ABSTRACT
The MtrC-MtrD-MtrE multidrug efflux pump of Neisseria gonorrhoeae confers resistance to a diverse array of antimicrobial agents by transporting these toxic compounds out of the gonococcus. Frequently in gonococcal strains, the expression of the mtrCDE operon is differentially regulated by both a repressor, MtrR, and an activator, MtrA. The mtrR gene lies 250 bp upstream of and is transcribed divergently from the mtrCDE operon. Previous research has shown that mutations in the mtrR coding region and in the mtrR-mtrCDE intergenic region increase levels of gonococcal antibiotic resistance and in vivo fitness. Recently, a C-to-T transition mutation 120 bp upstream of the mtrC start codon, termed mtr120, was identified in strain MS11 and shown to be sufficient to confer high levels of antimicrobial resistance when introduced into strain FA19. Here we report that this mutation results in a consensus −10 element and that its presence generates a novel promoter for mtrCDE transcription. This newly generated promoter was found to be stronger than the wild-type promoter and does not appear to be subject to MtrR repression or MtrA activation. Although rare, the mtr120 mutation was identified in an additional clinical isolate during sequence analysis of antibiotic-resistant strains cultured from patients with gonococcal infections. We propose that cis-acting mutations can develop in gonococci that significantly alter the regulation of the mtrCDE operon and result in increased resistance to antimicrobials.
IMPORTANCE
Gonorrhea is the second most prevalent sexually transmitted bacterial infection and a worldwide public health concern. As there is currently no vaccine against Neisseria gonorrhoeae, appropriate diagnostics and subsequent antibiotic therapy remain the primary means of infection control. However, the effectiveness of antibiotic treatment is constantly challenged by the emergence of resistant strains, mandating a thorough understanding of resistance mechanisms to aid in the development of new antimicrobial therapies and genetic methods for antimicrobial resistance testing. This study was undertaken to characterize a novel mechanism of antibiotic resistance regulation in N. gonorrhoeae. Here we show that a single base pair mutation generates a second, stronger promoter for mtrCDE transcription that acts independently of the known efflux system regulators and results in high-level antimicrobial resistance.
Introduction
Neisseria gonorrhoeae, the causative agent of the sexually transmitted infection gonorrhea, is a Gram-negative diplococcus and is strictly a human pathogen. Clinical isolates of N. gonorrhoeae frequently exhibit high levels of antimicrobial resistance mediated by multiple mechanisms, including active efflux of antimicrobials by four known efflux pumps (1–5). The MtrC-MtrD-MtrE efflux pump is a well-characterized system that recognizes and exports a wide variety of antimicrobial agents, including macrolide and β-lactam antibiotics, detergents, and host antimicrobial factors (1, 6). Transcription of the mtrCDE operon is differentially regulated by a repressor, MtrR, and an activator, MtrA (7–9). The mtrR gene is located 250 bp upstream of and is transcribed divergently from mtrCDE (3). MtrR represses the expression of mtrCDE via binding of two homodimers to pseudodirect repeats within the mtrCDE promoter (10); the MtrR helix-turn-helix (HTH) DNA-binding motif resides between residues 32 and 53 (7, 11).
A variety of mutations in mtrR and in the mtrR-mtrCDE intergenic region have been identified in antibiotic-resistant gonococcal strains recovered from outbreak investigations (12). Strain MS11, originally isolated in the 1960s from a patient with an uncomplicated cervical infection (13) and since used extensively by many researchers, exhibits higher levels of intrinsic in vitro resistance to MtrC-MtrD-MtrE substrates than do other laboratory strains (12). Sequence analysis of the MS11 mtr locus revealed that MS11 is a natural mtr mutant, containing an alanine-to-threonine substitution at position 39 in the MtrR DNA-binding domain, as well as a novel C-to-T transition mutation located 120 bp upstream of the mtrC start codon (mtr120) (12). Introduction of the mtr120 mutation into laboratory strain FA19 yielded one of the highest reported levels of MtrC-MtrD-MtrE-based antimicrobial resistance (12). Additionally, this mutation increased resistance to the host-derived antimicrobial compounds progesterone and CRAMP-38, the murine homologue of the human cathelicidin LL-37, suggesting that the mtr120 mutation facilitates resistance to host defense mechanisms (12). In agreement with this hypothesis, the mtr120 mutation increased in vivo fitness in a female mouse model of lower genital tract infection by nearly 3 logs compared to wild-type strain FA19 during competitive infection in the absence of antibiotic treatment (12).
Here we demonstrate that the mechanism of mtr120-based antimicrobial resistance is the generation of a consensus −10 element (14) that acts as a second, stronger promoter for mtrCDE transcription, resulting in substantially increased pump expression and enhanced resistance to antimicrobials. This promoter appears to function independently of MtrR and MtrA regulation. Additionally, we report that while the mtr120 mutation is rarer than other mutations that enhance mtrCDE expression, it was found in an additional multidrug-resistant strain that has been included in the 2008 WHO N. gonorrhoeae reference strain panel (15).
RESULTS
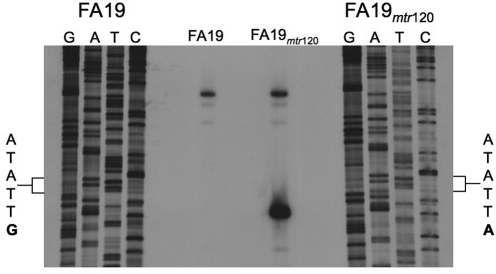
Analysis of the mtr120 locus in MS11 revealed that this C-to-T transition creates the consensus sequence for a −10 element (TATAAT) (Fig. 1) (14). A near-consensus −35 element sequence (TTGAGA) was located upstream of the potential −10 element; however, this putative −35 hexamer was separated from the potential −10 element by 24 bp rather than the optimal 17 bp (14). To determine if this −10 element could act to promote transcription, primer extension of mtrC was performed using total RNA from FA19 and DW120 cultures at mid-log phase (Fig. 2). As expected, primer extension of RNA from FA19 yielded a single mtrC transcript, which mapped to the previously identified transcription start site (Fig. 1) (1). The presence of the mtr120 mutation, however, resulted in a second, shorter mtrC transcript that was more intense than the wild-type mtrC transcript, suggesting a higher concentration of the shorter transcript. Importantly, the start site for this second transcript mapped to a site 7 bp downstream of the −10 consensus sequence generated by mtr120, a reasonable distance to suggest that this −10 sequence could act to promote the expression of this transcript.
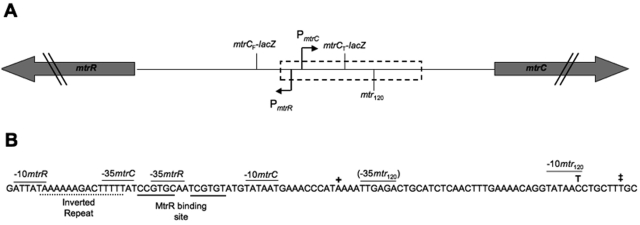
FIG 1 .
The mtr locus in N. gonorrhoeae. (A) Organization of the mtr locus. Bent arrows mark the mtrR and mtrCDE promoters (P). mtrR and mtrCDE are divergently transcribed on opposite strands. The locations of the mtr120 mutation and the mtrC-lacZ fusion start sites are indicated. The hatched box represents the location of the expanded sequence. (B) Sequence of the mtrR-mtrCDE intergenic region. The previously characterized mtrR and mtrCDE promoter elements, the consensus −10 sequence generated by the mtr120 mutation, and the putative −35 element for the mtr120 promoter are indicated in the expanded sequence. The transcription start site from the previously characterized mtrCDE promoter is marked by a single cross; the transcription start point from the mtr120 promoter is marked by a double cross.
FIG 2 .
Primer extension analysis of mtrC from FA19 and DW120. Primer extension products were generated using an mtrC-specific oligonucleotide (Table 4) hybridized to 50 µg of total RNA harvested from each strain. The DNA sequence was produced using the same oligonucleotide and is complementary to the mRNA. The wild-type and mutant sequences at the mtr120 locus are expanded, with the mutated nucleotide in bold.
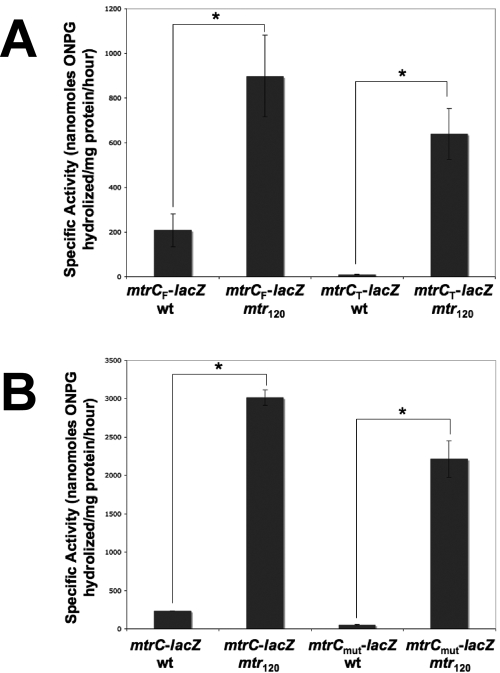
To rule out the possibility that the mutant transcript was a result of differential mRNA processing due to the mtr120 mutation, β-galactosidase assays were carried out with FA19 containing a promoterless lacZ gene translationally fused to either the entire mtrC promoter region (mtrCF-lacZ) or a truncated region lacking the wild-type mtrC promoter (mtrCT-lacZ) (Fig. 3A) (16). The mtr120 mtrCF-lacZ fusion showed significantly higher β-galactosidase activity than the wild-type mtrCF-lacZ fusion. As expected, the wild-type mtrCT-lacZ fusion showed no β-galactosidase activity. However, the mtr120 mtrCT-lacZ fusion showed levels of β-galactosidase activity significantly higher than those of the wild-type mtrCF-lacZ fusion, demonstrating that the −10 element generated by the mtr120 mutation is sufficient for transcription in the absence of the wild-type promoter and is stronger than the wild-type promoter, in agreement with the more intense band seen in the primer extension from mtr120. As further verification, we prepared mtrC-lacZ translational fusions containing a mutated −10 sequence (TATAAT to TGTCAC) of the wild-type mtrCDE promoter (mtrCmut-lacZ). We observed that mtrC-lacZ expression was abrogated when the wild-type promoter was mutated and the sequence at position 120 was wild type (Fig. 3B). However, mtrC-lacZ expression remained high when the mtr120 mutation was present. On the basis of these findings, we propose that the mtr120 mutation defines a new and highly active promoter for mtrCDE transcription.
FIG 3 .
Expression of β-galactosidase from the mtr120 locus. The β-galactosidase activities per milligram of total protein in cell extracts of FA19 containing translational mtrC-lacZ fusions are shown. Assays were performed in triplicate. Error bars represent 1 standard deviation. Asterisks correspond to a P value of <0.01 (Student’s t test). (A) The mtr120 locus is sufficient for mtrC-lacZ expression in the absence of the wild-type (wt) promoter. (B) The mtr120 locus promotes mtrC-lacZ expression when the wild-type mtrC promoter is inactivated.
To characterize the resistance phenotype of the mtr120 mutation and determine if MtrR and MtrA, the known regulators of the mtrCDE wild-type promoter, affect the resistance levels conferred by this mutation, we determined the MICs of antimicrobials against strains FA19 and DW120 and mtrR and mtrA knockout derivatives of each strain (genotypes described in Table 1, results shown in Table 2). In agreement with the increased mRNA levels detected in the primer extension experiment and high levels of β-galactosidase activity from the mtrC-lacZ fusions carrying the mtr120 mutation, the presence of mtr120 resulted in increased resistance to the MtrC-MtrD-MtrE pump substrates erythromycin (Erm), rifampin (Rif), crystal violet (CV), and Triton X-100 (TX-100). In contrast, the presence of mtr120 did not affect resistance to the nonpump substrate kanamycin (Km) in strains isogenic for mtrR or mtrA; increased Km resistance in mtrR and mtrA knockout strains is due to the presence of the aphA-3 cassette within these genes, and differences in resistance between strains with mtrR disruption and mtrA disruption may be attributed to differences in promoter strength for these genes. Importantly, the absence of MtrR and MtrA did not affect levels of resistance to pump substrates in strains with mtr120, suggesting that, unlike the wild-type promoter (17), the mtr120 promoter is not subject to MtrR or MtrA regulation.
TABLE 1 .
Strains used in this study
TABLE 2 .
Sensitivity to substrates of the MtrC-MtrD-MtrE efflux system
| Strain | Genotype | MIC (µg/ml) |
||||
|---|---|---|---|---|---|---|
| Erm | Rif | CV | TX-100 | Km | ||
| FA19 | Wild type | 0.25 | 0.06 | 0.6 | 125 | 30 |
| KH9 | FA19 mtrR::Kmr | 1 | 0.12 | 1.25 | 250 | 480 |
| CR1 | FA19 mtrA::Kmr | 0.25 | 0.06 | 0.6 | 125 | 240 |
| DW120 | FA19 mtr120 | 2 | 0.25 | 2.5 | >16,000 | 30 |
| EO1 | FA19 mtr120 mtrR::Kmr | 2 | 0.25 | 2.5 | >16,000 | 480 |
| EO2 | FA19 mtr120 mtrA::Kmr | 2 | 0.25 | 2.5 | >16,000 | 240 |
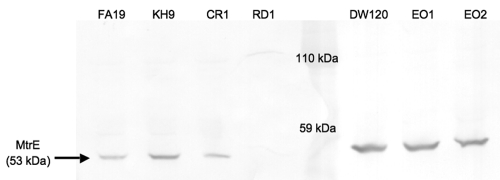
To verify that the observed increased antimicrobial resistance was due to increased levels of MtrC-MtrD-MtrE pump production, Western blot analysis was conducted to determine the effect of the mtr120 mutation on MtrE production. In agreement with the results of the MIC assays, strains bearing the mtr120 mutation produced much greater amounts of MtrE than strains with a wild-type sequence at this locus (Fig. 4). Additionally, the absence of MtrR and MtrA did not appear to affect MtrE levels in the presence of mtr120, further suggesting that these regulators do not act on the promoter generated by this mutation.
FIG 4 .
Expression of MtrE by wild-type and mtr120 mutant strains. Western blot analysis of whole-cell lysates from late-log-phase cultures was conducted using polyclonal rabbit MtrE-specific antibodies, followed by goat anti-rabbit IgG-alkaline phosphatase. Strain RD1 (44) contains a Kmr insertion in mtrE and was used as a negative control. The total protein from all strains was equally loaded, as assessed by Coomassie blue staining of a separate SDS-PAGE gel (data not shown).
The mtr120 mutation was originally identified in strain MS11 (12), which is a commonly utilized laboratory strain that has been used in gonococcal research for many years. Thus, to determine if the mtr120 mutation is present in strains isolated during recent clinical infection, as well as to compare its frequency to other mtr locus mutations in a clinical setting, the mtrR gene and the mtrR-mtrC intergenic region of 113 clinical isolates and 8 WHO reference strains were selected on the basis of their azithromycin MICs and sequenced. The azithromycin MIC range for the N. gonorrhoeae isolates sequenced, including the eight 2008 WHO reference strains (15), was 0.125 to 8 µg/ml, with 76% of the strains found to be resistant to azithromycin by the European Committee on Antimicrobial Susceptibility Testing (EUCAST; http://www.eucast.org) standard (MIC, >0.5 µg/ml). Among these strains, only one, the 2008 WHO reference strain WHO L (15), was found to contain the mtr120 mutation. This strain was also found to contain the previously described G45D mutation in the HTH DNA-binding domain of MtrR (11). In contrast to the low frequency of the mtr120 mutation, the previously defined single nucleotide (A) deletion in the 13-bp inverted repeat located between the −10 and −35 sequences of the mtrR promoter was found in 86 isolates (71%), of which 8 (7%) also had the G45D amino acid alteration in the coding region of mtrR (7). Moreover, five additional mutations in the promoter region of mtrR were found in a total of 12 isolates. Alteration of G45 (G45D [n = 11] and G45S [n = 3]) in the mtrR coding region alone was present in 14 isolates (12%). Other frequently occurring amino acid alterations found in the coding region of mtrR were A86T, which was found in 109 isolates (90%); Y105H, which was found in 25 isolates (21%); D79N, which was found in 9 isolates (7%); A39T, which was found in 7 isolates (6%); and L99G/H, which was found in 3 isolates (2%) (Table 3).
TABLE 3 .
The MIC of azithromycin and frequency of recovery of mutations in the promoter region of mtrR and the coding region of mtrR and the mtr120 mutation in 113 N. gonorrhoeae clinical isolates from 2002 to 2009 and 8 WHO N. gonorrhoeae reference strains from 2008
| Azithromycin MIC (µg/ml) (no. of isolates) | No. of isolates with: |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
mtrR promoter mutation |
Nonsynonymous mutation in mtrR coding region |
mtr 120 | |||||||||||
| ΔAa | C→Ab | Tc | Gd | Te | A→Cf | G45D/G45S | A86T | Y105H | D79N | A39T | L99G/L99H | ||
| 0.125 (2) | 1 | ||||||||||||
| 0.25 (4) | 4 | 2 | 4 | ||||||||||
| 0.38 (14) | 7 | 3 | 2 | 9 | 2 | 5 | 2 | 2 | |||||
| 0.5 (9) | 4 | 2 | 1 | 7 | 2 | 2 | 1 | 1 | 1 | ||||
| 0.75 (43) | 34 | 1 | 1 | 2 | 10 | 41 | 12 | 1 | 2 | ||||
| 1 (30) | 26 | 1 | 3 | 29 | 3 | 1 | 2 | ||||||
| 1.5 (8) | 7 | 1 | 8 | 1 | |||||||||
| 2 (4) | 2 | 1 | 1 | 4 | 1 | ||||||||
| 4 (2) | 2 | 2 | |||||||||||
| 6 (4) | 1 | 3 | 3 | 3 | |||||||||
| 8 (1) | 1 | 1 | 1 | ||||||||||
| Total (121) | 86 | 6 | 2 | 1 | 2 | 1 | 24 | 109 | 25 | 9 | 7 | 3 | 1 |
Deletion of A in 13-bp inverted repeat in the mtrR promoter.
Transversion from C to A 19 nucleotides upstream of where the A deletion occurs.
Insertion of one T 10 nucleotides downstream of where the A deletion occurs.
Deletion of one G 34 nucleotides upstream of where the A deletion occurs.
Deletion of one T 21 nucleotides upstream of where the A deletion occurs.
Transversion from A to C in the inverted repeat 3 nucleotides upstream of where the A deletion occurs.
DISCUSSION
The mtr120 mutation is novel in its mechanism of providing antimicrobial resistance in that it creates an entirely new promoter for mtrCDE transcription and acts independently of the MtrR and MtrA transcription regulatory proteins. To our knowledge, this is the first report of such a mechanism of efflux pump regulation. Two precedents exist in N. gonorrhoeae, however, for the upregulation of efflux pumps through cis-acting point mutations at existing promoters. First, the expression of norM, a gene encoding a multidrug and toxic compound extrusion family exporter that contributes to quaternary ammonium compound, norfloxacin, and ciprofloxacin resistance, is upregulated by point mutations in the −35 hexamer (C to T) of the norM promoter or in the ribosome-binding site (A to G) (5). Second, a point mutation in the −10 hexamer (G to T) of the promoter for macAB, which encodes an ABC transporter family efflux system that contributes to macrolide resistance, results in the increased expression of this efflux pump (4). Like the mtr120 mutation, these point mutations bring their respective promoter elements closer to the consensus sequences (TTGACA for the −35 element; AGGAGG for the ribosome-binding site), thereby enhancing recognition by RNA polymerase or, in the case of norM, the ribosome (14, 18). The mtr120 mutation is novel, however, in that there is no expression from the wild-type sequence at this locus, and the consensus −10 element generated by the mtr120 mutation acts as a second, independent promoter for mtrCDE transcription.
The mtr120 mutation appears to offer the gonococcus a relatively simple and convenient mechanism of antibiotic resistance. The change of this single base pair significantly increases mtrCDE expression and confers high-level antimicrobial resistance without disrupting other components of the efflux system, including the regulators MtrR and MtrA. Both MtrR and MtrA are global regulators in N. gonorrhoeae, controlling a multitude of genes outside the mtrCDE operon, many of which are important for virulence and in vivo fitness (19, 20). Thus, the ability to upregulate mtrCDE without affecting the regulation of other genes needed for infection and survival would be a highly efficient and minimally disruptive mechanism of developing antimicrobial resistance. It is therefore somewhat surprising that this mutation is so rare, especially compared to the frequency of mtrR promoter and coding region mutations in the clinical isolates examined in this study.
The reason for the rarity of the mtr120 mutation is thus a matter of speculation. The production of efflux pumps is an energy-expensive process, and it is therefore possible that the high levels of MtrC-MtrD-MtrE production stimulated by this mutation stress the gonococcus, resulting in slower or defective growth. In this respect, Eisenstein and Sparling noted that a single base pair deletion in the inverted repeat in the mtrR promoter, a mutation which also confers high-level antibiotic resistance through increased transcription of mtrCDE, results in a lower growth rate in vitro (7, 21). However, unlike mtr120, this mutation was recovered with high frequency in the strains sequenced in this study, and with the more recent finding that MtrR acts as a global regulator in the gonococcus, it is possible that the observed growth defect was at least in part due to lack of mtrR expression. Additionally, we have noticed no difference in the in vitro growth kinetics of strains carrying mtr120 and strains wild type at this site (data not shown), and Warner et al. found that strain DW120 has a fitness advantage in vivo over FA19 in a female mouse model of lower genital tract infection (12).
Another possibility to explain the rarity of the mtr120 mutation is that the mutational event required for this change occurs less frequently than those required for other mutations, particularly the deletion in the inverted repeat. A specific nucleotide change at a single base pair locus is required to generate the mtr120 phenotype. However, one of any five base pairs may be deleted in the inverted repeat to cause high-level resistance. Thus, it may be that the mtr120 mutation is simply less likely to occur, which would account for its scarcity in the isolates sequenced. Further analysis of the mtr120 mutation and its overall effects is required to elucidate the reason for this mutation’s relative infrequency.
It is important to note that the mtr120 mutation was originally identified in strain MS11 (12). This strain has been used extensively in the laboratory for studies on neisserial pili (22–25), Opa proteins (26, 27), in vitro cell infection (28, 29), antimicrobial resistance (30), and in vivo pathogenesis in male volunteers (31–34). The possession of this rare mutation, however, makes MS11 uncommon compared to other gonococcal strains, enhancing its resistance not only to antibiotics but also to host antimicrobial compounds that are MtrC-MtrD-MtrE substrates, such as the antimicrobial peptide LL-37 (6). MS11 has been found to be more infectious than another commonly studied N. gonorrhoeae strain, FA1090, in experimental infection of male volunteers (35), and it is likely that increased resistance to host antimicrobial compounds due to the mtr120 mutation plays an important role in this increased infectivity. It is therefore important to consider this mutation when interpreting findings from previous studies using strain MS11, particularly those involving antimicrobial resistance and pathogenesis and human volunteer studies in which virulence factors important in the evasion of innate defenses were assessed (35).
It is also important to note that WHO reference strain WHO L carries the mtr120 mutation. Although its levels of resistance to Erm and azithromycin were found to be slightly higher than those of reference strains with the single nucleotide deletion in the 13-bp inverted repeat of the mtrR promoter, WHO L does not contain this deletion, and its resistance was thus considered to be attributable to the G45D mutation in the HTH domain of MtrR (15). However, as mutations in the HTH domain of MtrR generally confer only low levels of antimicrobial resistance, it is far more likely that the higher level of azithromycin resistance of WHO L is due to the presence of mtr120, which is an important factor to consider in its use as a reference strain.
This study demonstrates the significant impact single base pair mutations may have on gene expression and the development of antimicrobial resistance and characterizes a novel cis-regulatory mechanism for efflux pump expression. Sequence analysis of the promoter regions of other efflux pumps, both in Neisseria and in other pathogenic organisms, will determine if this mechanism is widely used among pathogens or is unique to N. gonorrhoeae and mtrCDE. Additionally, the mtr120 mutation provides a unique opportunity for study of the physiological consequences of efflux pump overexpression on bacterial cells. This single point mutation in a noncoding region allows overproduction of the MtrC-MtrD-MtrE efflux pump without disruption or altered expression of local or global regulatory proteins. Thus, the direct phenotypic consequences of high-level efflux pump production can be examined without the introduction of confounding effects on cell physiology due to altered regulation of other genes, a challenge which has been difficult to overcome. Further study of the mtr120 mutation will help advance our understanding of antimicrobial resistance mechanisms, as well as elucidate the physiological consequences of efflux pump overexpression on bacterial cells.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Gonococci were routinely grown on GCB agar (Difco Laboratories, Detroit, MI) containing defined supplements I and II (36) at 37°C under 4% CO2 or in GCB broth (Difco Laboratories, Detroit, MI) containing defined supplements I and II and 0.048% (vol/vol) sodium bicarbonate with shaking at 37°C. E. coli DH5α was routinely grown on LB agar or in LB broth (Difco Laboratories, Detroit, MI).
The N. gonorrhoeae strains used in this study are described in Table 1. The oligonucleotide primers used are listed in Table 4. Strains DW120, KH9, and CR1 were previously described (1, 8, 12). Strain EO1 was constructed by transformation of DW120 with the mtrR::Kmr gene from KH9 chromosomal DNA by PCR amplification using primers KH9#10B, which anneals 10 bp downstream of the mtrR translational start site, and CEL1, which anneals 120 bp downstream of the mtrR translational stop site (1). Strain EO2 was constructed by transformation of DW120 with the mtrA::Kmr gene from CR1 PCR amplified using primers C6, which anneals 255 bp upstream of the mtrA translational stop site, and C7, which anneals 232 bp downstream of the mtrA translational start site. PCR products were purified using the QIAquick PCR purification kit (Qiagen Inc., Valencia, CA). Purified products were transformed into FA19, and transformants were selected on GCB agar supplemented with 50 µg/ml Km. Transformations were performed as previously described (37). All transformants were confirmed by PCR.
TABLE 4 .
Oligonucleotide primers used in this study
| Name | Sequence | Purpose |
|---|---|---|
| KH9#10B | 5′ CCAAAACCGAAGCCTTGAAAACCAA 3′ |
mtrR::Kmr amplification |
| CEL1 | 5′ GACAATGTTCATGCGATGATAGG 3′ |
mtrR::Kmr amplification |
| C6 | 5′ CGACATTCCATTCGTCTTCCGG 3′ |
mtrA::Kmr amplification |
| C7 | 5′ GCCACGACGGAAAATGCGGAG 3′ |
mtrA::Kmr amplification |
| PEmtrC181 | 5′ CCTTAGAAGCATAAAAAGCCAT 3′ | Primer extension of mtrC |
| mtrC_3 | 5′ AGTCGGATCCGGTTTGACGAGGGCGGAT 3′ | Full mtrC-lacZ fusion |
| mtrC_4 | 5′ AGTCGGATCCAATTGAGACTGCATCT CAACT 3′ | Truncated mtrC-lacZ fusion |
| PmtrCmut |
5′ AGTGGATCCGTTTCGGGTCGGTTTGACGAGGG CGGATTATAAAAAAGACTTTTTATCCGTGCAA TCGTGTATGTAGCACGAAACCCA 3′ |
Wild-type mtrC promoter −10 mutation for lacZ fusion |
| mtrC_7 | 5′ AGTCGGATCCGAAGCATAAAAAGCC 3′ | Reverse mtrC promoter primer |
| mtrC_F | 5′ CGTTTCGGGTCGGTTTGACG 3′ |
mtrR-mtrC intergenic region amplification |
| mtrC_R | 5′ CATCGCCTTAGAAGCATAAAAAGCC 3′ |
mtrR-mtrC intergenic region amplification |
| MTR1 | 5′ AACAGGCATTCTTATTTCAG 3′ |
mtrR
amplification |
| MTR2 | 5′ TTAGAAGAATGCTTTGTGTC 3′ |
mtrR
amplification |
Primer extension of mtrC.
Total RNA was prepared from gonococci by the method of Baker and Yanofsky (38). Primer extension analysis of mtrC was performed using SuperScript II reverse transcriptase (Invitrogen Co., Carlsbad, CA) according to the manufacturer’s instructions. Briefly, 50 µg of total RNA was reverse transcribed using the 5′-end 32P-labeled oligonucleotide primer PEmtrC181, which anneals to the first seven codons of mtrC (Table 4). To determine transcription start sites, primer extension products were electrophoresed on a 6% sequencing gel alongside reference sequencing reaction products derived from the PEmtrC181 primer using an mtrCDE promoter region PCR product amplified using primers mtrC_F and mtrC_R as templates. Sequencing was performed using the SequiTherm EXCEL II DNA sequencing kit (Epicenter Biotechnologies, Madison, WI) by following the manufacturer’s instructions. The dried gel was exposed to Kodak XAR film overnight at −70°C and developed using a Kodak X-Omat 1000A film processor.
Construction of mtrC-lacZ fusions.
Translational lacZ fusions were constructed as previously described (16). Briefly, the mtrC promoter region from FA19 or DW120 was amplified using primers that introduce a BamHI restriction site at the end of the PCR products; primer sequences are listed in Table 4. For all fusions, mtrC_7, which anneals to the first six codons of mtrC, was used as the reverse primer. Forward primer mtrC_3 was used to amplify the mtrC promoter sequence beginning 239 bp upstream of the mtrC start codon, encompassing all mtrC promoter elements, to make fusions wild-type mtrCF-lacZ and mtr120 mtrCF-lacZ. Forward primer mtrC_4 was used to PCR amplify a region beginning 157 bp upstream of the mtrC start codon from FA19 or DW120, excluding the previously identified mtrC promoter and transcription start site (1), to make the wild-type mtrCT-lacZ and mtr120 mtrCT-lacZ fusions. Forward primer PmtrCmut was used to PCR amplify a region beginning 254 bp upstream of the mtrC start codon from FA19 or DW120 and mutate the −10 region of the wild-type mtrC promoter from TATAAT to TGTCAC. PCR products were digested with BamHI, and the resulting DNA fragments were inserted into the BamHI site of pLES94 (16). Recombinant plasmids were transformed into E. coli DH5α. Transformants were selected on LB agar containing 100 µg/ml ampicillin. Correct insertion and orientation were confirmed by PCR analysis and DNA sequencing. The plasmids were transformed into FA19 to allow insertion into the chromosomal proAB locus. Transformants were selected on GCB agar containing 1 µg/ml chloramphenicol.
Preparation of cell extracts and β-galactosidase assays.
Strains containing translational mtrC-lacZ fusions were grown overnight on GCB agar plates containing 1 µg/ml chloramphenicol. Cells were scraped, washed once with phosphate-buffered saline (pH 7.4), and resuspended in lysis buffer (24 mM Na2HPO4, 16 mM NaH2PO4, 4 mM KCl, 0.4 mM MgSO4 ⋅ 7H2O). Cells were lysed by repeated freeze-thaw cycles, and cell debris was removed by centrifugation at 9,300 × g for 10 min at 4°C. β-Galactosidase assays were performed as previously described (39).
MIC assays.
The MICs of Erm, Rif, CV, TX-100, and Km were determined by 2-fold agar dilution assay (36). Strains were grown on GCB agar and resuspended in GCB broth to an optical density at 600 nm of 0.1, and 5-µl samples of these suspensions were inoculated onto GCB agar plates containing 2-fold serial dilutions of antibiotics. Plates were incubated overnight at 37°C under 4% CO2. Differences in MIC values greater than 2-fold were considered significant.
Western blot analysis of MtrE expression.
Whole-cell lysates from late-log-phase cultures (approximately 108 cells per sample) were subjected to 10% SDS-PAGE and transferred to nitrocellulose. The membrane was probed with a 1:10,000 dilution of rabbit polyclonal antibodies against amino acids 110 to 120 of MtrE (RQGSLSGGNVS) (20). Detection was performed with a 1:10,000 dilution of goat anti-rabbit IgG secondary antibody conjugated to alkaline phosphatase (Bio-Rad Laboratories, Hercules, CA) exposed to 5-bromo-4-chloro-3-indolylphosphate (BCIP) and Nitro Blue Tetrazolium (Roche Applied Science, Indianapolis, IN) according to the manufacturer’s instructions.
Detection of the mtr120 mutation in clinical isolates.
All examined clinical N. gonorrhoeae isolates (n = 113) were obtained at the National Reference Laboratory for Pathogenic Neisseria, Örebro University Hospital, Örebro, Sweden, from 2002 through 2009. Isolates were cultured from patients exposed to infection in many countries worldwide and were included based on having an azithromycin MIC of ≥0.38 µg/ml. Furthermore, the 2008 WHO N. gonorrhoeae reference strains (n = 8) were included for examination and quality control in all assays (15). The identities of all isolates were confirmed to the species level by the sugar utilization test and/or the Phadebact GC Monoclonal Test (Boule Diagnostics AB, Huddinge, Sweden), and they were preserved as previously described (40).
The MIC (µg/ml) of azithromycin was determined using the Etest method (AB bioMérieux, Solna, Sweden) as previously described (41). The breakpoints used for susceptibility, intermediate susceptibility, and resistance were according to EUCAST; for azithromycin, susceptibility was ≤0.25 µg/ml and resistance was >0.5 µg/ml.
N. gonorrhoeae DNA was isolated in the NorDiag Bullet instrument (Nordiag ASA Company, Oslo, Norway) using the BUGS’n BEADS STI-fast kit (Nordiag ASA Company), according to the manufacturer’s instructions. To identify putative mutations that cause enhanced expression of the MtrC-MtrD-MtrE efflux pump, the mtrR-mtrC intergenic region was amplified in a LightCycler 1.2 real-time PCR system (Roche Molecular Biochemicals, Mannheim, Germany) using primers mtrC_F, which anneals 11 bp upstream of the mtrR start codon and 249 bp upstream of the mtrC start codon (17), and mtrC_R, which anneals 24 nucleotides downstream of the mtrC translational start (1). Additionally, the promoter and coding region of mtrR was amplified using primers MTR1 and MTR2 (42) as previously described (43). All PCR amplification products were purified prior to sequencing using the High Pure PCR Product Purification Kit (Roche Diagnostics GmbH, Mannheim, Germany). Both DNA strands of amplicons were sequenced using the same primers as in the PCR amplification described above using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) on an ABI 3120 Genetic Analyzer (Applied Biosystems) according to the manufacturer’s instructions. Nucleotide and amino acid multiple-sequence alignments were performed using the BioEdit (version 5.0.9) software.
ACKNOWLEDGMENTS
This work was supported by NIH grants AI021150-26 (W.M.S.) and AI031496-21 (P. F. Sparling, University of North Carolina), by a VA Merit Review Grant to W.M.S., and by grants from the Örebro County Council Research Committee and the Foundation for Medical Research at Örebro University Hospital, Örebro, Sweden (M.U.). W.M.S. is the recipient of a Senior Research Career Scientist Award from the VA Medical Research Service. E.A.O. was supported in part by NIH training grant 2T32AI007470.
We thank L. Pucko for help in manuscript preparation.
Footnotes
Citation Ohneck EA, et al. 2011. A novel mechanism of high-level, broad-spectrum antibiotic resistance caused by a single base pair change in Neisseria gonorrhoeae. mBio 2(5):e00187-11. doi:10.1128/mBio.00187-11.
REFERENCES
- 1. Hagman KE, et al. 1995. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology 141:611–622 [DOI] [PubMed] [Google Scholar]
- 2. Lee EH, Shafer WM. 1999. The farAB-encoded efflux pump mediates resistance of gonococci to long-chained antibacterial fatty acids. Mol. Microbiol. 33:839–845 [DOI] [PubMed] [Google Scholar]
- 3. Pan W, Spratt BG. 1994. Regulation of the permeability of the gonococcal cell envelope by the mtr system. Mol. Microbiol. 11:769–775 [DOI] [PubMed] [Google Scholar]
- 4. Rouquette-Loughlin CE, Balthazar JT, Shafer WM. 2005. Characterization of the MacA-MacB efflux system in Neisseria gonorrhoeae. J. Antimicrob. Chemother. 56:856–860 [DOI] [PubMed] [Google Scholar]
- 5. Rouquette-Loughlin C, Dunham SA, Kuhn M, Balthazar JT, Shafer WM. 2003. The NorM efflux pump of Neisseria gonorrhoeae and Neisseria meningitidis recognizes antimicrobial cationic compounds. J. Bacteriol. 185:1101–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shafer WM, Qu X-D, Waring AJ, Lehrer RI. 1998. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. U. S. A. 95:1829–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hagman KE, Shafer WM. 1995. Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J. Bacteriol. 177:4162–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rouquette C, Harmon JB, Shafer WM. 1999. Induction of the mtrCDE-encoded efflux pump system of Neisseria gonorrhoeae requires MtrA, an AraC-like protein. Mol. Microbiol. 33:651–658 [DOI] [PubMed] [Google Scholar]
- 9. Shafer WM, Folster JP, Nicholas RA. 2010. Molecular mechanisms of antibiotic resistance expressed by the pathogenic Neisseria . In Genco C, Wetzler L, Neisseria: molecular mechanisms of pathogenesis. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 10. Hoffmann KM, Williams D, Shafer WM, Brennan RG. 2005. Characterization of the multiple transferable resistance repressor, MtrR, from Neisseria gonorrhoeae. J. Bacteriol. 187:5008–5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shafer WM, Balthazar JT, Hagman KE, Morse SA. 1995. Missense mutations that alter the DNA-binding domain of the MtrR protein occur frequently in rectal isolates of Neisseria gonorrhoeae that are resistant to faecal lipids. Microbiology 141:907–911 [DOI] [PubMed] [Google Scholar]
- 12. Warner DM, Shafer WM, Jerse AE. 2008. Clinically relevant mutations that cause derepression of the Neisseria gonorrhoeae MtrC-MtrD-MtrE efflux pump system confer different levels of antimicrobial resistance and in vivo fitness. Mol. Microbiol. 70:462–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Swanson J. 1973. Studies on gonococcus infection IV. Pili: their role in attachment of gonococci to tissue culture cells. J. Exp. Med. 137:571–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Browning DF, Busby SJW. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2:57–65 [DOI] [PubMed] [Google Scholar]
- 15. Unemo M, Fasth O, Fredlund H, Limnios A, Tapsall J. 2009. Phenotypic and genetic characterization of the 2008 WHO Neisseria gonorrhoeae reference strain panel intended for global quality assurance and quality control of gonococcal antimicrobial resistance surveillance for public health purposes. J. Antimicrob. Chemother. 63:1142–1151 [DOI] [PubMed] [Google Scholar]
- 16. Silver LE, Clark VL. 1995. Construction of a translational lacZ fusion system to study gene regulation in Neisseria gonorrhoeae. Gene 166:101–104 [DOI] [PubMed] [Google Scholar]
- 17. Lucas CE, Balthazar JT, Hagman KE, Shafer WM. 1997. The MtrR repressor binds the DNA sequence between the mtrR and mtrC genes of Neisseria gonorrhoeae. J. Bacteriol. 179:4123–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shine J, Dalgarno L. 1975. Determinant of cistron specificity in bacterial ribosomes. Nature 254:34–38 [DOI] [PubMed] [Google Scholar]
- 19. Folster JP, et al. 2009. MtrR modulates rpoH expression and levels of antimicrobial resistance in Neisseria gonorrhoeae. J. Bacteriol. 191:287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Warner DM, Folster JP, Shafer WM, Jerse AE. 2007. Regulation of the MtrC-MtrD-MtrE efflux-pump system modulates the in vivo fitness of Neisseria gonorrhoeae. J. Infect. Dis. 196:1804–1812 [DOI] [PubMed] [Google Scholar]
- 21. Eisenstein BI, Sparling PF. 1978. Mutations to increased antibiotic sensitivity in naturally-occurring gonococci. Nature 271:242–244 [DOI] [PubMed] [Google Scholar]
- 22. Chaussee MS, Wilson J, Hill SA. 1999. Characterization of the recD gene of Neisseria gonorrhoeae MS11 and the effect of recD inactivation on pilin variation and DNA transformation. Microbiology 145:389–400 [DOI] [PubMed] [Google Scholar]
- 23. Haas R, Meyer TF. 1986. The repertoire of silent pilus genes in Neisseria gonorrhoeae: evidence for gene conversion. Cell 44:107–115 [DOI] [PubMed] [Google Scholar]
- 24. Haas R, Veit S, Meyer TF. 1992. Silent pilin genes of Neisseria gonorrhoeae MS11 and the occurrence of related hypervariant sequences among other gonococcal isolates. Mol. Microbiol. 6:197–208 [DOI] [PubMed] [Google Scholar]
- 25. Manning PA, et al. 1991. L-pilin variants of Neisseria gonorrhoeae MS11. Mol. Microbiol. 5:917–926 [DOI] [PubMed] [Google Scholar]
- 26. Bhat KS, et al. 1991. The opacity proteins of Neisseria gonorrhoeae strain MS11 are encoded by a family of 11 complete genes. Mol. Microbiol. 5:1889–1901 [DOI] [PubMed] [Google Scholar]
- 27. Bos MP, Kuroki M, Krop-Watorek A, Hogan D, Belland RJ. 1998. CD66 receptor specificity exhibited by neisserial Opa variants is controlled by protein determinants in CD66 N-domains. Proc. Natl. Acad. Sci. U. S. A. 95:9584–9589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Criss AK, Seifert HS. 2008. Neisseria gonorrhoeae suppresses the oxidative burst of human polymorphonuclear leukocytes. Cell. Microbiol. 10:2257–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bergman P, et al. 2005. Neisseria gonorrhoeae downregulates expression of the human antimicrobial peptide LL-37. Cell. Microbiol. 7:1009–1017 [DOI] [PubMed] [Google Scholar]
- 30. Carbonetti N, Simnad V, Elkins C, Sparling PF. 1990. Construction of isogenic gonococci with variable porin structure: effects on susceptibility to human serum and antibiotics. Mol. Microbiol. 4:1009–1018 [DOI] [PubMed] [Google Scholar]
- 31. Schneider H, et al. 1991. Expression of paraglobiside-like lipooligosaccharides may be a necessary component of gonococcal pathogenesis in men. J. Exp. Med. 174:1601–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Swanson J, et al. 1987. Gonococcal pilin variants in experimental gonorrhea. J. Exp. Med. 165:1344–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ramsey KH, et al. 1995. Inflammatory cytokines produced in response to experimental human gonorrhea. J. Infect. Dis. 172:186–191 [DOI] [PubMed] [Google Scholar]
- 34. Schmidt KA, et al. 2001. Experimental gonococcal urethritis and reinfection with homologous gonococci in male volunteers. Sex. Transm. Dis. 28:555–564 [DOI] [PubMed] [Google Scholar]
- 35. Hobbs MM, et al. 2011. Experimental gonococcal infection in male volunteers: cumulative experience with Neisseria gonorrhoeae strains FA1090 and MS11mkC. Front. Microbiol. 2:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shafer WM, Guymon LF, Lind I, Sparling PF. 1984. Identification of an envelope mutation (env-10) resulting in increased antibiotic susceptibility and pyocin resistance in a clinical isolate of Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 25:767–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gunn JS, Stein DC. 1996. Use of a non-selective transformation technique to construct a multiply restriction/modification-deficient mutant of Neisseria gonorrhoeae. Mol. Gen. Genet. 251:509–517 [DOI] [PubMed] [Google Scholar]
- 38. Baker RF, Yanofsky C. 1968. The periodicity of RNA polymerase initiations: a new regulatory feature of transcription. Proc. Natl. Acad. Sci. U. S. A. 60:313–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Snyder LAS, Shafer WM, Saunders NJ. 2003. Divergence and transcriptional analysis of the division cell wall (dcw) gene cluster in Neisseria spp. Mol. Microbiol. 47:431–442 [DOI] [PubMed] [Google Scholar]
- 40. Unemo M, Olcén P, Berglund T, Albert J, Fredlund H. 2002. Molecular epidemiology of Neisseria gonorrhoeae: sequence analysis of the porB gene confirms presence of two circulating strains. J. Clin. Microbiol. 40:3741–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Berglund T, Unemo M, Olcen P, Giesecke J, Fredlund H. 2002. One year of Neisseria gonorrhoeae isolates in Sweden: the prevalence study of antibiotic susceptibility shows relation to the geographic area of exposure. Int. J. STD AIDS 13:109–114 [DOI] [PubMed] [Google Scholar]
- 42. Mavroidi A, et al. 2001. Multidrug-resistant strains of Neisseria gonorrhoeae in Greece. Antimicrob. Agents Chemother. 45:2651–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lindberg R, Fredlund H, Nicholas R, Unemo M. 2007. Neisseria gonorrhoeae isolates with reduced susceptibility to cefixime and ceftriaxone: association with genetic polymorphisms in penA, mtrR, porB1b, and ponA. Antimicrob. Agents Chemother. 51:2117–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Delahay RM, Robertson BD, Balthazar JT, Shafer WM, Ison CA. 1997. Involvement of the gonococcal MtrE protein in the resistance of Neisseria gonorrhoeae to toxic hydrophobic agents. Microbiology 143:2127–2133 [DOI] [PubMed] [Google Scholar]
- 45. Sarubbi FA, Jr, Blackman E, Sparling PF. 1974. Genetic mapping of linked antibiotic resistance loci in Neisseria goneorrhoeae. J. Bacteriol. 120:1284–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]