Introduction
The introduction of conjugate vaccination against seven serotypes of Streptococcus pneumoniae (the pneumococcus) has had profound consequences for the population structure of this pathogen, both in asymptomatic carriage and invasive disease. The prevalence of vaccine serotypes has, in vaccinated populations, fallen to the point that they are barely detectable (1). Nevertheless, the prevalence of overall pneumococcal carriage has shown no such decrease, remaining roughly constant. This is believed to be because the removal of the vaccine serotypes (4, 6B, 9V, 14, 18C, 19F and 23F) has enabled non-vaccine serotypes (of which more than 80 are known) to take their place: this phenomenon is known as serotype replacement.
Along with serotype replacement in carriage, an increase in invasive disease due to non-vaccine serotypes has been observed. Thus far in the US this has been small, and the net benefits of vaccination in terms of preventing disease have not been eroded to any great extent (2). This may be because of the serotypes which have to date been involved in replacement in carriage, only 19A is associated with a comparatively higher propensity to cause invasive disease (3). We do not know if the increased carriage prevalence of other serotypes will lead to their emergence as a cause of invasive disease.
The pneumococcus is a naturally transformable organism, with the ability to take up DNA from other strains (and species) and incorporate it into its own genome. This has been documented in many cases at the cps locus which contains the genes which determine serotype (4). Pneumococci which previously expressed vaccine serotypes can gain a non-vaccine serotype by this method (5). Uncertainty remains about the population level impact of this ‘serotype switching’ following vaccination; while it has been noted in surveillance of IPD (5, 6) the importance of clones produced by serotype switching in the population as a whole is not known. In the pre-vaccine era, antibiotic resistance was concentrated among the serotypes selected for inclusion in the vaccine. One of the advantages of vaccination has been a marked reduction in high level penicillin resistance, as a result of the removal of these highly drug resistant vaccine serotype strains (7). There are concerns that serotype switching could lead to the re-emergence of resistant clones in association with non-vaccine serotypes.
Possible cases of serotype switching are readily detected by a combination of serotyping and multi-locus sequence typing (MLST), a method for typing of bacterial strains which collects sequence data at seven housekeeping loci (8). Evidence of serotype switching is identified when isolates are identical at all MLST loci, yet have different serotypes. MLST also allows for the identification of clones associated with resistance or particular disease manifestations. We have applied this technique to a large sample of pneumococci isolated from carriage in generally healthy Massachusetts children during the fall and winter of 2006/2007.
Methods
Data collection
Carried pneumococcal strains were collected by nasopharyngeal swab from children 3 months <7 years of age, between October 2006 and April 2007 from children attending pediatric and family medicine practices for well-child or sick visits in 8 Massachusetts communities. Parental consent was obtained by study staff and nasopharyngeal swabs were obtained by trained nurses. All study procedures were approved by the Harvard Pilgrim Health Care institutional review board. Samples were processed for S. pneumoniae growth, antibiotic susceptibility, and serotype using the quellung reaction as previously described (9). Strains were maintained as glycerol stocks at -80°C and DNA was purified using DNeasy tissue kits (Qiagen, Valencia, CA). Following the initial strain collection, a new pneumococcal serotype, 6C was recognized and documented (10). Samples initially recorded as 6A were therefore tested to determine whether they expressed 6A or 6C capsules, as previously reported (11). To maintain consistency with our previous work (12), we consider 15B and 15C to be a single, rapidly interconverting serotype, 15B/C. The results from the 2006-7 dataset were compared with samples collected in 2001 and 2004 in 16 Massachusetts communities (which include those sampled in 2006-7), and previously described (12).
Multilocus Sequence Typing (MLST)
Sequence types (STs) of isolates were determined by MLST as previously described (13). Sequences of each of the seven gene fragments used in the pneumococcal MLST scheme were obtained on both DNA strands with an ABI 3700 or ABI 3730xl DNA analyzer. The sequences were aligned and trimmed to defined start and end positions using MEGA version 4 (14). Allele and ST assignments were made using the MLST website (www.spneumoniae.mlst.net). All alleles not already present in the pneumococcal MLST database were verified by re-sequencing the gene fragment on both strands. For STs found among isolates of more than one serotype the MLST loci were resequenced and serotypes were confirmed by quellung reaction.
Analysis
eBURSTv3 software was used in the analysis (15). This program groups related STs into clonal complexes, identifies the probable ancestor of each clonal complex as the ST with the largest number of single locus differences, and outputs a graphical representation of these relationships. This enabled us to assess the relative frequency of clonal complexes (CCs) of related strains in the 2004 sample and compare it with the sample collected in 2004 (12). To assess whether the difference in composition of the pneumococcal populations from the two time periods was statistically significant we used a permutation test based upon the Classification Index, as previously described (16), coded in R.
Serotype switching
Cases of serotype switching were identified by comparing MLST and serotype data. Switching will lead to isolates which are the same ST (or very closely related ones), but which have different serotypes. We approach this by first looking for examples of this within the 2007 dataset, and then by comparison with previously published datasets collected in Massachusetts in 2001 and 2004 (12) that have also been typed by MLST. Finally, we have compared the results reported here with the MLST database, which at the time of writing contains ST and serotype data from 7260 isolates contributed by a global network of researchers, to find any further cases of multiple serotype/MLST combinations. To assess whether serotype switching within serogroups was more common than expected by chance, we adopted a Monte Carlo approach: pairs of ancestral and new of serotypes, identified as above, were shuffled to produce 10000 random datasets, and for each the number of cases of switching within serogroups were calculated. The test is described in more detail in the supplemental digital content (SDC 1. Distribution of within-serogroup changes in 10000 pseudoreplicates).
Results
ST diversity
296 isolates were available for typing. One non-typable strain was excluded on the grounds that its MLST loci had sequences typical of the related species S. pseudopneumoniae (17). One 6B strain was excluded due to contamination with S. pseudopneumoniae DNA, leaving 294 in the final dataset. We found 86 distinct STs, 20 of which were new to the database. The classification index demonstrated that the ST composition of the 2006-7 dataset was significantly different from that of the previously reported 2004 dataset (p=0.006 from 1000 permutations). Despite this, there was relatively little change in the relative frequency of the major clonal complexes, shown in figure 1A and B. Fig 1a shows eBURST analysis of the population, compared with that found in 2004. eBURST groups related STs into clonal complexes (CCs), identifies the probable ancestor (shown in blue) of each CC as the ST with the largest number of minor differences, and outputs a graphical representation of these relationships. STs shown in green were only found in the 2007 dataset. STs in black were only found in the 2004 dataset and have not been recorded in 2007. Those in pink were found in both.
Figure 1.
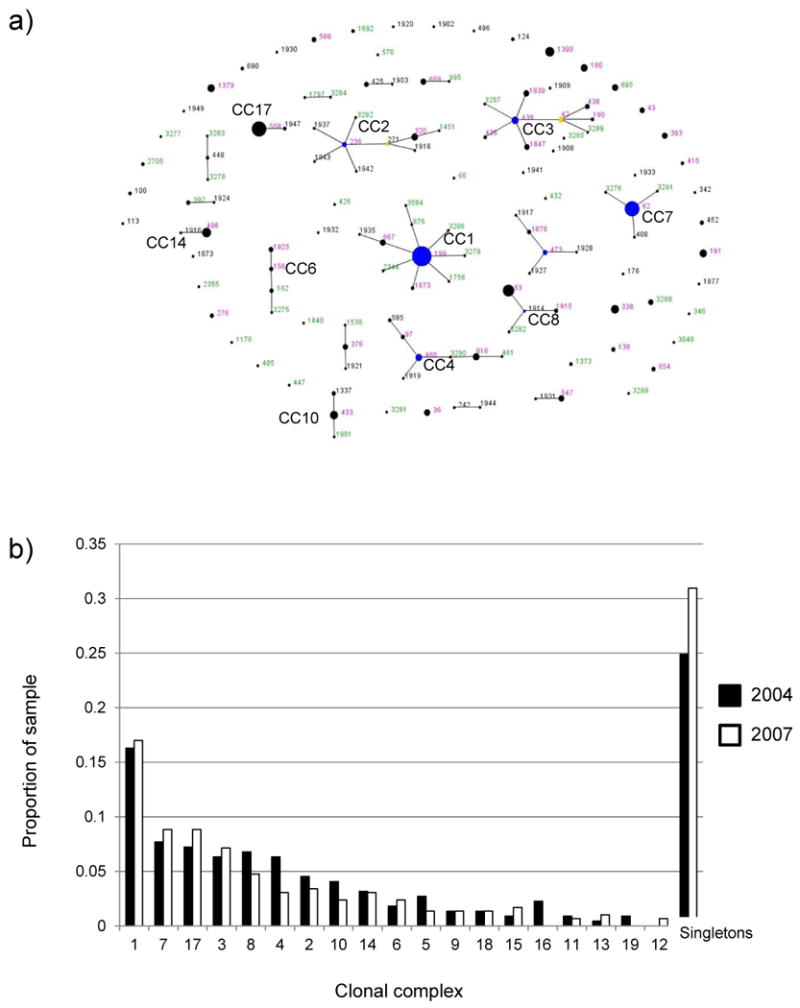
a) Comparative eBURST diagram showing the combined 2004 and 2007 samples. Each ST is represented by a point, the size of which is determined by the number of isolates with that ST in the combined dataset. STs differing at a single genetic locus are shown linked by a straight line. A clonal complex (CC) is a group of STs sharing 6 of 7 alleles with at least one other member of the group. Those STs which cannot be linked to any other in the sample are termed singletons and appear as unlinked points. For more information see http://spneumoniae.mlst.net/eburst/. The ST numbers are colored according to whether they were found only in 2004 (black), only in 2007 (green) or in both (pink). The 10 most common CCs in the combined sample are indicated.
b) bar chart showing changes in the relative frequency of the 10 most common CCs in 2004 and 2007.
The results are summarized in Table 1. This shows the numbers and serotype composition of all STs which were found ≥ 4 times, which taken together make up 70% of the sample. The full results including antibiograms (MICs for amoxicillin, benzylpenicillin, ceftriaxone, clindamycin, erythromycin, levofloxacin, rifampin, trimethoprim/sulfamthoxazole and vancomycin) are presented as supplemental digital content 5 (excel table).
Table 1.
Summary of the most common STs in the 2007 dataset, with serotype data.
| Sequence Type | N (%) | Serotypes (no.) |
|---|---|---|
| 199 | 40 (13.6) | 19A (16), 15B/C (21), 15F (3) |
| 558 | 26 (8.8) | 35B (26) |
| 62 | 24 (8.2) | 11A (23), 6A (1) |
| 63 | 11 (3.7) | 15A (10), 19A (1) |
| 338 | 9 (3.1) | 23A (9) |
| 498 | 9 (3.1) | 35F (9) |
| 1390 | 9 (3.1) | 6C (9) |
| 191 | 8 (2.7) | 7F (8) |
| 1379 | 8 (2.7) | 6C (8) |
| 320 | 7 (2.4) | 19A (7) |
| 695 | 7 (2.4) | 19A (7) |
| 433 | 6 (2.0) | 22F (6) |
| 36 | 5 (1.7) | 19A (1), 23B (4) |
| 439 | 5 (1.7) | 23B (5) |
| 3280 | 5 (1.7) | 15B/C (5) |
| 393 | 4 (1.4) | 38 (4) |
| 43 | 4 (1.4) | 19F (3), NT (1) |
| 547 | 4 (1.4) | 6C (2), 34 (2) |
| 659 | 4 (1.4) | 16F (4) |
| 816 | 4 (1.4) | 10A (4) |
| 1847 | 4 (1.4) | 23B (4) |
| Others | 91 (31.0) | See supplementary digital content |
Serotype switching
There are several cases in Table 1 of the same ST expressing more than one serotype. These presumably result from a history of serotype switching. Some of these are previously known combinations of ST and serotype (eg ST 199 has been previously shown to exist in 19A and 15B/C variants (18)) whereas others are not (eg ST 199 associated with a 15F capsule). In order to further explore the extent of serotype switching we have compared the results presented here with those from our previous surveys of carried pneumococci in the MA pediatric population (12), shown in Table 2. We have also compared the ST/serotype combinations reported here with those recorded in the MLST database (Table 3).
Table 2.
Serotype switching in the 2007 dataset detected by comparison with previous samples
| Sequence type | New serotype* (number) | Previous serotype†(s) |
|---|---|---|
| 36 | 23B (4), 19A (1) | 23F |
| 62 | 6A (1) | 11A |
| 66 | 11A (1) | 9V |
| 199 | 15F (3) | 19A, 15B/C |
| 138 | 6C (1) | 6B |
| 156 | 11A (1) | 9A, 9V |
| 447 | 37 (1) | 9N |
| 547 | 6C (2) | 34, 38 |
| 473 | 6C (3) | 6A |
All ST/serotype combinations shown above have not been previously recorded in MA.
Serotype(s) previous associated with the ST, based on sampling in 2001 and 2004. Serotype/ST combinations which have not been previously reported in the MLST database are underlined.
Table 3.
Serotype switching in the 2007 dataset detected by comparison with the MLST database.
| Sequence type | Current serotype | Expected serotype | Notes |
|---|---|---|---|
| 320 (1451) | 19A | 19F | See [5]. 1451 is a single locus variant of ST320 |
| 695 | 19A | 4 | see [5, 27] |
| 162 (3275) | 15B/C | 9V | SLV (and DLV) of ST 156 (Spain 9V-3 clone) [20] see [24] |
| 1925 | 19A | 9V | SLV of ST 156 (Spain 9V-3 clone) PMEN [20] |
| 338 | 23A | 23F | Columbia 23F-26 clone [25] |
The ST/serotype combinations shown above have been previously recorded in MA, in carriage or disease, but based on the records in the MLST database are apparent cases of serotype switching from vaccine serotypes. Serotype/ST combinations which have not been previously reported in the MLST database are underlined. SLV and DLV respectively refer to single and double locus variants in eBURST analysis [15]
Of the 10 inferred switching events shown in Table 2, 3 are between serotypes in the same serogroup (excluding the case of ST199 with a 15F capsule, because we cannot tell whether it arose from the 19A or 15B/C variant of this clone). To ask whether this was more than expected, we produced 10000 datasets in which pairs of serotypes were randomly shuffled, and calculated for each pseudoreplicate sample how many changes were within serogroups. 3 or more changes within serogroups were found in 430 pseudoreplicates (ie. p=0.043). The method is described in more detail in supplementary online material.
Changes among clones within individual serotypes
For certain replacing serotypes, we present the results from the MLST analysis of this dataset alongside those obtained previously in 2004. This enables us to track long term trends in the STs which compose these serotypes. For each of the four most common serotypes in 2007, we show a histogram illustrating the relative frequency of STs within that serotype in the total sample and the relative proportions of each ST with MICs for penicillin ≤ 0.06 μg/mL; ≥.0.06 μg/mL ≤ 2 μg/mL, and ≥ 2 μg/mL corresponding to the three classes of penicillin susceptibility presently in use (19)(Figure 2). To allow readers to visualize the relationships among STs within serotypes we also present eBURST diagrams comparing the ST composition of each serotype in 2004 and 2007. (Figure 3).
Figure 2.
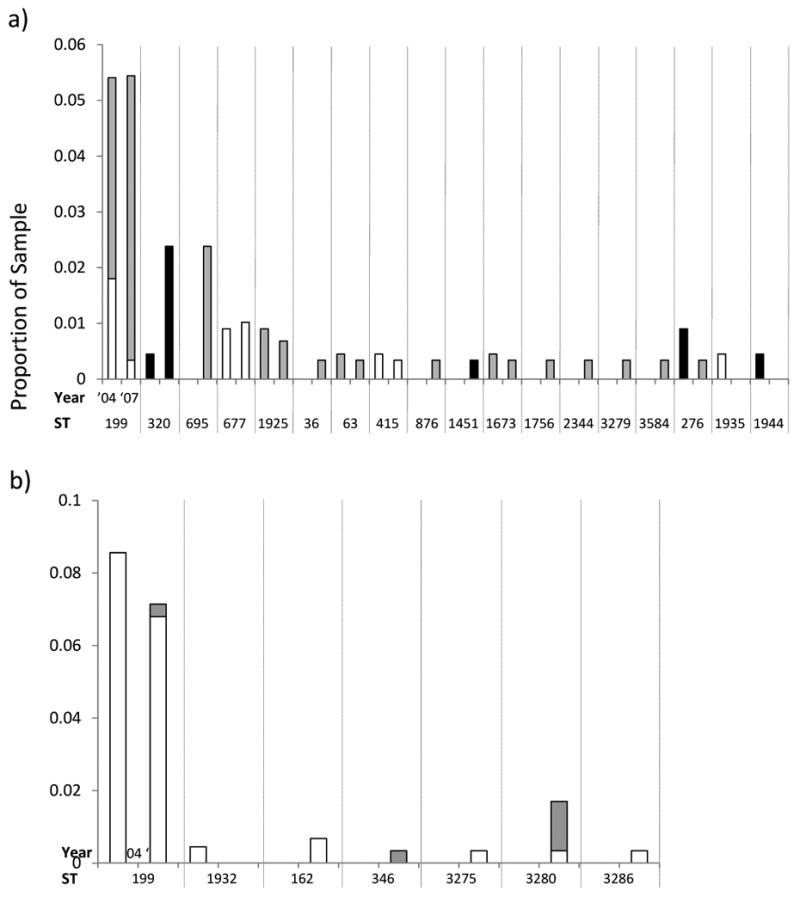
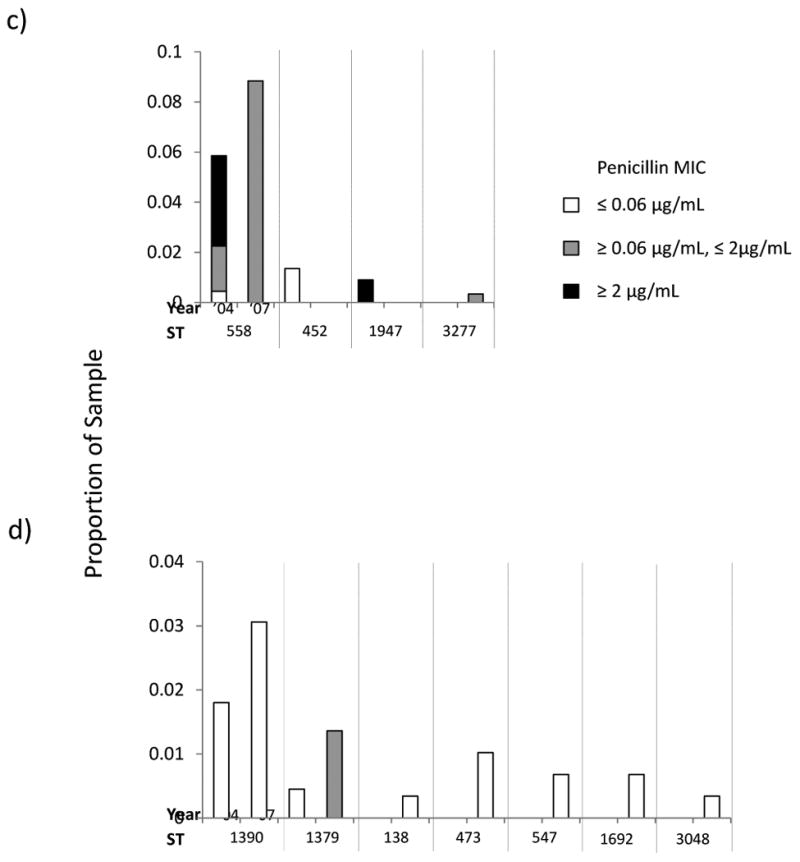
Histograms showing changes between 2004 and 2007 in the proportions of the sample for STs in the four most common serotypes: a) 19A, b) 15B/C, c) 35B and d) 6C. The levels of penicillin resistance are shown by the shading as indicated. To aid in comparing the results for the two years, isolates with the same ST are grouped between vertical dotted lines.
Figure 3.
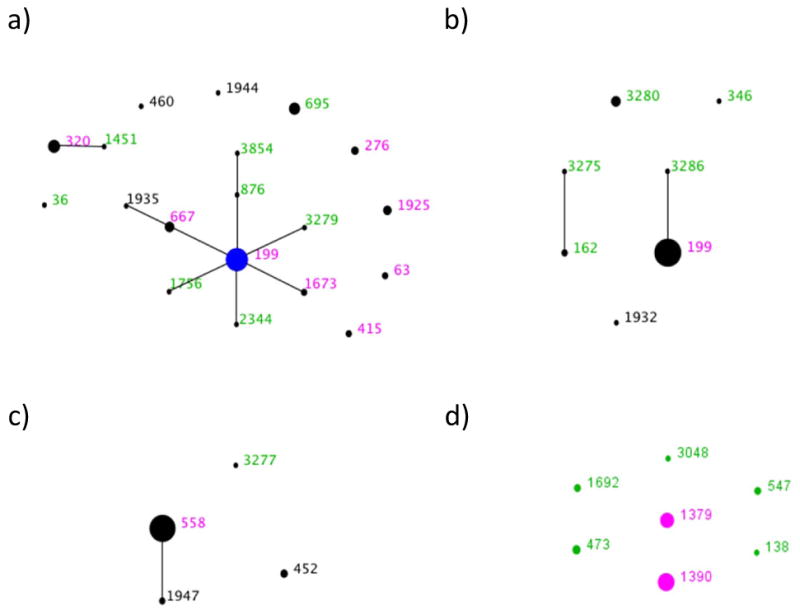
Comparative eBURST diagrams showing the combined 2004 and 2007 samples for the four most common serotypes: a) 19A, b) 15B/C, c) 35B and d) 6C. For details of interpreting eBURST diagrams please see the legend to Figure 1.
19A is a subject of considerable concern as an important replacement serotype in both carriage and invasive disease. Figs 2 a and 3a compare the 19A population in 2007 with 2004. The most common ST remains 199, but its overall frequency in the population has remained constant since 2004 (and indeed 2001, when it made up 5.4% of the sample). The increase in 19A is due to other sequence types, notably STs 320 and 695. Based on historical data, both of these STs would be expected to be associated with vaccine serotypes (see above and Table 3) and represent strains with the genetic structure of the MDR 19F (ST 320) and the PNSP 4 (ST 695) that previously circulated in the community. ST 320 is of specific concern as it is associated with high level penicillin resistance (see figure 2a) and also strongly associated with high level erythromycin resistance (all but one isolate MIC >256μg/ml. See SDC 2) and frequently with intermediate or high level ceftriaxone resistance.
Similarly to 19A, the most common ST in 15B/C strains remains 199, but the continued emergence of 15B/C has been due to multiple unrelated clones (fig 2b and 3b). These include ST 162, which is a part of the same clonal complex as ST 156 (the Spain 9V-3 clone (20)). It is interesting to note the apparent emergence of ST 3280 between 2004 and 2007, which exhibits diminished susceptibility to penicillin (Figure 2b). Contrastingly, the resistance found among serotype 35B isolates in 2004 is not to be found in 2007, although ST 558 continues to be the most common ST (figs 2c and 3c). ST 558 is a close relative of the Utah35B-24 clone, which has been noted to as an increasingly common cause of antimicrobial resistant IPD in the US (21).
Since this study commenced, a new serotype has been described. 6C may now be distinguished from 6A (10, 11), and now makes up 8.8% of the sample (26 isolates). Figs 2d and 3d show the clonal composition and relative clonal frequencies in this emerging serotype. In contrast with the other serotypes, the 6C population is made up of multiple unrelated STs. As found for 19A and 15B/C, a marked increase in clonal diversity is evident between 2004 and 2007. In 2004, only two 6C STs were observed (STs 1379 and 1390) but by 2007 five others are present. Of these, ST 138 appears to be a case of serotype switching from the vaccine serotype 6B. A single ST 473 with a 6A capsule was found in 2004.
Discussion
Between 2004 and 2007, the population of pneumococci isolated from asymptomatic nasopharyngeal carriage among Massachusetts children continued to change in response to pressure from a 7-valent vaccine (PCV7) (as shown by the significant classification index). As previously noted (1) the serotypes included in PCV7 have almost disappeared from the population, but the overall prevalence of pneumococcal carriage has remained constant as others have replaced them. By applying MLST to this dataset we are able to define the clones responsible, and identify isolates generated by serotype switching.
Prior to implementation of PCV7 in 2000, it was expected that under vaccine pressure, some clones expressing vaccine serotypes would undergo homologous recombination with non vaccine serotypes at the capsular locus, and thereby change their capsular type. Such serotype switching has not been in evidence in previous carriage samples from this population or others (12, 22, 23). However, it has been described in at least two cases of 19A STs previously associated with vaccine serotypes, and isolated from invasive disease (STs 320 and 695) (5). We have now found both these clones in carriage in MA, along with a number of other apparent cases of serotype switching, shown in tables 2 a and b. Of 86 distinct STs found in the sample, 11 expressed >1 serotype (Table 2), and 7 more expressed a serotype which was not in accordance with those expected from the samples collected in 2001 and 2004, or the MLST database (Table 3).
The role of serotype switching in the changing clonal composition of individual serotypes is best illustrated by 19A (Figs 2a and 3a). The dominant ST remains 199, which has diversified since 2004, producing a number of close relatives that were not previously observed and can be seen in fig 3a shown in green. These were all found in 2007 but not 2004. Those which are part of the same clonal complex as ST 199 show its diversification, which has also been noted in large studies of invasive disease (6, 24) but here is seen at the level of local carriage. However the STs showing the largest increase in prevalence are 320 and 695, both of which are unrelated to ST 199, and were generated by serotype switching (5). Our evidence that carriage prevalence of ST 199 has remained relatively static in MA, is in contrast with evidence that it has increased in invasive disease elsewhere (6). It is not clear whether this is due regional variation in carriage populations or conceivably, increased virulence.
It is reasonable to suggest that serotype switching has also produced the 19A variants of STs 1925 (a single locus variant of the Spain 9V-3 clone), 63 (the Sweden 15A-23 clone), and ST 36 (previously 23F). Again, some of these have been previously noted in invasive disease (6, 24, 25), and this work demonstrates their success in carriage. Of the 19A clones generated by serotype switching, ST 320 is notable for its high level of resistance to multiple antibiotic classes (Fig 2a and SDM). Our sample suggests a marked increase in carriage prevalence, showing the potential of switching to produce vaccine escape mutants with the capacity for considerable success in carriage, which is consistent with a marked increase in IPD due to this ST (21, 26). However other 19A STs with much lower levels of resistance (for example ST 695) have also increased, which suggests strongly that resistance is only one of many factors contributing to the success of individual clones.
While we have found 10 combinations of serotype and ST which were not previously noted in MA (table 2 and 3), it is important to note that it is not possible to tell from the data presented here whether these variants were generated prior or subsequent to vaccination, nor whether the switching event occurred in MA or the new variant migrated from elsewhere. However, their presence and prevalence is in marked contrast with our previous samples.
While 19A is of particular interest because of its importance in invasive disease (27), other serotypes have also been involved in switching. Before vaccination, the majority of serotype switching was documented in the vaccine serotypes (which were common and hence more likely to be detected). Examples of STs previously associated with vaccine serotypes which have generated escape variants by this method are STs 36 (23F to 23A and 19A), 66 (9V to 11A), 138 (6B to 6C), and 473 (again 6B to 6C). Table 2 also shows several examples of switching between non vaccine serotypes, which we expect to detect as they become more common. The future role of these strains in carriage or disease cannot be predicted and it must be emphasized that they remain, mostly, rare.
Another observation is that changes of serotype within serogroup are more common. 3 of 9 putative switching events shown in Table 2 were between serotypes in the same serogroup. Using a permutation approach to test whether this is more than would be expected by chance, we estimated p=0.043. It should be noted that we excluded ST199 from our analysis because both 19A and 15B/C variants of this clone are prevalent, and it is reasonable to suggest either could have given rise to the new 15F variant observed in this study. However the 19A variant of this clone shows consistently elevated resistance to penicillin, while the 15B/C variant does not. The resistance profile of the three 15F isolates reported here appears more similar to the 15B/C strains (see SDC 2) and so the number of changes within serogroups may be higher than the conservative assumptions we make here. While some serotypes, for example 6A and B, are known to interconvert by single nucleotide changes (28), this is not so for the within-serogroup changes we have found, which require the loss or acquisition of additional genes (29). Two broad classes of explanation for this phenomenon are possible: either transformation events are more frequent between serotypes within a serogroup (perhaps due to greater homology of capsular biosynthesis genes), or the genetic background of the strains involved in some way is more fit when combined with serotypes within that serogroup, than are other genetic backgrounds.
We hypothesise that the response of the pneumococcal population to vaccination can be divided into two stages. First, following the removal of vaccine serotypes, there is rapid outgrowth of clones of non-vaccine serotypes already present in the population, leading to serotype replacement in carriage. During this process, some clones of vaccine serotypes produce non-vaccine type variants through serotype switching. While these are initially rare, in time they emerge and become detectable, as we have found here. Whether their re-emergence is due to any selective advantage is not clear, but we note the continued representation of clones with lowered susceptibility to antibiotics (ST 320 in particular); in some cases this is because of former vaccine-type clones which maintain their resistance phenotype with non-vaccine serotype. Examples of this include ST 320, ST 338, and multiple different serotype variants of ST 156. It was also noted that ST 695, in changing its serotype from 4 to 19A acquired an enhanced non-susceptibility to penicillin (5). Among other clones, which are not produced by switching from vaccine serotypes, ST 558 and the 19A variant of ST 199 show diminished sensitivity to some antibiotics. Clinicians should continue to be aware of the presence of highly resistant and/or multiple drug resistant strains that continue to circulate even after the elimination of vaccine serotypes in the community.
Supplementary Material
SDC 1. Distribution of within serogroup changes within 10000 Monte-Carlo pseudoreplicates. (text)
SDC 2 (figure)
Possible routes by which a single 23F ST can generate two alternative serotype variants. A) illustrates two separate acquisitions of novel serotypes by a 23F background. B) and C) show sequential acquisition of serotypes. A) and B) both contain one case of serotype switching within a serogroup, whereas C) contains none.
SDC 3 (table) Cases of serotype switching
SDC 4 (figure) Frequency distribution of the number of switches within serogroups in 10000 random combinations of ancestral and derived serotypes drawn from SDC 3 (table).
SDC 5. Complete results, including serotype, ST and antibiogram.
Results of MLST analysis of 294 pneumococcal carriage isolates, with MICs to amoxicillin, benzylpenicillin, ceftriaxone, clindamycin, erythromycin, levofloxacin, rifampin, trimethoprim/sulfamthoxazole and vancomycin.
(excel table)
Acknowledgments
We thank Christophe Fraser for helpful discussions.
Funding: WPH acknowledges funding from the Royal Society. The work of the remaining authors and data collection for the samples from 2001 and beyond were supported by a grant (R01 AI066304, J Finkelstein) from the US National Institute of Allergy and Infectious Diseases.
References
- 1.Huang SS, Hinrichsen VL, Stevenson AE, et al. Continued impact of pneumococcal conjugate vaccine on carriage in young children. Pediatrics. 2009;124:e1–11. doi: 10.1542/peds.2008-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hicks LA, Harrison LH, Flannery B, et al. Increase in Non-vaccine-Type Pneumococcal Disease in the Era of Widespread Pneumococcal Conjugate Vaccination, United States, 1998-2004. Journal of Infectious Diseases. 2007;196:1346–1354. doi: 10.1086/521626. [DOI] [PubMed] [Google Scholar]
- 3.Hanage WP, Kaijalainen TH, Syrjanen RK, et al. Invasiveness of serotypes and clones of Streptococcus pneumoniae among children in Finland. Infect Immun. 2005;73:431–435. doi: 10.1128/IAI.73.1.431-435.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coffey TJ, Daniels M, Enright MC, Spratt BG. Serotype 14 variants of the Spanish penicillin-resistant serotype 9V clone of Streptococcus pneumoniae arose by large recombinational replacements of the cpsA-pbp1a region. Microbiology. 1999;145:2023–2031. doi: 10.1099/13500872-145-8-2023. [DOI] [PubMed] [Google Scholar]
- 5.Brueggemann AB, Pai R, Crook DW, Beall B. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog. 2007;3:e168. doi: 10.1371/journal.ppat.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore MR, Gertz RE, Jr, Woodbury RL, et al. Population snapshot of emergent Streptococcus pneumoniae serotype 19A in the United States, 2005. J Infect Dis. 2008;197:1016–1027. doi: 10.1086/528996. [DOI] [PubMed] [Google Scholar]
- 7.Kyaw MH, Lynfield R, Schaffner W, et al. Effect of introduction of the pneumococcal conjugate vaccine on drug-resistant Streptococcus pneumoniae. N Engl J Med. 2006;354:1455–1463. doi: 10.1056/NEJMoa051642. [DOI] [PubMed] [Google Scholar]
- 8.Enright MC, Spratt BG. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144:3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 9.Huang SS, Platt R, Rifas-Shiman SL, et al. Post-PCV7 changes in colonizing pneumococcal serotypes in 16 Massachusetts communities, 2001 and 2004. Pediatrics. 2005;116:e408–413. doi: 10.1542/peds.2004-2338. [DOI] [PubMed] [Google Scholar]
- 10.Park IH, Pritchard DG, Cartee R, et al. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J Clin Microbiol. 2007;45:1225–1233. doi: 10.1128/JCM.02199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nahm MH, Lin J, Finkelstein JA, Pelton SI. Increase in the prevalence of the newly discovered pneumococcal serotype 6C in the nasopharynx after introduction of pneumococcal conjugate vaccine. J Infect Dis. 2009;199:320–325. doi: 10.1086/596064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanage WP, Huang SS, Lipsitch M, et al. Diversity and antibiotic resistance among nonvaccine serotypes of Streptococcus pneumoniae carriage isolates in the post-heptavalent conjugate vaccine era. J Infect Dis. 2007;195:347–352. doi: 10.1086/510249. [DOI] [PubMed] [Google Scholar]
- 13.Enright MC, Spratt BG. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144:3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 14.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 15.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jolley KA, Wilson DJ, Kriz P, McVean G, Maiden MC. The influence of mutation, recombination, population history, and selection on patterns of genetic diversity in Neisseria meningitidis. Mol Biol Evol. 2005;22:562–569. doi: 10.1093/molbev/msi041. [DOI] [PubMed] [Google Scholar]
- 17.Hanage WP, Kaijalainen T, Herva E, et al. Using multilocus sequence data to define the pneumococcus. J Bacteriol. 2005;187:6223–6230. doi: 10.1128/JB.187.17.6223-6230.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gertz RE, Jr, McEllistrem MC, Boxrud DJ, et al. Clonal distribution of invasive pneumococcal isolates from children and selected adults in the United States prior to 7-valent conjugate vaccine introduction. J Clin Microbiol. 2003;41:4194–4216. doi: 10.1128/JCM.41.9.4194-4216.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weinstein MP, Klugman KP, Jones RN. Rationale for revised penicillin susceptibility breakpoints versus Streptococcus pneumoniae: coping with antimicrobial susceptibility in an era of resistance. Clin Infect Dis. 2009;48:1596–1600. doi: 10.1086/598975. [DOI] [PubMed] [Google Scholar]
- 20.Zhou J, Enright MC, Spratt BG. Identification of the major Spanish clones of penicillin-resistant pneumococci via the Internet using multilocus sequence typing. J Clin Microbiol. 2000;38:977–986. doi: 10.1128/jcm.38.3.977-986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richter SS, Heilmann KP, Dohrn CL, et al. Changing epidemiology of antimicrobial-resistant Streptococcus pneumoniae in the United States, 2004-2005. Clin Infect Dis. 2009;48:e23–33. doi: 10.1086/595857. [DOI] [PubMed] [Google Scholar]
- 22.Sa-Leao R, Nunes S, Brito-Avo A, et al. Changes in pneumococcal serotypes and antibiotypes carried by vaccinated and unvaccinated day-care centre attendees in Portugal, a country with widespread use of the seven-valent pneumococcal conjugate vaccine. Clin Microbiol Infect. 2009;15:1002–1007. doi: 10.1111/j.1469-0691.2009.02775.x. [DOI] [PubMed] [Google Scholar]
- 23.Vestrheim DF, Hoiby EA, Aaberge IS, Caugant DA. Impact of a pneumococcal conjugate vaccination program on carriage among children in Norway. Clin Vaccine Immunol. 2010;17:325–334. doi: 10.1128/CVI.00435-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pai R, Moore MR, Pilishvili T, et al. Postvaccine genetic structure of Streptococcus pneumoniae serotype 19A from children in the United States. J Infect Dis. 2005;192:1988–1995. doi: 10.1086/498043. [DOI] [PubMed] [Google Scholar]
- 25.Beall B, McEllistrem MC, Gertz RE, Jr, et al. Pre- and postvaccination clonal compositions of invasive pneumococcal serotypes for isolates collected in the United States in 1999, 2001, and 2002. J Clin Microbiol. 2006;44:999–1017. doi: 10.1128/JCM.44.3.999-1017.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hulten KG, Kaplan SL, Lamberth LB, Mason EO. Diferences in MLST types in Streptococcus pneumoniae 19A from 8 US medical centers before and after introductions of the 7-valent pneumococcal conjugate vaccine [Abstract LB-36]. 47th Annual Meeting of the Infectious Diseases Society of America; Philadelphia, PA. Arlington VA: IDSA; 2009. [Google Scholar]
- 27.Pelton SI, Huot H, Finkelstein JA, et al. Emergence of 19A as virulent and multidrug resistant Pneumococcus in Massachusetts following universal immunization of infants with pneumococcal conjugate vaccine. Pediatr Infect Dis J. 2007;26:468–472. doi: 10.1097/INF.0b013e31803df9ca. [DOI] [PubMed] [Google Scholar]
- 28.Mavroidi A, Godoy D, Aanensen DM, et al. Evolutionary genetics of the capsular locus of serogroup 6 pneumococci. J Bacteriol. 2004;186:8181–8192. doi: 10.1128/JB.186.24.8181-8192.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bentley SD, Aanensen DM, Mavroidi A, et al. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2006;2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SDC 1. Distribution of within serogroup changes within 10000 Monte-Carlo pseudoreplicates. (text)
SDC 2 (figure)
Possible routes by which a single 23F ST can generate two alternative serotype variants. A) illustrates two separate acquisitions of novel serotypes by a 23F background. B) and C) show sequential acquisition of serotypes. A) and B) both contain one case of serotype switching within a serogroup, whereas C) contains none.
SDC 3 (table) Cases of serotype switching
SDC 4 (figure) Frequency distribution of the number of switches within serogroups in 10000 random combinations of ancestral and derived serotypes drawn from SDC 3 (table).
SDC 5. Complete results, including serotype, ST and antibiogram.
Results of MLST analysis of 294 pneumococcal carriage isolates, with MICs to amoxicillin, benzylpenicillin, ceftriaxone, clindamycin, erythromycin, levofloxacin, rifampin, trimethoprim/sulfamthoxazole and vancomycin.
(excel table)


