Summary
Polg mtDNA mutator mice are important models for investigating the role of acquired mtDNA mutations in aging. Despite extensive study, there remains little consensus on either the etiology of the progeroid phenotype or the mtDNA mutation spectrum induced by disrupted polymerase-γ function. To investigate the latter we have developed a novel, pragmatic approach we term “Mito-seq”, applying next generation sequencing to enriched, native mtDNA. Regardless of detection parameters we observed an increase of at least two orders of magnitude in the number of mtDNA single nucleotide variants in Polg mutator mice compared to controls. We found no evidence for the accumulation of canonical mtDNA deletions but multimers of the mtDNA control region were identified in brain and heart. These control region multimers (CRMs) contained heterogeneous breakpoints and formed species that excluded the majority of mtDNA genes. CRMs demonstrate that polymerase-γ 3′–5′ exonuclease activity is required for preserving mtDNA integrity.
Introduction
A central tenet of the mitochondrial theory of aging is that the accumulation of somatic mtDNA mutations plays a causative role in aging. DNA polymerase-γ is the only DNA polymerase in mitochondria and provides an attractive target for genetic manipulation of mtDNA mutation rates. The PolgD257A mtDNA mutator mouse (Kujoth et al., 2005; Trifunovic et al., 2004) carries a missense mutation which drastically reduces 3′–5′ exonuclease activity required for proof-reading. Homozygous PolgD257A/D257A mice have a progeroid phenotype, with alopecia, loss of body fat, kyphosis, anemia and osteoporosis present by six months, and a life-span of around 12 months. Understanding how somatic mtDNA mutations contribute to a progeroid phenotype has been hindered by a lack of consensus on the somatic mtDNA mutation spectrum of PolgD257A/D257A mice (Kraytsberg et al., 2009; Vermulst et al., 2009; Edgar et al., 2010). To overcome the limitations of conventional mtDNA mutation screening techniques we have applied next generation sequencing to native mtDNA enriched via organelle purification (see Methods). This approach, that we term Mito-seq, differs from other applications of next generation sequencing to mtDNA as it requires no a priori assumption of the mutation spectrum present in a sample before sequencing. Thus it enables detection of rearrangements not represented in PCR-derived libraries (He et al., 2010) and it removes the risk of enrichment of nuclear-mtDNA pseudogenes during homology-dependent array capture (Vasta et al., 2009). In addition, the application of paired-end sequencing facilitates the detection of rearrangements (see Supplemental Data). We have evaluated Mito-seq in tissue from Polgwt/wt and PolgD257A/D257A mice and murine mtDNA-less LMTK- ρ0 cells. Our results expand the mutation spectrum of PolgD257A mice and demonstrate that polymerase-γ 3′–5′ exonuclease activity is required for maintenance of mtDNA integrity.
Results
Mito-Seq is not influenced by nuclear-mtDNA pseudogene sequences
Mito-seq involves enrichment of mitochondria from a tissue homogenate or cells, followed by total DNA extraction, and library preparation (see Methods). As total DNA derived from purified mitochondria invariably contains nuclear DNA, interference from the hundreds of nuclear-mtDNA pseudogene sequences (NUMTS) present in mammalian genomes is a concern (Woischnik and Moraes, 2002). To determine whether NUMTS affect Mito-seq analysis, we sequenced whole-cell DNA from murine LMTK-ρ0 cells that do not contain any mtDNA. Of 5,304,393 reads that aligned to the murine whole genome, only 104 (0.002% of aligned reads) aligned to mtDNA (Supplemental Table S1) and no evidence of read clustering was observed (data not shown). The low level of interference is likely due to a number of factors. Most importantly, NUMTS are not present in highly repetitive genomic DNA, thus no single region of mtDNA is highly represented in the nuclear genome. As they constitute less than 0.15% of the nuclear genome (Woischnik and Moraes, 2002) and our whole genome coverage was less than 10%, the probability of any given NUMTS being represented in native DNA libraries of this size is very low. On the basis of such low levels of interference we conclude that NUMTS have no confounding influence on Mito-seq analysis.
Characteristics of Mito-seq assemblies
Paired-end Mito-seq libraries were prepared from brain and heart of 44 and 27 week old PolgD257A/D257A mice (P1 and P2 respectively), and 43 and 12 week old Polgwt/wt mice (W1 and W2 respectively). Assemblies were built using quality-trimmed reads and a gapped, local alignment with CLCbio Genomics Workbench (GWB). This approach protected against assembly errors due to short insertion/deletions (indels) and mismatches at the ends of reads. Average coverage depth was between 1,717x and 6,876x and a typical GC-biased coverage pattern was observed (Bentley et al., 2008). For tissues from P1 and W1, the percentage of total reads aligning to mtDNA ranged from 3.6–9.0% and for P2 and W2 78.3–81.0% (Supplemental Table S1). The differences in mtDNA content in these libraries being due to differences in nuclear DNA carryover as a result of protocol optimization. In all cases over 97% of reads aligned to the mouse whole genome, thus confirming library quality.
PolgD257A/D257A mtDNA contains elevated levels of heteroplasmic single nucleotide variants and small indels
Total unfiltered variance in PolgD257A/D257A assemblies was higher than in Polgwt/wt assemblies, where 84.5% and 84.9% of sequence reads assembled without conflict in W1 brain and heart respectively, and only 75.7% and 73.0% of reads assembled without conflict in P1 brain and heart. Prior to filtering, total conflicts exceeded 12,800–16,400 in all assemblies, including controls. To reduce the influence of sequencing and assembly errors in single nucleotide variant (SNV) analysis, detection of heteroplasmic SNVs was carried out using stringent read quality, call quality, call environment and call frequency filtering of total conflicts using the GWB SNP detection algorithm. Importantly, the use of call frequency filtering excluded detection of single copy SNVs restricting analysis to absolute quantitation of low frequency clonal SNVs. Two orders of magnitude more SNVs were detected in PolgD257A/D257A assemblies than controls under all conditions (Supplemental Table S2 and Workbook S1). To provide a point of reference, assembly and detection parameters that called zero SNVs in W1 assemblies were used to select PolgD257A/D257A variants for analysis. Using these parameters when minimum call frequencies were adjusted to control for differences in average coverage, between 100 and 712 SNVs were detected in PolgD257A/D257A assemblies (Table 1). An alternate normalization assembling partial data sets from PolgD257A/D257A samples to build assemblies with the same coverage as Polgwt/wt assemblies detected between 514 and 1,070 SNVs in brain and heart respectively. Heteroplasmy levels and the number of SNVs over 1% abundance were similar in all PolgD257A/D257A assemblies at 5.6% – 8.3% and 33 – 46 in brain, 7.2% and 58 in heart respectively. Of the 100 most abundant SNVs in brain and heart of P1, 76–84% were present in both tissues. In contrast the identity of SNVs in brain between the PolgD257A/D257A littermates P1 and P2 was only 4–12%.
Table 1. Overview of Mito-seq data.
SNV and indel data is corrected for differences in coverage using differences in minimum counts or assembly of partial data sets (parenthesis). Normalized read pair data and BLAST data refers to normalization of read pair frequencies to the ratio of total aligned read pairs within each assembly.
| Subject information | Tissue | Brain | Brain | Brain | Heart | Heart | Brain |
| Mouse ID | P1 | W1 | P1 | W1 | P2 | W2 | |
| Genotype | PolgD257A/D257A | Polgwt/wt | PolgD257A/D257A | Polgwt/wt | PolgD257A/D257A | Polgwt/wt | |
| Age (weeks) | 44 | 48 | 44 | 48 | 27 | 12 | |
| SNV and indel data | Total SNPs | 253 (514) | 0 | 712 (1070) | 0 | 100 | 15 |
| % identity vs brain P1 | X | - | 76 | - | 12 | - | |
| % identity vs heart P1 | 84 | - | X | - | 8 | - | |
| % identity vs brain P2 | 4 | - | 0 | - | X | - | |
| Total Indels | 15 (17) | 4 | 21 (23) | 4 | 8 | 4 | |
| Indels in OL | 2 | 1 | 2 | 1 | 2 | 2 | |
| Indels in N[>=4] | 11 | 2 | 16 | 3 | 5 | 2 | |
| Read pair data | Total aligned pairs | 747,583 | 236,629 | 381,669 | 289,841 | 1,091,157 | 543,069 |
| Pairs with extended read distance (F---R) | 80 | 39 | 33 | 97 | 47 | 14 | |
| Pairs with R-F orientation | 10,827 | 42 | 2,518 | 84 | 1,993 | 13 | |
| Normalized (F---R) | 80 | 123 | 65 | 250 | 32 | 28 | |
| Normalized (R-F) | 10,827 | 133 | 4,932 | 217 | 1,993 | 26 | |
| BLAST data | Reads with F-R oriented segments | 47 | 40 | 32 | 31 | - | - |
| Reads with R-F oriented segments | 3,307 | 35 | 767 | 16 | - | - | |
| Normalized (F-R) | 47 | 115 | 61 | 81 | - | - | |
| Normalized (R-F) | 3,307 | 100 | 1,475 | 42 | - | - |
The use of gapped, local sequence alignment enabled screening for heteroplasmic indels of ≤3 bp. Heteroplasmic indels in the poly-A tract of the light-strand origin of replication (OL) were the most abundant in every assembly, present at 1.0–1.7% abundance in Polgwt/wt assemblies and 2.6%–4.9% in PolgD257A/D257A assemblies. OL was a region of low coverage in all Mito-seq assemblies probably due to the fact that it forms a strong stem-loop structure, an observation supported by the accumulation of unpaired forward-oriented sequence reads 5′ of the stem loop and reverse-oriented sequence reads 3′ of it (data not shown). Outside of OL most indels also fell in poly-N[>=4] homopolymer tracts (Table 1).
PolgD257A/D257A mitochondria contain abundant control region multimers
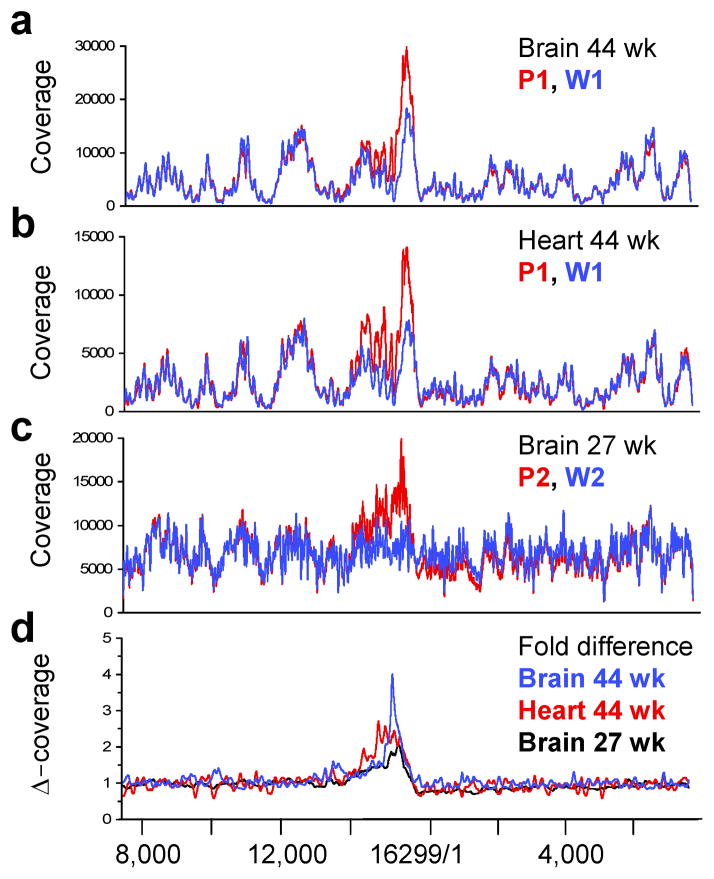
Large mtDNA deletions have been suggested by some (Vermulst et al., 2008), but not all studies (Kraytsberg et al., 2009; Edgar et al., 2009), to contribute to the pathology of PolgD257A/D257A mice. We screened for canonical deletions that do not remove the nt1/16,299 origin of the circular mtDNA reference sequence by extracting read pairs with a standard F-R read orientation (forward 5′ – reverse 3′) but extended read distances (see Supplemental Data). Surprisingly, very few such pairs were identified in any assembly (Table S1). However, normalized coverage maps of Polgwt/wt and PolgD257A/D257A assemblies identified a complex copy number variation in PolgD257A/D257A mice extending from ~nt14,000–16,200 (Fig. 1a–c). Peak increases in coverage of 2 – 4-fold were seen in brain and 2.5-fold in heart (Fig. 1d). Since the copy number variation extended into the mtDNA control region (nt15,423–16,299) we refer to these alterations as Control Region Multimers (CRMs). Within this region we identified a large number of read pairs that aligned with a R-F read orientation (reverse 5′ – forward 3′) indicative of non-canonical deletions or tandem duplications, which are indistinguishable by this analysis (see Supplemental Data). When normalized to total aligned pairs, between 1,993 and 10,827 such pairs were identified in brain and heart from PolgD257A/D257A mice, but only 26 – 217 in brain and heart from Polgwt/wt mice. Moreover, 90% of these events were clustered within the CRM region (Fig. 2a, c).
Figure 1. CRMs in PolgD257A/D257A Mito-seq assemblies.
Coverage overlay of P1 (red, PolgD257A/D257A) and W1 (blue, Polgwt/wt) libraries from brain (a) and heart (b) aligned to a control region-centric mtDNA reference sequence. (c) Coverage overlay of P2 (red, PolgD257A/D257A) and W2 (blue, Polgwt/wt) libraries from brain aligned to the same reference. Overlays were normalized to average coverage between nt8,000–10,000. (d) Fold differences in coverage for each pair (Δ-coverage) using a 100 bp rolling average. P1-W1 brain shown in blue, P1-W1 heart in red and P2-W2 brain in black. Numbering below figure corresponds to standard murine mtDNA reference sequence (NC_005089).
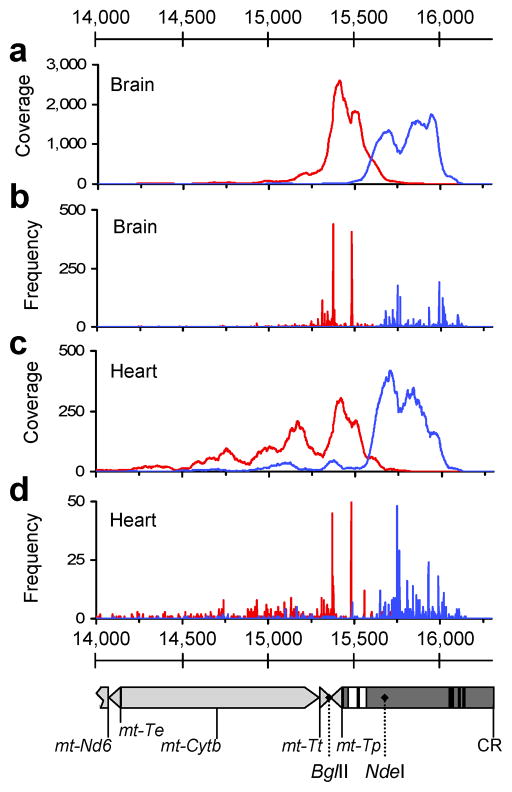
Figure 2. Localization of R-F oriented read pairs and R-F breakpoints within the CRM region in PolgD257A/D257A assemblies.
(a, c) Coverage maps of R-F oriented read pairs extracted from brain and heart assemblies from P1 (PolgD257A/D257A). Forward oriented reads are shown in blue and reverse oriented reads in red. (b, d) Frequencies of breakpoints positions in brain and heart libraries from P1. Positions of 5′ bases (F) shown in blue and 3′ bases (R), in red. Detail of mtDNA nt14,000–16,299 aligned to panels a-d is shown below. Genes (light grey) and control region (dark grey) are depicted. ETAS1 and -2 (extended termination sequence) are represented as white boxes and CSBI, -II and -III (conserved sequence blocks) as black boxes. Relevant restriction sites are labeled with dotted lines.
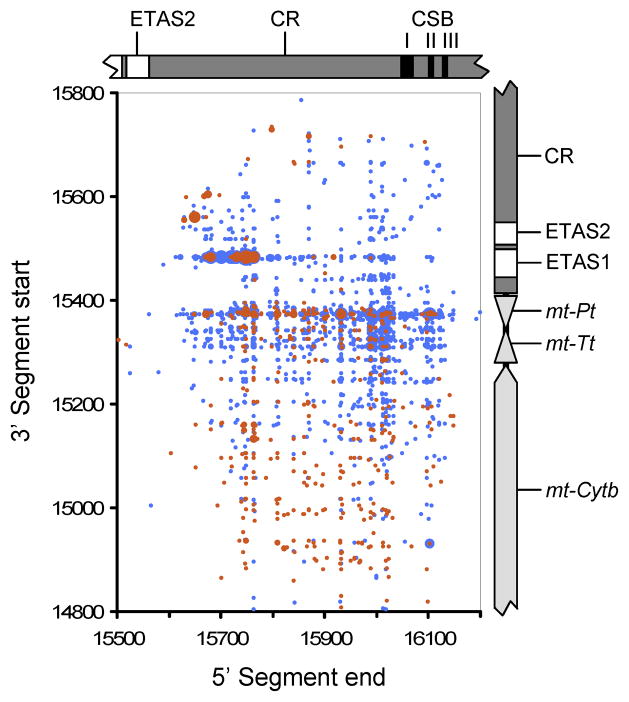
We were able to examine breakpoints at base-level resolution by using gapped BLAST to align unassembled reads against the mtDNA reference sequence and then parsing results to extract reads with high scoring segment pairs. Identical to the paired-end alignments above, only few F-R breakpoints, but a large number of R-F breakpoints were identified in PolgD257A/D257A mice. After normalizing, 1,475 – 3,307 R-F breakpoints were present in brain and heart of PolgD257A/D257A mice, compared to 42 – 100 in Polgwt/wt mice. Dot-plots visualizing R-F breakpoint locations showed linear clustering with a predominantly non-clonal distribution (Fig. 3). There did not appear to be any clear association between breakpoint locations and control region elements such as ETAS1/2 or CSBI/II/III involved in mtDNA transcription and replication. Breakpoint locations indicated that rearrangements had a unit length of 200–800 bp with an average of 566 bp. Coverage maps of CRM units derived from sequenced breakpoint locations from P1 brain and heart demonstrated that regions around ETAS1/2 were retained in most CRMs with peak coverage around nt15,600 present in 96.5 % of CRMs from brain and 82.8 % from heart. In contrast sequences around CSBI/II/III were rarely present (10.9 % and 5.8 % respectively, Fig S1). The pool of 4,074 R-F breakpoints that we sequenced in PolgD257A/D257A brain and heart contained little evidence of recombination at direct repeats. Only 11.4–12.6% of breakpoints had overlap of 3–14 bp and 10–11.2% of 2 bp. In contrast, 74.2–77.2% had an overlap of 0–1 bp. When considering all F-R breakpoints not associated with CRMs from both Polgwt/wt brain and heart 95% contained direct repeats of >=2 bp, whereas in PolgD257A/D257A mice this figure dropped to 55%.
Figure 3. Dot-plot of breakpoints from PolgD257A/D257A brain and heart (P1).
For each breakpoint, the position of the 5′ base (F) is plotted against the position of the 3′ base (R), equivalent to the blue and red frequencies respectively in Figures 2b and 2d. The frequency of each unique breakpoint is indicated by dot size with frequencies binned into nine equally sized bins from 1 (
 ) to 119 (
) to 119 (
 ) occurrences in brain and six bins from 1 (
) occurrences in brain and six bins from 1 (
 ) to 24 (
) to 24 (
 ) occurrences in heart. Genetic maps of mtDNA are aligned opposite each axis with detail as in Figure 2.
) occurrences in heart. Genetic maps of mtDNA are aligned opposite each axis with detail as in Figure 2.
CRMs are distinct from wild-type mtDNA
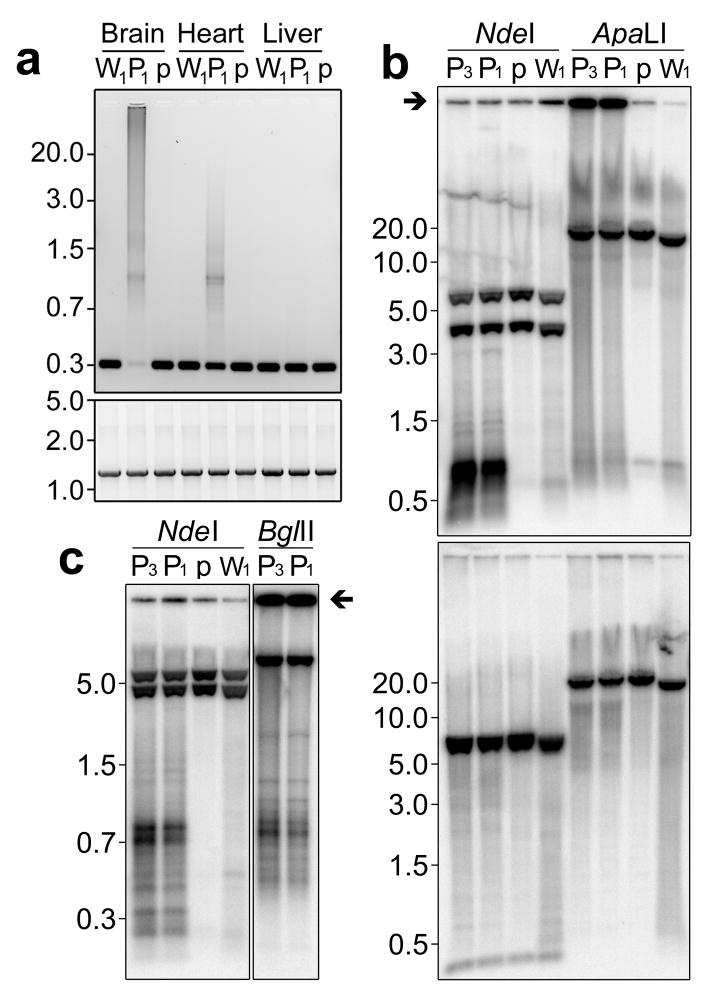
Amplification using long-extension PCR with primers located inside the CRM region resulted in large smeared products in PolgD257A/D257A brain and heart samples (Fig. 4a). In contrast, brain and heart of Polgwt/wt and PolgD257A/wt mice and also liver of PolgD257A/D257A mice produced single bands indicating an absence or very low levels of CRMs. Only by increasing the number of cycles in the PCR we were able to detect CRMs in liver. This analysis also detected CRMs in skeletal muscle, and to a lower extent in intestine, lung and testis. (Fig S2). Using primers outside the CRM region, we amplified only wild-type sized fragments. (Fig. 4a). This was confirmed using PCR with only one of each primer pair inside the CRM region and by Southern blotting where no fragment length polymorphism was seen for a fragment spanning the control region (data not shown). To determine whether CRMs accumulate in normal murine aging, total DNA from cortex or whole brain of 9 mice from 108–147 weeks of age was amplified using primers within the CRM region. No evidence for the presence of CRMs was found.
Figure 4. Probing CRM structure using PCR and Southern blotting.
(a) Resolution of long-extension PCR products amplified using PCR primers within the CRM region (upper panel) and outside of it (lower panel) using DNA from brain, heart and liver of Polgwt/wt (W1), PolgD257A/D257A (P1) and PolgD257A/wt (p) mice. (b) Southern blots of total DNA from brain of mice described in panel (a) with an additional PolgD257A/D257A sample (P3). DNA was digested with NdeI or ApaLI and hybridized with a probe against the control region (upper panel) or an alternate probe against nt8,926 – 10,081 (lower panel). (c) Resolution of CRMs using high percentage agarose gels following NdeI or BglII digestion. Immobile control region signal is indicated with arrows in panels b and c. NdeI cleaves wild-type mtDNA at 5 sites. The control region probe binds wild-type NdeI fragments of 5.9 and 3.8 Kb and the alternate probe binds NdeI fragments of 6.3, 5.9 and 0.2 Kb.
To further resolve CRMs, total DNA from brain of PolgD257A/D257A, PolgD257A/wt and Polgwt/wt mice was investigated using Southern blotting with probes encompassing the control region (nt15,196–16,127) or an alternate probe spanning nt8,926–10,081 (Fig. 4b, c). Following digestion with ApaLI, which cleaves mtDNA at a site distal to the control region, substantial levels of immobile signal were seen in PolgD257A/D257A samples when blots were probed with the control region probe (upper panel Fig. 4b, see also Fig. S3) but not when probed with the alternate probe, which only detected the linearized full length mtDNA (lower panel Fig. 4b). Breakpoint mapping indicated that ~95% of CRM sequences retained the NdeI restriction site at nt15,665. Digestion with NdeI mobilized control region signal resulting in abundant banding below 1.5 Kb in PolgD257A/D257A samples (Fig 4c). Similar banding was also seen using BglII with the exception that a significant amount of signal remained immobile. This is consistent with the fact that only 26% of CRM sequences retain the BglII restriction site nt15,336 (Fig. 4c). Densitometry revealed that 74.6–73.5 % of total control region signal was immobile in PolgD257A/D257A samples when cleaved with ApaLI and that cleavage with NdeI released 65.1–71.6% of total control region signal as <1.5 Kb bands. These numbers match the peak CRM coverage seen in P1 which at 4-fold is equivalent to a 75% increase in control region DNA. Together these data demonstrate that CRMs form massive DNA species distinct from full length mtDNA.
CRMs are associated with mtDNA depletion and increased mitochondrial gene expression
Southern blotting using probes against the nuclear 18S rDNA and mtDNA was used to determine relative mtDNA copy number. mtDNA was detected using a probe hybridizing to nt1,210–3,364, a region not present in CRMs and outside the 11kb region duplicated in mtDNA theta structures resulting from stalled replication (Bailey et al., 2009). We found that in PolgD257A/D257A mice full length mtDNA levels were increased 2.29-fold in liver but reduced to 0.55-fold wild-type levels in brain (Table 2). Absolute steady state levels of the mitochondrial mRNAs encoding mt-Co1, mt-Nd1, mt-Nd5 and mt-Nd6 were increased in both tissues but when normalized to relative mtDNA copy number there was no overall increase in liver yet a 2.49 – 3.47-fold increase in transcripts in brain (Table 2). Absolute levels of control region RNA were increased in both PolgD257A/D257A liver and brain but again when normalized to relative mtDNA copy number, an increase was only observed in brain. The magnitude of the increase in control region RNA in PolgD257A/D257A brain (8.78-fold) suggests that CRMs are transcribed.
Table 2. Relative mitochondrial mRNA expression and relative mtDNA copy number.
qPCR derived ΔΔCT values for mitochondrial RNAs from age matched PolgD257A/D257A (n=4, mice P4–P7) and Polgwt/wt littermates (n=4, mice W3–W6). Two tail t-test indicated all ΔCT values to be significant (p=0.014 to 6.4E-05). Relative mtDNA copy number was determined using Southern blotting. Data is also presented normalized to relative mtDNA copy number “(normalized)”.
| mt-Co1 | mt-Nd1 | mt-Nd5 | mt-Nd6 | Control region | mtDNA | |
|---|---|---|---|---|---|---|
| Brain | 1.91 | 1.37 | 1.61 | 1.67 | 4.83 | 0.55 |
| Liver | 3.24 | 1.83 | 2.56 | 2.96 | 1.65 | 2.29 |
| Brain (normalized) | 3.47 | 2.49 | 2.93 | 3.04 | 8.78 | 1 |
| Liver (normalized) | 1.41 | 0.80 | 1.12 | 1.29 | 0.72 | 1 |
Discussion
mtDNA mutator mice are important models for interrogating mtDNA replication and the mitochondrial theory of aging. In PolgD257 strains, the loss of polymerase-γ 3′–5′ exonuclease activity required for proof-reading leads to the accumulation of heteroplasmic single nucleotide variants (Trifunovic et al., 2004). We investigated the abundance of heteroplasmic SNVs using high coverage next generation sequencing. In our study total unfiltered variance was higher in PolgD257A/D257A assemblies than Polgwt/wt assemblies, confirming their high mutant load. Applying call filtering, two orders of magnitude more heteroplasmic SNV loci were consistently detected in PolgD257A/D257A assemblies using a range of assembly and detection parameters. We also identified increased levels of heteroplasmic indels in homopolymer tracts in PolgD257A/D257A mice in agreement with previous reports (Stewart et al., 2008). Similar indels have been noted at high levels in certain and wild-type mouse strains (Sachadyn et al., 2008) implying that where tolerated, indels my occur relatively frequently, underlying the importance of gapped assembly of mtDNA sequencing data. As we sampled whole organs, the most abundant SNVs and indels we detected were those fixed early in development during either oogenesis (Wai et al., 2008) or early embryogenesis. This is supported by our finding of a higher identity of abundant SNVs between different tissues of the same mouse than between littermates.
Our data provided further evidence that PolgD257A/D257A mice do not accumulate canonical mtDNA deletions spanning the major arc between OH and OL. Stalled replication forks (Bailey et al., 2009) that would have created extended CNVs were also not observed in brain or heart. Instead we identified abundant CRMs comprising multiple copies of sequence around the 5′ end of the mtDNA control region in brain, heart and skeletal muscle but not other tissues. PCR and Southern blotting confirmed that CRMs comprised DNA species distinct from wild-type mtDNA and formed massive high-molecular structures that could be mobilized with specific restriction enzymes. This is compatible with the results obtained by Kasahara and co-workers in mtDNA of PolgD181A brain after BglII digestion (Kasahara et al., 2006). The large size and heterogeneity of CRMs meant that we were unable to determine their structure. Free single unit mini-circles can be excluded due to the immobility of undigested CRM DNA. Catenated single- or multiple-unit mini-circles, branched linear tandem duplications or combinations of these are possible structures.
PCR and Southern blotting showed that CRMs accumulated in PolgD257A/D257A mice and not in age matched PolgD257A/wt or Polgwt/wt mice suggesting an association with a progeroid phenotype. While the elevated levels of control region RNA present in PolgD257A/D257A brain implies CRMs may be transcriptionally active, mitochondrial mRNA levels in brain were increased relative to controls both in terms of absolute transcript levels and transcripts relative to mtDNA copy number. This indicates that CRMs do not have a negative influence on mitochondrial transcription. Increased mitochondrial transcript levels in PolgD257A/D257A brain are likely a compensatory mechanism to counteract the reduced mtDNA copy number, although unbalanced transcription could have functional consequences.
A mild mtDNA depletion was associated with the presence of CRMs in PolgD257A/D257A brain that was not apparent in PolgD257A/D257A liver where CRMs were not readily detectable. This suggests that CRMs may negatively influence mtDNA replication and maintenance. As mtDNA is thought to be attached to mitochondrial membranes via the control region (Albring et al., 1977) and CRMs contain multimers of part of the control region, it is possible that the presence of CRMs may affect mtDNA distribution within large cells such as neurons.
The mechanism by which CRMs accumulate is limited to both tissue and mtDNA location. The apparent restriction to brain and muscle may reflect the clearance of cells with high CRM loads in tissues with higher mitotic potential rather than differences in mtDNA metabolism. CRMs were not present at detectable levels in brains of aged wild-type mice indicating that they are consequence of the loss in Polg 3′–5′ exonuclease activity.
Current models of mtDNA replication include termination in the control region (Brown et al., 2005; Yasukawa et al., 2006). Similarly, the termination of replication of circular prokaryotic chromosomes is localized and carefully controlled (Schaeffer et al., 2005). Aberrant termination of chromosome replication leads to replichore collision and the induction of a process known as “pathological DNA replication”, the formation of massive branched structures containing multiple copies of replication origins and termini, that are immobile under electrophoresis (Rudolph et al., 2009b). These features bear a striking similarity to our findings as undigested CRMs are immobile and replication origins and termini are both present in the control region of mtDNA. Pathological DNA replication arises from uncontrolled replication initiated on 3′ displaced leading strands (Rudolph et al., 2009a), removal of which requires either 3′–5′ exonuclease activity or a RecG 5′–3′ helicase activity (Rudolph et al., 2009b). The manifest presence of polymerase-γ at sites of replichore collision makes this enzyme a good candidate for removal of 3′ displaced strands via 3′–5′ exonuclease activity (Johnson and Johnson, 2001). If so this would be analogous to the role of polymerase-δ 3′–5′ exonuclease activity in Okazaki fragment maturation (Jin et al., 2001). The loss of 3′–5′ exonuclease activity in PolgD257A/D257A mice may increase the life-time of 3′ displaced strands and thus may increase the likelihood of replicative cascades similar to pathological DNA replication initiating on mtDNA. While this might provide a model for multiplication of the control region it does not explain the recombination breakpoints sequenced in CRMs. The paucity of micro-homology within such sequences argues against homologous recombination or strand invasion. We have previously found evidence for non-homologous end joining (NHEJ) in some mtDNA deletion breakpoints in transgenic mice (Fukui and Moraes, 2009). NHEJ of double stranded products of aberrant replication initiation or fork reversal could satisfy the creation of such breakpoints. It is also interesting to the note the very low levels of direct repeats in PolgD257A/D257A breakpoints outside CRMs. This is in agreement with previous studies (Vermulst et al., 2008) and suggests that Polg 3′–5′ exonuclease activity may be required for exposing single stranded DNA for homologous recombination.
Human mutations in POLG are most often associated with progressive external ophthalmoplegia (PEO). PEO is also caused by mutations in PEO1, encoding a mitochondrial 5′–3′ helicase stimulated by 3′ displaced strands (Korhonen et al., 2003). Given the pathological association, it is possible that PEO1 may perform an analogous function to RecG in recovery from replichore collision. Indeed, breakpoints similar to those identified here, with one segment mapping to the 3′ segment (R) cluster around 15,370bp (Fig. 3), have been cloned in mice expressing mutant Twinl, the murine homologue of PEO1 (Tyynismaa et al., 2005). R-F breakpoints consistent with those in CRMs have also been sequenced in patients with Kearns Sayre syndrome and their family members (Brockington et al., 1993), patients with mitochondrial myopathy (Manfredi et al., 1995), large haplogroup screens (Torroni et al., 1994) and aged subjects (Wei et al., 1996). The breakpoints were ascribed to tandem duplications of the mtDNA control region within full length mtDNA despite the fact that amplification across complete duplication units using primers distal to the lesion was not possible (Brockington et al., 1993) just as we observed for CRMs. This raises the possibility that CRMs in PolgD257A/D257A mice may resemble abnormal mtDNA species that accumulate in mitochondrial myopathies and aging.
Methods
Mice
Identification (ID), age, sex, Polg genotype and background information: P1, 44 wk ♀, PolgD257A/D257A C57BL/6 (sib P2); P2, 27 wk ♀, PolgD257A/D257A C57BL/6 (sib P1); P3, 53 wk ♂, PolgD257A/D257A C57BL/6 (maternal sib P1 and P2); P4, 48 wk ♀, PolgD257A/D257A C57BL/6; P5, 47 wk ♂, PolgD257A/D257A C57BL/6; P6, 47 wk ♂, PolgD257A/D257A C57BL/6; P7, 47 wk ♀, PolgD257A/D257A C57BL/6; p, 55 wk ♂, PolgD257A/wt C57BL/6; W1, 48 wk ♀, Polgwt/wt C57BL/6 (non-PolgD257A line); W2, 12 wk ♂, Polgwt/wt BALB/c (heteroplasmic NZB/BALB/c mtDNA); W3, 47 wk ♂, Polgwt/wt C57BL/6 (PolgD257A line); W4, 47 wk ♀, Polgwt/wt C57BL/6 (PolgD257A line); W5, 47 wk ♀, Polgwt/wt C57BL/6 (PolgD257A line); W6, 46 wk ♂, Polgwt/wt C57BL/6 (PolgD257A line). PolgD257/+ founders and tissues were kindly donated by Professor Tomas Prolla (Kujoth et al., 2005).
Preparation of enriched mtDNA samples
Whole brains (~0.30–0.50g), were dissected and homogenized on ice using 12–24 strokes of a teflon-glass Dounce homogenizer in 3 ml of 200 mM mannitol, 50 mM sucrose, 10 mM HEPES (pH 7.0), 1 mM ETDA, 1x Roche Complete protease inhibitors. Whole heart (0.12–0.17g), was prepared identically except that the tissue was shredded prior to homogenization. Un-broken tissue was removed by centrifugation at 600 g and supernatants were centrifuged at 6,000–12,000 g to obtain mitochondria-enriched pellets. Pellets were re-suspended in 2 ml 250 mM sucrose, 10 mM HEPES (pH 7.0), 1 mM ETDA and diluted 1:1 with 50% Nycodenz (w/v), 10 mM HEPES (pH 7.0), 1 mM ETDA. The resulting sample in 25% Nycodenz was layered between equal volumes of 30% and 20% Nycodenz in 10 mM HEPES (pH 7.0) and 1 mM ETDA. Gradients were centrifuged for 120 minutes at 95,000 g and swollen, intact mitochondria were collected from the 25%–20% boundary. Fractions were diluted 1:1 with 250 mM sucrose, 10 mM HEPES (pH 7.0), 1 mM ETDA and mitochondrial pellets were obtained by centrifugation at 16,000 g. Aspirated pellets were re-suspended in 300 μl of 30 mM TRIS (pH 8.3), 100mM NaCl, 10 mM EDTA. Proteinase K (Qiagen) and SDS were added to 2 mAU/ml and 0.5% (w/v) respectively. Samples were incubated 8–12hrs at 55°C. Total nucleic acids were extracted using 50:48:2 phenol:cholorfom:isoamyl-alcohol (v/v/v) followed by two extractions with 24:1 cholorfom:isoamyl-alcohol (v/v). Nucleic acids were then pelleted and washed by ethanol precipitation (NaAc) and re-suspended in 50μl of 10mM TRIS (pH 8.4). RNA was digested by treatment with RNAse A and digestion products were removed using Sephadex G50 spin columns (GE, Roche).
Paired-end sequencing
MtDNA-enriched paired-end libraries were prepared for sequencing using first party kits according to manufacturers instructions (Illumina). Libraries were sequenced using an Illumina GAII sequencer and images were analyzed using pipeline 1.4, except for samples P2 and W2, which were analyzed using pipeline 1.3. 2x 52 bp paired-end reads were acquired for P1 brain and heart, W1 brain, and LMTK ρ0 cells.1x 52/1x 37 bp paired-end reads were acquired for W1 heart, and 2x 34 bp paired-end reads were acquired for P2 and W2. P1 and W1 brain and heart libraries were each run in single channels of the same chip; P2 and W2 libraries were each run in single channels of a second chip; and the LMTK-ρ0 cell library was run in a single channel of a third chip.
Sequence assembly, read extraction and analysis
All sequencing assemblies were prepared and analyzed using CLCbio Genomics Workbench (GWB). Read files were trimmed using default quality scores, Illumina sequencing primer sequence and a delete read cut-off of 40 bp for 2x 52 bp reads, 28 bp for W1 heart and 25 bp for 2x 34 bp reads. Single reads resulting from trimmed paired-end reads were used in analysis where appropriate. Excluding exceptions in table S2, all assemblies were built using local alignment with limits of 10 and scores of 2 for mismatch and 3 for insertion or deletion. For 2x 34 bp assemblies the limit was reduced to 8. Total coverage maps were derived from assemblies against the mtDNA reference sequence NC_005089 with the start position shifted to position 7401 without any read extraction. SNV and indel data were derived from assemblies against NC_005089 with the first 40 bp (2x 52 bp) or 20 bp (2x 34 bp) repeated at the end of the sequence to enable full circle read-through. Paired-end distances were set to 110–320 bp for 2x 52 bp reads and 75–160 bp for 2x 34 bp reads and all broken pairs were removed prior to analysis to remove duplicate calls from short overlapping pairs and the influence of rearrangement breakpoints. For SNV detection, default call quality settings were used with detection limits set to 1% or 8 reads in W1 heart and brain, 11 reads in P1 heart, 23 reads in P1 brain, 31 reads in P2 brain and 21 reads in W2 brain, to account for differences in coverage normalized to average coverage between nt8,000–10,000 after broken read removal. Data reported in table S2 was obtained from assemblies with minimum read counts stated for W1 and equivalent coverage-normalized minimum counts for other assemblies. Because of the coverage variation (Fig. 1), SNVs localized to regions of low coverage may be under represented. For indel detection, minimum read counts were the same as those for SNV detection. R-F oriented read pairs were extracted as specific pairs from assemblies against a reference sequence composed of a tandem duplication of NC_005089 with paired-end distances set to 720–16,299bp. F-R oriented read pairs spanning canonical deletions were extracted as non-specific pairs from the same assemblies. BLAST alignments were done using unassembled reads derived from mtDNA assemblies described above (~14–17M reads per sample) with a word length of 15. Stringent parsing extracted only results with two high scoring segments which extended to at least the read start and end positions minus 1 bp. Whole genome assemblies used the same unassembled reads files as BLAST alignments and assembled them against the mouse whole genome (NCBI m37) excluding mtDNA, using a local un-gapped alignment. Percentage alignment was calculated from the sum of total reads aligning to NC_00589 and NCBI m37.
PCR
For brain and heart samples, total DNA was extracted from residual 600 g homogenates using phenol/chloroform extraction as described above. For liver samples enriched mtDNA was prepared as described above. Roche LA Expand polymerase was used to amplify control region-spanning and control region fragments using buffer 2 and primers 14177F(+20 bp) and 225R(−23 bp) and 15720F(+20 bp) and 16022R(−22 bp) respectively. Extension times were 240 s per cycle and 120+2 s respectively.
Southern blotting
Total DNA from homogenates was extracted as above, was digested with 4–15 U/μg of restriction enzyme for 3–12 hrs. DNA was resolved using 0.5% or 1.3% agarose TTE or TAE gels with ethidium bromide as stated and blotted onto Zeta-probe membranes (Bio-Rad) according to manufacturers instructions. Blots were hybridized using α-[32P]-dCTP-labeled, random-primed probes synthesized against a PCR fragments spanning nt15,196–16,127 (control region probe), nt8,926–10,081 (alternate mtDNA probe) or nt1,210–3,364 (mtDNA copy number probe) of NC_005089 mtDNA reference sequence or nt4,509–5,522 of BK000964 murine rDNA repeat unit (18S). Signal was visualized using a Perkin Elmer Cyclone storage phosphor imager.
qPCR
Total RNA was extracted from snap frozen brain and liver using the Qiagen miRNeasy kit and treated with DNAse I digestion prior to cleanup using RNeasy columns. PCR confirmed the absence of mtDNA in RNA preps prior to reverse transcription. cDNA was made using Invitrogen Superscript III kit. Quantitative PCR was carried out in triplicate using Fermentas Sybr ROX mix on an ABI 7300 instrument using a standard 3 step cycle. ΔΔCT values were calculated relative to CT values for β-actin for n=4 Polgwt/wt vs n=4 PolgD257A/D257A samples for each tissue. mtDNA primers: mt-Co1, 6530F(+22):6647R(−22); mt-Nd1, 3281F(+20):3364B(−22); mt-Nd5, 12782F(+22):12880B(−20); mt-Nd6, 13578F(+22):13713B(−22); control region, 15664F(+22):15815B(−22).
Supplementary Material
Highlights.
Mito-Seq provides a sensitive, pragmatic approach to mtDNA mutation detection.
PolgD257A/D257A mice accumulate control region multimers (CRMS) distinct from mtDNA.
CRMs are associated with a mild mtDNA depletion and increased mtDNA gene expression.
Acknowledgments
Supported by PHS grants EY10804, AG036871, CA85700 (CTM), NS053767 (SZ), P50NS071674 (JV) and a fellowship from the United Mitochondrial Disease Foundation (SLW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Albring M, Griffith J, Attardi G. Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication. Proc Natl Acad Sci U S A. 1977;74:1348–1352. doi: 10.1073/pnas.74.4.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey LJ, Cluett TJ, Reyes A, Prolla TA, Poulton J, Leeuwenburgh C, Holt IJ. Mice expressing an error-prone DNA polymerase in mitochondria display elevated replication pausing and chromosomal breakage at fragile sites of mitochondrial DNA. Nucleic Acids Res. 2009;37:2327–2335. doi: 10.1093/nar/gkp091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockington M, Sweeney MG, Hammans SR, Morgan-Hughes JA, Harding AE. A tandem duplication in the D-loop of human mitochondrial DNA is associated with deletions in mitochondrial myopathies. Nat Genet. 1993;4:67–71. doi: 10.1038/ng0593-67. [DOI] [PubMed] [Google Scholar]
- Brown TA, Cecconi C, Tkachuk AN, Bustamante C, Clayton DA. Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes Dev. 2005;19:2466–2476. doi: 10.1101/gad.1352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar D, Larsson NG, Trifunovic A. Response: Point Mutations Are Causing Progeroid Phenotypes in the mtDNA Mutator Mouse. Cell Metab. 2010;11:93. doi: 10.1016/j.cmet.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Edgar D, Shabalina I, Camara Y, Wredenberg A, Calvaruso MA, Nijtmans L, Nedergaard J, Cannon B, Larsson NG, Trifunovic A. Random point mutations with major effects on protein-coding genes are the driving force behind premature aging in mtDNA mutator mice. Cell Metab. 2009;10:131–138. doi: 10.1016/j.cmet.2009.06.010. [DOI] [PubMed] [Google Scholar]
- Fukui H, Moraes CT. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum Mol Genet. 2009;18:1028–1036. doi: 10.1093/hmg/ddn437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, Diaz LA, Jr, Kinzler KW, Vogelstein B, Papadopoulos N. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010 doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YH, Obert R, Burgers PM, Kunkel TA, Resnick MA, Gordenin DA. The 3′-->5′ exonuclease of DNA polymerase delta can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc Natl Acad Sci U S A. 2001;98:5122–5127. doi: 10.1073/pnas.091095198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AA, Johnson KA. Exonuclease proofreading by human mitochondrial DNA polymerase. J Biol Chem. 2001;276:38097–38107. doi: 10.1074/jbc.M106046200. [DOI] [PubMed] [Google Scholar]
- Kasahara T, Kubota M, Miyauchi T, Noda Y, Mouri A, Nabeshima T, Kato T. Mice with neuron-specific accumulation of mitochondrial DNA mutations show mood disorder-like phenotypes. Mol Psychiatry. 2006;11:577–93. 523. doi: 10.1038/sj.mp.4001824. [DOI] [PubMed] [Google Scholar]
- Korhonen JA, Gaspari M, Falkenberg M. TWINKLE Has 5′ -> 3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J Biol Chem. 2003;278:48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- Kraytsberg Y, Simon DK, Turnbull DM, Khrapko K. Do mtDNA deletions drive premature aging in mtDNA mutator mice? Aging Cell. 2009;8:502–506. doi: 10.1111/j.1474-9726.2009.00484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- Manfredi G, Servidei S, Bonilla E, Shanske S, Schon EA, DiMauro S, Moraes CT. High levels of mitochondrial DNA with an unstable 260-bp duplication in a patient with a mitochondrial myopathy. Neurology. 1995;45:762–768. doi: 10.1212/wnl.45.4.762. [DOI] [PubMed] [Google Scholar]
- Rudolph CJ, Upton AL, Harris L, Lloyd RG. Pathological replication in cells lacking RecG DNA translocase. Mol Microbiol. 2009a;73:352–366. doi: 10.1111/j.1365-2958.2009.06773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph CJ, Upton AL, Lloyd RG. Replication fork collisions cause pathological chromosomal amplification in cells lacking RecG DNA translocase. Mol Microbiol. 2009b;74:940–955. doi: 10.1111/j.1365-2958.2009.06909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachadyn P, Zhang XM, Clark LD, Naviaux RK, Heber-Katz E. Naturally occurring mitochondrial DNA heteroplasmy in the MRL mouse. Mitochondrion. 2008;8:358–366. doi: 10.1016/j.mito.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer PM, Headlam MJ, Dixon NE. Protein--protein interactions in the eubacterial replisome. Iubmb Life. 2005;57:5–12. doi: 10.1080/15216540500058956. [DOI] [PubMed] [Google Scholar]
- Stewart JB, Freyer C, Elson JL, Wredenberg A, Cansu Z, Trifunovic A, Larsson NG. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 2008;6:e10. doi: 10.1371/journal.pbio.0060010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Lott MT, Cabell MF, Chen YS, Lavergne L, Wallace DC. mtDNA and the origin of Caucasians: identification of ancient Caucasian-specific haplogroups, one of which is prone to a recurrent somatic duplication in the D-loop region. Am J Hum Genet. 1994;55:760–776. [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly Y, Gidlof S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Tyynismaa H, Mjosund KP, Wanrooij S, Lappalainen I, Ylikallio E, Jalanko A, Spelbrink JN, Paetau A, Suomalainen A. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17687–17692. doi: 10.1073/pnas.0505551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasta V, Ng SB, Turner EH, Shendure J, Hahn SH. Next generation sequence analysis for mitochondrial disorders. Genome Med. 2009;1:100. doi: 10.1186/gm100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermulst M, Wanagat J, Kujoth GC, Bielas JH, Rabinovitch PS, Prolla TA, Loeb LA. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat Genet. 2008;40:392–394. doi: 10.1038/ng.95. [DOI] [PubMed] [Google Scholar]
- Vermulst M, Wanagat J, Loeb LA. On mitochondria, mutations, and methodology. Cell Metab. 2009;10:437. doi: 10.1016/j.cmet.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Wai T, Teoli D, Shoubridge EA. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat Genet. 2008;40:1484–1488. doi: 10.1038/ng.258. [DOI] [PubMed] [Google Scholar]
- Wei YH, Pang CY, You BJ, Lee HC. Tandem duplications and large-scale deletions of mitochondrial DNA are early molecular events of human aging process. Ann N Y Acad Sci. 1996;786:82–101. doi: 10.1111/j.1749-6632.1996.tb39054.x. [DOI] [PubMed] [Google Scholar]
- Woischnik M, Moraes CT. Pattern of organization of human mitochondrial pseudogenes in the nuclear genome. Genome Research. 2002;12:885–893. doi: 10.1101/gr.227202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa T, Reyes A, Cluett TJ, Yang MY, Bowmaker M, Jacobs HT, Holt IJ. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 2006;25:5358–5371. doi: 10.1038/sj.emboj.7601392. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.