Abstract
Most pathogenic mtDNA mutations are heteroplasmic and there is a clear correlation between high levels of mutated mtDNA in a tissue and pathology. We have found that in vivo double strand breaks (DSB) in mtDNA lead to digestion of cleaved mtDNA and replication of residual mtDNA. Therefore, if DSB could be targeted to mutations in mtDNA, mutant genomes could be eliminated and the wild-type mtDNA would repopulate the cells. This can be achieved by using mitochondria-targeted restriction endonucleases as a means to degrade specific mtDNA haplotypes in heteroplasmic cells or tissues. In the present work we investigated the potential of systemic delivery of mitochondria-targeted restriction endonucleases to reduce the proportion of mutant mtDNA in specific tissues. Using the asymptomatic NZB/BALB mtDNA heteroplasmic mouse as a model, we found that a mitochondria-targeted ApaLI (that cleaves BALB mtDNA at a single site and does not cleave NZB mtDNA) increased the proportion of NZB mtDNA in target tissues. This was observed in heart, using a cardiotropic adeno-associated virus type-6 (AAV6) and in liver, using the hepatotropic adenovirus type-5 (Ad5). No mtDNA depletion or loss of cytochrome c oxidase activity was observed in any of these tissues. These results demonstrate the potential of systemic delivery of viral vectors to specific organs for the therapeutic application of mitochondria-targeted restriction enzymes in mtDNA disorders.
Keywords: Mitochondrial, gene therapy, adenovirus, AAV6
Introduction
Recent epidemiological studies indicate that mtDNA-related diseases have a prevalence of 9.2 per 100,0001. In addition, these disorders constitute one of the most common neurometabolic diseases of childhood, with a risk of developing mitochondrial disease of 1 in 5,0002, making them a common cause of morbidity3. Mitochondrial diseases can affect multiple tissues, have variable severity and heterogeneous clinical expression, not only among individuals with different mutations, but also among individuals with the same mutation4. Cardiomyopathy is diagnosed in approximately 17–40% of pediatric patients with mitochondrial disorders5,6 and the presence of cardiomyopathy is indicative of poor prognosis, particularly among patients with cytochrome c oxidase (COX) deficiency6. Approximately 27% of patients with mitochondrial cardiomyopathy are found to carry heteroplasmic mtDNA point mutations7. Eighteen heteroplasmic point mutations have been associated with cardiomyopathies, 11 of which fall within in tRNALeu (UUR) or tRNAIle, the latter appearing to be a hotspot for mutations, leading to isolated or primary mitochondrial cardiomyopathy6. Hepatopathies are seen in between 10–20% of pediatric patients with mitochondrial disease8, being less frequently associated with heteroplasmic mtDNA point mutations than cardiomyopathies. Hepatopathy can be a feature of some common multisystem disorders caused by heteroplasmic mtDNA mutations, such as MERRF9.
Normal expression of the mitochondrial genome (mtDNA) is essential for the biogenesis of the oxidative phosphorylation system (OXPHOS). Tissues from patients with heteroplasmic mtDNA mutations contain mtDNA populations with variable proportions of mutant and wild-type mtDNA10. Disease phenotypes are only observed once the percentage of wild-type mtDNA drops below a threshold level, and because of this, increasing the proportion of wild-type mtDNA in affected tissues by inducing a so-called “heteroplasmy shift”, is seen as a therapeutic strategy for mtDNA disorders11,12. Ours and other groups have successfully induced heteroplasmy shift by delivering restriction endonucleases (RE) that target specific mtDNA sub-populations in mitochondria. Cleaved mtDNA is rapidly degraded allowing repopulation with uncleaved mtDNA13. Using this approach, levels of the pathogenic 8993T-G mutation (MT-ATP6) have been reduced in heteroplasmic cybrid cells using SmaI14 and XmaI15 that target the mutation. Working with asymptomatic heteroplasmic mice that carry NZB and BALB murine mtDNA haplotypes, we have previously shown that a localized heteroplasmy shift can be induced following intra-tissue injection of viral vectors expressing mitochondria-targeted RE (mito-RE)13,16. Heart and liver are both accessible to systemically administered viral vectors and this prompted us to investigate whether heteroplasmy shift could be induced in these two disease-relevant organs following systemic delivery of viral vectors expressing mitochondria-targeted RE.
To target mitochondrial RE to the heart we used AAV6 vectors that have a well characterized tropism for heart17 and to a lesser extent liver and skeletal muscle18. Recombinant adenoviral vectors were used to target the liver as they are highly hepatotropic when administered intravenously19. NZB/BALB heteroplasmic mice were used as a model of heteroplasmy and ApaLI (mito-ApaLI-HA) as the mito-RE.
Materials and methods
Mito-ApaLI–HA Constructs
A synthetic gene coding for the ApaLI restriction endonuclease with a C-terminal HA (Hemagglutinin antigen) tag was purchased from Integrated DNA Technologies (Coralville, IA). The codon usage was optimized for mammalian translation. An N-terminal mitochondrial import signal derived from COX8A was added and the resulting construct was cloned into pAd5-Track for production of rAd5[mito-ApaLI-HA] adenoviral vectors13. The same insert was cloned in an adenovirus associated viral vector (rAAV6[mito-ApaLI-HA]) for the production of viral particles (Fig. S2). In both cases expression was under the control of the cytomegalovirus (CMV) promoters. Respective CMV-driven control viral vectors expressing green fluorescent protein (rAd5[eGFP]), or alkaline phosphatase (rAAV6[AP]) were also produced.
NZB/BALB Mice
Female NZB/BALB heteroplasmic founders were a kind gift from Eric Shoubridge (McGill University)20. These mice carry variable levels of the NZB and BALB murine mtDNA haplotypes in a BALB genetic background and do not display any form of mitochondrial dysfunction.
Virus administration and sample preparation
To achieve systemic delivery, anesthetized adult female mice (6–8 weeks-old) were injected in the right external jugular vein. Viral vector concentration was determined by A260 measurements21,22. For adenoviral vectors 3.0x10e10 viral genomes (vg) of rAd5[mito-ApaLI-HA] or rAd5[eGFP] diluted in 30–50 μl of PBS were injected using a 30-gauge needle attached to a 100 μl gas tight Hamilton syringe via 30-gauge tygon tubing. Identical injections of 1.0 or 5.0 x 10e11 vg of rAAV6[mito-ApaLI-HA] or 1.0 x 10e11 vg rAAV6[AP] diluted in 100 μl of PBS were also performed. For adenovirus studies, pre-injection liver biopsies were obtained from open surgeries after anesthesia on day 0. At 1 or 2 weeks post-injection, liver, kidney and spleen samples were taken following perfusion with chilled PBS. For AAV6 studies, a muscle biopsy was performed on day 0 before injection and multiple tissue-samples were obtained at 2, 8 or 12 weeks post-injection following perfusion with chilled PBS. All the samples were obtained after anesthesia. The University of Miami IACUC approved the studies reported here.
Heteroplasmy analysis and Southern blotting
Total DNA from tissue samples was obtained after phenol-chloroform-extraction. mtDNA heteroplasmy was determined by ‘Last-cycle hot’ PCR23 using mtDNA primers (5,228–5,250; 5,665–5,690). The PCR product was digested with ApaLI, which digests BALB mtDNA at position 5,461 and subjected to electrophoreses in an 8% polyacrylamide gel. The radioactive signal was quantified using a Cyclone phosphorimaging system (Perkin-Elmer, Waltham, MA, USA) as described16. For Southern blotting analysis, five microgram of total DNA was digested with SacI, which linearized mouse mtDNA by cleavage at position 9,047. Digested samples were electrophoresed through 0.8% agarose gels and transferred to zeta-probe membranes (Bio-Rad, USA). mtDNA was detected by hybridization with a probe corresponding to positions 10–10,145 of the mouse mtDNA (accession number AJ512208). To quantify nuclear DNA, the blots were hybridized with a probe corresponding to positions (502–1,515) of the mouse 18S rDNA (accession number BK000964). The radioactive signal was quantified using a Cyclone phosphor-imager system as previously described16.
Western blotting
Fifty micrograms of total protein from liver or cell homogenates was electrophoresed using 4–20% Tris-HCl polyacrylamide gels (Bio-Rad) and transferred to nitrocellulose membranes (Bio-Rad). Rat anti-HA antibody was obtained from Roche Biochemicals and a donkey anti-rat IgG IRDye 800-conjugated secondary antibody from Rockland (Gilbertsville, PA, USA). The Odyssey Infrared Imaging System (LI-COR) was used to quantitate western blots.
Histological and immunohistochemical studies
NZB/BALB mouse hepatocytes were plated onto coverslips and infected with 1x10e3 viral genomes/cell of rAd5[mito-ApaLI-HA] or rAAV6[mito-ApaLI-HA] for 48 h. Cells were then incubated for 30 min at 37°C with 200 nM Mito Tracker Red CMXRos (Invitrogen, Carlsbad, CA) and fixed with 2% paraformaldehyde (PFA) in PBS for 20 min. After a brief treatment with methanol (5 min), a primary anti-HA antibody (Roche) (in 2% bovine serum albumin (BSA) in phosphate-buffered saline (PBS)) was left overnight at 4°C followed by 2 h incubation at room temperature with an Alexa Fluor 488-conjugated secondary antibody (Molecular Probes, Invitrogen). Images were recorded using a confocal microscope LSM510 from Carl Zeiss.
Tissue samples were fixed in 4% PFA and cryopreserved using 30% sucrose in PBS prior to freezing in liquid nitrogen-cooled isopentane and stored at 80°C. For staining samples were sectioned using a cryostat (20 μm) and mounted on Superfrost Plus microscope slides (Thermo Fisher Scientific). Samples from rAd5-injected mice were checked for eGFP expression. Tissue samples from rAAV6-injected mice were frozen in isopentane and sectioned (20 μm). Slices were fixed in 4% PFA, pre-heated for 90 min in PBS at 67°C to eliminate the endogenous alkaline phosphatase and stained for alkaline phosphatase. Chromogenic staining was achieved by incubation with an NBT/BCIP solution (Sigma-Aldrich). Samples for immunofluorescence were treated with 0.5% Triton X-100 in PBS with 2% BSA for 30 min and then stained using anti-HA, anti-SDHA or anti-MTCO1-Alex 594 conjugated as above. For histological studies, samples were frozen in isopentane, fixed in PFA 4% and stained for COX or COX/SDH activities as described24.
Results
mtDNA segregation patterns in NZB/BALB heteroplasmic mice
Previous characterization of mice harboring two types of mtDNA (NZB and BALB) mice has demonstrated an uneven segregation of mtDNA haplotypes in liver, spleen and kidney but not in muscle20. We confirmed the skewed natural segregation of the NZB mtDNA haplotype over 22 months in this heteroplasmic mouse model showing increases in NZB mtDNA in liver and kidney, and decreases in spleen, relative to the amount of NZB mtDNA from ear samples of the same mice obtained at the time of genotyping (3–4 weeks). No changes in the levels of NZB mtDNA were detected in ear samples over time (Fig. S1).
Mito-ApaLI-HA expressed from AAV6 vectors is targeted to mitochondria
We have previously shown that mito-ApaLI-HA expressed from Ad5 vectors is directed to mitochondria13. To confirm that mito-ApaLI-HA expressed from AAV6 vectors (Fig. S2) is also targeted to mitochondria, cultured mouse hepatocytes were infected with with rAAV6[mito-ApaLI-HA] and analyzed 1 week post-transduction by immunocytochemistry. Cells transduced with rAAV6[mito-ApaLI-HA], showed HA expression that colocalized with the mitochondrial dye Mito Tracker Red CMXRos (Fig. S3).
Cardiac expression of rAAV6[mito-ApaLI-HA]
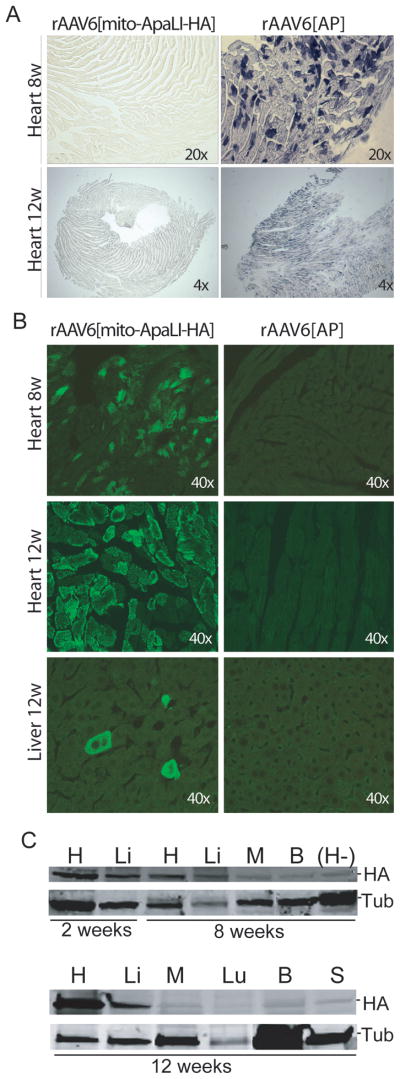
Following intra-jugular injection of 1–5x10e11 vector genome particles (vg)/mouse, the expression of the rAAV6[mito-ApaLI-HA] and the control rAAV6[AP] were analyzed at 2, 8 and 12 weeks post-injection (2 animals per group). In animals injected with the alkaline phosphatase vector, activity was detectable mainly in heart, and only at 8 and 12 weeks (Fig. 1A). Expression of mito-ApaLI-HA in heart was detectable by immunocytochemistry staining with an anti-HA antibody after 8, and 12 weeks, while liver (Fig. 1B) and skeletal muscle (not shown) only showed focal positive staining at 12 weeks. Expression of mito-ApaLI-HA was positive in heart and liver by western blot at 2 weeks post-injection and remained detectable at 8 and 12 weeks post-injection (Fig. 1C). On the other hand, expression of mito-ApaLI-HA was barely detectable in skeletal muscle (M), brain (B), lung (Lu), and spleen (S) at 12 weeks post-injection (Fig. 1C).
Figure 1. Expression of the rAAV6[mito-ApaLI-HA] in targeted tissues.
(A) Alkaline phosphatase (AP) staining performed in heart showed expression of AP after 8 weeks of injection of rAAV6[AP] (1x10e11vg). The staining persisted in heart after 12 weeks (1x10e11vg (not shown) and 5x10e11vg). (B) HA expression post-delivery of the transgenes. We observed high expression in the heart after 8 and 12 weeks, but few cells expressing the transgene in liver, and no expression in the tissues targeted with control rAAV6[AP] . (C) The expression of the rAAV6[mito-ApaLI-HA] was also analyzed by western blotting using the same anti-HA antibody as in figures 1B. Positive expression was detected in heart (H) and liver (Li) after 2 weeks, 8 weeks (1x10e11vg) and 12 weeks (5x10e11vg) post-delivery of the rAAV6[mito-ApaLI-HA], with reduced expression to undetectable levels in other tissues analyzed such as quadriceps muscle (M), brain (B), lung (Lu), spleen (S). A negative control of a heart sample from a non-injected animal (H-) was also analyzed, with no detectable expression of HA. Samples were compared to tubulin used as control.
Liver expression of rAd5[mito-ApaLI-HA]
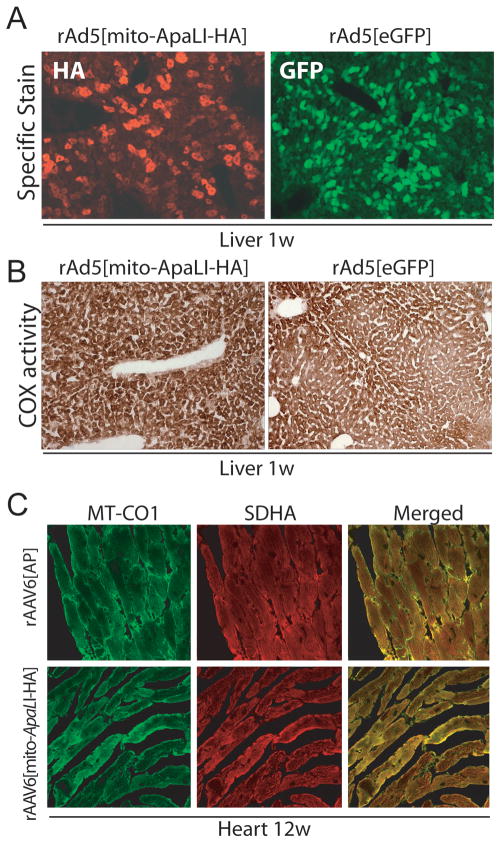
Similar injections were carried out using 3 x 10e10 vg/mouse of the rAd5[mito-ApaLI-HA] (6 animals) or control rAd5[eGFP] (3 animals). Using immunofluorescence, expression of mito-ApaLI-HA was visible from 1 week post-injection (Fig. 2A). In animals that received control vector, direct visualization of eGFP expression was also possible at 1 week post-injection (Fig. 2A).
Figure 2. [Mito-ApaLI-HA] expression in liver or heart is not associated with COX deficiencies.
(A) Expression of Mito-ApaLI-HA in liver after 7 days of the systemic delivery of the transgene rAd[Mito-ApaLI-HA]. Animals were sacrificed and liver samples frozen in liquid nitrogen-cooled isopentane as described in Methods. Twenty-μm liver sections were immunostained for HA (magnification 20x). eGFP liver staining was observed after 7 days systemic delivery of rAd5[eGFP] (magnification 20x). (B) COX staining in liver samples after 1 week post-delivery of the rAd5[mito-ApaLI-HA] samples did not show Cox deficiency when compared to rAd5[eGFP] transduced liver samples (magnification 20x). (C) Transduction of rAAV6[mito-ApaLI-HA] in heart showed no changes in the levels of subunit I of COX (MT-CO1)(green)/SDH (red) immunostaining after 12 weeks post-delivery of the transgene when compared to rAAV6[AP] control samples (magnification 40x).
Cardiac expression of rAAV6[mito-ApaLI-HA] - No mtDNA depletion nor cytochrome c oxidase (COX) deficiency observed in heteroplasmic mice
One concern related to the use of mito-RE is the potential for the rapid mtDNA cleavage to cause a mtDNA depletion. For this reason Southern blots were used to determine mtDNA/nDNA ratios in vector-positive tissues. No evidence of mtDNA depletion (as a ratio to the nuclear 18S rDNA) was observed in any cardiac samples at 2, 8 or 12 weeks post-injection of rAAV6[mito-ApaLI-HA] when compared to samples from mice injected with rAAV6[AP] (Fig. 3A).
Figure 3. Expression of mito-ApaLI-HA in heart and liver does not lead to mtDNA depletion.

(A) MtDNA levels in heart were analyzed by Southern blot in samples obtained 2, 8 and 12 weeks after rAAV6[mito-ApaLI-HA] or rAAV6[AP] injections. No changes in the ratios of mtDNA/nuclear DNA were observed (B) rAd5[mito-ApaLI-HA] or controls rAd5[eGFP] transduced liver samples showed no changes in the ratios of mtDNA/nuclear DNA when comparing pre-injection liver samples to the post-injection samples after 1 or 2 weeks. Each double bar represents one animal sample before and after the delivery of the transgene. mtDNA/nDNA is expressed as ratios of arbitrary densitometric units.
Heart sections from mice injected with rAAV6[mito-ApaLI-HA] were evaluated for cytochrome c oxidase (COX) deficiency using immuno-fluorescence with an antibody against MT-CO1, a mtDNA encoded catalytic subunit of COX. No evidence of loss of MT-CO1 staining or an increase in SDHA staining, indicative of mtDNA-related OXPHOS dysfunction, was seen in any of the transduced fibers at 12 weeks post-injection (Fig. 2C). No alterations in MT-CO1 or SDHA expression were observed in samples of mice injected the control rAAV6[AP] vector (Fig. 2C). No evidence of morphological abnormalities after H&E staining was observed in mice injected with either AAV vector at 12 weeks post injection (data not shown).
Liver expression of rAd5[mito-ApaLI-HA] - No mtDNA depletion nor cytochrome c oxidase (COX) deficiency observed in heteroplasmic mice
Similar analysis of liver DNA taken from mice injected with either rAd5[mito-ApaLI-HA] or rAd5[eGFP] found no evidence of mtDNA depletion in the samples at 2 weeks post-injection. There was a transient reduction in mtDNA levels in mice injected with rAd5[mito-ApaLI-HA] at 1 week post-injection, but normal levels were observed by 2 weeks (Fig. 3B).
Fixed liver sections from mice injected with the adenoviral vectors were evaluated histochemically for COX activity. No COX deficient areas were visible in samples from mice injected with rAd5[mito-ApaLI-HA] taken at 1 week (Fig. 2B) or 2 weeks (data not shown) post injection . In addition, no morphological abnormalities were observed with H&E staining at 2 weeks post injection (data not shown).
Cardiac expression of rAAV6[mito-ApaLI-HA] - Mito-ApaLI-HA expression induces a shift in mtDNA heteroplasmy
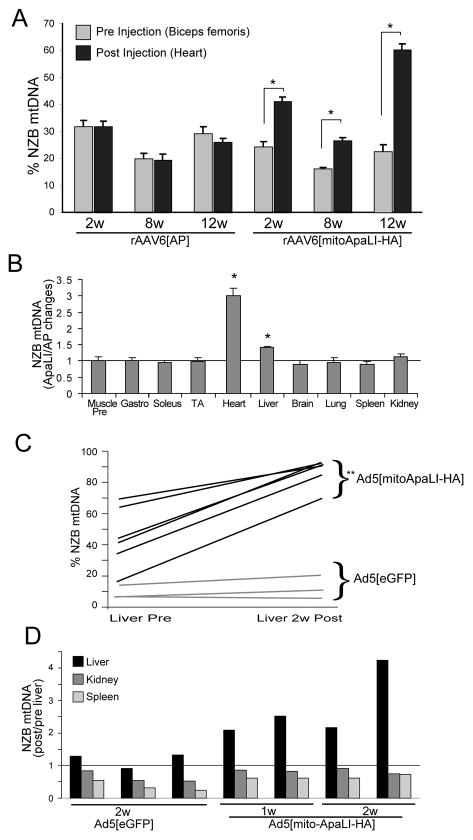
Changes in mtDNA heteroplasmy were evaluated using radiolabeled RFLP after amplification using a RE ApaLI to investigate the levels of NZB mtDNA relative to BALB mtDNA (NZB mtDNA is not cleaved by mito-ApaLI-HA). For mice injected with the AAV6 vectors, changes in NZB levels were determined by comparing the levels in skeletal muscle biopsies taken before injection on day 0 to levels in cardiac muscle samples taken at 2, 8 or 12 weeks post injection (Fig. 4A). We used skeletal muscle as the tissue of choice for baseline mtDNA heteroplasmy because, in this model, mtDNA heteroplasmy is stable over time in skeletal muscle, which has many functional and structural similarities to cardiac muscle20. A significant increase in the levels of NZB mtDNA was observed in all cardiac samples from mice injected with rAAV6[mito-ApaLI-HA] with the highest change seen at 12 weeks post-injection. No change was observed in mice receiving rAAV6[AP] at any time point (Fig. 4A).
Figure 4. [mito-ApaLI-HA] induces a significant shift in mtDNA heteroplasmy in targeted tissues after systemic injection of the transgenes.
The percentage of NZB mtDNA genotype was quantified by the last-cycle hot PCR/RFLP analysis (as described in Methods). (A) DNA samples from injected animals with rAAV6[AP] or rAAV6[mito-ApaLI-HA] were evaluated for the increase of NZB mtDNA after 2 weeks (2w), 8 weeks (8w) and 12 weeks (12w) systemic delivery of the transgenes (one animal per group, 6 animals total). Significant increase of the NZB mtDNA was observed in heart after injection of rAAV6[mito-ApaLI-HA] over time, with no change when the control rAAV6[AP] virus was delivered (3 independent samples from the same mouse *p<0.05). (B) Because of the natural increase of NZB mtDNA in liver and kidney, and decrease in spleen (S1)20, the NZB mtDNA ratio from samples obtained after delivery of the rAAV6[mito-ApaLI-HA] were normalized to the rAAV6[AP] injected samples, and the ratios were compared to muscle samples before the injection. Only liver and heart showed significantly increased NZB mtDNA above 1, when compared to muscle-pre-injected tissue after 12 weeks (3 independent samples from the same mouse *p<0.05). (C) NZB mtDNA was increased in liver of injected mice 1 or 2 weeks after delivery of rAd5[mito-ApaLI-HA] (n=6) with no changes in the level of NZB mtDNA in the control animals after systemic delivery of the rAd5[eGFP] (n=3). Each line connects “Liver Pre” (as values obtained from liver DNA samples before injection) and “Liver 2w Post” as post-injection liver samples after 2 weeks of delivery of the transgenes. Each line represents one animal (**p<0.01). (D) Ratios of NZB percentages after/before treatment with rAd5[mito-ApaLI-HA]. No changes in NZB mtDNA percentages were observed in kidney and spleen. However, liver samples showed a marked increase in the percentages of NZB mtDNA after treatment. Each cluster of bars represents one animal (n=3 for rAd5[eGFP], n=2 for rAd5[mito-ApaLI-HA] 1 week and, n=2 for rAd5[mito-ApaLI-HA] 2 weeks).
At 12 weeks post delivery of rAAV6[mito-ApaLI-HA], changes in NZB levels were also examined in other tissues (Fig. 4B). No significant change was seen in the percentage of NZB mtDNA at 12 weeks post-delivery of the transgene in spleen, brain, lung, kidney, gastrocnemius, triceps, quadriceps, soleus nor tibialis anterior muscles. A significant but small increase was observed in liver (Fig. 4B).
Liver expression of rAd5[mito-ApaLI-HA] - Mito-ApaLI-HA expression induces a shift in mtDNA heteroplasmy
For the mice injected with adenoviral vectors, liver samples for DNA extraction were taken on day 0 prior to injection and at 1 or 2 weeks post-injection. As shown in Fig. 4C, the initial proportion of NZB mtDNA ranged from 4% to 70%. Samples from mice that received the rAd5[eGFP] control vector showed no change in the level of NZB mtDNA at 2 weeks post-injection. However, samples obtained from mice injected with rAd5[mito-ApaLI-HA] showed an increase in the levels of NZB mtDNA at 1 or 2 weeks post-injection. Note that the age-related increase in relative levels of NZB mtDNA seen in liver in NZB/BALB mice is far less than observed here (compare the induced change with the natural drift shown in Fig. S1). We also determined whether there were any change in the percentage of NZB mtDNA in kidney and spleen that are not targeted by adenovirus. No increase in the relative levels of NZB mtDNA was seen in these tissues in any samples (Fig. 4D).
Apparent reduction in ApaLI cleavage in post injection samples is not caused by mutation of ApaLI sites
Because ApaLI was used both to induce heteroplasmy shift in vivo and to diagnose heteroplasmy levels, it is possible that apparent changes in heteroplasmy could be related to mutations of ApaLI sites caused by cleavage and defective repair rather than by a heteroplasmy shift. To investigate this we sequenced the ApaLI site in pre- and post-injection samples from liver of an rAd5[mito-ApaLI-HA]-injected mouse. The only differences visible in electropherograms of the region in both the pre- and post-injection samples corresponded to the sequeces expected from NZB or BALB haplotypes and not to novel mutations (Fig. S4). This confirms that the observed changes in ApaLI RFLP reflect changes in heteroplasmy.
DISCUSSION
Human cells contain on average 1,000 copies of mtDNA compared to the 2n copies of nuclear genome25. The concurrent existence of variable proportions of both wild-type and mutated mtDNA in cells of patients with heteroplasmic mtDNA mutations offers the potential for unique approaches to gene therapy26. The multicopy nature of the mitochondrial genome means that, rather than inserting wild-type versions of mutant genes or silencing them (as it is done with nuclear genes), the selective removal of mutant genomes is a viable strategy for gene therapy. This approach is termed heteroplasmy-shift. It is now well established that restriction endonucleases targeted to mitochondria (mito-RE) are able to bring about heteroplasmy shift by stimulating the digestion of mtDNA with double-strand breaks (DSB) and replication of residual intact mtDNA27,13. In mtDNA heteroplasmic models where a pathogenic mutation creates a unique restriction site not present on co-existent wild-type mtDNA, this cycle enables marked decreases in mutant load and recovery of OXPHOS function14,15. Similarly, we have shown that local injection of rAd5[mito-ApaLI-HA] which targets only BALB mtDNA in the NZB/BALB mouse, leads to a localized heteroplasmy shift towards the NZB haplotype13. We have now explored the systemic delivery of mito-RE using tissue tropic viral vectors.
MtDNA point mutations cause clinical symptoms when the percentage of wild-type DNA drops below a critical threshold, typically 10–40%28. The absolute levels of NZB mtDNA, representing wild-type mtDNA in our model, ranged from 6–70% with a mean of 30%, being a reasonable model for the levels of heteroplasmy seen in patients. Following expression of mito-ApaLI-HA, NZB mtDNA levels in liver ranged from 70–93% (mean 87%). Sequencing through the mitochondrial ApaLI restriction site region confirmed that the observed heteroplasmy shift was not due to cleavage-induced mutation. Heteroplasmy shift of this magnitude would typically be sufficient to reduce the mutant load and protect against a biochemical defect in different cell or tissue types. Nevertheless, in the cases that overall mutant load is very high, the wild-type levels would have to increase relatively fast to compensate for the loss of mutant mtDNA and restore function.
The degree and efficiency of heteroplasmy shift depends on the presence of a residual population of uncut mtDNA. In a “differential multiple cleavage site model”, employing a restriction endonuclease that recognized multiple sites on both mtDNA haplotypes in the NZB/BALB mouse16, we observed only slight changes in heteroplasmy and a transient mtDNA depletion caused by cutting of the entire mtDNA population in cells or tissue. As the mito-ApaLI-HA restriction endonuclease used in this study only cut BALB mtDNA, we observed larger changes in mtDNA heteroplasmy and no mtDNA depletion. There was a small mtDNA depletion in mice injected with rAd5[mito-ApaLI-HA] and sampled at 7 days, but no depletion was present at 14 days, a time where the vector would have been cleared29. The lack of observable COX deficiency in the samples with depletion is most likely due to the transient nature of the depletion and to the fact that the depletion was not severe enough to impair OXPHOS assembly. This is in agreement with the observation that it can take weeks for a detectable mtDNA depletion to manifest as an observable biochemical defect30. No evidence of partially-deleted mtDNA species was found on either Southern blots or using PCR (data not shown). In systems where we have used mitochondria-targeted restriction endonucleases that cleave all the mtDNA in cells or tissues we have observed double strand break-induced recombination and the production of partially-deleted mtDNAs, albeit at low levels31–33. The absence of such mtDNA species in this study mirrors findings of earlier work with mito-ApaLI-HA and is probably due to the efficient repletion of mtDNA via replication of the pool of uncut NZB mtDNA blocking the fixation of rare BALB recombination events13.
Numerous serotypes of AAV have been identified that have different tissue tropisms and binding characteristics17. An elegant analysis of AAV serotypes 1–9 was performed using bioluminescence imaging to compare spatial and temporal differences in reporter gene expression from the different serotypes following systemic administration18. AAV6 was found to transduce heart, skeletal muscle, liver and lung with expression reaching plateau at about 8 weeks when 1x 10e11 viral particles were administered through the tail vein18. We were unable to detect AP expression in liver and lung. Expression in heart was barely detectable at 2 weeks but strong at 8 and 12 weeks post administration of rAAV6[AP]. Heteroplasmy shift was already detectable in heart at 2 weeks post-injection of rAAV6[mito-ApaLI-HA] and increased until 12 weeks. A small change in heteroplasmy was seen in skeletal muscle, only detectable at 8 weeks post-injection. HA expression in skeletal muscle was weak at 8 and 12 weeks post-injection, only detected by immunohistochemistry. Apparent variations in spatial and temporal expression of AP and mito-ApaLI-HA most likely reflect differences in expression assays as AP is detected as total activity at a given time point whereas mito-ApaLI-HA is detected as a cumulative heteroplasmy shift over the course of an experiment. The tissue tropism of rAAV6[mito-ApaLI-HA] as determined by heteroplasmy shift is in accordance with results obtained by Yang et al34 that showed high reporter gene expression in the heart following tail vein injection of AAV6. Similar results were obtained by Palomeque et al17 who showed AAV6 vectors efficiently transduced rat myocardium reaching a plateau at 4 weeks post injection. Skeletal muscle is clearly a relevant tissue for mtDNA gene therapy. High levels of transduction of skeletal muscle have been achieved using very high titers of AAV6 vectors35 or in combination with VEGF as a vasodilator36.
Adenovirus vectors are currently used in more clinical trials than any other viral vector [clinicaltrials.gov at NIH] due to their minimal toxic effect and lack of integration37–39. In rodents and non-human primates, systemic administration of rAd5 results in high transduction in liver19,39–41. In addition, jugular vein injection appears to increase survival compared to tail vein injection42.
Hepatotropism of Ad vectors is mediated via interactions with blood29 and hepatocyte cell surface receptors. Reporter gene analysis and the observed heteroplasmy shift in liver but not kidney or spleen in this study are in accordance with the high hepatotropism of rAd5 vectors. Because we did find evidence of a transient mtDNA depletion in liver at 1 week, one could argue that the change in heteroplasmy could be related to the death of cells with a respiratory defect (mtDNA depletion) and subsequent regeneration of cells with a different mtDNA heteroplasmy state. However, we do not believe this is the case because we did not observe COX deficiency at any time point after injection. Although the robust change in mtDNA heteroplasmy observed in liver is likely to be long lasting, we have not performed long term studies after rAd5[mito-ApaLI-HA].
The ease with which we were able to induce mtDNA heteroplasmy shift in different tissues, using unmodified vector systems with expression driven by simple CMV promoters, reflects the wide expression profile of mito-ApaLI-HA that is capable of inducing heteroplasmy shift. This implies that partial removal of mutant mtDNA genomes can be achieved either through a short burst of high expression, as demonstrated by rAd5[mito-ApaLI-HA] in liver or by longer-term/lower-level expression of rAAV6[mito-ApaLI-HA] in the heart. There is no good evidence of any form of replicative advantage or selection toward mtDNA with pathogenic point mutations in vivo thus a shift in mtDNA heteroplasmy is likely to be long lasting. Off-site transduction of viral vectors (e.g. AAV6 expression in skeletal muscle and liver) probably would be desirable in the case of mitochondrial disease, which are commonly multisystemic disorders.
Among potential therapies to treat mitochondrial diseases associated with mtDNA mutations, changes in mtDNA heteroplasmy is arguably the most promising approach. The use of bacterial restriction nucleases is limited by the requirement that a unique site must be created by the mutation. However, we have shown that under controlled conditions, mtDNA with multiple sites can be differentially susceptible to degradation if one molecule harbors more sites than the other16. A related but broader approach would be the use of mitochondria-targeted zinc finger nucleases with designed recognition sites43. The current work should stimulate research into the therapeutic potential of mitochondrial nucleases in heteroplasmic mtDNA disorders.
Supplementary Material
Figure S1. NZB mitochondrial DNA segregates over time in certain tissues. Ear DNA samples were obtained from heteroplasmic mice [BALB/cByJ (BALB) and NZB/BINJ (NZB) at the time of genotyping (3–4 weeks). DNA samples from liver, kidney and spleen were also obtained from different animals sacrificed at different time points over a 22 months period and tested for the levels of NZB by RFLP-PCR as described in methods. NZB levels in different tissues were normalized by subtracting the NZB mtDNA levels measured in the ear DNA.
Figure S2. Construction and characterization of a rAAV6[Mito-ApaLI-HA]. The genetic code of a synthetic ApaLI gene was optimized for mammalian translation and a HA-tag coding sequence added to the 3’-end. The gene was cloned downstream of the cytochrome c oxidase subunit 8 (COX 8) mitochondrial targeting sequence. The recombinant rAAV6[mito-ApaLI-HA] contained a mitochondrial targeting sequence (cytochrome c oxidase subunit 8 (Cox8)), a gene coding for a mammalianized form of ApaLI restriction endonuclease synthesized in vitro and an HA tag for immunological detection. The construct was cloned in a recombinant adeno-associated serotype 6 backbone, under the control of cytomegalovirus promoter (CMV).
Figure S3. Expression of the recombinant [mito-ApaLI-HA] in cells. To test for the capacity of the rAAV6[mito-ApaLI-HA] to infect cells and localize to mitochondria with Mito Tracker Red CMXRos, hepatocytes derived from heteroplasmic mice were transduced with rAAV6[mito-ApaLI-HA] (5x10e4 vg/cell) for 7 days. Using an anti-HA antibody, we detect colocalization of the transgene in mitochondria of the cells that express the recombinant restriction endonuclease, but not the ones infected with the control (not shown).
Figure S4. Electropherograms of the ApaLI restriction site region. Heteroplasmic mtDNA sequence is shown and pre- and post-injection reads with consensus base-calling below. The ApaLI restriction site [GTGCA`C] on BALB mtDNA (Pre-injection) is marked. The only altered peaks visible post-injection correspond to NZB mtDNA and no novel peaks corresponding to ApaLI site mutations are present.
Acknowledgments
We are grateful to James M. Allen and Jeffrey S. Chamberlain for help preparing high titers of AAV6 and advice. This work was supported by PHS grant EY10804, NS041777, CA85700 and the Muscular Dystrophy Association to CTM.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Schaefer AM, McFarland R, Blakely EL, He L, Whittaker RG, Taylor RW, et al. Prevalence of mitochondrial DNA disease in adults. Ann Neurol. 2008;63:35–39. doi: 10.1002/ana.21217. [DOI] [PubMed] [Google Scholar]
- 2.Haas RH, Parikh S, Falk MJ, Saneto RP, Wolf NI, Darin N, et al. Mitochondrial disease: a practical approach for primary care physicians. Pediatrics. 2007;120:1326–1333. doi: 10.1542/peds.2007-0391. [DOI] [PubMed] [Google Scholar]
- 3.Horvath R, Gorman G, Chinnery PF. How can we treat mitochondrial encephalomyopathies? Approaches to therapy. Neurotherapeutics. 2008;5:558–568. doi: 10.1016/j.nurt.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeviani M, Carelli V. Mitochondrial disorders. Curr Opin Neurol. 2007;20:564–571. doi: 10.1097/WCO.0b013e3282ef58cd. [DOI] [PubMed] [Google Scholar]
- 5.Holmgren D, Wahlander H, Eriksson BO, Oldfors A, Holme E, Tulinius M. Cardiomyopathy in children with mitochondrial disease; clinical course and cardiological findings. Eur Heart J. 2003;24:280–288. doi: 10.1016/s0195-668x(02)00387-1. [DOI] [PubMed] [Google Scholar]
- 6.Towbin JA. Mitochondrial Cardiology. In: Di Maurom SHM, Schon EA, editors. Mitochondrial Medicine. Informa Healthcare; Oxon: 2006. pp. 75–103. [Google Scholar]
- 7.Scaglia F, Towbin JA, Craigen WJ, Belmont JW, Smith EO, Neish SR, et al. Clinical spectrum, morbidity, and mortality in 113 pediatric patients with mitochondrial disease. Pediatrics. 2004;114:925–931. doi: 10.1542/peds.2004-0718. [DOI] [PubMed] [Google Scholar]
- 8.Lee WS, Sokol RJ. Mitochondrial hepatopathies: advances in genetics and pathogenesis. Hepatology. 2007;45:1555–1565. doi: 10.1002/hep.21710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vallance HD, Jeven G, Wallace DC, Brown MD. A case of sporadic infantile histiocytoid cardiomyopathy caused by the A8344G (MERRF) mitochondrial DNA mutation. Pediatr Cardiol. 2004;25:538–540. doi: 10.1007/s00246-003-0446-y. [DOI] [PubMed] [Google Scholar]
- 10.Wallace DC, Brown MD, Lott MT. Mitochondrial DNA variation in human evolution and disease. Gene. 1999;238:211–230. doi: 10.1016/s0378-1119(99)00295-4. [DOI] [PubMed] [Google Scholar]
- 11.Chomyn A, Martinuzzi A, Yoneda M, Daga A, Hurko O, Johns D, et al. MELAS mutation in mtDNA binding site for transcription termination factor causes defects in protein synthesis and in respiration but no change in levels of upstream and downstream mature transcripts. Proc Natl Acad Sci U S A. 1992;89:4221–4225. doi: 10.1073/pnas.89.10.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanna MG, Nelson IP, Morgan-Hughes JA, Harding AE. Impaired mitochondrial translation in human myoblasts harbouring the mitochondrial DNA tRNA lysine 8344 A-->G (MERRF) mutation: relationship to proportion of mutant mitochondrial DNA. J Neurol Sci. 1995;130:154–160. doi: 10.1016/0022-510x(95)00022-t. [DOI] [PubMed] [Google Scholar]
- 13.Bayona-Bafaluy MP, Blits B, Battersby BJ, Shoubridge EA, Moraes CT. Rapid directional shift of mitochondrial DNA heteroplasmy in animal tissues by a mitochondrially targeted restriction endonuclease. Proc Natl Acad Sci U S A. 2005;102:14392–14397. doi: 10.1073/pnas.0502896102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka M, Borgeld HJ, Zhang J, Muramatsu S, Gong JS, Yoneda M, et al. Gene therapy for mitochondrial disease by delivering restriction endonuclease SmaI into mitochondria. J Biomed Sci. 2002;9:534–541. doi: 10.1159/000064726. [DOI] [PubMed] [Google Scholar]
- 15.Alexeyev MF, Venediktova N, Pastukh V, Shokolenko I, Bonilla G, Wilson GL. Selective elimination of mutant mitochondrial genomes as therapeutic strategy for the treatment of NARP and MILS syndromes. Gene Ther. 2008;15:516–523. doi: 10.1038/gt.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bacman SR, Williams SL, Hernandez D, Moraes CT. Modulating mtDNA heteroplasmy by mitochondria-targeted restriction endonucleases in a 'differential multiple cleavage-site' model. Gene Ther. 2007;14:1309–1318. doi: 10.1038/sj.gt.3302981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palomeque J, Chemaly ER, Colosi P, Wellman JA, Zhou S, Del Monte F, et al. Efficiency of eight different AAV serotypes in transducing rat myocardium in vivo. Gene Ther. 2007;14:989–997. doi: 10.1038/sj.gt.3302895. [DOI] [PubMed] [Google Scholar]
- 18.Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 19.Huard J, Lochmuller H, Acsadi G, Jani A, Massie B, Karpati G. The route of administration is a major determinant of the transduction efficiency of rat tissues by adenoviral recombinants. Gene Ther. 1995;2:107–115. [PubMed] [Google Scholar]
- 20.Jenuth JP, Peterson AC, Shoubridge EA. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat Genet. 1997;16:93–95. doi: 10.1038/ng0597-93. [DOI] [PubMed] [Google Scholar]
- 21.Segura MM, Monfar M, Puig M, Mennechet F, Ibanes S, Chillon M. A real-time PCR assay for quantification of canine adenoviral vectors. J Virol Methods. 163:129–136. doi: 10.1016/j.jviromet.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 22.Sommer JM, Smith PH, Parthasarathy S, Isaacs J, Vijay S, Kieran J, et al. Quantification of adeno-associated virus particles and empty capsids by optical density measurement. Mol Ther. 2003;7:122–128. doi: 10.1016/s1525-0016(02)00019-9. [DOI] [PubMed] [Google Scholar]
- 23.Moraes CT, Ricci E, Petruzzella V, Shanske S, DiMauro S, Schon EA, et al. Molecular analysis of the muscle pathology associated with mitochondrial DNA deletions. Nat Genet. 1992;1:359–367. doi: 10.1038/ng0892-359. [DOI] [PubMed] [Google Scholar]
- 24.Diaz F, Thomas CK, Garcia S, Hernandez D, Moraes CT. Mice lacking COX10 in skeletal muscle recapitulate the phenotype of progressive mitochondrial myopathies associated with cytochrome c oxidase deficiency. Hum Mol Genet. 2005;14:2737–2748. doi: 10.1093/hmg/ddi307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Satoh M, Kuroiwa T. Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell. Exp Cell Res. 1991;196:137–140. doi: 10.1016/0014-4827(91)90467-9. [DOI] [PubMed] [Google Scholar]
- 26.Koene S, Smeitink J. Mitochondrial medicine: entering the era of treatment. J Intern Med. 2009;265:193–209. doi: 10.1111/j.1365-2796.2008.02058.x. [DOI] [PubMed] [Google Scholar]
- 27.Srivastava S, Moraes CT. Manipulating mitochondrial DNA heteroplasmy by a mitochondrially targeted restriction endonuclease. Hum Mol Genet. 2001;10:3093–3099. doi: 10.1093/hmg/10.26.3093. [DOI] [PubMed] [Google Scholar]
- 28.Gardner JL, Craven L, Turnbull DM, Taylor RW. Experimental strategies towards treating mitochondrial DNA disorders. Biosci Rep. 2007;27:139–150. doi: 10.1007/s10540-007-9042-3. [DOI] [PubMed] [Google Scholar]
- 29.Baker AH, McVey JH, Waddington SN, Di Paolo NC, Shayakhmetov DM. The influence of blood on in vivo adenovirus bio-distribution and transduction. Mol Ther. 2007;15:1410–1416. doi: 10.1038/sj.mt.6300206. [DOI] [PubMed] [Google Scholar]
- 30.Sorensen L, Ekstrand M, Silva JP, Lindqvist E, Xu B, Rustin P, et al. Late-onset corticohippocampal neurodepletion attributable to catastrophic failure of oxidative phosphorylation in MILON mice. J Neurosci. 2001;21:8082–8090. doi: 10.1523/JNEUROSCI.21-20-08082.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srivastava S, Moraes CT. Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Hum Mol Genet. 2005;14:893–902. doi: 10.1093/hmg/ddi082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukui H, Moraes CT. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum Mol Genet. 2009;18:1028–1036. doi: 10.1093/hmg/ddn437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bacman SR, Williams SL, Moraes CT. Intra- and inter-molecular recombination of mitochondrial DNA after in vivo induction of multiple double-strand breaks. Nucleic Acids Res. 2009;37:4218–4226. doi: 10.1093/nar/gkp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang L, Jiang J, Drouin LM, Agbandje-McKenna M, Chen C, Qiao C, et al. A myocardium tropic adeno-associated virus (AAV) evolved by DNA shuffling and in vivo selection. Proc Natl Acad Sci U S A. 2009;106:3946–3951. doi: 10.1073/pnas.0813207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blankinship MJ, Gregorevic P, Allen JM, Harper SQ, Harper H, Halbert CL, et al. Efficient transduction of skeletal muscle using vectors based on adeno-associated virus serotype 6. Mol Ther. 2004;10:671–678. doi: 10.1016/j.ymthe.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 36.Gregorevic P, Blankinship MJ, Allen JM, Crawford RW, Meuse L, Miller DG, et al. Systemic delivery of genes to striated muscles using adeno-associated viral vectors. Nat Med. 2004;10:828–834. doi: 10.1038/nm1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Everett RS, Hodges BL, Ding EY, Xu F, Serra D, Amalfitano A. Liver toxicities typically induced by first-generation adenoviral vectors can be reduced by use of E1, E2b-deleted adenoviral vectors. Hum Gene Ther. 2003;14:1715–1726. doi: 10.1089/104303403322611737. [DOI] [PubMed] [Google Scholar]
- 38.Nicklin SA, Wu E, Nemerow GR, Baker AH. The influence of adenovirus fiber structure and function on vector development for gene therapy. Mol Ther. 2005;12:384–393. doi: 10.1016/j.ymthe.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 39.Kishi Y, Kuba K, Nakamura T, Wen J, Suzuki Y, Mizuno S, et al. Systemic NK4 gene therapy inhibits tumor growth and metastasis of melanoma and lung carcinoma in syngeneic mouse tumor models. Cancer Sci. 2009 doi: 10.1111/j.1349-7006.2009.01184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sullivan DE, Dash S, Du H, Hiramatsu N, Aydin F, Kolls J, et al. Liver-directed gene transfer in non-human primates. Hum Gene Ther. 1997;8:1195–1206. doi: 10.1089/hum.1997.8.10-1195. [DOI] [PubMed] [Google Scholar]
- 41.Shayakhmetov DM, Li ZY, Ni S, Lieber A. Analysis of adenovirus sequestration in the liver, transduction of hepatic cells, and innate toxicity after injection of fiber-modified vectors. J Virol. 2004;78:5368–5381. doi: 10.1128/JVI.78.10.5368-5381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boquet MP, Wonganan P, Dekker JD, Croyle MA. Influence of method of systemic administration of adenovirus on virus-mediated toxicity: focus on mortality, virus distribution, and drug metabolism. J Pharmacol Toxicol Methods. 2008;58:222–232. doi: 10.1016/j.vascn.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minczuk M, Papworth MA, Miller JC, Murphy MP, Klug A. Development of a single-chain, quasi-dimeric zinc-finger nuclease for the selective degradation of mutated human mitochondrial DNA. Nucleic Acids Res. 2008;36:3926–3938. doi: 10.1093/nar/gkn313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. NZB mitochondrial DNA segregates over time in certain tissues. Ear DNA samples were obtained from heteroplasmic mice [BALB/cByJ (BALB) and NZB/BINJ (NZB) at the time of genotyping (3–4 weeks). DNA samples from liver, kidney and spleen were also obtained from different animals sacrificed at different time points over a 22 months period and tested for the levels of NZB by RFLP-PCR as described in methods. NZB levels in different tissues were normalized by subtracting the NZB mtDNA levels measured in the ear DNA.
Figure S2. Construction and characterization of a rAAV6[Mito-ApaLI-HA]. The genetic code of a synthetic ApaLI gene was optimized for mammalian translation and a HA-tag coding sequence added to the 3’-end. The gene was cloned downstream of the cytochrome c oxidase subunit 8 (COX 8) mitochondrial targeting sequence. The recombinant rAAV6[mito-ApaLI-HA] contained a mitochondrial targeting sequence (cytochrome c oxidase subunit 8 (Cox8)), a gene coding for a mammalianized form of ApaLI restriction endonuclease synthesized in vitro and an HA tag for immunological detection. The construct was cloned in a recombinant adeno-associated serotype 6 backbone, under the control of cytomegalovirus promoter (CMV).
Figure S3. Expression of the recombinant [mito-ApaLI-HA] in cells. To test for the capacity of the rAAV6[mito-ApaLI-HA] to infect cells and localize to mitochondria with Mito Tracker Red CMXRos, hepatocytes derived from heteroplasmic mice were transduced with rAAV6[mito-ApaLI-HA] (5x10e4 vg/cell) for 7 days. Using an anti-HA antibody, we detect colocalization of the transgene in mitochondria of the cells that express the recombinant restriction endonuclease, but not the ones infected with the control (not shown).
Figure S4. Electropherograms of the ApaLI restriction site region. Heteroplasmic mtDNA sequence is shown and pre- and post-injection reads with consensus base-calling below. The ApaLI restriction site [GTGCA`C] on BALB mtDNA (Pre-injection) is marked. The only altered peaks visible post-injection correspond to NZB mtDNA and no novel peaks corresponding to ApaLI site mutations are present.