure 7D). The ability of DBP, in addition to C/EBP-α and C/EBP-β, to interact with the Sftpb −339/−316 region raises the possibility that other transcription factors or bZIP family members could bind to the same site.
DISCUSSION
SFTPB is critical in maintaining lung function, because SFTPB-deficient mice die of respiratory failure shortly after birth (3–6). One of the key events in acute lung injury is the loss of functional surfactant. Thus, surfactant has long been investigated as a treatment for acute lung injury. Surfactant replacement, however, has yet to be demonstrated as an effective therapy for acute lung injury because of immense hurdles in its administration (44). An alternative strategy may involve the development of treatments that maintain or restore endogenous surfactant production. We previously reported that in a murine model of acute lung injury, the induction of JUN was associated with a diminution of SFTPB expression (7). SFTPB expression was maintained in resistant, compared with sensitive, murine strains, and inducible SFTPB expression increased the survival of mice (7). The present work focused on analyses of the core Sftpb promoter, to identify transcription factor recognition sites that contribute to the JUN-mediated inhibition of SFTPB.
The murine Sftpb promoter, spanning nucleotides −653 to +42, contains two functionally distinguishable proximal (−132 to −1) and distal (−382 to −283) promoter regions (8). The distal promoter region can be transcriptionally active in the absence of the proximal promoter region. The distal promoter region contains a putative initiator sequence (CATTCTG) at nucleotides −286 to −280. This initiator element, which was originally identified in the TATA-less murine terminal deoxynucleotidyl transferase gene, encompasses the transcription start site and can direct basal transcription (45). As determined by primer extension (8) and expressed-sequence Tag (EST) database analyses, the SFTPB mRNA 5′ untranslated region contains ≤14–16 nucleotides. However, basal promoter activity was lost with the deletion of promoter sequences from −415 to −353 (34), demonstrating the critical role of the distal promoter region. Although the occurrence of an alternative SFTPB transcriptional initiation site is unknown, the functionality of the distal region as a promoter provided a useful tool for analyzing the role of its putative transcription factor recognition sites, independent of those in the proximal region.
The Sftpb promoter contains AP-1, NKX2–1, trans-acting transcription factors 1 and 3 (SP1, SP3), hepatocyte nuclear factor 3 (HNF3), retinoic acid receptor, and other recognition elements (8, 34, 38, 40, 46). Although JUN can activate numerous genes by binding to AP-1 recognition elements, it can also inhibit the induction of other genes by a different mechanism. JUN inhibits the induction of the bone γ-carboxyglutamate protein (osteocalcin) gene by retinoic acid and vitamin D3 (35) and the induction of the kallikrein-related peptidase 3 (prostate-specific antigen) gene by androgen (47). Direct interactions between JUN and the receptors for vitamin D3 or androgen inhibit the induction of target genes.
The Sftpb proximal and distal promoter regions contain putative AP-1 sites. The proximal AP-1 site is identical to the optimal consensus AP-1 site (TGACTCA), whereas the distal AP-1 site (TGCGTCA) differs by two nucleotides. In addition, the distal CREB site differs from the consensus CREB site (TGACGTCA). However, these nucleotide sequence differences could not explain JUN's ability to inhibit the distal, but not the proximal, promoter region. The patterns of JUN inhibition of the distal AP-1 wild-type and point mutant reporters were comparable. To rule out the possibility that the introduced mutations may not have sufficiently altered the mode of protein–DNA interactions, we investigated the effects of JUN co-expression on reporter constructs that lacked the distal putative AP-1 site. Despite the absence of the putative AP-1 site, JUN inhibited reporter activity. These results suggest that the presence of the putative AP-1 site in the distal promoter region is not required for the inhibition by JUN of Sftpb promoter activity, raising the possibility of protein–protein interactions as a mediator of JUN's inhibitory effect.
Our conclusion about the role of the Sftpb AP-1 sites contrasts with those of previous studies (34). Sever-Chroneos and colleagues (34) observed that mutation in the distal AP-1 binding site increased basal promoter activity fivefold in MLE-15 cells. In addition, they concluded that the distal AP-1 element is involved in, but is not sufficient for, the inhibition by JUN of promoter activity. In our analysis, although deletion of the distal element containing the overlapping AP1/NKX2–1 recognition sites reduced promoter activity, mutation of the AP-1 site did not induce basal promoter activity or reverse JUN inhibition. It is unclear whether the use of the luciferase reporter (half-life of approximately 0.84 hours) (48) versus the CAT reporter (half-life of approximately 16 hours) (49) and other experimental variations (e.g., the use of different promoter fragments) contributed to the discrepancies observed. In support of our observations, a genome-wide analysis of the frequency and distribution of AP-1 sites indicated that the number of AP-1–regulated genes identified is far smaller than the number of genes containing potential AP-1 sites, and that not all AP-1 sites are activated in a given cell under a given condition (50, 51).
In addition, the interaction of JUN with NKX2–1 appears unlikely to inhibit Sftpb promoter activity. Point mutations at the NKX2–1 sites in the distal promoter reduced reporter activity, but the mutants remained sensitive to inhibition by JUN, suggesting that other recognition sites or regulatory factors mediated the JUN-mediated inhibition of Sftpb.
Mobility shift assays indicated that JUN may target the Sftpb promoter by binding to a site within the −339/−316 region. Nucleotide sequence analysis for potential transcription factor binding sites in the −339/−316 Sftpb promoter region predicted recognition sites for DBP and C/EBP. This was a pertinent finding because DBP and the C/EBPs, like JUN, belong to the bZIP protein family. The contribution of the identified site to the regulation of Sftpb promoter activity was demonstrated in MLE-15 cells transfected with pGL4−653/+42 wild-type and mutant reporters. Mutagenesis of the putative recognition site reduced Sftpb promoter activity and sensitivity to JUN-mediated inhibition.
Endogenous DBP or C/EBP binding to the Sftpb promoter was investigated by chromatin immunoprecipitation. Immunoprecipitation of MLE-15 chromatin sonicate with anti-DBP antibodies did not enrich the Sftpb promoter PCR amplification product, suggesting that DBP does not associate with Sftpb promoter in vivo, or that the abundance of DBP–Sftpb promoter complexes formed in vivo does not permit detection by this method. In contrast, C/EBP-α and C/EBP-β binding to the Sftpb promoter was evidenced by the enrichment of chromatin immunoprecipitation.
JUN modulates gene transcription by forming homodimeric or heterodimeric complexes (52, 53). Our results indicate that JUN may not inhibit Sftpb promoter activity by forming homodimers. The recombinant JUN protein formed a complex with the consensus AP-1 oligonucleotide, but with none of the Sftpb-derived oligonucleotides spanning the Sftpb promoter region −375 to −275. However, in the presence of MLE-15 nuclear protein extract, the addition of JUN increased the intensity of DNA–protein complexes formed with the −339/−316 oligonucleotide probe. Further analysis, using point mutant oligonucleotides, suggested that multiple transcription factors bound to the −339/−316 probe.
Previous studies demonstrated that C/EBP-β and JUN interacted through their bZIP region in the absence of DNA. Such interactions altered the specificity of DNA binding (54). In our EMSA analyses, C/EBP binding to the Sftpb −339/−316 region, to which JUN by itself cannot bind, was shifted in the presence of purified recombinant JUN protein. These results suggest that the Sftpb −339/−316 region is recognized by C/EBP-α and/or C/EBP-β dimers or C/EBP-α and/or C/EBP-β/JUN heterodimers, and that the formation of heteromeric complexes is key to Sftpb promoter targeting by JUN. The PAR-bZIP DBP also bound to the −339/−316 probe, raising the possibility that other transcription factors or bZIP proteins could bind to the same site.
In conclusion, we analyzed the murine Sftpb promoter encompassing nucleotides −653 to +42. Deletion and site-directed Sftpb promoter reporter mutants and EMSAs indicate that the distal Sftpb promoter region −339 to −316 is a critical regulatory element. Mutagenesis in this region, which contains a C/EBP recognition site, reduced Sftpb promoter activity and sensitivity to JUN-mediated inhibition. Endogenous C/EBP-α and C/EBP-β bind to the Sftpb promoter. The transcription factor JUN can partner with C/EBP-α or c/EBP-β, bind to the identified cis-acting regulatory DNA site, and inhibit Sftpb promoter activity. Thus, the inhibition by JUN of the Sftpb promoter is likely indirect and dependent on heteromeric complex formation.
Supplementary Material
This study was supported by National Institutes of Health grants ES015675, HL077763, and HL085655 (G.D.L.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2010-0260OC on December 10, 2010
Author Disclosure: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Metcalfe IL, Enhorning G, Possmayer F. Pulmonary surfactant–associated proteins: their role in the expression of surface activity. J Appl Physiol 1980;49:34–41. [DOI] [PubMed] [Google Scholar]
- 2.Curstedt T, Jornvall H, Robertson B, Bergman T, Berggren P. Two hydrophobic low-molecular-mass protein fractions of pulmonary surfactant: characterization and biophysical activity. Eur J Biochem 1987;168:255–262. [DOI] [PubMed] [Google Scholar]
- 3.Beers MF, Hamvas A, Moxley MA, Gonzales LW, Guttentag SH, Solarin KO, Longmore WJ, Nogee LM, Ballard PL. Pulmonary surfactant metabolism in infants lacking surfactant protein B. Am J Respir Cell Mol Biol 2000;22:380–391. [DOI] [PubMed] [Google Scholar]
- 4.Clark JC, Wert SE, Bachurski CJ, Stahlman MT, Stripp BR, Weaver TE, Whitsett JA. Targeted disruption of the surfactant protein B gene disrupts surfactant homeostasis, causing respiratory failure in newborn mice. Proc Natl Acad Sci USA 1995;92:7794–7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melton KR, Nesslein LL, Ikegami M, Tichelaar JW, Clark JC, Whitsett JA, Weaver TE. SP-B deficiency causes respiratory failure in adult mice. Am J Physiol Lung Cell Mol Physiol 2003;285:L543–L549. [DOI] [PubMed] [Google Scholar]
- 6.Tokieda K, Whitsett JA, Clark JC, Weaver TE, Ikeda K, McConnell KB, Jobe AH, Ikegami M, Iwamoto HS. Pulmonary dysfunction in neonatal SP-B–deficient mice. Am J Physiol 1997;273:L875–L882. [DOI] [PubMed] [Google Scholar]
- 7.Bein K, Wesselkamper SC, Liu X, Dietsch M, Majumder N, Concel VJ, Medvedovic M, Sartor MA, Henning LN, Venditto C, et al. Surfactant-associated protein B is critical to survival in nickel-induced injury in mice. Am J Respir Cell Mol Biol 2009;41:226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruno MA, Bohinski RJ, Carter JE, Foss KA, Whitsett JA. Structure and function of the mouse surfactant protein B gene. Am J Physiol 1995;268:L381–L389. [DOI] [PubMed] [Google Scholar]
- 9.Curran T, Franza BR Jr. Fos and Jun: the AP-1 connection. Cell 1988;55:395–397. [DOI] [PubMed] [Google Scholar]
- 10.Johnson RS, van Lingen B, Papaioannou VE, Spiegelman BM. A null mutation at the c-Jun locus causes embryonic lethality and retarded cell growth in culture. Genes Dev 1993;7:1309–1317. [DOI] [PubMed] [Google Scholar]
- 11.Eferl R, Sibilia M, Hilberg F, Fuchsbichler A, Kufferath I, Guertl B, Zenz R, Wagner EF, Zatloukal K. Functions of c-Jun in liver and heart development. J Cell Biol 1999;145:1049–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rebollo A, Dumoutier L, Renauld JC, Zaballos A, Ayllon V, Martinez AC. Bcl-3 expression promotes cell survival following interleukin-4 deprivation and is controlled by AP1 and AP1-like transcription factors. Mol Cell Biol 2000;20:3407–3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castellazzi M, Dangy JP, Mechta F, Hirai S, Yaniv M, Samarut J, Lassailly A, Brun G. Overexpression of avian or mouse c-Jun in primary chick embryo fibroblasts confers a partially transformed phenotype. Oncogene 1990;5:1541–1547. [PubMed] [Google Scholar]
- 14.Schutte J, Viallet J, Nau M, Segal S, Fedorko J, Minna J. Jun-B inhibits and c-Fos stimulates the transforming and trans-activating activities of c-Jun. Cell 1989;59:987–997. [DOI] [PubMed] [Google Scholar]
- 15.Eferl R, Ricci R, Kenner L, Zenz R, David JP, Rath M, Wagner EF. Liver tumor development: c-Jun antagonizes the proapoptotic activity of p53. Cell 2003;112:181–192. [DOI] [PubMed] [Google Scholar]
- 16.Hilberg F, Aguzzi A, Howells N, Wagner EF. c-Jun is essential for normal mouse development and hepatogenesis. Nature 1993;365:179–181. [DOI] [PubMed] [Google Scholar]
- 17.Angel P, Allegretto EA, Okino ST, Hattori K, Boyle WJ, Hunter T, Karin M. Oncogene Jun encodes a sequence-specific trans-activator similar to AP-1. Nature 1988;332:166–171. [DOI] [PubMed] [Google Scholar]
- 18.Rauscher FJ III, Voulalas PJ, Franza BR Jr, Curran T. Fos and Jun bind cooperatively to the AP-1 site: reconstitution in vitro. Genes Dev 1988;2:1687–1699. [DOI] [PubMed] [Google Scholar]
- 19.Harshman KD, Moye-Rowley WS, Parker CS. Transcriptional activation by the SV40 AP-1 recognition element in yeast is mediated by a factor similar to AP-1 that is distinct from GCN4. Cell 1988;53:321–330. [DOI] [PubMed] [Google Scholar]
- 20.Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos–Jun interactions that mediate transcription regulatory specificity. Oncogene 2001;20:2438–2452. [DOI] [PubMed] [Google Scholar]
- 21.Newman JR, Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 2003;300:2097–2101. [DOI] [PubMed] [Google Scholar]
- 22.Cassel TN, Nord M. C/EBP transcription factors in the lung epithelium. Am J Physiol Lung Cell Mol Physiol 2003;285:L773–L781. [DOI] [PubMed] [Google Scholar]
- 23.Cassel TN, Nordlund-Moller L, Andersson O, Gustafsson JA, Nord M. C/EBPalpha and C/EBPdelta activate the Clara cell secretory protein gene through interaction with two adjacent C/EBP-binding sites. Am J Respir Cell Mol Biol 2000;22:469–480. [DOI] [PubMed] [Google Scholar]
- 24.Li F, Rosenberg E, Smith CI, Notarfrancesco K, Reisher SR, Shuman H, Feinstein SI. Correlation of expression of transcription factor C/EBP alpha and surfactant protein genes in lung cells. Am J Physiol 1995;269:L241–L247. [DOI] [PubMed] [Google Scholar]
- 25.Rosenberg E, Li F, Reisher SR, Wang M, Gonzales LW, Ewing JR, Malek S, Ballard PL, Notarfrancesco K, Shuman H, et al. Members of the C/EBP transcription factor family stimulate expression of the human and rat surfactant protein A (SP-A) genes. Biochim Biophys Acta 2002;1575:82–90. [DOI] [PubMed] [Google Scholar]
- 26.Sugahara K, Sadohara T, Sugita M, Iyama K, Takiguchi M. Differential expression of CCAAT enhancer binding protein family in rat alveolar epithelial cell proliferation and in acute lung injury. Cell Tissue Res 1999;297:261–270. [DOI] [PubMed] [Google Scholar]
- 27.Flodby P, Barlow C, Kylefjord H, Ahrlund-Richter L, Xanthopoulos KG. Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein alpha. J Biol Chem 1996;271:24753–24760. [DOI] [PubMed] [Google Scholar]
- 28.Sugahara K, Iyama KI, Kimura T, Sano K, Darlington GJ, Akiba T, Takiguchi M. Mice lacking CCAAT/enhancer-binding protein–alpha show hyperproliferation of alveolar Type II cells and increased surfactant protein mRNAs. Cell Tissue Res 2001;306:57–63. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka T, Yoshida N, Kishimoto T, Akira S. Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J 1997;16:7432–7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wikenheiser KA, Vorbroker DK, Rice WR, Clark JC, Bachurski CJ, Oie HK, Whitsett JA. Production of immortalized distal respiratory epithelial cell lines from surfactant protein C/simian virus 40 large tumor antigen transgenic mice. Proc Natl Acad Sci USA 1993;90:11029–11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Field-Corbett C, English K, O'Dea S. Regulation of surfactant protein B gene expression in bone marrow–derived cells. Stem Cells 2009;27:662–669. [DOI] [PubMed] [Google Scholar]
- 32.Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. Curr Protoc Bioinform 2008;21:2.6.1-2.6.15. [DOI] [PubMed]
- 33.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res 2004;32:D91–D94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sever-Chroneos Z, Bachurski CJ, Yan C, Whitsett JA. Regulation of mouse SP-B gene promoter by AP-1 family members. Am J Physiol 1999;277:L79–L88. [DOI] [PubMed] [Google Scholar]
- 35.Schule R, Umesono K, Mangelsdorf DJ, Bolado J, Pike JW, Evans RM. Jun–Fos and receptors for vitamins A and D recognize a common response element in the human osteocalcin gene. Cell 1990;61:497–504. [DOI] [PubMed] [Google Scholar]
- 36.Yang-Yen HF, Chambard JC, Sun YL, Smeal T, Schmidt TJ, Drouin J, Karin M. Transcriptional interference between c-Jun and the glucocorticoid receptor: mutual inhibition of DNA binding due to direct protein–protein interaction. Cell 1990;62:1205–1215. [DOI] [PubMed] [Google Scholar]
- 37.Minoo P, Hu L, Zhu N, Borok Z, Bellusci S, Groffen J, Kardassis D, Li C. SMAD3 prevents binding of NKX2.1 and FOXA1 to the SPB promoter through its MH1 and MH2 domains. Nucleic Acids Res 2008;36:179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naltner A, Ghaffari M, Whitsett JA, Yan C. Retinoic acid stimulation of the human surfactant protein B promoter is thyroid transcription factor 1 site-dependent. J Biol Chem 2000;275:56–62. [DOI] [PubMed] [Google Scholar]
- 39.Yan C, Naltner A, Conkright J, Ghaffari M. Protein–protein interaction of retinoic acid receptor alpha and thyroid transcription factor–1 in respiratory epithelial cells. J Biol Chem 2001;276:21686–21691. [DOI] [PubMed] [Google Scholar]
- 40.Yan C, Sever Z, Whitsett JA. Upstream enhancer activity in the human surfactant protein B gene is mediated by thyroid transcription factor 1. J Biol Chem 1995;270:24852–24857. [DOI] [PubMed] [Google Scholar]
- 41.Falvey E, Marcacci L, Schibler U. DNA-binding specificity of PAR and C/EBP leucine zipper proteins: a single amino acid substitution in the C/EBP DNA-binding domain confers PAR-like specificity to C/EBP. Biol Chem 1996;377:797–809. [PubMed] [Google Scholar]
- 42.Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J 1990;9:1897–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sawaya PL, Stripp BR, Whitsett JA, Luse DS. The lung-specific CC10 gene is regulated by transcription factors from the AP-1, octamer, and hepatocyte nuclear factor 3 families. Mol Cell Biol 1993;13:3860–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spragg R. Surfactant for acute lung injury. Am J Respir Cell Mol Biol 2007;37:377–378. [DOI] [PubMed] [Google Scholar]
- 45.Smale ST, Baltimore D. The “initiator” as a transcription control element. Cell 1989;57:103–113. [DOI] [PubMed] [Google Scholar]
- 46.Bohinski RJ, Di Lauro R, Whitsett JA. The lung-specific surfactant protein B gene promoter is a target for thyroid transcription factor 1 and hepatocyte nuclear factor 3, indicating common factors for organ-specific gene expression along the foregut axis. Mol Cell Biol 1994;14:5671–5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato N, Sadar MD, Bruchovsky N, Saatcioglu F, Rennie PS, Sato S, Lange PH, Gleave ME. Androgenic induction of prostate-specific antigen gene is repressed by protein–protein interaction between the androgen receptor and AP-1/c-Jun in the human prostate cancer cell line LNCaP. J Biol Chem 1997;272:17485–17494. [DOI] [PubMed] [Google Scholar]
- 48.Leclerc GM, Boockfor FR, Faught WJ, Frawley LS. Development of a destabilized Firefly luciferase enzyme for measurement of gene expression. Biotechniques 2000;29:590–591, 594–596, 598 passim. [DOI] [PubMed] [Google Scholar]
- 49.Krebsbach PH, Harrison JR, Lichtler AC, Woody CO, Rowe DW, Kream BE. Transgenic expression of COL1A1–chloramphenicol acetyltransferase fusion genes in bone: differential utilization of promoter elements in vivo and in cultured cells. Mol Cell Biol 1993;13:5168–5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu SL, Waha A, Vogt PK. Identification and characterization of genes upregulated in cells transformed by v-Jun. Oncogene 2000;19:3537–3545. [DOI] [PubMed] [Google Scholar]
- 51.Zhou H, Zarubin T, Ji Z, Min Z, Zhu W, Downey JS, Lin S, Han J. Frequency and distribution of AP-1 sites in the human genome. DNA Res 2005;12:139–150. [DOI] [PubMed] [Google Scholar]
- 52.Nakabeppu Y, Nathans D. The basic region of Fos mediates specific DNA binding. EMBO J 1989;8:3833–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryseck RP, Bravo R. C-JUN, JUN B, and JUN D differ in their binding affinities to AP-1 and CRE consensus sequences: effect of FOS proteins. Oncogene 1991;6:533–542. [PubMed] [Google Scholar]
- 54.Hsu W, Kerppola TK, Chen PL, Curran T, Chen-Kiang S. Fos and Jun repress transcription activation by NF-IL6 through association at the basic zipper region. Mol Cell Biol 1994;14:268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.