Abstract
WRKY transcription factors regulate diverse plant processes including responses to biotic stresses. Our previous studies indicate that OsWRKY62, an OsWRKY IIa subfamily member, functions as a negative regulator of the rice defense against Xanthomonas oryzae pv. oryzae. Here, we report that a large inverted repeat construct designed to knock down the expression of the four OsWRKY IIa subfamily members (OsWRKY62, OsWRKY28, OsWRKY71, and OsWRKY76) leads to overexpression of all four genes and disease resistance in some transgenic plants. These phenotypes are stably inherited as reflected by progeny analysis. A pathogenesis-related gene, PR10, is up-regulated in plants overexpressing the OsWRKY IIa genes. These results suggest that OsWRKY IIa proteins interact functionally to modulate plant innate immunity.
Keywords: Rice, OsWRKY IIa, Xoo, Innate immunity
Introduction
WRKY proteins constitute a family of mostly plant-specific zinc-finger transcription factors composed of 74 and 109 members in Arabidopsis and rice, respectively (Eulgem and Somssich 2007; Ross et al. 2007). A common feature of WRKY proteins is the WRKY domain, which is defined by a region of approximately 60 amino acids containing a conserved WRKYGQK amino acid sequence adjacent to a zinc-finger-like motif (Eulgem et al. 2000). Both regions are necessary for high-affinity binding to a canonical W-box motif containing a consensus sequence (C/T)TGAC(C/T). Based on primary structure, WRKY proteins have been classified into three groups (I, II, and III) and various subgroups (e.g., IIa, IIb, etc.).
Recent studies have shown that WRKY transcription factors are involved in various biological processes but most notably respond to biotic and abiotic stresses (Pandey and Somssich 2009). WRKY transcription factors from various plant species function as both positive and negative regulators in plant innate immunity. Compared with the gain-of-function analysis which has been a powerful tool to reveal defense-related phenotype of individual WRKY transcription factor, the success of loss-of-function analysis has been limited due to functional redundancy among structurally related WRKY proteins (Wang et al. 2006; Xu et al. 2006). For example, Piffanelli and colleagues screened 20 Oswrky insertion mutants but observed no phenotypic change for host and non-host pathogen interaction (Berri et al. 2009). Only a few WRKY genes have been reported to show an altered defense response in knockout or knock-down plants. For example, loss of AtWRKY70 function compromises both basal defense response to bacterial and fungal pathogens and RPP4-mediated race-specific resistance to Hyaloperonospora parasitica (Li et al. 2004; Li et al. 2006a; Knoth et al. 2007). Knock-down of AtWRKY23 expression results in decreased susceptibility to the cyst nematode Heterodera schachtii (Grunewald et al. 2008). OsWRKY89 suppression lines show reduced wax content and increased susceptibility to rice blast fungus (Wang et al. 2007).
To study the function of WRKY family genes with functional redundancies, one strategy is to generate lines in which multiple WRKY genes are knocked-out. For example, knock-outs of AtWRKY IIa subfamily genes AtWRKY18, AtWRKY40 and AtWRKY60 individually does not affect the plant response to the hemibiotrophic bacterial pathogen Pseudomonas syringae and the necrotrophic fungal pathogen Botrytis cinerea. However, wrky18 wrky40 and wrky18 wrky60 double mutants and the wrky18 wrky40 wrky60 triple mutant are more resistant to P. syringae but more susceptible to B. cinerea (Xu et al. 2006). The Arabidopsis WRKY IId subfamily member wrky11 but not wrky7 or wrky17 mutants show enhanced basal resistance P. syringae. Further enhanced resistance is observed in wrky11 wrky17 double mutant plants that are generated and tested (Journot-Catalino et al. 2006).
We have previously reported that overexpression of OsWRKY62 compromises resistance to Xanthomonas oryzae pv. oryzae (Xoo) in the presence and absence of the Xa21 pattern recognition receptor (Peng et al. 2008; Lee et al. 2009). OsWRKY62, together with OsWRKY28, OsWRKY71 and OsWRKY76, comprise the rice WRKY IIa subfamily. It has been shown that transcripts of OsWRKY62 and OsWRKY76 accumulate in response to benzothiadiazole, salicylic acid (SA) and the rice fungal pathogen Magnaporthe grisea (Ryu et al. 2006; Shimono et al. 2007). In addition, OsWRKY71 is induced by SA and rice bacterial pathogen treatment (Liu et al. 2007). To understand the complex of this phylogenetically related gene family in the rice innate immunity, we designed a large inverted repeat construct to knock-down expression of all four members. Intriguingly, this silencing construct led to over-expression of all four genes in some transgenic plants. Those lines showing this “super-overexpression” phenotype displayed resistance against Xoo. In these super-overexpression resistant lines, pathogenesis-related (PR) gene expression was activated. Although the mechanism of the super-overexpression phenotype induced by the gene silencing construct is unknown, our results indicate functional interactions between OsWRKY IIa subfamily members in regulating plant innate immunity.
Results
Generation of a super long inverted repeat construct
To probe the role of the OsWRKY IIa subfamily genes in plant innate immunity, we attempted to knock down all four family members simultaneously with one “super” long inverted repeat (SIR) construct. SIR constructs have been used to successfully silence various numbers of a family of four Rac GTPases (Miki et al. 2005).
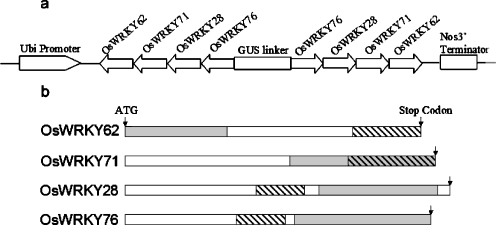
To design the OsWRKY IIa SIR construct, WRKYssRNAi, we used the maize ubiquitin promoter to drive the expression of both sense and antisense sequences of a segment (ranging from 165 to 265 bp) of each family member, separated by a 979-bp ß-glucuronidase (GUS) spacer (Fig. 1a). This construct is predicted to induce formation of hairpin mRNAs carrying the fragments of OsWRKY IIa genes in the stem and the GUS spacer in the loop. The segments chosen for OsWRKY62, OsWRKY76 and OsWRKY28 are gene-specific regions (Fig. 1b) and do not contain a contiguous stretch of over 16 bases of identical nucleotides to the other annotated rice genes. The segment of OsWRKY71 contains contiguous stretches of 20–24 bases identical nucleotides to OsWRKY28.
Fig. 1.
Schemetic representation of the WRKYssRNAi construct. a The structure of the WRKYssRNAi construct. The OsWRKY IIa sequences (OsWRKY62, 228 bp; OsWRKY71, 265 bp; OsWRKY28, 186 bp; OsWRKY76, 165 bp) were placed in the antisense-GUS linker-sense orientation. b Diagrams of the OsWRKY IIa coding regions. Regions used to generate the WRKYssRNAi construct are labeled by diagonal lines. The grey boxes indicate the regions which were amplified by RT-PCR using the gene specific primers for endogenous gene expression analysis.
Endogenous OsWRKY IIa gene expression in the WRKYssRNAi transgenic lines
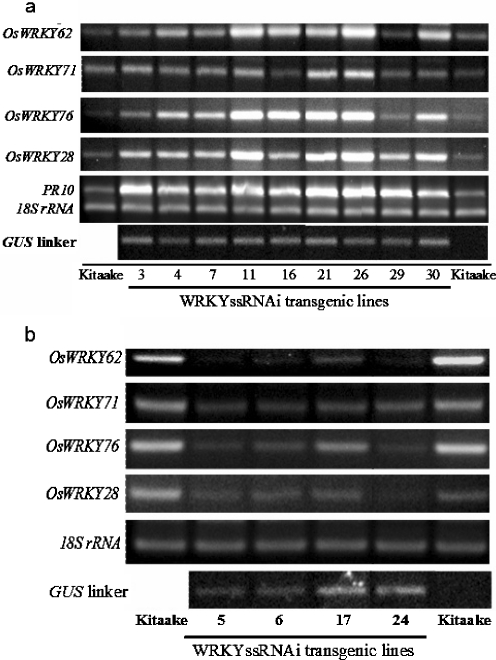
The WRKYssRNAi construct was introduced into Kitaake plants by Agrobacterium-mediated transformation. The presence of the transgene in individually transformed plants was confirmed by PCR (data not shown). To determine the RNA levels of the endogenous OsWRKY IIa genes, real-time polymerase chain reaction (RT-PCR) was performed on RNA samples from 17 transgenic lines using gene-specific primers (Fig. 1b). Surprisingly, super-overexpression of all four genes was observed in nine transgenic lines when compared to the wild type controls (Fig. 2a). Four transgenic lines showed silencing of all four genes, called super-silencing, as expected (Fig. 2b), and four lines showed little change in gene expression relative to the controls (data not shown). To ensure that the inserted construct is transcribed as expected, the gus linker expression was examined using RT-PCR. As shown in Fig. 2, the gus linker expression was detected in all 17 transgenic lines.
Fig. 2.
RNA analysis of WRKYssRNAi T0 lines. RNA samples were extracted from non-inoculated leaves. 18S ribosomal RNA was used as an internal control. a Lines showing super-overexpression of OsWRKY IIa genes. b Lines showing super-silencing of OsWRKY IIa genes.
Super-overexpression of OsWRKY IIa subfamily genes enhances rice basal defense against Xoo
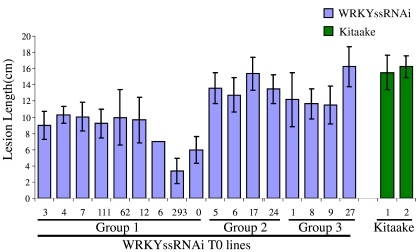
To study the possible function of OsWRKY IIa genes in basal defense response to pathogens, WRKYssRNAi transgenic T0 lines were challenged with Xoo at 5 weeks of age. The nine plants that displayed the super-overexpressed OsWRKY IIa molecular phenotype also displayed resistance to Xoo. Figure 3 indicates that lesions developed on the plants super-overexpressing OsWRKY IIa genes were significantly shorter than those of wild-type Kitaake controls. The four plants that showed super-silencing of OsWRKY IIa genes did not exhibit alterations in lesion length and thus were still susceptible. To ensure that our observations were not somehow related to the one set of transformations, we also performed a second transformation of the OsWRKYIIa SIR construct into Kitaake. Many of these plants also exhibited a disease resistance phenotype (data not shown).
Fig. 3.
Xoo inoculation results of WRKYssRNAi T0 lines. The numbers listed below the bar represent individual transgenic plants. Group 1 includes plants super-overexpressing OsWRKY IIa genes. Group 2 includes plants super-silencing OsWRKY IIa genes. Group 3 includes plants with no change in OsWRKY IIa gene expression. Kitaake plants were used as controls. Leaves were inoculated when the plants were 5 weeks old. Lesion lengths were measured 14 days afterwards. Each bar represents the average and standard deviation of at least three leaves.
To elucidate the molecular basis of the disease resistance phenotype developed on plants super-overexpressing OsWRKY IIa genes, we carried out RT-PCR analysis to compare the expression of PR10, a defense-related gene, in un-inoculated leaves. As seen in Fig. 2a, PR10 was highly induced in the lines super-overexpressing OsWRKY IIa members when compared to the Kitaake controls. These results indicate that super-overexpression of OsWRKY IIa genes activates pathogenesis-related gene expression, which likely contributes to enhanced basal defense against Xoo in rice plants.
The disease resistance and super-overexpression phenotypes are stably inherited
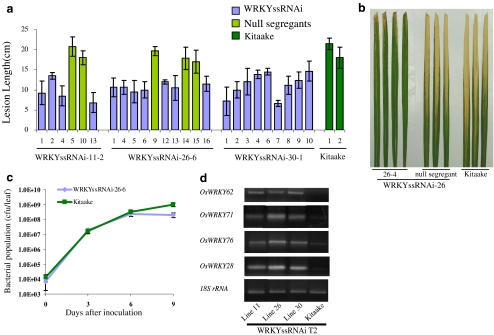
To examine whether the disease resistance phenotype can be maintained in the next generation, progeny from the WRKYssRNAi T1 lines, WRKYssRNAi-11-2, -26-6, and -30-1, were tested for co-segregation between the WRKYssRNAi construct and the disease resistance phenotype. The presence of the WRKYssRNAi construct in each plant was examined by PCR (data not shown). Five-week old plants were subjected to Xoo inoculation. As shown in Fig. 4a, progeny from three independently transformed lines carrying the WRKYssRNAi construct showed shorter lesions than the null segregants as well as the Kitaake controls. Figure 4b shows typical leaf samples after Xoo infection from segregating progeny of WRKYssRNAi-26 and Kitaake. The null segregants developed long water-soaked lesions similar to those observed in the Kitaake controls. By contrast, lesion development was restricted in the WRKYssRNAi transgenic plant (26-4). To quantify the Xoo population in the leaves, we carried out growth curve analysis. We found that that the shorter lesions of WRKYssRNAi line 26 correlated with reduced populations of Xoo in rice leaves (Fig. 4c).
Fig. 4.
Xoo inoculation results and RNA analysis of WRKYssRNAi progeny. a Leaf lesion lengths of WRKYssRNAi T2 segregating progeny. Kitaake plants were used as controls. Leaves were inoculated when the plants were 5 weeks old; lesion lengths were measured 14 days afterwards. Each bar represents the average and standard deviation of at least three leaves. b A picture of lesions developed on leaves from T1 progeny of WRKYssRNAi line 26 and Kitaake taken 14 days after Xoo inoculation. c Xoo growth curve analysis of progeny of WRKYssRNAi-26-6 and Kitaake lines. For each time point, the bacterial population size was determined separately for three leaves of similar growth stage from three plants. Error bars represents the standard deviation of three leaves. d RNA expression levels of OsWRKY IIa genes in un-inoculated leaves of WRKYssRNAi T2 progeny. RT-PCR was performed using gene-specific primers targeted to the regions not represented in the WRKYssRNAi construct. 18S ribosomal RNA was used as an internal control.
To verify that the disease resistance phenotype correlated with super-overexpression of OsWRKY IIa genes, we analyzed the expression of OsWRKY IIa genes in the T2 progeny from WRKYssRNAi T1 lines, 11-2, -26-6, and -30-1. RNA samples were extracted from un-inoculated leaves. RT-PCR results in Fig. 4d shows that the progeny that inherited the WRKYssRNAi construct maintained the elevated expression level of OsWRKY IIa genes. Together, these results indicate that super-overexpression of OsWRKY IIa genes caused by the WRKYssRNAi construct can by stably inherited. Moreover, super-overexpression of OsWRKY IIa genes enhances resistance to Xoo in rice.
Discussion
OsWRKY IIa subfamily members play both positive and negative regulatory roles in regulating plant innate immunity. Overexpression of OsWRKY62 compromises rice basal defense and Xa21-mediated resistance to Xoo (Peng et al. 2008). By contrast, overexpression of OsWRKY71 results in elevated expression of defense-related genes and enhanced resistance to Xoo (Liu et al. 2007). The work presented here demonstrates that super-overexpression of OsWRKY IIa subfamily members enhances basal defense against Xoo. These data indicate functional interactions between OsWRKY IIa proteins in regulating plant innate immunity. Yeast two-hybrid and bimolecular fluorescence complementation experiments provide support for physical interactions among the OsWRKYIIa proteins (Y. Seo et al., in preparation).
Similarly, Arabidopsis WRKY IIa subfamily members interact both physically and functionally in plant defense (Xu et al. 2006). AtWRKY IIa proteins form both homocomplexes and heterocomplexes. Association of different WRKY IIa members can alter their sequence-specific DNA binding activities. In addition, constitutive expression of AtWRKY18 enhances resistance to P. syringae, while plants overexpressing AtWRKY18 with either AtWRKY40 or AtWRKY60 are more susceptible to both P. syringae and B. cinerea. These observations indicate that association of different WRKY IIa members may alter defense gene regulation and ultimately affect the plant defense response.
A recent report on interactions of two transcriptional repressors and two transcriptional activators in modulating gibberellin signaling in aleurone cells provides an intriguing example of how different transcription factor complex affects the expression of target genes (Zou et al. 2008). HvWRKY38, a barley ortholog of OsWRKY71, functions as a repressor of GA-induced expression of Amy32b a-amylase. When present individually, the inductive activities of SAD, a DNA-binding with one finger (DOF) protein and HvGAMYB (a R2R3 MYB protein) on Amy32b expression are blocked by HvWRKY38. However, SAD and HvGAMYB together overcome the inhibitory effect of HvWRKY38. Interestingly, the synergistic effect of SAD and HvGAMYB transcriptional activators is almost abolished when repressor BPBF, a barley prolamin-box binding factor which is another DOF protein, is present with HvWRKY38. Further investigation shows that HvWRKY38 interacts with HvBPBF in the nuclei of aleurone cells. Moreover, HvWRKY38 interferes with the binding of HvGAMYB to the Amy32b promoter. Taken together, these studies suggest that relative amounts of activator and repressor proteins in a complex can modulate the expression of target genes.
OsWRKY IIa proteins contain putative leucine zipper motifs at the N-terminus, which suggests potential homodimerization or heterodimerization between proteins. It has been shown that leucine zipper motifs are critical for the physical interaction of AtWRKY IIa proteins and homodimerization of HvWRKY38 (Xu et al. 2006; Zou et al. 2008). OsWRKY71 has also recently been reported to function as a transcriptional repressor (Chujo et al. 2008). As a positive regulator of plant defense response, OsWRKY71 may play an indirect role in transcriptional activation of defense-related gene expression by interacting with other regulatory proteins. Taken together, we hypothesize that different combinatorial dimers formed by OsWRKY IIa proteins may exhibit different functions in regulating target gene expression. Moreover, changes in the participants within protein complexes caused by mutation, overexpression or environmental stress can also affect the expression of target genes, leading to altered plant defense response. This may explain the data observed with rice lines overexpressing OsWRKY62 alone (susceptible) compared with plants overexpressing all four OsWRKY IIa proteins (resistant).
While ineffective gene silencing by short interference RNA (siRNA) is commonly observed (Rose et al. 2005), it has not previously been observed that a siRNA construct can lead to super-overexpression of the target genes. Inverted repeats are usually found to be very efficient for silencing (Miki et al. 2005), although different levels of efficiency of silencing are not uncommon (Chern et al. 2005a). We have searched the literature for an explanation of the super-overexpression phenomenon and found reports of RNAs that are able to stimulate transcription; however, these examples are directed toward promoter sequences, not the translated region of a gene. For instance, Dahiya and associates discovered that exogenous dsRNAs designed against human promoter regions increase expression of the corresponding coding region (Li et al. 2006b). Our results indicate that simultaneous over-expression of all the members of the rice WRKY IIa complex reveals a previously unobserved outcome of attempted gene silencing. Although the mechanism remains unknown at this point, one possibility is that self-regulation by WRKY genes or regulation via other transcription factors and proteins may contribute to this phenotype.
Methods
Plant materials and growth conditions
Rice (Oryza sativa L.) plants were grown in the greenhouse until 5 weeks of age and transferred to the growth chamber before Xoo inoculation. In the greenhouse, the light intensity in photosynthetic photon flux across the spectrum from 400 to 700 nm was approximately 250 μmol m−2 s−1 in spring. The growth chamber was set on a 14-hour daytime period, a 28/26°C temperature cycle and at 90% humidity. The chamber was equipped with metal halide and incandescent lights. The light intensity in the growth chamber was approximately 100 μmol m−2 s−1.
Xoo inoculation and quantification of bacterial populations
For Xoo inoculation, a bacterial suspension (OD600 of 0.5) of Xoo stain PX099 (Philippine race 6, PR6), which carries ax21 activity, was used to inoculate rice by the scissors-dip method (Kauffman et al. 1973; Lee et al. 2009).
To determine the bacterial populations, three inoculated leaves from each genotype were ground up and resuspended in 10 mL H2O to harvest bacteria separately. Diluted extract was plated on peptone sucrose agar plates containing 15 mg/L cephalexin.
Plasmid construction and rice transformation
To generate the WRKYssRNAi construct, the fragments of OsWRKY62 and OsWRKY71 were fused by overlapping PCR using primers 5′-TCGCCGACCAAGGATTGAGCACGATCACACTCGAC-3′/5′-TTTGGATCCCAGGAATGTGTGGGAT-3′ (for OsWRKY62); 5′-TCCAAGCGCTTCGTCCAGAACAGCGACGGCTCCG-3′/5′-GTCGAGTGTGATCGTGCTCAATCCTTGGTCGGC-3′ (for OsWRKY71); The fragments of OsWRKY28 and OsWRKY76 were fused by overlapping PCR using primers 5′-TTTGAATTCCTGCGACGCCAA-3′/5′-GAGGCTGTCGACGCTAATTGGTACCCGTCCTTGA-3′ (for OsWRKY76); 5′-TCAAGGACGGGTACCAATTAGCGTCGACAGCCTCGA-3′/5′-CGGAGCCGTCGCTGTTCTGGACGAAGCGCTTGGAG-3′ (for OsWRKY28). Both PCR products were purified and fused by overlapping PCR using primers 5′-TTTGATTCCTGCCGACGCCAA-3′/5′-TTTGGATCCCAGGAATGTGTGGGAT-3′. The final product was cloned into the pGEM T Easy vector (Promega) and confirmed by sequencing. The insert was then excised with BamHI and EcoRI and ligated with a 1 kb, EcoRI-digested GUS fragment into BamHI pre-cut pBlueScript II SK-, yielding a 2.6 kb insert containing fragments from four WRKY IIa member genes in tandem in antisense and sense orientations with GUS linker in the center. This insert was excited with BamHI and cloned into the Ubi-NC1300 vector (Chern et al. 2005b). The insert sequence is available at GenBank under accession number 1288449.
Rice transformation was as described previously (Chern et al. 2001). Agrobacterium EHA105 was used to infect rice callus. For transformation of Kitaake, hygromycin was used for selection.
RNA extraction and RT-PCR analysis
Total RNA was extracted from leaves using the Trizol reagent (Invitrogen). Total RNA was converted into cDNA using M-MLV reverse transcriptase (EC 2.7.7.49; Invitrogen). PCR analyses were performed with gene-specific primer sets: 5′-ATGGACGACGACGGCGACGGCT-3′/5′- GCGGTCGGCGGCTGCTGTCTC-3′ (for OsWRKY62); 5′- AATGGAGGAAGTATGGTCAGAAGGT-3′/5′- CGTCGATCAGTAGCAATCGTCAAT-3′ (for OsWRKY71); 5′- CATCGTGCCCGGTGAAGAAGAA-3′/5′- AATTCGGGCAGCTTCTGGAGGAT-3′ (for OsWRKY76); 5′- AGAAGGTGACGAAGGACAACCC-3′/5′- GTGACGAGCGCCGCCTTGAAC-3′ (for OsWRKY28); 5′-CGCAGCTCACATTATCAAGTCAGA-3′/5′-GAAGCAGCAATACGGAGATGGATG-3′ (for PR10); 5′- CATGAAGATGCGGACTTACG-3′/5′- ATCCACGCCGTATTCGG-3′ (for GUS). 18S ribosomal RNA was used as an internal control following the QuantumRNA 18S Internal Standards manual (Ambion, Austin, TX, USA). The amplified products were then resolved by gel electrophoresis.
Acknowledgement
Thanks to Maw-Sheng Chern for valuable discussions and to Brian Conlin for his technical assistance. This work was supported by the National Institute of Health grant #GM59962 to PCR. LEB was supported in part by a University of California Office of the President Postdoctoral Fellowship.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Berri S, Abbruscato P, Faivre-Rampant O, Brasileiro AC, Fumasoni I, Satoh K, et al. Characterization of WRKY co-regulatory networks in rice and Arabidopsis. BMC Plant Biol. 2009;9:120. doi: 10.1186/1471-2229-9-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern MS, Fitzgerald HA, Yadav RC, Canlas PE, Dong X, Ronald PC. Evidence for a disease-resistance pathway in rice similar to the NPR1-mediated signaling pathway in Arabidopsis. Plant J. 2001;27:101–113. doi: 10.1046/j.1365-313x.2001.01070.x. [DOI] [PubMed] [Google Scholar]
- Chern MS, Canlas PE, Fitzgerald HA, Ronald PC. Rice NRR, a negative regulator of disease resistance, interacts with Arabidopsis NPR1 and rice NH1. Plant J. 2005;43:623–635. doi: 10.1111/j.1365-313X.2005.02485.x. [DOI] [PubMed] [Google Scholar]
- Chern M, Fitzgerald HA, Canlas PE, Navarre DA, Ronald PC. Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol Plant Microbe Interact. 2005;18:511–520. doi: 10.1094/MPMI-18-0511. [DOI] [PubMed] [Google Scholar]
- Chujo T, Kato T, Yamada K, Takai R, Akimoto-Tomiyama C, Minami E, et al. Characterization of an elicitor-induced rice WRKY gene, OsWRKY71. Biosci Biotechnol Biochem. 2008;72:240–245. doi: 10.1271/bbb.70553. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol. 2007;10:366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- Grunewald W, Karimi M, Wieczorek K, Van de Cappelle E, Wischnitzki E, Grundler F, et al. A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiol. 2008;148:358–368. doi: 10.1104/pp.108.119131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journot-Catalino N, Somssich IE, Roby D, Kroj T. The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell. 2006;18:3289–3302. doi: 10.1105/tpc.106.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman HE, Reddy APK, Hsieh SPV, Marca SD. An improved technique for evaluation of resistance of rice varieties to Xanthomonas oryzae. Plant Dis Rep. 1973;57:537–541. [Google Scholar]
- Knoth C, Ringler J, Dangl JL, Eulgem T. Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol Plant Microbe Interact. 2007;20:120–128. doi: 10.1094/MPMI-20-2-0120. [DOI] [PubMed] [Google Scholar]
- Lee SW, Han SW, Sririyanum M, Park CJ, Seo YS, Ronald PC. A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science. 2009;326:850–853. doi: 10.1126/science.1173438. [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET. The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 2004;16:319–331. doi: 10.1105/tpc.016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Brader G, Kariola T, Palva ET. WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 2006;46:477–491. doi: 10.1111/j.1365-313X.2006.02712.x. [DOI] [PubMed] [Google Scholar]
- Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, et al. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Bai X, Wang X, Chu C. OsWRKY71, a rice transcription factor, is involved in rice defense response. J Plant Physiol. 2007;164:969–979. doi: 10.1016/j.jplph.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Miki D, Itoh R, Shimamoto K. RNA silencing of single and multiple members in a gene family of rice. Plant Physiol. 2005;138:1903–1913. doi: 10.1104/pp.105.063933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009;150:1648–1655. doi: 10.1104/pp.109.138990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Bartley LE, Chen X, Dardick C, Chern M, Ruan R, et al. OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol Plant. 2008;1:446–458. doi: 10.1093/mp/ssn024. [DOI] [PubMed] [Google Scholar]
- Rose SD, Kim DH, Amarzguioui M, Heidel JD, Collingwood MA, Davis ME, et al. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005;33:4140–4156. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Liu Y, Shen QJ. The WRKY Gene Family in Rice (Oryza sativa) J Integr Plant Biol. 2007;49:827–842. doi: 10.1111/j.1744-7909.2007.00504.x. [DOI] [Google Scholar]
- Ryu HS, Han M, Lee SK, Cho JI, Ryoo N, Heu S, et al. A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Rep. 2006;25:836–847. doi: 10.1007/s00299-006-0138-1. [DOI] [PubMed] [Google Scholar]
- Shimono M, Sugano S, Nakayama A, Jiang CJ, Ono K, Toki S, Takatsuji H. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell. 2007;19:2064–2076. doi: 10.1105/tpc.106.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2006;2:e123. doi: 10.1371/journal.ppat.0020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hao J, Chen X, Hao Z, Wang X, Lou Y, et al. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol Biol. 2007;65:799–815. doi: 10.1007/s11103-007-9244-x. [DOI] [PubMed] [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell. 2006;18:1310–1326. doi: 10.1105/tpc.105.037523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X, Neuman D, Shen QJ. Interactions of two transcriptional repressors and two transcriptional activators in modulating gibberellin signaling in aleurone cells. Plant Physiol. 2008;148:176–186. doi: 10.1104/pp.108.123653. [DOI] [PMC free article] [PubMed] [Google Scholar]