Abstract
As transcriptional regulators, circadian genes have the potential to influence a variety of biological pathways, including many cancer-related processes. Cryptochrome 2 (CRY2) is essential for proper circadian timing, and is a key component of the circadian regulatory feedback loop. Here, we report findings from genetic, epigenetic, loss-of-function, and transcriptional profiling analyses of CRY2 in breast cancer. Six SNPs in CRY2 were identified for genotyping in a case-control population (N=441 cases and N=479 controls), and three SNPs (rs11038689, rs7123390, and rs1401417) were significantly associated with postmenopausal breast cancer risk, with significant effect modification by menopausal status (dominant model for rs11038689: odds ratio (OR) = 0.71, 95% confidence interval (CI): 0.51–0.99, P for trend = 0.028; homozygous variants for rs7123390: OR = 0.44, 95% CI: 0.22–0.86, P for trend = 0.028; and rs1401417, OR=0.44, 95% CI: 0.21–0.92, P for trend = 0.017). Interestingly, this association was only evident in women with estrogen and progesterone receptor (ER/PR) negative breast tumors, but not with ER/PR positive tumors. Breast cancer patients also had significantly higher levels of CRY2 promoter methylation relative to controls, which is consistent with tissue array data showing lower levels of CRY2 expression in tumor tissue relative to adjacent normal tissue. Furthermore, in vitro analyses identified a number of breast cancer-relevant genes which displayed altered expression following CRY2 knockdown. These findings suggest a role for CRY2 in breast tumorigenesis, and provide further evidence that the circadian system may be an important modulator of hormone-related cancer susceptibility.
Keywords: CRY2, Breast Cancer, Circadian Biomarker, Methylation, Microarray
Introduction
The human circadian rhythm is a fundamental aspect of human physiology, and a wide range of biological processes are influenced by the circadian clock, including body temperature, energy metabolism, hormone secretion, and sleep-wake cycles (1). Circadian disruption is associated with a variety of adverse affects including metabolic disruption, the promotion of oxidative stress, and alterations in immune function (2). Increasing evidence also suggests that the circadian system may play a critical role in various cancer-related processes (3, 4), and the International Agency for Research on Cancer (IARC) recently concluded that shift work that involves circadian disruption is “probably carcinogenic to humans” (5).
At the molecular level, the maintenance of circadian rhythm involves a complicated interplay between environmental and endogenous cues, and many circadian processes are regulated by a relatively small set of core circadian genes (6). These genes form a negative and positive feedback system, which allows for fairly tight control of circadian gene expression, thereby ensuring that circadian gene products will be active during the appropriate phase of the circadian cycle (7–9). Recent evidence indicates that several of the core circadian genes are involved in transcriptional regulation, and as many as 2–10% of all mammalian genes may be regulated to some degree by the circadian oscillatory mechanism, suggesting the potential for broad consequences of disruptions to this system (10–12).
One such core circadian gene, CRY2, is essential to the maintenance of circadian rhythm through its role in the negative arm of the feedback loop, and may function more broadly as a transcriptional repressor (13–17). CRY2 has also been shown to be involved in cellular proliferation, including roles in DNA damage checkpoint control (18) and regulation of genes important for cell cycle progression (19, 20). The role of cryptochromes in DNA damage response and susceptibility to carcinogenesis remains controversial, however, as Cry1−/−, Cry2−/− transgenic mice did not display a cancer-prone phenotype in response to ionizing radiation exposure (19). Here, we report findings from genetic and epigenetic epidemiological analyses of CRY2 in breast cancer, as well as a loss-of-function investigation into the effects of CRY2 knockdown on breast cancer-relevant gene expression.
Materials and Methods
Study subjects
The study population consists of subjects enrolled in a previous breast cancer case-control study conducted in Connecticut. Details regarding subject recruitment and participant characteristics have been described in previous publications (21, 22). Briefly, cases were incident, histologically confirmed breast cancer patients (International Classification of Diseases for Oncology, 174.0 –174.9) between the ages of 30 and 80 with no previous diagnosis of cancer other than non-melanoma skin cancer, who were alive at the time of the interview. Cases were either identified from computerized patient information at Yale-New Haven Hospital (YNHH) in New Haven County, Connecticut, or from nearby Tolland County, Connecticut via hospital records by the Rapid Case Ascertainment Shared Resource at the Yale Cancer Center. YNHH controls were patients at YNHH who underwent breast-related surgery for histologically confirmed benign breast diseases. Tolland county controls were identified either through random digit dialing (for subjects younger than 65), or through the Health Care Finance Administration files (for subjects age 65 and older). After obtaining approval from each participant’s hospital and physician, potential subjects were contacted by letter and then by telephone, and those who agreed to participate were interviewed by a trained interviewer. Among YNHH subjects, participation rates were 71% for controls and 77% for cases, and among Tolland County subjects, participation rates were 61% for controls and 74% for cases. A standardized, structured questionnaire was used to obtain a number of participant characteristics including family history of cancer, reproductive history, diet, and demographic factors. At the conclusion of the interview, blood was drawn into sodium-heparinized tubes and DNA was isolated from peripheral blood lymphocytes (PBLs) for use in genotyping and methylation analyses. ER and PR status were determined immunohistochemically at YNHH, as previously described (23), with an H-score greater than 75 considered receptor positive. A total of 441 cases (93%) and 479 controls (95%) had DNA samples available for genetic analysis in the current study. The mean age of cases was 57, while controls had a mean age of 55. Both cases and controls were predominantly Caucasian (91% and 92%, respectively) with very few African Americans (6% and 6%) or other races (3% and 2%). With the exception of menopausal status (cases were 76.6% post-menopausal, compared to 65.6% in controls; P<0.001), no measured factor differed significantly by case-control status.
SNP selection and genotyping
SNPs were identified in the Haploview interface of the HapMap genome browser, Release 22 (http://www.hapmap.org/cgi-perl/gbrowse/hapmap22_B36), using the Tagger algorithm (24). Applying the pairwise tagging approach, which provides a list of the minimum number of SNPs which can represent all of the markers in a given region at a specified level of correlation, five SNPs (rs11038689, rs7123390, rs2292912, rs10838524, and rs11605924) with minor allele frequencies greater than 0.1 were identified as representative of all variation found within the exonic and intronic regions of the CRY2 gene with r2 ≥ 0.8. One additional intronic SNP (rs1401417) which had been previously identified as significantly associated with prostate cancer risk (25) was also included in the genotyping pool.
Genotyping for all SNPs was performed at Yale University’s W.M. Keck Foundation Biotechnology Research Laboratory using the Sequenom MassARRAY multiplex genotyping platform (Sequenom, Inc., San Diego, CA) according to the manufacturer’s protocol. Duplicate samples from 100 study subjects and 40 replicate samples from each of two blood donors were interspersed throughout each batch for all genotyping assays. The concordance rates for QC samples were over 95% for all assays. All genotyping calls, including quality control data, were re-checked by different laboratory personnel and genotyping scores were reproduced with 100% accuracy.
CpG island identification and methylation analysis
Using the CpG Island Searcher web tool (http://www.cpgislands.com/), one CpG island was identified that spans the promoter region, the first exon, and part of the first intron of the CRY2 gene (−450 to +750). The MethPrimer program (www.urogene.org//methprimer) was then used to design methylation specific PCR primers within the identified CpG island region. To distinguish methylated and unmethylated DNA sequences, genomic DNA samples isolated from PBLs were bisulfite treated using the EZ DNA Methylation Kit (Zymo Research, Orange, CA) according to the manufacturer’s protocol. Upon bisulfite treatment, unmethylated cytosines are converted into uracil whereas methylated cytosines remain unchanged. After the conversion, the presence of methylation was determined by quantitative PCR using primers specific to the methylated or unmethylated sequence, and the Power SYBR Green Kit (Applied Biosystems) according to the manufacturer’s protocol. The primer sequences used to detect unmethylated DNA were L: GTTTGTGGATAGTTTTAGTTTGT and R: CACCTAACAATTAACCCAAAAACA, and the primers for methylated DNA were L: GTTTGCGGATAGTTTTAGTTTGC R: CCTAACGATTAACCCAAAAACG. A methylation index was determined for each subject using the following formula: MI = [1 / (1+2−(CTu-CTme)] × 100%, as previously described (26), where CTu = the average cycle threshold obtained from duplicate qPCRs using the unmethylated primers, and CTme = the average cycle threshold obtained using the methylated primers. Since radio- and chemotherapy can affect DNA methylation, only patients from the breast cancer population who had not undergone these treatments were included in this portion of the analysis (n=80), along with an equal number of age-matched controls. Each PCR reaction was performed in duplicate using both the unmethylated and methylated primers, for a total of four reactions per subject. One or more of these reactions failed (no amplification or standard deviation greater than 1 across replicates) in samples from four of the cases, and were thus excluded, leaving a final sample of 76 cases and 80 controls.
Cell culture and treatments
Human breast adenocarcinoma cells (MCF-7) were obtained from American Type Culture Collection (Manassas, VA). Cells were maintained in Dulbecco’s modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Invitrogen), 0.01 mg/ml bovine insulin, and 1% penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO). siRNA oligos targeting CRY2 (Sense: 5’-UGCUUCAUUCGUUCAAUGUUAAGCCGG-3’ Antisense: 5’-GGCUUAACAUUGAACGAAUGAAGCA-3’) and a scrambled sequence negative control siRNA (Sense: 5’-CUUCCUCUCUUUCUCUCCCUUGUGA-3’, Antisense: 5’-UCACAAGGGAGAGAAAGAGGGAAGGA-3’) were designed and manufactured by Integrated DNA Technologies (IDT, Coralville, IA). Each oligo was diluted in OPTI-MEM serum-free medium (Invitrogen), complexed with Lipofectamine RNAiMax transfection reagent (Invitrogen), and reverse transfected with approximately 100,000 cells in 12 well plates at a final concentration of 10nM. 48 hours after transfection, cells were harvested and RNA was isolated for determination of CRY2 knockdown and subsequent whole genome expression microarray.
RNA isolation, knockdown assessment, and whole genome expression microarray
RNA was extracted from cells treated with negative control and CRY2-targeting siRNA oligos using the RNA Mini Kit (Qiagen, Valencia, CA), with on-column DNA digestion, according to the manufacturer’s instructions for mammalian cells. The purified RNA was then used as a template for first-strand cDNA synthesis using the AffinityScript cDNA kit (Stratagene, La Jolla, CA) with oligo-dT primers. Quantitative real-time PCR was performed using the Power SYBR Green PCR master mix (Applied BioSystems, Foster City, CA) with gene-specific primers, and a standard thermal cycling procedure on an ABI 7500 instrument (Applied BioSystems). The primers used for CRY2 amplification were: (L: ACCGGGGACTCTGTCTACTG, R: GCCTGCACTGCTCATGCT). RNA quantity was normalized using HPRT1 content, and CRY2 silencing was quantified according to the 2−ΔΔCt method. Biological replicates of RNA extracted from CRY2 knockdown and normal cells were interrogated by whole genome microarray (Agilent, Inc 44k chip, performed by MoGene, LC, St Louis, MO). All microarray data has been uploaded to the Gene Expression Omnibus (GEO) database, and this data can be accessed by referencing accession #GSE14617. As the focus of the current study was on breast cancer and hormone signaling, we performed a search for breast cancer-related genes using two online resources: The Breast Cancer & Estrogen Receptor Signaling pathway gene list from SABiosciences (cat#PAHS-005), and the hormone-related genes identified by the Breast and Prostate Cancer Cohort Consortium (http://epi.grants.cancer.gov/BPC3/gene.html) in order to compile a targeted list of transcripts for analysis.
Expression analysis of CRY2 in breast tumor tissues
We searched the Array Express database (27) (www.ebi.ac.uk/arrayexpress) using the Atlas of Gene Expression function to identify an expression array comparison involving breast tumor tissue and adjacent normal tissue. Using the keywords: Gene, “CRY2”; Conditions, “Breast Cancer”; and setting the species filter to Homo Sapiens, one array was identified (accession number E-TABM-276). The investigators of this study collected samples from breast tumors, and the surrounding zones (1cm, 2cm, 3 cm and 4 cm away from the boundary of the tumor) (28). For the current analysis, we focused on patients for which expression data was available for both tumor samples and distant normal tissue (3–4cm away), and conducted a paired analysis of CRY2 expression levels. Further details regarding tissue collection and the experimental protocol of this array are available at the Array Express database, or from the primary publication (28).
Statistical analysis
All statistical analyses were performed using the SAS statistical software, version 9.1 (SAS Institute, Cary, NC). CRY2 knockdown was assessed using the 2−ΔΔCt method with RNA content normalized to the housekeeping gene HPRT1. For the case-control analyses, allelic distributions for all SNPs were tested by goodness-of-fit chi-square for compliance with Hardy-Weinberg equilibrium (HWE). Odds ratios and 95% confidence intervals were determined for each disease associations by unconditional multivariate logistic regression. Covariates included in the final model were: age (continuous), race (Caucasian, African-American, Other), family history of breast cancer in a first-degree relative (no or yes; in mother, sister, or daughter), BMI (<21, 21–24.9, 25+ kg/m2), smoking (ever or never), family income per person 10 years prior to diagnosis or interview (<$20,000, $20,000–24,999, $25,000+, or unknown), age at menarche (<13, 13–14, 15+ years), lifetime months of lactation (0, 1–10, 11+), number of live births (0, 1–3, 4+), and study site (New Haven County or Tolland County). Information on hormone use was not available for this population and thus could not be controlled for. Other factors, including alcohol consumption and age at first birth did not influence the effect estimates and were thus excluded from the final models. An interaction term was added to the model and the P-value associated with the Wald test comparing the parameter estimates for each association in pre- and post-menopausal women was used to assess whether the effect of each SNP was modified by menopausal status. Haplotypes were determined using the PHASE program (29), and haplotypes with a frequency of less than 3% were not individually analyzed, but were maintained in the analysis population as controls. Of the 12 haplotypes identified, 98% of the sample chromosomes came from the 4 most frequent haplotypes, which were then analyzed by unconditional multivariate logistic regression, using all other haplotypes as the comparison group, and including the same group of covariates as the main effects models. The sign rank test was used for comparison of CRY2 expression in paired samples of tumor tissue and adjacent normal tissue, and expression level by ER/PR status was compared using the Wilcoxon two-sample test. Due to the multiple comparisons involved in the microarray analysis, adjustments were made to control for false positive findings. A correction was applied to each observation using the Benjamini-Hochberg false discovery rate adjustment, as previously described (30), to obtain a new adjusted P-value (Q).
Results
SNPs in CRY2 are associated with postmenopausal breast cancer risk
Six SNPs were identified for genotyping, including five internal tagging SNPs identified through HaploView, and one SNP which had previously been identified as significantly associated with prostate cancer risk (25). Genotyping failed for one SNP (rs10838524), which was thus excluded from further analysis. For each of the five remaining SNPs, compliance with Hardy-Weinberg equilibrium was examined among the controls, and no significant departures were observed (P<0.10). Odds ratios and confidence intervals were determined for each of the five remaining SNPs. A test of homogeneity indicated significant (P<0.05) effect modification by menopausal status for three of these SNPs when comparing homozygous genotypes. As such, overall associations are stratified by menopausal status. For the three SNPs for which menopausal status was an effect modifier, significant associations were observed with post-menopausal breast cancer risk (rs11038689: dominant model odds ratio (OR)=0.71, 95% confidence interval (CI):0.51–0.99, rs7123390: homozygous variant OR=0.44, 95% CI:0.22–0.86, rs1401417: homozygous variant OR=0.44, 95% CI:0.21–0.92) (Table 1). A haplotype analysis of the five variants demonstrated that only one haplotype, that containing the three significant variants from the main effects model, was significantly associated with breast cancer risk in postmenopausal women (frequency=22.5%; OR=0.72, 95% CI:0.56–0.94), while no haplotypes were significantly associated with pre-menopausal breast cancer (data not shown). Note that despite the fact that each SNP was chosen using the haplotype-based Tagger algorithm (24), we found significant correlation among the three significant SNPs (Supplementary Table 1). As such, each of these SNPs may be serving as markers for the same causal SNP, and therefore the three significant associations may be interpreted as approximating one single association.
Table 1.
Association of CRY2 SNPs with breast cancer risk by menopausal status
| Pre-Menopausal | Post-Menopausal | ||||||
|---|---|---|---|---|---|---|---|
| Genotype | Cases | Controls | OR* | Cases | Controls | OR* | Interaction |
| N | N | (95% CI) | N | N | (95% CI) | P-Value† | |
| rs11038689 | |||||||
| A/A | 55 | 97 | Ref. | 197 | 167 | Ref. | |
| A/G | 37 | 53 | 1.31 (0.74–2.29) | 101 | 116 | 0.74 (0.52–1.04) | 0.100 |
| G/G | 7 | 5 | 2.95 (0.82–10.66) | 13 | 20 | 0.54 (0.26–1.13) | 0.044 |
| A/G or G/G | 44 | 58 | 1.43 (0.83–2.46) | 114 | 136 | 0.71 (0.51–0.99) | 0.038 |
| Trend | P = 0.101 | P = 0.028 | 0.018 | ||||
| rs11605924 | |||||||
| A/A | 31 | 35 | Ref. | 90 | 90 | Ref. | |
| A/C | 43 | 76 | 0.64 (0.33–1.25) | 138 | 145 | 0.91 (0.62–1.34) | 0.316 |
| C/C | 24 | 44 | 0.69 (0.33–1.44) | 86 | 68 | 1.22 (0.77–1.93) | 0.097 |
| A/C or C/C | 67 | 120 | 0.66 (0.36–1.22) | 224 | 213 | 1.00 (0.69–1.45) | 0.158 |
| Trend | P = 0.334 | P = 0.410 | 0.104 | ||||
| rs2292912 | |||||||
| C/C | 60 | 90 | Ref. | 183 | 188 | Ref. | |
| C/G | 28 | 57 | 0.71 (0.40–1.29) | 104 | 95 | 1.17 (0.82–1.67) | 0.254 |
| G/G | 11 | 8 | 1.75 (0.56–5.44) | 28 | 22 | 1.67 (0.85–3.28) | 0.495 |
| C/G or G/G | 39 | 65 | 0.80 (0.45–1.41) | 132 | 117 | 1.23 (0.88–1.74) | 0.437 |
| Trend | P = 0.950 | P = 0.129 | 0.437 | ||||
| rs7123390 | |||||||
| G/G | 53 | 90 | Ref. | 168 | 144 | Ref. | |
| G/A | 35 | 58 | 1.03 (0.59–1.81) | 125 | 126 | 0.84 (0.60–1.18) | 0.503 |
| A/A | 8 | 7 | 2.05 (0.65–6.44) | 15 | 28 | 0.44 (0.22–0.86) | 0.029 |
| G/A or A/A | 43 | 65 | 1.13 (0.66–1.94) | 140 | 154 | 0.77 (0.56–1.07) | 0.215 |
| Trend | P = 0.396 | P = 0.028 | 0.065 | ||||
| rs1401417 | |||||||
| G/G | 55 | 100 | Ref. | 198 | 169 | Ref. | |
| G/C | 35 | 55 | 1.27 (0.72–2.23) | 104 | 116 | 0.77 (0.55–1.09) | 0.180 |
| C/C | 7 | 5 | 2.87 (0.79–10.44) | 12 | 23 | 0.44 (0.21–0.92) | 0.017 |
| G/C or C/C | 42 | 60 | 1.39 (0.81–2.40) | 116 | 139 | 0.72 (0.52–1.00) | 0.056 |
| Trend | P = 0.122 | P = 0.017 | 0.016 | ||||
Adjusted for age, race, family history of breast cancer, BMI, parity, years of breastfeeding, age at menarche, and study site.
Wald test of equality of the parameter estimates for each association in pre- and post-menopausal women.
In an attempt to further explore these associations in a separate population, we also analyzed a genome-wide association study (GWAS) of post-menopausal breast cancer initiated by NCI’s Division of Cancer Epidemiology and Genetics (DCEG) in collaboration with the Nurses’ Health Study (31). This dataset (available at: http://cgems.cancer.gov/data/), termed the Cancer Genetic Markers of Susceptibility (CGEMS) study, contains four markers within the genomic region of CRY2. None of these markers are significantly associated with breast cancer risk in this population. However, only one of the four markers, rs11605924, was also included in our study. The CGEMS estimated odds ratio for homozygotes (1.02), was similar to our estimate among post-menopausal women (1.22), and both found no significant association with breast cancer risk. In addition, the minor allele frequency (MAF) among the controls for this SNP matches very closely with the MAF among controls in our population (46.5% and 46.4%, respectively).
Association of CRY2 and postmenopausal breast cancer risk by hormone receptor status
Since estrogen and progesterone receptor (ER/PR) status is an important clinical factor in predicting breast cancer prognosis and dictating treatment options, we performed a separate analysis of each SNP with the population stratified according to joint estrogen and progesterone receptor status, where known. Interestingly, the homozygous or combined heterozygous/homozygous variant genotypes for four of the five SNPs (rs11038689, rs11605924, rs7123390, and rs1401417) were significantly associated with breast cancer risk in women with ER/PR negative tumors, but none were associated with ER/PR positive disease (H score > 75; Table 2). Similarly, the P-value for trend associated with increasing number of variant alleles was statistically significant (P<0.05) for each of these four SNPs in the ER/PR negative stratum, while no significant associations were observed among ER/PR positive tumors, despite the approximately equal number of subjects in each category. This data suggest that the effect of CRY2 on breast cancer risk may be significantly influenced by the presence of endogenous hormones, and CRY2 may be particularly relevant for ER and PR negative tumorigenesis. Note that due to sample size limitations, this analysis was performed for post-menopausal women only.
Table 2.
Association of CRY2 SNPs with postmenopausal breast cancer by ER/PR status
| ER/PR Positive | ER/PR Negative | ||||
|---|---|---|---|---|---|
| Genotype | Controls | Cases | OR* | Cases | OR* |
| N | N | (95% CI) | N | (95% CI) | |
| rs11038689 | |||||
| A/A | 167 | 38 | Ref. | 38 | Ref. |
| A/G | 116 | 21 | 0.82 (0.44–1.53) | 13 | 0.48 (0.23–0.99) |
| G/G | 20 | 2 | 0.36 (0.08–1.70) | 1 | 0.21 (0.03–1.73) |
| A/G or G/G | 136 | 23 | 0.74 (0.40–1.36) | 14 | 0.44 (0.22–0.89) |
| Trend | 0.200 | 0.018 | |||
| rs11605924 | |||||
| A/A | 90 | 20 | Ref. | 12 | Ref. |
| A/C | 145 | 24 | 0.73 (0.36–1.51) | 20 | 1.15 (0.50–2.63) |
| C/C | 68 | 16 | 1.12 (0.49–2.55) | 21 | 2.49 (1.03–5.99) |
| A/C or C/C | 213 | 40 | 0.84 (0.43–1.65) | 41 | 1.55 (0.72–3.30) |
| Trend | 0.834 | 0.035 | |||
| rs2292912 | |||||
| C/C | 188 | 37 | Ref. | 31 | Ref. |
| C/G | 95 | 18 | 0.88 (0.45–1.73) | 19 | 1.15 (0.57–2.31) |
| G/G | 22 | 7 | 2.03 (0.64–6.48) | 3 | 1.05 (0.22–4.94) |
| C/G or G/G | 117 | 25 | 1.00 (0.53–1.90) | 22 | 1.14 (0.58–2.26) |
| Trend | 0.558 | 0.765 | |||
| rs7123390 | |||||
| G/G | 144 | 29 | Ref. | 35 | Ref. |
| G/A | 126 | 29 | 1.30 (0.70–2.42) | 16 | 0.53 (0.27–1.06) |
| A/A | 28 | 2 | 0.30 (0.06–1.40) | 2 | 0.27 (0.06–1.27) |
| G/A or A/A | 154 | 31 | 1.08 (0.59–1.97) | 18 | 0.48 (0.25–0.93) |
| Trend | 0.530 | 0.023 | |||
| rs1401417 | |||||
| G/G | 169 | 39 | Ref. | 40 | Ref. |
| G/C | 116 | 21 | 0.79 (0.42–1.48) | 14 | 0.50 (0.25–1.01) |
| C/C | 23 | 2 | 0.31 (0.07–1.42) | 2 | 0.39 (0.08–1.87) |
| G/C or C/C | 139 | 23 | 0.70 (0.38–1.29) | 16 | 0.49 (0.25–0.95) |
| Trend | 0.129 | 0.040 | |||
Adjusted for age, race, family history of breast cancer, BMI, parity, years of breastfeeding, age at menarche, and study site.
CRY2 promoter methylation is associated with breast cancer
To determine whether CRY2 promoter methylation was associated with breast cancer risk, we performed methylation-specific PCR for all patients who had not undergone radio- or chemotherapy, as well as an equal number of controls frequency matched by age. Although only untreated cases could be used for methylation analysis, no differences were detected among several characteristics between untreated and treated cases (Supplementary Table 2). In both pre- and post-menopausal women, cases had higher methylation indices, although the difference was only statistically significant among post-menopausal women, where the mean MI among controls was 22.32% and the mean MI among cases was 30.59% (P=0.049; Table 3). Although it is unclear how CRY2 promoter methylation influences gene expression, the fact that cases had a higher degree of methylation in this region implies that CRY2 may operate as a tumor suppressor.
Table 3.
Methylation of the CRY2 promoter by case/control and menopausal status
| Pre-Menopausal | Post-Menopausal | |||||
|---|---|---|---|---|---|---|
| Methylation Index (%) | Methylation Index (%) | |||||
| Mean | SEM | P-value* | Mean | SEM | P-value* | |
| Controls | 29.34 | 5.27 | 22.32 | 2.24 | ||
| Cases | 34.93 | 8.16 | 0.552 | 30.59 | 3.50 | 0.049 |
t-test comparing mean methylation index in cases and controls
SEM: standard error of the mean
Decreased expression of CRY2 in breast tumor tissue, especially in ER/PR positive cancers
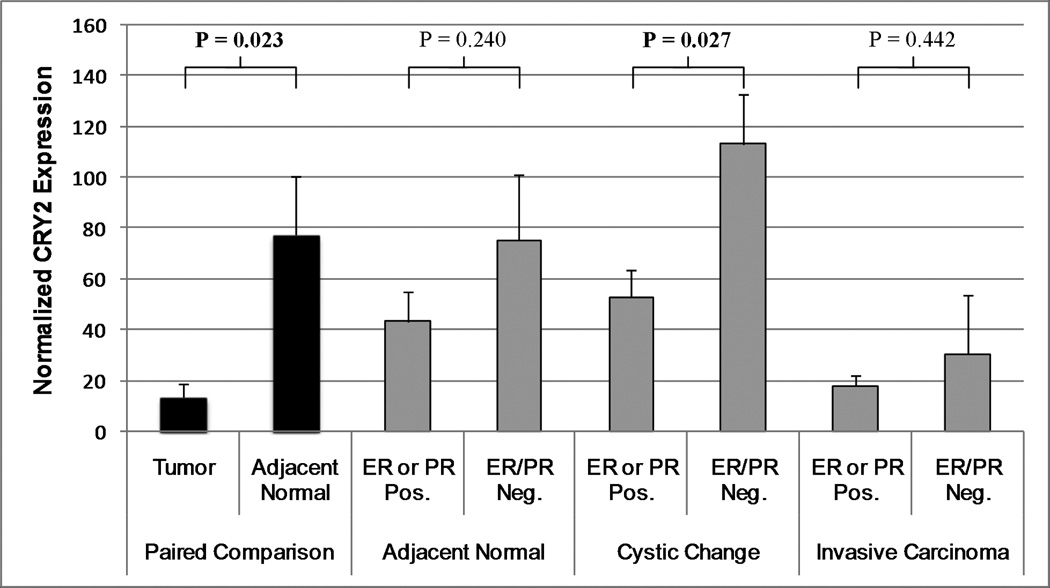
In order to determine whether CRY2 expression is altered in breast tumor samples, we identified a transcriptional profiling array from the Atlas database which examined gene expression in breast tumor tissue and adjacent normal breast tissue. As expected based on our methylation findings, CRY2 levels were significantly lower in tumor tissue (mean normalized expression=13.4) compared to adjacent normal tissue (mean normalized expression=77.2; P=0.023; Figure 1). The tissue samples were also categorized according to their histology (“invasive carcinoma”, “cystic change”, and “normal” tissue), and hormone receptor status (ER and PR, positive and negative). For each of the histological subgroups, we also compared CRY2 expression in ER/PR negative tumors, to that in tumors with either ER or PR expressed. In each case, ER/PR negative tumors had higher CRY2 levels, although this difference was only statistically significant in the “cystic change” subgroup (mean CRY2 expression in ER or PR positive samples=52.8, mean CRY2 expression in ER/PR negative samples=113.1; P=0.027).
Figure 1.
CRY2 expression in breast tissue taken at mastectomy; from a publicly available breast tumor tissue array. CRY2 expression was compared in paired samples of tumor tissue and adjacent normal tissue (3–4cm from the tumor boundary) extracted from the same patient. In addition, CRY2 expression was compared in tissue expressing either ER or PR, versus ER/PR negative samples from each of three histological subtypes. CRY2 expression was lower in tumor tissue than in adjacent normal tissue, and ER/PR negative samples had consistently higher CRY2 levels than those expressing either ER, PR, or both; although this difference was only statistically significant in the “cystic change” histological subtype.
Breast cancer relevant transcripts are influenced by CRY2 knockdown in vitro
A whole genome expression microarray revealed several breast cancer-related genes which displayed significantly altered expression in MCF-7 cells treated with CRY2-targeting siRNA oligos, relative to cells treated with negative control siRNAs. In order to focus our analysis on the most highly relevant transcripts, we identified 131 unique breast cancer and hormone-related transcripts using online resources. Of these, 43 appeared at low intensity in one or both biological replicates of the microarray, and were thus deemed unreliable estimates of gene expression, and were excluded from further analysis. 22 of the remaining 88 transcripts (25%) had significantly altered expression following CRY2 knockdown, representing a variety of biological pathways including regulation of cell differentiation, proliferation, motility, angiogenesis, and apoptosis, in addition to sex hormone regulation and estrogen signaling (Table 4). Although it remains unclear why variants in CRY2 might be especially relevant for ER/PR negative tumors in post-menopausal women, this data represent an intriguing set of observations which warrant further investigation.
Table 4.
Breast cancer-relevant transcripts which displayed significantly altered expression following CRY2 knockdown. Fold change and false discovery rate-adjusted P-values (Q) represent the mean of biological replicate microarray assays.
| Gene ID | RefSeq | Description | Fold Change | Q-Value |
|---|---|---|---|---|
| AKR1C3 | NM_003739 | Sex Hormone Metabolism, Proproliferative Signalling | 1.95 | 0.0028 |
| BCL2 | NM_000633 | Regulation of Apoptosis, Prognosis and Therapeutic Response | −1.47 | 0.0487 |
| C3 | NM_000064 | Estrogen Signalling | 2.09 | 0.0007 |
| CCND1 | NM_053056 | Cell Cycle Regulation, Tumor Progression | 1.51 | 0.0087 |
| CD44 | NM_000610 | Cellular Adhesion, Tumor Metastasis | 1.58 | 0.0028 |
| CD47 | NM_198793 | Induction of Apoptosis | 1.59 | 0.0255 |
| CDKN1A | NM_000389 | Cell Cycle Regulation | 1.71 | 0.0001 |
| FAS | NM_000043 | Regulation of Apoptosis, Tumor Progression and Prognosis | 1.81 | 0.0003 |
| GNAS | NM_080425 | Regulation of Apoptosis, Proliferation, and Tumor Progression | 1.90 | 0.0261 |
| HSPB1 | NM_001540 | Regulation of Estrogen Signalling | 1.79 | 0.0000 |
| IL6 | NM_000600 | Promotion of Tumor Growth and Invasion | 8.17 | 0.0000 |
| IL6R | NM_000565 | Promotion of Tumor Growth and Invasion | 6.06 | 0.0385 |
| INHA | NM_002191 | Regulation of Cell Differentiation and Proliferation | 3.61 | 0.0000 |
| INHBA | NM_002192 | Regulation of Cell Differentiation and Proliferation | 2.00 | 0.0004 |
| KIT | NM_000222 | Proto-oncogene, Cell Growth and Differentiation | −2.03 | 0.0000 |
| LHB | NM_000894 | Sexual Reproduction, Promotes Mammary Tumorigenesis | −1.85 | 0.0050 |
| RAC2 | NM_002872 | Cell Growth Regulation, Immune Response, Chemotaxis | −1.87 | 0.0016 |
| SERPINA3 | NM_001085 | Estrogen Signalling, Serine Protease Inhibitor | 1.63 | 0.0126 |
| TFF1 | NM_003225 | Estrogen Inducible, Promotes Cell Migration | 3.48 | 0.0000 |
| TFF3 | NM_003226 | Estrogen Inducible, Promotes Cell Migration | −1.58 | 0.0023 |
| THBS1 | NM_003246 | Cell-Cell Interactions, Control of Tumor Metastisis | 2.21 | 0.0000 |
| TNFAIP2 | NM_006291 | Induced by Tumor Necrosis Factor, Angiogenesis | 1.70 | 0.0086 |
Discussion
Although recent evidence suggests an important role for circadian genes in transcriptional regulation and cancer-related processes, the extent to which individual genes may serve as risk or prognostic biomarkers for cancer has yet to be fully elucidated. Together with CRY1 and the Period genes, CRY2 forms the negative arm of the circadian feedback loop, and is essential for proper maintenance of circadian timing. As such, CRY2 has the potential to influence, directly or indirectly, the expression and availability of gene products in a variety of biological pathways (13, 32, 33). The results presented here suggest that variants in CRY2 may significantly influence breast cancer susceptibility, which is consistent with recent observations from genetic association studies which showed that SNPs in CRY2 are associated with risk of non-Hodgkin lymphoma (34), and functional genetic variations in two other circadian genes, PER3 and NPAS2, were significantly associated with breast cancer risk (35, 36).
An interesting aspect of the epidemiological portion of this study was the impact of menopausal and hormone receptor status on the association between CRY2 SNPs and breast cancer risk. Since ER/PR negative tumors are more common among pre-menopausal women, the implication from these data are that menopausal status is likely a primary modifier of the gene-disease association, independent from the effects of ER/PR status. However, the further observation that CRY2 variants were most strongly associated with ER/PR negative tumors may be of particular interest for future investigations, given that ER/PR negative tumors tend to be more aggressive, are not generally treatable with selective estrogen receptor modulators such as tamoxifen, and are thus associated with decreased survival (37). While several previous studies have suggested that hormones, including estrogen, may influence the expression of genes in the circadian system (38–40), the mechanisms underlying the observed differences in the effect of CRY2 SNPs on breast cancer risk remain unclear, and warrant further investigation. Of note, we did not have information on hormone replacement therapy (HRT) use in our population, and were thus unable to control for it in the analysis. While HRT use is associated with elevated risk of breast cancer, in order for this exposure to operate as a confounder in our study, it would have to be associated with CRY2 genotypes. Although this is possible by chance, we feel that confounding by HRT use is unlikely to explain our results.
Compared to genetic studies of circadian genes in tumorigenesis, epigenetic changes, such as promoter methylation, remain a relatively unexplored area in the field of cancer research. The findings from our current study of CRY2 are among the first evidence suggesting that promoter methylation status in circadian genes could be a potential biomarker for cancer susceptibility. Our results demonstrate that CRY2 promoter methylation, which may lead to decreased gene expression, was elevated in breast cancer cases relative to controls. Several previous studies have also noted significant down-regulation of circadian genes associated with various cancers. For example, the expression level of several circadian genes, including CRY2, was significantly diminished among those suffering from chronic myeloid leukemia compared to healthy individuals (41). Decreased expression of CRY2 was also observed in hepatocellular carcinomas, and this reduction was not caused by genetic mutations, but by several factors, including promoter methylation (42). Together, these findings suggest that methylation changes, in addition to genetic variants in circadian genes, could serve as novel cancer biomarkers. However, despite these promising initial findings, further study is needed to confirm this association, as very few cases in our sample were untreated, leaving a fairly small sample size available for this portion of the study.
Since the methylation analysis was performed using DNA samples isolated from PBLs, it remains unclear whether epigenetic changes measured in this surrogate tissue are an accurate reflection of the methylation status in breast tissue. A previous study has demonstrated strong correlation between methylation in PBLs and colon tissue (kappa statistic = 86.5%, p < 0.0001) (43), and a large proof-of-principle study found significant associations between the level of PBL methylation in a number of ER-α target (ERT) genes and breast cancer risk, highlighting the potential for epigenotyping to be useful in estimating risk, even when measured in peripheral blood cell DNA (44). If hypermethylation in the CRY2 promoter region does in fact lead to diminished gene expression, our data indicate that CRY2 may operate as a tumor suppressor, as increased methylation was associated with decreased breast cancer risk. This suggestion is corroborated by the transcriptional profiling array showing that CRY2 expression was lower in tumor tissue than in adjacent normal tissue. This array also demonstrated that CRY2 was elevated in ER/PR negative breast tissue samples, which is in keeping with the general suggestion that variants in CRY2 had the biggest impact on breast cancer risk in women with ER/PR negative tumors.
Among the microarray findings, the strong induction of trefoil factor 1 (TFF1) in cells with reduced CRY2 (fold change=3.5, Q<0.0001) is of particular interest. TFF1 is strongly estrogen regulated, and aberrant expression of this transcript can influence a number of cancer-related pathways, including proliferation, apoptosis, anoikis, angiogenesis, and migration and invasion (45). In addition, while it has been suggested that TFF1 encourages cell cycle delay at the G1-S transition, it has also been shown to increase the levels of cyclin D1 (CCND1), a key stimulator of cell cycle progression (46). Interestingly, CCND1 was also significantly induced following CRY2 knockdown (fold change=1.51, Q=0.009). While the nature of these relationships remains unclear, this data is consistent with the overall suggestion that CRY2 may have tumor suppressor properties, and provide the intriguing suggestion that CRY2 may be involved in hormone signaling, with the potential to regulate, directly or indirectly, transcripts with well-established relevance for breast tumorigenesis.
In summary, our study demonstrates a genetic association of the circadian gene CRY2 with breast cancer risk, which is potentially modified by menopausal and ER/PR status. Moreover, an epigenetic association between CRY2 promoter hypermethylation and increased breast cancer risk was also detected, suggesting that CRY2 may operate as a tumor suppressor. This role was further supported by an observed decreased expression of CRY2 in breast tumor tissues and an in vitro loss-of-function analysis, which demonstrated that a number of breast cancer relevant transcripts displayed altered expression following CRY2 knockdown. These data suggest a novel role for CRY2 in hormone signaling and breast tumorigenesis, and provide evidence supportive of the hypothesis that the circadian system may be an important modulator of breast cancer risk. However, additional large studies will be required in order to further elucidate the role of CRY2 in breast cancer susceptibility.
Supplementary Material
Acknowledgments
Financial Support: This work was supported by the US National Institutes of Health (grants CA122676 and CA110937).
References
- 1.Kondratov RV, Gorbacheva VY, Antoch MP. The role of mammalian circadian proteins in normal physiology and genotoxic stress responses. Curr Top Dev Biol. 2007;78:173–216. doi: 10.1016/S0070-2153(06)78005-X. [DOI] [PubMed] [Google Scholar]
- 2.Navara KJ, Nelson RJ. The dark side of light at night: physiological, epidemiological, and ecological consequences. J Pineal Res. 2007;43:215–224. doi: 10.1111/j.1600-079X.2007.00473.x. [DOI] [PubMed] [Google Scholar]
- 3.Lamont EW, James FO, Boivin DB, Cermakian N. From circadian clock gene expression to pathologies. Sleep Med. 2007;8:547–556. doi: 10.1016/j.sleep.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Stevens RG, Blask DE, Brainard GC, et al. Meeting report: the role of environmental lighting and circadian disruption in cancer and other diseases. Environ Health Perspect. 2007;115:1357–1362. doi: 10.1289/ehp.10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.IARC. IARC monographs on the evaluation of carcinogenic risks to humans; 2007. Lyon: International Agency for Research on Cancer; 2007. Shift-work, painting and fire-fighting. in press. [PMC free article] [PubMed] [Google Scholar]
- 6.Oster H. The genetic basis of circadian behavior. Genes Brain Behav. 2006;5 Suppl 2:73–79. doi: 10.1111/j.1601-183X.2006.00226.x. [DOI] [PubMed] [Google Scholar]
- 7.Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 8.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 9.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 10.Le Minh N, Damiola F, Tronche F, Schutz G, Schibler U. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. Embo J. 2001;20:7128–7136. doi: 10.1093/emboj/20.24.7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Storch KF, Lipan O, Leykin I, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 12.Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12:551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 13.Griffin EA, Jr, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- 14.van der Horst GT, Muijtjens M, Kobayashi K, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 15.Yu W, Nomura M, Ikeda M. Interactivating feedback loops within the mammalian clock: BMAL1 is negatively autoregulated and upregulated by CRY1, CRY2, and PER2. Biochem Biophys Res Commun. 2002;290:933–941. doi: 10.1006/bbrc.2001.6300. [DOI] [PubMed] [Google Scholar]
- 16.Sancar A. Cryptochrome: the second photoactive pigment in the eye and its role in circadian photoreception. Annu Rev Biochem. 2000;69:31–67. doi: 10.1146/annurev.biochem.69.1.31. [DOI] [PubMed] [Google Scholar]
- 17.Vitaterna MH, Selby CP, Todo T, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci U S A. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol. 2005;25:3109–3116. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gauger MA, Sancar A. Cryptochrome, circadian cycle, cell cycle checkpoints, and cancer. Cancer Res. 2005;65:6828–6834. doi: 10.1158/0008-5472.CAN-05-1119. [DOI] [PubMed] [Google Scholar]
- 20.Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 21.Zheng T, Holford TR, Mayne ST, et al. Risk of female breast cancer associated with serum polychlorinated biphenyls and 1,1-dichloro-2,2'-bis(p-chlorophenyl)ethylene. Cancer Epidemiol Biomarkers Prev. 2000;9:167–174. [PubMed] [Google Scholar]
- 22.Zheng T, Holford TR, Tessari J, et al. Breast cancer risk associated with congeners of polychlorinated biphenyls. Am J Epidemiol. 2000;152:50–58. doi: 10.1093/aje/152.1.50. [DOI] [PubMed] [Google Scholar]
- 23.McCarty KS, Jr, Miller LS, Cox EB, Konrath J, McCarty KS., Sr Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Arch Pathol Lab Med. 1985;109:716–721. [PubMed] [Google Scholar]
- 24.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nature Genetics. 2005;37:1217–1223. doi: 10.1038/ng1669. [see comment] [DOI] [PubMed] [Google Scholar]
- 25.Chu LW, Zhu Y, Yu K, et al. Variants in circadian genes and prostate cancer risk: a population-based study in China. Prostate Cancer Prostatic Dis. 2008;11:342–348. doi: 10.1038/sj.pcan.4501024. [DOI] [PubMed] [Google Scholar]
- 26.Lu L, Katsaros D, de la Longrais IA, Sochirca O, Yu H. Hypermethylation of let-7a-3 in epithelial ovarian cancer is associated with low insulin-like growth factor-II expression and favorable prognosis. Cancer Res. 2007;67:10117–10122. doi: 10.1158/0008-5472.CAN-07-2544. [DOI] [PubMed] [Google Scholar]
- 27.Parkinson H, Kapushesky M, Kolesnikov N, et al. ArrayExpress update--from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng AS, Culhane AC, Chan MW, et al. Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res. 2008;68:1786–1796. doi: 10.1158/0008-5472.CAN-07-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 31.Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmesser S, Tallone T, Bordon A, Rusconi S, Albrecht U. Interaction of circadian clock proteins PER2 and CRY with BMAL1 and CLOCK. BMC Mol Biol. 2008;9:41. doi: 10.1186/1471-2199-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kume K, Zylka MJ, Sriram S, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 34.Hoffman AE, Zheng T, Stevens RG, et al. Clock-cancer connection in non-Hodgkin's lymphoma: a genetic association study and pathway analysis of the circadian gene cryptochrome 2. Cancer Res. 2009;69:3605–3613. doi: 10.1158/0008-5472.CAN-08-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Y, Brown HN, Zhang Y, Stevens RG, Zheng T. Period3 structural variation: a circadian biomarker associated with breast cancer in young women. Cancer Epidemiol Biomarkers Prev. 2005;14:268–270. [PubMed] [Google Scholar]
- 36.Zhu Y, Stevens RG, Leaderer D, et al. Non-synonymous polymorphisms in the circadian gene NPAS2 and breast cancer risk. Breast Cancer Res Treat. 2008;107:421–425. doi: 10.1007/s10549-007-9565-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruder AM, Lubin F, Wax Y, Geier A, Alfundary E, Chetrit A. Estrogen and progesterone receptors in breast cancer patients. Epidemiologic characteristics and survival differences. Cancer. 1989;64:196–202. doi: 10.1002/1097-0142(19890701)64:1<196::aid-cncr2820640134>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura TJ, Shinohara K, Funabashi T, Kimura F. Effect of estrogen on the expression of Cry1 and Cry2 mRNAs in the suprachiasmatic nucleus of female rats. Neurosci Res. 2001;41:251–255. doi: 10.1016/s0168-0102(01)00285-1. [DOI] [PubMed] [Google Scholar]
- 39.Nakamura TJ, Sellix MT, Menaker M, Block GD. Estrogen directly modulates circadian rhythms of PER2 expression in the uterus. Am J Physiol Endocrinol Metab. 2008;295:E1025–E1031. doi: 10.1152/ajpendo.90392.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mostafaie N, Kallay E, Sauerzapf E, et al. Correlated downregulation of estrogen receptor beta and the circadian clock gene Per1 in human colorectal cancer. Mol Carcinog. 2009 doi: 10.1002/mc.20510. [DOI] [PubMed] [Google Scholar]
- 41.Yang MY, Chang JG, Lin PM, et al. Downregulation of circadian clock genes in chronic myeloid leukemia: alternative methylation pattern of hPER3. Cancer Sci. 2006;97:1298–1307. doi: 10.1111/j.1349-7006.2006.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin YM, Chang JH, Yeh KT, et al. Disturbance of circadian gene expression in hepatocellular carcinoma. Mol Carcinog. 2008;47:925–933. doi: 10.1002/mc.20446. [DOI] [PubMed] [Google Scholar]
- 43.Cui Y, Liao YC, Lo SH. Epidermal growth factor modulates tyrosine phosphorylation of a novel tensin family member, tensin3. Mol Cancer Res. 2004;2:225–232. [PubMed] [Google Scholar]
- 44.Widschwendter M, Apostolidou S, Raum E, et al. Epigenotyping in peripheral blood cell DNA and breast cancer risk: a proof of principle study. PLoS One. 2008;3:e2656. doi: 10.1371/journal.pone.0002656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perry JK, Kannan N, Grandison PM, Mitchell MD, Lobie PE. Are trefoil factors oncogenic? Trends Endocrinol Metab. 2008;19:74–81. doi: 10.1016/j.tem.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 46.Bossenmeyer-Pourie C, Kannan R, Ribieras S, et al. The trefoil factor 1 participates in gastrointestinal cell differentiation by delaying G1-S phase transition and reducing apoptosis. J Cell Biol. 2002;157:761–770. doi: 10.1083/jcb200108056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.