Abstract
Tissue-specific stem cell (TSC) behavior is determined by the stem cell niche. However, delineation of the TSC–niche interaction requires purification of both entities. We reasoned that the niche could be defined by the location of the TSC. We demonstrate that a single CD49fbright/Sca1+/ALDH+ basal cell generates rare label-retaining cells and abundant label-diluting cells. Label-retaining and label-diluting cells were located in the rimmed domain of a unique clone type, the rimmed clone. The TSC property of self-renewal was tested by serial passage at clonal density and analysis of clone-forming cell frequency. A single clone could be passaged up to five times and formed only rimmed clones. Thus, rimmed clone formation was a cell-intrinsic property. Differentiation potential was evaluated in air–liquid interface cultures. Homogenous cultures of rimmed clones were highly mitotic but were refractory to standard differentiation signals. However, rimmed clones that were cocultured with unfractionated tracheal cells generated each of the cell types found in the tracheal epithelium. Thus, the default niche is promitotic: Multipotential differentiation requires adaptation of the niche. Because lung TSCs are typically evaluated after injury, the behavior of CD49fbright/Sca1+/ALDH+ cells was tested in normal and naphthalene-treated mice. These cells were mitotically active in the normal and repaired epithelium, their proliferation rate increased in response to injury, and they retained label for 34 days. We conclude that the CD49fbright/Sca1+/ALDH+ tracheal basal cell is a TSC, that it generates its own niche in vitro, and that it participates in tracheal epithelial homeostasis and repair.
Clinical Relevance
Identification of the dual-purpose stem and in vitro niche cell and development of an in vitro model system will enable analysis of signals that regulate the tissue-specific stem cell–niche interaction.
A tissue-specific stem cell (TSC) is defined as a cell that self-renews and has a differentiation potential that is equivalent to the cellular diversity of its home tissue (1). These functions are regulated by a specialized microenvironment termed the stem cell niche (2). Classical studies in the hematopoietic system and gut indicate that the niche serves a protective role, possibly by shielding the stem cell from injurious agents or by limiting its exposure to proliferation or differentiation signals. Current paradigms suggest that the niche provides signals that regulate self-renewal of TSC, production of highly mitotic transit-amplifying cells (TACs), and differentiation of TAC into terminally differentiated cell types. A precise understanding of the TSC–niche interaction and its role in tissue homeostasis awaits purification of the TSC and characterization of the niche.
The origin of the stem cell niche is unknown. Because the niche is frequently located at the junction between disparate cell types within a single tissue or at the interface between tissues or involves interactions between the TSC and the extracellular matrix, distinct origins for the niche and the TSC have been suggested (3). However, analysis of dermal stem cells suggests that the TSC and the niche-forming cell are a single entity (4). The possibility of a dual-purpose cell that acts as the TSC and niche-forming cell is also supported by the finding that purified mammary, prostate, and neural TSCs do not require a preexisting niche for appropriate proliferation and differentiation (5). These data suggest that TSCs may generate their own niche.
Several assays were developed to distinguish the TSC from its progeny, the TAC population. Cells can be classified on the basis of mitotic frequency. The duration of the cell cycle varies among known TSC types, but a generally accepted guideline is that TSCs proliferate rarely, whereas TACs proliferate repeatedly. Mitotic frequency is analyzed by pulse-chase analysis using labeled nucleotides (6) or the histone 2B:green fluorescent protein reporter (7). In this label retention assay, TSCs are identified as cells that retain the label, and TACs are identified as those that dilute the label. Distinctive colony morphology, maximal cell generation number, and colony-forming efficiency have also been used to discriminate between TSCs and TACs (8). Clonal origin is established using the statistically rigorous limiting dilution assay (9). Self-renewal and cell autonomous function are tested by serial passage at clonal density. Individually, these assays are not sufficient to identify the TSC. However, they can be used collectively to distinguish TSCs and TACs.
The existence of a tracheobronchial TSC and a specific niche has been postulated (10). However, in vivo analyses of tracheobronchial stem and progenitor cells did not combine the three standards that are critical to the demonstration of a TSC. Label-retention analysis indicated that TSC-like basal cells reside in tracheal gland–duct junction or intercartilaginous zones (11). Lineage tracing studies suggested that a subset of human (12) or mouse (13–15) tracheobronchial basal cells were multipotential progenitors for the differentiated tracheal cell types: basal, secretory, and ciliated cells. However, other basal cell subtypes were identified (14), and the lineage relationships among these populations was not assessed.
The lung's predilection for particle (i.e., cells) expulsion makes in vivo analysis of putative TSCs difficult and prone to failure. Consequently, ex vivo and in vitro assays are preferred for the initial evaluation of putative TSCs. Analysis of human nasal polyp or bronchial cells (16–18), rat tracheal cells (19), and mouse tracheal cells (20) in air–liquid interface (ALI) cultures demonstrated that these cultures support basal cell self-renewal as well as ciliated and secretory differentiation. The ALI method was used to demonstrate that a population of basal cells defined by their cell surface phenotype generated a differentiated epithelium (18). Similar results were reported using tracheal xenografts, indicating equivalence of the in vitro and ex vivo methods. The differentiation potential of mouse keratin 5+ basal cells was tested in ALI (21) or spheroid cultures (13). However, these mouse studies reported only ciliated cell differentiation. The limited differentiation of spheroids suggested that an additional component, possibly another cell type, regulated secretory differentiation (13).
In the present study, we investigated the stem cell–niche interaction using a prospective purification strategy and a novel rim–clone forming assay. A subpopulation of mouse tracheal basal cells, defined by expression of CD49f, Sca1, and aldehyde dehydrogenase activity (CD49fbright/Sca1+/ALDH+), was purified by flow cytometry. Limiting dilution culture and statistical analysis of clonality demonstrated that individual cells exhibited the stem cell characteristics of label retention, self-renewal, and multipotential differentiation. This cell type participated in epithelial homeostasis and repair in vivo. Culture of TSCs at clonal density generated a three-dimensional in vitro niche that reformed in clonal cultures at each of five successive generations. This niche promoted generation of numerous basal cells but prevented differentiation even in the presence of appropriate stimuli. The default niche could be modified by tracheal cells or their secretions to a niche that was permissive for differentiation. These data support the conclusion that the CD49fbright/Sca1+/ALDH+ cell exhibits the properties of a TSC that generates its own niche in vitro.
Materials and Methods
Additional information is supplied in the online supplement.
Reagents
Reagents and sources are noted in Table E1. Immunostaining and flow cytometry antibodies are noted in Tables E2 and E3.
Mouse Strains
Range-finding experiments were conducted using 40 to 45 pooled trachea derived from male or female 129, C57Bl/6, and Fvb/n mice. All analyses were repeated in FVB/n mice. Mice were maintained in an AAALAC-approved facility and screened for pathogens on a quarterly basis. All procedures involving animal use were reviewed and approved by the National Jewish Health Institutional Animal Care and Use Committee.
Naphthalene and Bromodeoxyuridine Treatment
Female Fvb/n mice were treated with 300 mg naphthalene (NA) in corn oil as previously detailed (22). Bromodeoxyuridine (BrdU) was dissolved at 5 mg/ml in saline and delivered at 10 μl/g body weight (intraperitoneally) on a daily basis for 6 consecutive days.
Isolation and Culture of Cells
Slight modifications to previously published tracheal cell preparation and culture methods (20) are described in the online supplement. Hydrocortisone (MP Biomedicals, Solon, OH) 25 ng/ml was added to the culture medium. For flow cytometric analysis, cells were recovered by digestion with 0.3% dispase/0.2% collagenase VIII/0.05% trypsin at 37°C for 1 hour (23). Sorted epithelial cells were cultured on lethally irradiated (5,000 rad) NIH3T3 feeders in modified mMTEC+ (20). Feeder cell density was 3 × 104/cm2. Cultures were grown at 37°C in 5% CO2 for 14 to 30 days. For clonal and subculturing studies, individual rimmed clones were recovered with 0.25% trypsin/0.53 mM EDTA using a cloning cylinder.
Flow Cytometry
Epithelial cells were analyzed and sorted using a Moflo high-speed cell sorter (Dako Cytomation, Carpinteria, CA) equipped with CyCLONE automated cloner. Gates for fluorescence fractionations were set with unstained and isotype controls (see Figure E2 in the online supplement). Single color positive and negative compensation beads were used to compensate for spectral overlap. Aldehyde dehydrogenase (ALDH) activity was evaluated using the ALDEFLUOR assay kit. The negative control was set by inhibiting ALDH activity by preincubation of cells with competitive inhibitor (di-ethyl-amino benzaldehyde) followed by addition of the fluorescent substrate (bodipy-aminoacetate). For clonogenic frequency analysis, cells of interest were directly sorted into 96-well plates containing irradiated 3T3-feeders. Cell cycle analysis was performed by incubation of fixed rimmed clone cells overnight with a 0.5 mg/ml propidium iodide (24). Cell cycle position was analyzed on a FC-500 cytometer (Beckman-Coulter, Brea, CA) and data was processed with the Mod-fit program (Verity Software House, Topsham, ME).
Immunofluorescence
Fluorescence-activated cell–sorted tracheal epithelial cells were deposited on glass slides using a cytospin and stained using standard methods. Images were captured using a microscope (Axiovision; Carl Zeiss Microimaging, Jena, Germany), and data were analyzed with AxioVision digital imaging software or an inverted Zeiss 200M microscope using Intelligent Imaging Innovations Inc. software (3I, Inc., Denver, CO).
Quantitative RT-PCR
Total RNA was prepared from the flow-sorted cells using RNA isolation kit following the manufacturer's instructions. cDNA was prepared and assayed as previously described (25) using Assays-on-Demand (Applied Biosystems, Foster City, CA). mRNA expression was quantified using an ABI Prism 7000 (Applied Biosystems) Sequence Detection System, and values were calculated by the ΔΔCT method (26).
Statistical Analysis
Results are presented as mean ± SD. Differences were evaluated by Students t test using Graph-pad prism (Graph-pad Software Inc., San Diego, CA).
Results
Rimmed Clones Contain Label-Retaining and Label-Diluting Cells
Analysis of mouse tracheal epithelial colony-forming cells identified two types of colonies: nonrimmed and rimmed (Figures 1A and 1B). The frequency of nonrimmed colony-forming cells was approximately 860 colonies in 105. The frequency of rimmed colony-forming cells was approximately 2.5 colonies in 105 cells. Both colony types were derived from a single cell (Figures 1C–1E). Only rimmed clones contained BrdU+ cells on culture Days 12 to 14 (Figure 1I).
Figure 1.
Rimmed clones contain label-retaining and label-diluting cells. (A and B) Colony morphology. Bright-field images of a nonrimmed (A) and rimmed (B) colony generated by unfractionated trach0065al cells. (C–E) Analysis of clonality. Tracheal epithelial cells expressing green flourescent protein (GFP) or red flourescent protein (RFP) were mixed, and endogenous fluorescence was examined on culture Day 14. Merged green and red images illustrate monochromatic GFP+ (C and D) and RFP+ (E) colonies. (F–J) Label retention analysis. Tracheal cell cultures were pulsed with bromodeoxyuridine (BrdU) on culture Days 0 to 3, 3 to 5, 5 to 7, or 12 to 14 and fixed on culture Day 14. BrdU was detected by immunofluorescence analysis (red, arrows). (G) Cells that proliferated during the 0- to 3-day interval (F, J) or the 3- to 5-day interval retained label to Day 14. (H) Cells labeled between Days 5 and 7 did not retain label to Day 14. (I) Numerous cells were labeled with BrdU between Days 12 and 14. (K and L) Molecular phenotype of label-retaining cells. (K) BrdU label–retaining cells (red) coexpress keratin (K)5 (green) or (L) K14 (green). Dual-positive cells are indicated by arrows. Original magnification is noted in the upper right hand corner of each image.
Label retention was used as the initial indicator of a cell that proliferated infrequently relative to its progeny. Cultures were pulsed with BrdU and chased for 2 weeks (Figures 1F–1L). Nonrimmed clones did not contain label-retaining cells (LRCs). In contrast, rare LRCs were present in rimmed clones pulsed between Days 0 and 3 and chased to Day 14 (Figures 1F and 1J). LRCs were located in the rim domain. LRCs were also detected in cultures that were pulsed on Days 3 to 5 and 5 to 7 and chased to Day 14 (Figures 1G and 1H) but were less frequent than those detected on Days 0 to 3.
Dual immunofluorescence analysis was used to determine the cellular phenotype of rimmed clone cells. Cells in the central and rim domains were positive for keratin (K) 5, a basal cell marker (Figure 1K). In contrast, K14+ cells were present only in the rim domain (Figure 1L). BrdU label–retaining cells coexpressed K5 or K14 (Figures 1K and 1L). Rimmed clones did not express differentiation markers associated with secretory cell types, Clara cell secretory protein (CCSP) and Muc5AC, or a marker associated with ciliated cells, acetylated tubulin (ACT) (data not shown).
Prospective Purification of Tracheal-Rimmed Clone-Forming Cells
Existing data suggest that the putative tracheal TSCs were a subset of the basal cell population (11, 12, 14, 15, 27, 28). Consequently, tracheal cells were tested for expression of surface markers that were used to subset basal cells from the epidermis, esophagus, prostate, and mammary gland. Tracheal cell recovery was optimized to retain these markers (Figure E1).
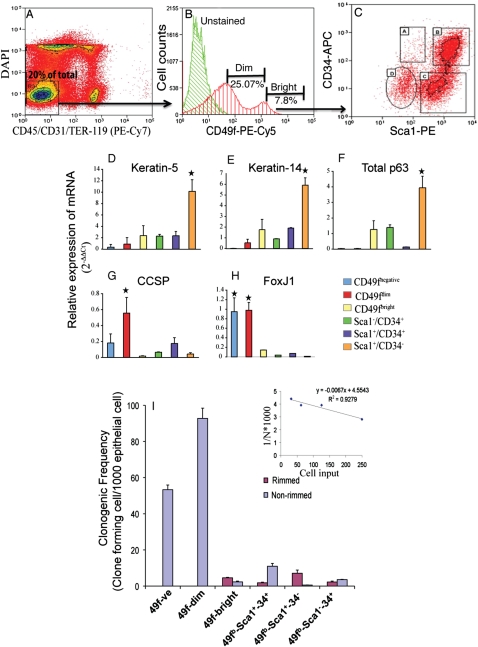
Tracheal cells were fractionated iteratively using flow cytometry. Isotype and postsorting controls are presented in Figure E2. Nonviable and nonepithelial cells were excluded by staining for DAPI, CD45, Ter119, and CD31 (Figure 2A). The remaining cells were 20 ± 3.26% (n = 10) of total and were termed tracheal epithelial cells (TECs). A population of CD49f+ cells was detected and conservatively separated into dim and bright subsets (Figure 2B). CD49fdim cells were 25.07 ± 4.18% (n = 7) of the TECs and expressed CCSP+ and ACT+ predominantly (Figures E3A and E3B; Table 1). CD49fbright cells were 7.96 ± 1.20% (n = 7) of the TECs, and 57 ± 3.8% expressed K5+. Approximately 10% of CD49fbright or CD49fdim K5+ cells coexpressed K14.
Figure 2.
Prospective purification of tracheal rimmed clone-forming cells. All quantitative data are presented as mean ± SD. (A) Contour plot indicating the exclusion markers used in this study: DAPI (dead cells), CD45 (hematopoietic), CD31 (endothelial), and TER-119 (erythrocytes). Viable, CD45−, CD31−, and TER119− cells (R3) were selected for further analysis (arrow). (B) Fractionation according to the expression CD49f (α6 integrin). Green line, unstained cells; red line, CD49f+ cells. The CD49f+ population was divided into two subsets: CD49fdim and CD49fbright. (C) Flow cytometric analysis Sca1 and CD34 expression in CD49fbright cells. The abundance of each subpopulation within the total tracheal cell preparation was (A) 0.1 ± 0.02%, (B) 1.18 ± 0.3%, (C) 0.5 ± 0.05%, and (D) 0.2 ± 0.08% (n = 5). (D–H) Gene expression profile of tracheal epithelial cell subsets. Quantitative RT-PCR was used to determine the abundance of basal cell markers: K5 (D), K14 (E), total p63 (F), Clara cell secretory protein (CCSP) (G), and the ciliated cell marker FoxJ1 (H) relative to a tracheal RNA calibrator (n = 4). *Significance at P < 0.005. (G) Colony-forming cell frequency of tracheal epithelial cell subsets. Predefined numbers of cells from various subpopulations were sorted into 96-well plates containing irradiated NIH-3T3 feeders and cultured for 14 days. Two types of clones were detected: nonrimmed (blue bars) or rimmed (pink bars). Clonogenic frequency for each clone type was calculated by linear regression (inset), where N = colony-forming cell frequency, y = 1/N, and x = cell input. The equation was solved for y = 37%, and the colony forming cell frequency presented as 1,000N. Data are presented as the mean ± SD (n = 3).
TABLE 1.
MOLECULAR PHENOTYPE OF CELLS WITHIN CD49F-POSITIVE POPULATION*
| CD49f+ Subsets |
||
| Cell Type | CD49fdim | CD49fbright |
| K5 | 43 ± 1.57 | 57 ± 3.8 |
| K14 | 10 ± 0.93 | 11.73 ± 1.96 |
| CCSP | 17 ± 0.63 | 0.1 ± 0.023 |
| ACT | 5 ± 0.22 | 0 |
Definition of abbreviations: ACT = acetylated tubulin; CCSP = Clara cell secretory protein; K5 = keratin 5; K14 = keratin 14.
To determine the molecular phenotype of cells present in these populations, cells were cytospun and stained for the basal cell marker K5, the secretory cell marker CCSP, and the nuclear stain DAPI. Similar preparations were stained for the ciliated cell marker ACT, K5, and DAPI. Representation of cells expressing cell-type specific markers was calculated as 100(no. positive cells/no. total cells) (n = 4).
The CD49fbright subset was fractionated on the basis of CD34 and Sca1 expression (Figure 2C). Sca1−/CD34+ (population A), Sca1+/CD34+ (population B), Sca1+/CD34− (population C), and Sca1−/CD34− (population D) accounted for 2.84 ± 1.50%, 52.64 ± 5.82%, 25.6 ± 3.18%, and 7.85 ± 2.62%, respectively, of the CD49fbright population (n = 5). Only population C was enriched for K5+ basal cells (Figure E3C; Table 2).
TABLE 2.
QUANTIFICATION OF CELL TYPES WITHIN CD49FBRIGHT CELL SUBSETS DEFINED BY EXPRESSION OF SCA1 AND CD34*
| Cell Type | Subset A: CD49fb/Sca1−/CD34+ | Subset B: CD49fb/Sca1+/CD34+ | Subset C: CD49fb/Sca1+/CD34− |
| K5 | 10.5 ± 2.18 | 0.656 ± 0.346 | 79.36 ± 5.151 |
| K14 | 3.37 ± 1.28 | 0.124 ± 0.039 | 13.95 ± 1.96 |
| CCSP | 0 | 0.257 ± 0.032 | 0 |
| ACT | 0 | 0 | 0 |
Definition of abbreviations: ACT = acetylated tubulin; CCSP = Clara cell secretory protein; K5 = keratin 5; K14 = keratin 14.
Representation of cells expressing cell type–specific markers were calculated as mentioned in Table 1 (n = 4).
Because some of the markers used for immunofluorescence analysis may be secreted in response to cell isolation (CCSP) or are found in structures susceptible to mechanical stress (ACT), expression of epithelial cell–specific mRNAs was determined by quantitative real-time PCR (Figures 2D–H). The CD49fbright and CD49fbright/Sca1+/CD34− populations were enriched for cells that express the basal cell markers K5, K14, and p63. CCSP and FoxJ1 mRNAs were depleted in these populations. Because Sca1 cell type specificity is debated (29, 30), vimentin was used as a mesenchymal cell marker. Vimentin mRNA was not detected in the CD49fbright population or in its subpopulations.
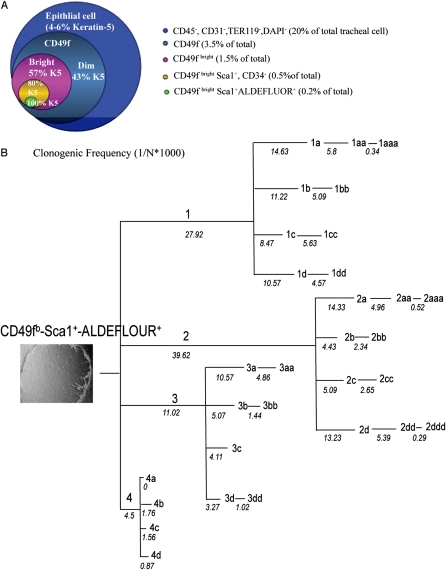
The functional properties of individual cells within each TEC subset were evaluated using a limiting dilution assay (9). A prespecified number of cells (2-fold decrements from 200 to 1 cell/well) were directly deposited into 96-well plates. Linear regression was used to determine clone-forming cell frequency (Figure 2I, inset). The CD49f− and CD49fdim populations formed nonrimmed clones exclusively (Figure 2I, blue bar). In contrast, the CD49fbright population and three of its derivatives, the Sca1+/CD34+, Sac1+/CD34−, Sca1−/CD34+cells, gave rise to nonrimmed and rimmed clones (Figure 2I, magenta bar). CD49fbright/Sca1+/CD34− basal cells generated rimmed clones with the highest frequency (95 ± 3.2%; n = 4). Rimmed clone–forming cell frequency within this population was 74-fold greater than that of the CD49fbright/Sca1+/CD34+ population.
Aldehyde Dehydrogenase Activity Subsets CD49fbright/Sca1+ Basal Cells
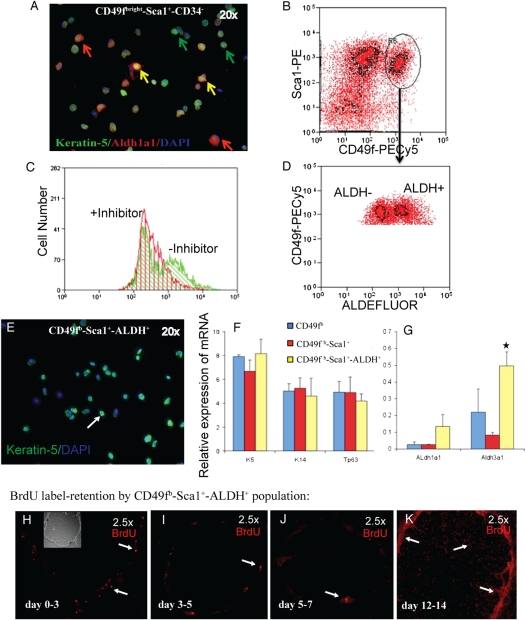
To determine whether aldehyde dehydrogenase (ALDH) activity, a marker for primitive hematopoietic, neural, and mammary epithelial stem cells (31), could distinguish tracheal basal cell subsets, tracheal cells were costained for K5 and ALDH1A1 (Figure 3A). ALDH1A1 immunopositive cells were 52.00 ± 5.32% of CD49fbright/Sca1+/CD34− cells and 43.35 ± 6.17% of CD49fbright/Sca1+/CD34+ cells. These data demonstrate that CD34 did not separate tracheal epithelial cells into discrete ALDH1A1 expressing subsets.
Figure 3.
Subsetting of CD49fbright/Sca1+ basal cells by aldehyde dehydrogenase (ALDH) activity. (A) Dual-immunofluorescence analysis of K5 (green) and ALDH1A1 (red, red arrows) in the CD49fbright/Sca1+/CD34− population. Yellow arrows indicate dual-positive cells. (B) Flow cytometric identification of CD49fbright/Sca1 dual-positive tracheal epithelial cells. Cells selected for further analysis are indicated as R6 (arrow). (C) Flow cytometric analysis of ALDEFLUOR activity in the presence of the diethylaminobenzaldehyde (DEAB) inhibitor (+inhibitor, red) and in the absence of the DEAB inhibitor (−inhibitor, green). (D) Separation of CD49fbright/Sca1+ cells into ALDH-negative (ALDH−) and -positive (ALDH+) fractions. (E) Immunofluorescence analysis of cytospin preparations from CD49fbright/Sca1+/ALDH+ cells. Cells were stained for K5 (green) and DAPI (blue). Arrow indicates a typical K5+ cell. (F and G) Quantitative RT-PCR analysis of gene expression in the CD49fbright (blue), CD49fbright/Sca1+ (red), and CD49fbright/Sca1+/ALDH+ (yellow) subsets. (F) The basal cell markers K5, K14, and Trp63 were analyzed. (G) Expression of ALDH1a1 and ALDH3a1. Mean ± SD (n = 4). *Significance at P < 0.003. (H–K) Proliferation of CD49fbright/Sca1+/ALDH+ cells as a function of time in culture. BrdU label (red) retention was evaluated as described in Figure 1. (H) Labeling Days 0 to 3, (I) 3 to 5, (J) 5 to 7, and (K) 12 to 14. Original magnification: 2.5×. The inset in H is a bright-field image of this clone.
Flow cytometric analysis of ALDEFLUOR activity demonstrated that 0.25 ± 0.021% (n = 8) of TECs possessed diethylaminobenzaldehyde-inhibited ALDH activity (Figure 3C). CD49fbright/Sca1+ cells (Figure 3B, R6) were divided into ALDH+ and ALDH− populations (Figure 3D). CD49fbright/Sca1+/ALDH+ TECs had low side scatter, indicating that they were agranular cells (data not shown). Immunofluorescence analysis indicated that 100% of CD49fbright/Sca1+/ALDH+ cells were K5+ (Figure 3E). Gene expression analysis (Figure 3F) demonstrated that expression of basal-specific mRNAs did not vary between ALDH+ and ALDH− populations. However, ALDH1A1 and ALDH3A1 mRNAs were enriched in ALDH+ cells (Figure 3G).
When cultured in standard mouse TEC culture medium (20) on irradiated NIH3T3 fibroblasts, the CD49fbright/Sca1+/ALDH− population generated only nonrimmed clones. The colony-forming cell frequency (CFCF) was 3.563 ± 0.831 (n = 3). This CFCF is equivalent to that of terminal rimmed clones (see Figure 6). CD49fbright/Sca1+/ALDH−–derived nonrimmed clones did not retain label and could not be serially passaged. This cell type was not pursued further.
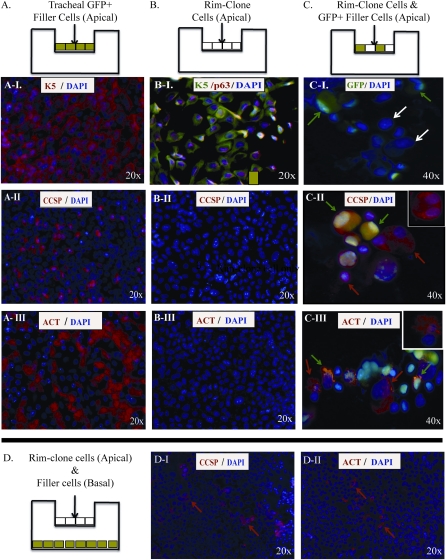
Figure 6.
Differentiation of rimmed clone-forming cells. (A–C) Schematic representing the cell types used to generate air–liquid interface (ALI) cultures. (A) Cultures generated with tracheal epithelial cells from GFP mice (GFP-filler). (B) Cultures generated with rimmed clone cells. (C) Cultures generated using a combination of GFP+ filler cells and rimmed clone cells. (A-I–A-III) Immunofluorescence analysis of differentiated cell types in ALI cultures generated from GFP+ filler cells. (A-I) Expression of the basal cell marker K5 (red) and DAPI (blue). (A-II) Clara cell marker CCSP (red) and DAPI (blue). (A-III) Ciliated cell marker ACT (red) and DAPI (blue). All images are en face. Original magnification: 20×. (B-I–B-III) Immunofluorescence analysis of differentiated cell types in ALI cultures generated from rimmed clone cells. (B-I) K5 (green), p63 (red), and DAPI (blue). (B-II) CCSP (red) and DAPI (blue). (B-III) ACT (red) and DAPI (blue). All images are en face. Original magnification: 20×. (C-I–C-III) Immunofluorescence analysis of differentiated cell types in ALI cultures generated from rimmed clone cells plus GFP+ filler cells. Cells were recovered by trypsinizatin, and cytospin preparations were generated. (C-I) Endogenous GFP fluorescence (green) and DAPI (blue). Green arrows indicate GFP+ filler cells. White arrows indicate GFP− clone cells. (C-II) Endogenous GFP fluorescence (green) and CCSP (red). Arrows indicate filler-derived GFP+/CCSP+ cells (green/red) and rimmed clone-derived GFP−/CCSP+ cells (red). Inset illustrates a rimmed clone-derived GFP−/CCSP+ cell focused on the optical plane of CCSP, a cytosolic protein. (C-III) Endogenous GFP fluorescence (green) and ACT (red). Arrows indicate filler-derived GFP+/ACT+ (green/red) cells and rimmed clone-derived GFP−/ACT+ cells (red). Inset illustrates a clone-derived GFP−/ACT+ focused on the optical plane ACT, an extracellular protein localized to motile cilia on the cell surface. Original magnification: 40×. (D) Analysis of paracrine signaling. Rimmed clone cells were cultured in the apical compartment and GFP+ filler cells were cultured in the basal compartment. Differentiation was evaluated 10 days after establishment of the ALI. Immunofluorescence analysis of differentiation markers in cells present on the luminal surface of the Transwell membrane, which contains the rimmed cells. All images are en face. Original magnification: 20×. (D-I) CCSP (red, arrows), DAPI (blue). (D-II) ACT (red, arrows), DAPI (blue).
The CD49fbright/Sca1+/ALDH+ population was 0.2% of total tracheal cells and 1% of the TECs. All of these cells were positive for K5 and generated only rimmed clones (Figure 3H; data not shown). Selection of this subpopulation resulted in a 100-fold enrichment of the rimmed clone–forming cell compared with unfractionated TECs. Pulse-chase analysis demonstrated that CD49fbright/Sca1+/ALDH+–derived rimmed clones contained rare LRCs and abundant label-diluting cells (LDCs) that were localized to the clone rim (Figures 3I and 3J). A similar distribution of LRCs and LDCs in the K5/TRE-histone 2B model (Figure E4) indicated that BrdU toxicity did not affect proliferation of central region cells (32).
Molecular and Mitotic Phenotype of CD49fbright/Sca1+/ALDH+–Derived Rimmed Clones
CD49fbright/Sca1+/ALDH+ cell–derived rimmed clones were round and suggested that clone growth was radially symmetric (Figures 4A–4D). Immunofluorescence analysis showed that cells within the rim and central regions expressed E-cadherin (Figures 4E and 4F). K5 and K14 were coexpressed in the rim, but central region cells expressed K5 only. CCSP, ACT, and Muc5Ac were not detected (data not shown). These data indicate that CD49fbright/Sca1+/ALDH+–derived rimmed clones were composed of basal cells.
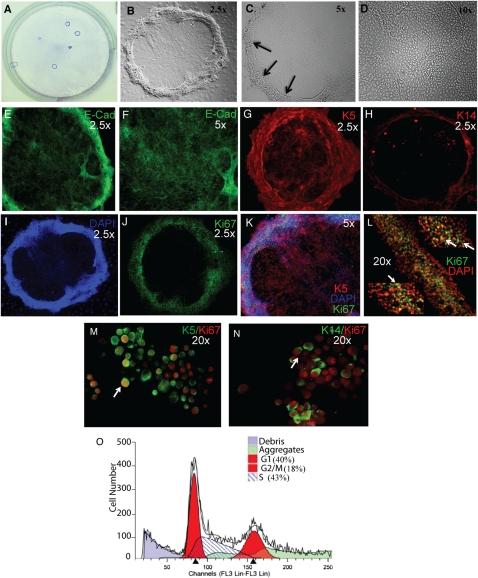
Figure 4.
Phenotypic characterization of rimmed clones generated by CD49fbright/Sca1+/ALDH+ cells. (A) Giemsa stain of rimmed clones in a 6-well plate. Brightfield; original magnification 1×. (B–D) Phase contrast images of (B) a rimmed clone, (C) the three-dimensional rim (arrows), and (D) the cobblestone central region. Original magnifications vary for each panel and are indicated in the upper right corner. (E–H) Immunofluorescence analysis of epithelial markers. (E) E-cad (green) expression in the rim. (F) E-cad expression in the cobblestone region. (G) K5 (red). (H) K14 (red). (I–N) Immunofluorescence analysis of proliferation. Single-color images: (I) DAPI (blue) and (J) Ki67 (green). (K) Merged image for K5 (red), Ki67 (green), and DAPI (blue). (L) Higher magnification of Ki67 (green) and DAPI (red) at three positions within the rimmed region of a rimmed clone. Arrows indicate dual-positive cells. (M and N) Immunofluorescence analysis of (M) K5 (green) and Ki67 (red) or (N) K14 (green) Ki67 (red) on a cytospin preparation of a rimmed clone. Arrows indicate dual-positive cells. (O) Flow cytometric analysis of cell cycle position in cells derived from an individual rimmed clone. Colors are defined in the panel. Representative of 10 analyses.
Ki67 analysis also demonstrated that mitotic cells were abundant and highly localized to the rim domain of rimmed clones on culture Days 12 to 14 (Figures 4I–4L). Rare Ki67+ cells were detected in the central domain (Figure 4K). Cell density, as evaluated by DAPI staining, was very high in the rim but low in the central region (Figures 4I and 4K). Cytospin preparations of rimmed clone isolates were stained for K5, K14, and Ki67 (Figures 4M and 4N). Quantification demonstrated that 77.71 ± 4.12% of K5+ cells were positive for Ki67 and that 76.13 ± 3.45% K14+ cells coexpressed Ki67. The contribution of each cell type to the mitotic pool was 86.06 ± 5.12% for K5+ cells and 73.85 ± 2.66% for K14+ cells. These data, in combination with the demonstration that the majority of rim domain cells were dual positive for K5 and K14 (Figures 4G and 4H), suggest that clone expansion was driven by K5/14 dual-positive cells.
Propidium iodide staining and flow cytometric analysis (24) were used to determine the cell cycle characteristics of passage 1 rimmed clone cells. Evaluation of 10 rimmed clones on culture Day 14 demonstrated that 40 ± 2.37% of cells were in G1 phase, 43 ± 3.8% were in S phase, and 18 ± 1.79% were in G2/M phase (Figure 5O). Only debris was detected to the left of the G1 peak, indicating that few if any cells were apoptotic.
Figure 5.
Self-renewal properties of rimmed clone-forming cells. (A) Summary of the prospective purification strategy for tracheal basal cells. The diameter of each circle represents the abundance of each subpopulation within the original tracheal cell preparation. K5 staining of each population indicates enrichment of basal cells within the population. (B) Self-renewal of individual rimmed clone cells as a function of passage. Colony-forming cell frequency (CFCF) was determined as indicated in Figure 2. Rimmed clone (left panel), phase contrast image; original magnification 2.5×. Only rimmed clones were detected. Passage number for each clone is depicted above the line, and CFCF at each passage is noted below the line. The length of each line is proportional to the CFCF.
Self-Renewal Properties of the Rimmed-Cloned Forming Cell
Self-renewal of individual CD49fbright/Sca1+/ALDH+ cells was evaluated as a function of passage using the limiting dilution assay and the statistical method detailed in Figure 2I. A single clone and its derivatives could be passaged five times (Figure 5B). For example, a single CD49fbright/Sca1+/ALDH+ cell (passage [p]0) resulted in the generation of a clone that could be passaged to generate a second clone, clone 2 (p1). Clone 2 had a CFCF of 39.62, and its derivatives could be passaged three times to generate clones 2a (p2), 2aa (p3), and 2aaa (p4). The CFCF for clone 2 derivatives decreased as a function of passage and was 0.3 to 0.5 at termination. In contrast, clone 4 had an initial CFCF of 4.5 and terminated at p2. Because the average number of cells per clone was 8 × 103 ± 2 × 103 cells (n = 15) on Day 14, these data indicate that the most clonogenic p0 cells generated approximately 40 million progeny.
Differentiation Potential of Rimmed Clone Cells
The differentiation potential of CD49fbright/Sca1+/ALDH+ rimmed clone cells was tested using ALI cultures (Figure 6) (20). Control cultures were generated from green flourescent protein (GFP)+ tracheal cells (Figure 6A and E5) and served as differentiation controls. On differentiation Day 9, numerous K5+ basal cells, CCSP+ Clara-like cells, and ACT+ ciliated cells were detected (Figures 6A-I–6A-III). Enface and cytospin analysis demonstrated that all cells derived from GFP+ cells retained endogenous fluorescence as they proliferated and differentiated in vitro (Figure E5).
ALI cultures were also generated from passage 1 rimmed clones. These cultures were termed “homogenous ALI” (Figure 6B). Homogenous ALI polarized on culture Day 5, demonstrating functional epithelial barrier formation. However, homogenous ALI did not differentiate into CCSP+ or ACT+ cells when subjected to the differentiation protocol (Figures 6B-II and 6B-III). DAPI staining confirmed that cells were present at high density throughout the culture (Figures 6B-II and 6B-III). Immunostaining for K5 and p63 (Figures 6B–6I) demonstrated that homogenous ALI culture cells retained the basal cell characteristics that were previously shown for rimmed clones grown at clonal density on irradiated feeders (Figure 4).
ALI cultures composed of GFP− rimmed clones, and GFP+ “filler cells” were termed “heterogeneous ALI.” These cultures polarized on Day 5. At this time, the epithelium was composed of rimmed clones that contained GFP+ filler cells in in the central cobblestone region and in the rim (data not shown). The high cell density within the rimmed region prevented phenotypic analysis of rimed clone cells and filler cells. Consequently, differentiation was evaluated in cytospin preparations. Identification of GFP+/CCSP+ and GFP+/ACT+ indicated successful differentiation of the filler cells (Figures 6C-II and 6C-III; green arrows). The frequency of these differentiated cell types was similar to that achieved in control ALI cultures (Figures 6A-I–6A-III). GFP− rimmed clone cells that expressed CCSP or ACT were also identified. The frequency of these rimmed clone–derived differentiated cells was similar to that noted for the filler cells.
The hypothesis that tracheal cells induced rimmed clone cell differentiation via secreted factors was tested by culturing rimmed clone cells and tracheal cells in separate compartments of the Transwell culture system (Figure 6D). Differentiation of rimmed clone cells was evaluated on Day 10 and demonstrated numerous CCSP+ (Figure 6D-I) or ACT+ (Figure 6D-II) cells. Green cells were not detected, indicating integrity of the Transwell membrane and its ability to separate the two cell types.
LRCs In Vivo
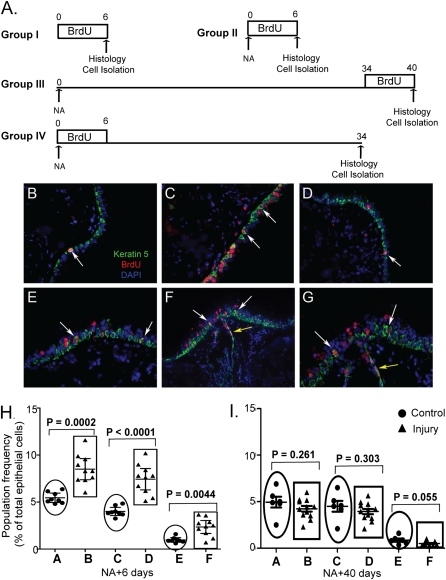
TSCs are typically identified after severe epithelial injury and identified as LRCs or as cells that generate multipotential clones (11, 13–15). Because tracheal LRCs have not been evaluated after naphthalene (NA) injury, the distribution of K5+/BrdU+ cells was evaluated in mice treated as indicated in Figure 7A. In the steady state, K5+/BrdU+ cells were detected throughout the trachea (Figure 7B). An increased number of K5+/BrdU+ cells were detected on NA recovery Day 6 (Figure 7C). The distribution of these cells was similar to that detected in the steady state. Mice treated with NA and recovered after 40 days also contained K5+/BrdU+ cells that were distributed as shown for control and Day 6 (Figure 7D). A rare population of K5+/BrdU+ cells were located below the basement membrane in intercartilaginous regions that did not contain submucosal glands (data not shown). Tissues from mice that were treated with NA, labeled on recovery Days 1 to 6, and chased for 34 days contained K5+ and K5− LRCs (Figures 7E–7G). These cells were more frequent in intercartilaginous regions than in midcartilaginous regions and were also located in submucosal gland duct junctions.
Figure 7.
Contribution of CD49fbright/Sca1+/ALDH+ cells in epithelial homeostasis and repair: (A) Mitotic index of CD49fbright/Sca1+/ALDH+ cells at steady state and after Naphthalene (NA) injury. Four experimental groups were used. Group I (no-injury and 6 consecutive days of BrdU), Group II (NA-injury at day 0 and 6 consecutive days of BrdU), Group III (NA-injury at day 0 and BrdU from day 34-40) and Group IV (NA-injury at day 0 followed by BrdU for 6 consecutive days and chased up to day 34). CD49fbright/Sca1+/ALDH+ cells were isolated by flow-cytometry at the end of each time-period mentioned above, cytospin preparations were generated and immunostained for BrdU. BrdU positive and negative cells were counted and expressed as percentages of total number of cells detected by DAPI staining. (B) Histological analysis of mouse trachea (group I) by immunofluorescence microscopy with anti-BrdU (red) and K5 (green) antibodies. Arrow showed a BrdU/K5 double positive cell. (C) Histology of Group II, (D) Histology of Group III. (E) Immunostaining of tracheal tissue (Group IV) showing the presence of label-retaining cells in the intercartlagenious region. Arrow showed label-retaining K5 positive cells. (F) Anatomical location of BrdU label-retaining cells (Group IV) on the surface epithelium (white arrows) and also in the submucosal gland (yellow arrow). (G) Higher magnification of the picture shown in F. (H) Participation of CD49fbright/Sca1+/ALDH+ cells in epithelial repair in response to NA-injury was assessed at day 6 post-injury by flow cytometry. Epithelial cells expressing CD49fbright (population A&B), CD49fbright/Sca1+ (population C&D) and CD49fbright/Sca1+/ALDH+ (population E&F) were analyzed. Individual animals, control group (n = 8, closed circles) and NA-treated group (n = 10, closed triangles) were evaluated and the representation of various cell subsets is presented as percentage of total epithelial cells. Differences between control and treated group were assessed by non-parametric two-tailed t test. (I) Participation of CD49fbright/Sca1+/ALDH+ cells in epithelial homeostasis on recovery day 40. Experimental design was similar to that explained in (H). Control (n = 6) and treated (n = 12).
CD49fbright/Sca1+/ALDH+ Cells Participate in Tracheal Homeostasis and Repair
To determine whether CD49fbright/Sca1+/ALDH+ cells were NA resistant, the frequency of this cell type was compared in control and NA-treated mice. On recovery Day 6, CD49fbright/Sca1+/ALDH+ cells were 2.35 ± 0.30% of epithelial cells (Figure 7H, group F). This frequency was 3-fold greater than control (Figure 7H, group E; P = 0.004). Similar enrichment of basal cells (Figure 7H, groups A versus B and groups C versus D) suggested that this difference could be accounted for by depletion of CD49fdim Clara and ciliated cells. On recovery Day 40, CD49fbright/Sca1+/ALDH+ cells were 0.51 ± 0.07% of epithelial cells (Figure 7I, group F), and this frequency was similar to control (Figure 7I, group E; P = 0.06). The frequency of CD49fbright and CD49fbright/Sca1+ cells was similar in control and treated mice on Day 40. These data further supported the conclusion that changes in the number of Clara-like and ciliated cells within the total epithelial cell population accounted for the increased frequency of CD49fbright/Sca1+/ALDH+ cells on recovery Day 6.
To determine whether CD49fbright/Sca1+/ALDH+ cells participated in tracheal homeostasis and repair, mitotically active cells were labeled with BrdU in vivo (Figure 7A). CD49fbright/Sca1+/ALDH+ cells were isolated by flow cytometry, and the frequency of BrdU+ cells was determined by immunostaining of cytospin preparations (Figure E6). The mitotic fraction of CD49fbright/Sca1+/ALDH+ cells was 10.3 ± 1.2% in control mice, 56.9 ± 2.18% in mice treated with NA and recovered after 6 days, and 21.2 ± 1.84% in mice treated with NA and recovered after 40 days. This study indicated that CD49fbright/Sca1+/ALDH+ cells were mitotically active under homeostatic conditions before and after repair. The 6-fold increase in their mitotic index indicated recruitment into the mitotic pool after NA treatment.
To determine whether CD49fbright/Sca1+/ALDH+ cells were label-retaining cells, mice were treated with NA, labeled with BrdU on recovery Days 1 to 6, and chased to Day 40 (Figure 7A, Group IV). After a 34-day chase, 17.8 ± 1.7% of CD49fbright/Sca1+/ALDH+ cells were BrdU+, indicating that they had proliferated after injury, survived 5 weeks, and divided infrequently. Of the BrdU+/CD49fbright/Sca1+/ALDH+ cells, 4.68 ± 0.6% had BrdU levels that were equivalent to that detected in cells labeled immediately after NA treatment (Figure E6). These BrdUbright/CD49fbright/Sca1+/ALDH+ cells were 28.30 ± 3.83% of the mitotic fraction.
Discussion
We demonstrated that a specific mouse tracheal epithelial cell type, defined by its surface phenotype and biochemical properties, functions as a self-renewing multipotential progenitor and as an in vitro niche-forming cell. The CD49fbright/Sca1+/ALDH+ cell is a basal cell subtype. It generates LRCs and LDCs that are sequestered within the rim domain of a specific clone type, the rimmed clone. These clones can be serially passaged, at clonal density, for five generations. At each generation, only rimmed clones are produced. When cultured under homogenous conditions, this cell produced a niche that favored production of LDCs. Cells sequestered within this niche were refractory to differentiation stimuli. However, coculture of rimmed clone cells with unfractionated tracheal cells (UTCs) or exposure to the soluble factors produced by UTCs resulted in differentiation. BrdU labeling demonstrated that CD49fbright/Sca1+/ALDH+ proliferated in the normal epithelium in response to NA injury and in the repaired epithelium. Cells that proliferated after NA injury retained label to recovery Day 40, demonstrating the slow-cycling phenotype and longevity associated with TSCs.
Purification of the TSC
We present evidence that CD49fbright/Sca1+/ALDH+ cells are TSCs. The CD49fbright/Sca1+/ALDH+ cell is a K5/14-expressing basal cell (see Figure 2). This molecular phenotype is consistent with a previous analysis of multipotential cells from the human bronchus (12) and the mouse trachea (11). Single CD49fbright/Sca1+/ALDH+ cells could be cultured at clonal density over five passages (see Figure 5), indicating that label-retaining cells are not merely a long-lived cell type. When exposed to differentiation stimuli in the presence of tracheal filler cells, the CD49fbright/Sca1+/ALDH+ cell self-renews the basal cell population and generates differentiated Clara-like and ciliated cells (Figure 6C). Thus, the CD49fbright/Sca1+/ALDH+ cell is multipotential. Previous analyses of putative tracheal TSCs were unable to demonstrate differentiation to the Clara-like cell phenotype (13, 21). Consequently, the present study is the first to demonstrate multipotential differentiation of a single prospectively purified mouse tracheal basal cell into all cell types found in its home epithelium. Collectively, these data indicate that CD49fbright/Sca1+/ALDH+ cells exhibited the essential characteristics of a TSC.
Architecture and Function of the Stem Cell Niche
We propose that the tracheal rimmed clone recapitulates the most basic function of a stem cell niche in vitro: It harbors TSCs. Because a single cell forms the in vitro niche over multiple passages, we suggest that the rimmed domain maintains TSCs in vitro. The importance of the rim as an in vitro TSC niche is indicated by the fact that the tracheal clones that lack the rimmed domain terminate at p0 and do not contain LRCs (see Figure 1). Furthermore, rimmed domain integrity decreases with age and passage. Loss of the rim is associated with a low colony-forming efficiency (see Figure 5). These data suggest that the rim is critical to the maintenance of tracheal TSCs in vitro. This niche is permissive for intermittent TSC cell division and promotes repeated TAC proliferation. This cell-selective effect may be a consequence of heterogeneity within the rim or interaction between rim and central domain cells.
The demonstration that rimmed clones are refractory to differentiation stimuli suggests that the in vitro niche also limits differentiation. Thus, we suggest that the in vitro niche maintains TSCs and TACs at the expense of a differentiated cell production. Coculture of rimmed clones with UTCs disrupts the rim edge, resulting in a mixture of rimmed clone and nonclone cells. These modified clones are responsive to differentiation signals (see Figure 6). The finding that differentiation was also induced by factors elaborated by UTCs residing in the basal compartment indicated that niche modification occurred via paracrine factors rather than direct cell–cell contact (see Figure 6D). Analysis of other epithelial systems suggests that the niche may exist in distinct proliferation and differentiation states. By analogy to these systems (7, 33), we suggest that the in vitro niche is also regulated by nonniche cells.
Analysis demonstrated that holoclones generated from hair follicle TSCs are bordered by a raised edge similar to the tracheal rimmed clone (4). Because holoclone-forming cells exhibited the defining features of TSCs (i.e., self-renewal and multipotency), Blanplain and colleagues indicated that the holoclone models the “bulge niche microenvironment and architecture” (4). An alternative interpretation of the holoclone analysis and our data is that the rimmed domain establishes conditions that are permissive for maintenance of a specific cell type with the properties of a TSC. Because the TSC niche remains an ambiguous entity in vivo, the present analysis serves as a starting point for identification and functional assessment of the TSC niche within in the respiratory epithelium.
Participation of TSCs in Homeostasis and Repair
The BrdU labeling study (Figure 7) indicated that the CD49fbright/Sca1+/ALDH+ cells participated in tracheal homeostasis and repair. These data are consistent with previous reports of label-retaining cells in the detergent and acid injury models (11). However, participation of tracheal TSCs in homeostasis is novel and contrasts with the quiescence of bronchiolar variant CCSP-expressing cell (vCE) in the normal epithelium (34, 35). The tracheal TSC is defined by two surface markers, a biochemical property, and the absence of four markers. Thus, the location of these cells could not be determined in vivo. However, the demonstration that a high percentage of CD49fbright/Sca1+/ALDH+ cells were also LRCs and the finding that LRCs were not restricted to a recognizable anatomical location suggested that TSCs were not sequestered in the gland duct junction or the intercartilaginous regions. This lack of spatial restriction is consistent with distribution of multipotential K14+ cells (M. Ghosh, unpublished) and may be related to the ability of the TSC to generate its own niche.
Supplementary Material
Acknowledgments
The authors thank Drs. Peter Henson and Gregory Downey for critical reading of the manuscript, Douglas Hicks for animal husbandry, Bilan Li for histology, Kaili Weil of Carl Zeiss MicroImaging, Inc., and Joshua Loomis of the NJH Microscopy Core for microscopy training. Drs. Dennis R. Roop, K. Dallaglio, and Susan Majka of the University of Colorado-Denver provided valuable reagents and advice.
Footnotes
This work was supported by NIH grants R01HL075585 and R01HL075585-04S1 (S.D.R.), by Cystic Fibrosis Foundation Research and Pilot Project grants (S.D.R.), and by grant 2T32HL007085 awarded to the Division of Pulmonary Science and Critical Care Medicine, University of Colorado-Denver (M.G.).
This article contains an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2010-0314OC on December 3, 2010
Author Disclosure: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Morrison SJ. Stem cell potential: can anything make anything?. Curr Biol 2001;11:R7–R9 [DOI] [PubMed] [Google Scholar]
- 2.Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells 1978;4:7–25 [PubMed] [Google Scholar]
- 3.Fuchs E. The tortoise and the hair: slow-cycling cells in the stem cell race. Cell 2009;137:811–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 2004;118:635–648 [DOI] [PubMed] [Google Scholar]
- 5.Sneddon JB, Werb Z. Location, location, location: the cancer stem cell niche. Cell Stem Cell 2007;1:607–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotsarelis G, Cheng SZ, Dong G, Sun TT, Lavker RM. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: implications on epithelial stem cells. Cell 1989;57:201–209 [DOI] [PubMed] [Google Scholar]
- 7.Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Defining the epithelial stem cell niche in skin. Science 2004;303:359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrandon Y, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci USA 1987;84:2302–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taswell C. Limiting dilution assays for the determination of immunocompetent cell frequencies: I. Data analysis. J Immunol 1981;126:1614–1619 [PubMed] [Google Scholar]
- 10.Engelhardt JF. Stem cell niches in the mouse airway. Am J Respir Cell Mol Biol 2001;24:649–652 [DOI] [PubMed] [Google Scholar]
- 11.Borthwick DW, Shahbazian M, Krantz QT, Dorin JR, Randell SH. Evidence for stem-cell niches in the tracheal epithelium. Am J Respir Cell Mol Biol 2001;24:662–670 [DOI] [PubMed] [Google Scholar]
- 12.Engelhardt JF, Schlossberg H, Yankaskas JR, Dudus L. Progenitor cells of the adult human airway involved in submucosal gland development. Development 1995;121:2031–2046 [DOI] [PubMed] [Google Scholar]
- 13.Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BL. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA 2009;106:12771–12775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. Basal cells are a multipotent progenitor capable of renewing the bronchial epithelium. Am J Pathol 2004;164:577–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. In vivo differentiation potential of tracheal basal cells: evidence for multipotent and unipotent subpopulations. Am J Physiol Lung Cell Mol Physiol 2004;286:L643–L649 [DOI] [PubMed] [Google Scholar]
- 16.Avril-Delplanque A, Casal I, Castillon N, Hinnrasky J, Puchelle E, Peault B. Aquaporin-3 expression in human fetal airway epithelial progenitor cells. Stem Cells 2005;23:992–1001 [DOI] [PubMed] [Google Scholar]
- 17.Hajj R, Baranek T, Le Naour R, Lesimple P, Puchelle E, Coraux C. Basal cells of the human adult airway surface epithelium retain transit-amplifying cell properties. Stem Cells 2007;25:139–148 [DOI] [PubMed] [Google Scholar]
- 18.Dupuit F, Gaillard D, Hinnrasky J, Mongodin E, de Bentzmann S, Copreni E, Puchelle E. Differentiated and functional human airway epithelium regeneration in tracheal xenografts. Am J Physiol Lung Cell Mol Physiol 2000;278:L165–L176 [DOI] [PubMed] [Google Scholar]
- 19.Randell SH, Comment CE, Ramaekers FC, Nettesheim P. Properties of rat tracheal epithelial cells separated based on expression of cell surface alpha-galactosyl end groups. Am J Respir Cell Mol Biol 1991;4:544–554 [DOI] [PubMed] [Google Scholar]
- 20.You Y, Richer EJ, Huang T, Brody SL. Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am J Physiol Lung Cell Mol Physiol 2002;283:L1315–L1321 [DOI] [PubMed] [Google Scholar]
- 21.Schoch KG, Lori A, Burns KA, Eldred T, Olsen JC, Randell SH. A subset of mouse tracheal epithelial basal cells generates large colonies in vitro. Am J Physiol Lung Cell Mol Physiol 2004;286:L631–L642 [DOI] [PubMed] [Google Scholar]
- 22.Cole BB, Smith RW, Jenkins KM, Graham BB, Reynolds PR, Reynolds SD. Tracheal basal cells: a facultative progenitor cell pool. Am J Pathol 2010;177:362–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niu Y, Epperly MW, Shen H, Smith T, Wang H, Greenberger JS. Intraesophageal mnsod-plasmid liposome enhances engraftment and self-renewal of bone marrow derived progenitors of esophageal squamous epithelium. Gene Ther 2008;15:347–356 [DOI] [PubMed] [Google Scholar]
- 24.Krishan A. Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide staining. J Cell Biol 1975;66:188–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reynolds SD, Zemke AC, Giangreco A, Brockway BL, Teisanu RM, Drake JA, Mariani TJ, Yp D, Taketo MM, Stripp BR. Conditional stabilization of beta-catenin expands the pool of lung stem cells. Stem Cells 2008;26:1337–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative pcr. Genome Res 1996;6:986–994 [DOI] [PubMed] [Google Scholar]
- 27.Rock JR, Randell SH, Hogan BL. Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis Model Mech 2010;3:545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engelhardt JF, Allen ED, Wilson JM. Reconstitution of tracheal grafts with a genetically modified epithelium. Proc Natl Acad Sci USA 1991;88:11192–11196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teisanu RM, Lagasse E, Whitesides JF, Stripp BR. Prospective isolation of bronchiolar stem cells based upon immunophenotypic and autofluorescence characteristics. Stem Cells 2009;27:612–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McQualter JL, Brouard N, Williams B, Baird BN, Sims-Lucas S, Yuen K, Nilsson SK, Simmons PJ, Bertoncello I. Endogenous fibroblastic progenitor cells in the adult mouse lung are highly enriched in the sca-1 positive cell fraction. Stem Cells 2009;27:623–633 [DOI] [PubMed] [Google Scholar]
- 31.Moreb JS, Zucali JR, Ostmark B, Benson NA. Heterogeneity of aldehyde dehydrogenase expression in lung cancer cell lines is revealed by aldefluor flow cytometry-based assay. Cytometry B Clin Cytom 2007;72:281–289 [DOI] [PubMed] [Google Scholar]
- 32.Masterson JC, O'Dea S. 5-bromo-2-deoxyuridine activates DNA damage signalling responses and induces a senescence-like phenotype in p16-null lung cancer cells. Anticancer Drugs 2007;18:1053–1068 [DOI] [PubMed] [Google Scholar]
- 33.Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine: Iii. Entero-endocrine cells. Am J Anat 1974;141:503–519 [DOI] [PubMed] [Google Scholar]
- 34.Reynolds SD, Giangreco A, Power JH, Stripp BR. Neuroepithelial bodies of pulmonary airways serve as a reservoir of progenitor cells capable of epithelial regeneration. Am J Pathol 2000;156:269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giangreco A, Arwert EN, Rosewell IR, Snyder J, Watt FM, Stripp BR. Stem cells are dispensable for lung homeostasis but restore airways after injury. Proc Natl Acad Sci USA 2009;106:9286–9291 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.