Abstract
Background
An optimal staging system to estimate the risk of recurrence following the complete resection of localized, primary gastrointestinal stromal tumor (GIST) has not been established. Recently, it has been shown that adjuvant imatinib mesylate prolongs recurrence-free survival (RFS) following the resection of localized, primary GIST. We sought to develop a nomogram to predict RFS after surgery in the absence of adjuvant therapy to help guide patient selection for adjuvant imatinib therapy.
Methods
A nomogram to predict RFS based on tumor size (in cm), location (stomach, small intestine, colon/rectum, or other), and mitotic index (<5 or ≥5 mitoses per 50 high power fields) was developed from 127 patients treated at Memorial Sloan-Kettering Cancer Center. The nomogram was tested in cohorts of patients from the Spanish national registry (GEIS; n=212) and the Mayo Clinic (Mayo; n=148). The nomogram was evaluated by calculating concordance probabilities as well as testing calibration of predicted RFS with observed RFS. Concordance probabilities were also compared with those of 3 commonly employed staging systems.
Findings
The nomogram had a concordance probability of 0.78 (±0.02) in the MSKCC dataset and 0.76 (±0.03) and 0.80 (±0.02) in the validation cohorts. The nomogram predictions were well calibrated. Remodeling the nomogram to include tyrosine kinase mutation status did not improve its discriminatory ability. Concordance probabilities of the nomogram were superior to those of the 2 NIH staging systems (0.76 (±0.03) versus 0.70 (±0.04) (p=0.04) and 0.66 (±0.04) (p=0.01) in the GEIS validation cohort; 0.80 (±0.02) versus 0.74 (±0.02) (p=0.04) and 0.78 (±0.02) (p=0.05) in the Mayo cohort) and equivalent to the AFIP-Miettinen staging system (0.76 (±0.03) versus 0.73 (±0.004) (p=0.28) in the GEIS cohort; 0.80 (±0.02) versus 0.76 (±0.003) (p=0.09) in the Mayo cohort). Nomogram predictions of RFS appeared better calibrated than predictions made using the AFIP-Miettinen system.
Interpretation
The nomogram accurately predicts RFS following the resection of localized, primary GIST and may be useful to select patients for adjuvant imatinib therapy.
Keywords: gastrointestinal stromal tumor, GIST, nomogram, recurrence, prognosis, survival, imatinib mesylate, staging, surgery
INTRODUCTION
GISTs typically arise from the gastrointestinal tract, but also can be found in the mesentery, omentum, and retroperitoneum.1,2 They commonly contain a mutation in the KIT proto-oncogene, or less frequently, in platelet-derived growth factor receptor alpha (PDGFRA).3–5 GISTs have received considerable attention due to their sensitivity to tyrosine kinase inhibitors. Imatinib mesylate (Gleevec™, Novartis Pharmaceuticals, Basel Switzerland) is a specific inhibitor of the KIT and PDGFRA proteins (as well as ABL and BCR-ABL). Imatinib achieves a partial response or stable disease in over 80% of patients with metastatic GIST with a median survival of 5 years.6 Unfortunately, acquired resistance to imatinib occurs at a median treatment duration of less than 2 years.6,7 The other FDA-approved targeted agent for advanced GIST is sunitinib maleate (Sutent™, Pfizer, New York), which inhibits KIT, PDGFRA, the vascular endothelial growth factor receptors, fms-like tyrosine kinase-3 receptor (FLT3), and the RET receptor. When used in patients who are intolerant to imatinib or have refractory disease, sunitinib achieves a median progression-free survival (PFS) of 6 months.8
The gold standard for localized, primary GIST is surgical resection. Unfortunately, tumor recurrence is common and usually occurs in the liver and/or the peritoneum.9 Nearly all GISTs, with the possible exception of very small tumors (<1 cm) found incidentally, seem to have the potential to recur following surgical resection. However, determination of the risk of recurrence in a particular patient has been difficult. The rarity of the tumor and the relatively recent recognition of GIST as a distinct pathologic entity among soft tissue tumors have limited the identification of prognostic variables and the establishment of staging systems. The widespread use of immunohistochemical staining for KIT (CD117) and the overexpression of KIT by virtually all GISTs have now enabled GIST to be diagnosed with precision.
Recently, the American College of Surgeons Oncology Group (ACOSOG) reported the results of study Z9001, an intergroup, randomized, double-blind, placebo controlled trial evaluating adjuvant imatinib for patients with primary GIST ≥3 cm. Adjuvant imatinib therapy prolonged recurrence-free survival (RFS) in comparison to placebo.10 With a median follow-up of 19 months, RFS at 1 year was 98% on the imatinib arm versus 83% on the placebo arm (hazard ratio 0.35, p<0.0001). Based on the trial results, the United States Food and Drug Administration (FDA) approved the use of adjuvant imatinib in December 2008, and EMEA approved its use in March 2009.11,12 In particular, the European Medicines Agency (EMEA) approval is restricted to patients at “significant risk” of relapse, without reference to what criteria should be used to make this determination. In light of the potential toxicity and the financial cost of the treatment, the ability to precisely calculate the risk of recurrence for individual patients is important.
Risk stratification in GIST based on tumor size, mitotic activity, and tumor location has been suggested, but an optimal staging system has not been established and validated.13–21 Two commonly used staging systems for prognosis were developed at a 2001 United States National Institutes of Health (NIH) Workshop and are shown in Table 1.14,15 A modification of one of these staging systems was suggested in 2006 and is also shown in Table 1.18 Notably, none of these staging systems provides a quantifiable risk of recurrence for individual patients. While an American Joint Committee on Cancer (AJCC) TNM staging system for sarcoma exists, it is not specific enough for GIST and therefore has not been used. Prognostic nomograms for assessing postoperative outcome in sarcomas and other malignancies have been developed at Memorial Sloan-Kettering Cancer Center (MSKCC) and elsewhere.22–26 A nomogram is a graphical interface for a statistical model utilizing variables with additive prognostic importance to predict outcome precisely for a given patient. Nomograms are typically more accurate than staging systems, such as AJCC groupings.24,25 The aim of the current study was to establish a nomogram to predict the risk of tumor recurrence after gross surgical resection of a localized, primary GIST in the absence of tyrosine kinase inhibitor therapy.
Table 1.
Commonly used staging systems for assessing behavior of GIST.
| NIH-Fletcher14 | |
|---|---|
| Risk | Features |
| Very low | <2 cm and <5 mitotic index |
| Low | 2–5 cm and <5 mitotic index |
| Intermediate | 5–10 cm and <5 mitotic index or <5 cm and 6–10 mitotic index |
| High | >5 cm and >5 mitotic index or >10 cm and any mitotic index or any size and >10 mitotic index |
| NIH-Miettinen15 | |
|---|---|
| Group | Features |
| Probably benign | gastric: ≤5 cm and ≤5 mitotic index intestinal: ≤2 cm and ≤5 mitotic index |
| Uncertain or low malignant potential |
gastric: >5 cm, ≤10 cm, and ≤5 mitotic index intestinal: >2 cm, ≤5 cm, and ≤5 mitotic index |
| Probably malignant | gastric: >10 cm or >5 mitotic index intestinal: >5 cm or >5 mitotic index |
| AFIP-Miettinen18 | |
|---|---|
| Group | Features |
| Very low, if any malignant potential |
≤2 cm and ≤5 mitotic index |
| Low malignant potential |
gastric: >2/ ≤10 cm and ≤5 mitotic index, ≤2 cm and >5 mitotic index intestinal: >2/ ≤5 cm and ≤5 mitotic index |
| Intermediate malignant potential |
gastric: >10 cm and ≤5 mitotic index, >2/ ≤5 cm and >5 mitotic index intestinal: >5/ ≤10 cm and ≤5 mitotic index |
| High malignant potential |
gastric: >5 cm and >5 mitotic index intestinal: >10 cm or >5 mitotic index |
Mitotic index = number of mitoses per 50 high power fields
METHODS
Patients and Clinicopathologic Variables
Three databases of patients who underwent complete gross resection of a localized, primary GIST without adjuvant therapy were used in this study. Within each dataset, an expert pathologist confirmed the diagnosis of GIST and calculated the mitotic index (number of mitoses per 50 randomly selected high power microscopic fields (HPF)). Tumor size was measured by the pathologist either before or after formalin fixation. The nomogram was constructed based on 127 patients treated at MSKCC between 1983 and 2002. The validation cohort from the Spanish Group for Research in Sarcomas (Grupo Español de Investigación en Sarcomas, GEIS) consisted of 212 patients with GIST diagnosed between 1994 and 2001 at 30 of the 80 member hospitals. The Mayo Clinic validation cohort included 148 patients who underwent surgery between 1978 and 2004. Demographic information and clinicopathologic variables of the patients in the 3 datasets are shown in Table 2. Individual analyses from these datasets have been published previously.27–29 This study was approved by the institutional review board at each institution.
Table 2.
Demographic information and clinicopathologic variables.
| Variable | N (%) or Median (Range) | |||
|---|---|---|---|---|
| MSKCC (n=127) |
GEIS (n=212) |
Mayo (n=148) |
||
| Sex | Female | 54 (43) | 116 (55) | 66 (45) |
| Male | 73 (57) | 96 (45) | 82 (55) | |
| Age | Years | 67 (10–94) | 66 (25–93) | 63 (13–91) |
| Tumor site | Stomach | 74 (58) | 125(59) | 79 (53) |
| Small intestine | 35 (28) | 74 (35) | 54 (37) | |
| Colon/rectum | 14 (11) | 3 (1) | 15 (10) | |
| Other | 4 (3) | 10 (5) | 0 (0) | |
| Tumor size | Centimeters | 6 (0.3–50) | 6 (0.4–27) | 6.5 (1–37) |
| Mitotic index | <5 | 94 (74) | 192 (91) | 89 (60) |
| ≥5 | 33 (26) | 20 (9) | 59 (40) | |
| Completeness of resection |
R0 | 108 (85) | 195 (92) | 144 (97) |
| R1 | 19 (15) | 8 (4) | 4 (3) | |
| Unknown | 0 (0) | 9 (4) | 0 (0) | |
| Tumor Rupture |
No | 123 (97) | 212 (100) | 147 (99) |
| Yes | 4 (3) | 0 (0) | 148 (1) | |
Mitotic index = number of mitoses per 50 high power fields
Statistical Analysis
RFS probabilities were estimated using the Kaplan-Meier method.30 Multivariate analysis was conducted using Cox proportional hazards regression models. The proportional hazards assumption was verified by tests of correlations with time and examination of residual plots. A restricted cubic spline was used to model the non-linear relationship between tumor size and recurrence.31 Only 4 variables (tumor size, mitotic index, site and type of tyrosine kinase mutation) were considered for this model due to the limited number of recurrences in the data. This Cox model (available online) was the basis for a nomogram, and our modeling and internal validation procedures are similar to those used previously.32
Nomogram performance was assessed in 2 ways. First, the concordance probability was calculated.33 The concordance probability is the chance that given 2 randomly drawn patients, the patient who recurs first had a higher nomogram-predicted probability of recurrence. If both patients recur at the same time, or if the patient with shorter follow-up does not recur, the probability does not apply to that pair of patients. The interpretation of the concordance probability is similar to that of the area under the receiver operating characteristic curve.34 For comparison, the concordance probability was also calculated for the 3 staging systems (Table 1) that are commonly used to predict the risk of tumor recurrence.14,15 Concordance probabilities were compared using 1000 bootstrap resamples and the method of asymptotic significance level (ASL).35 The second component of model assessment involved testing calibration. Nomogram-predicted probability of recurrence was compared against Kaplan-Meier observed RFS for 4 quartiles of patients stratified by nomogram score in each dataset. Calibration of the AFIP-Miettinen staging system was performed by assigning predictions of 2-year and 5-year RFS for each stage using the Kaplan-Meier predictions obtained using the MSKCC dataset. These predictions were then compared against the Kaplan-Meier observed RFS in the 2 validation cohorts. Calibration is assessed by plotting the predicted probabilities against the actual outcome. This graph should be close to the 45-degree line if the predictions are well calibrated. All analyses were performed using R version 2.3 (www.r-project.org) and SAS version 9.1 (Cary, NC).
Role of the Funding Sources
This work was supported by Public Health Service Grants CA102613 and P01 CA47179 from the National Cancer Institute. The sponsors did not have a role in study design; collection, analysis, or interpretation of the data; or writing this report. All authors had access to portions of the raw data. JSG, MG and RPD had access to all raw data and analysis. The sponsors did not have access to any of the raw data. The corresponding author had full access to all of the data and the final responsibility to submit for publication.
RESULTS
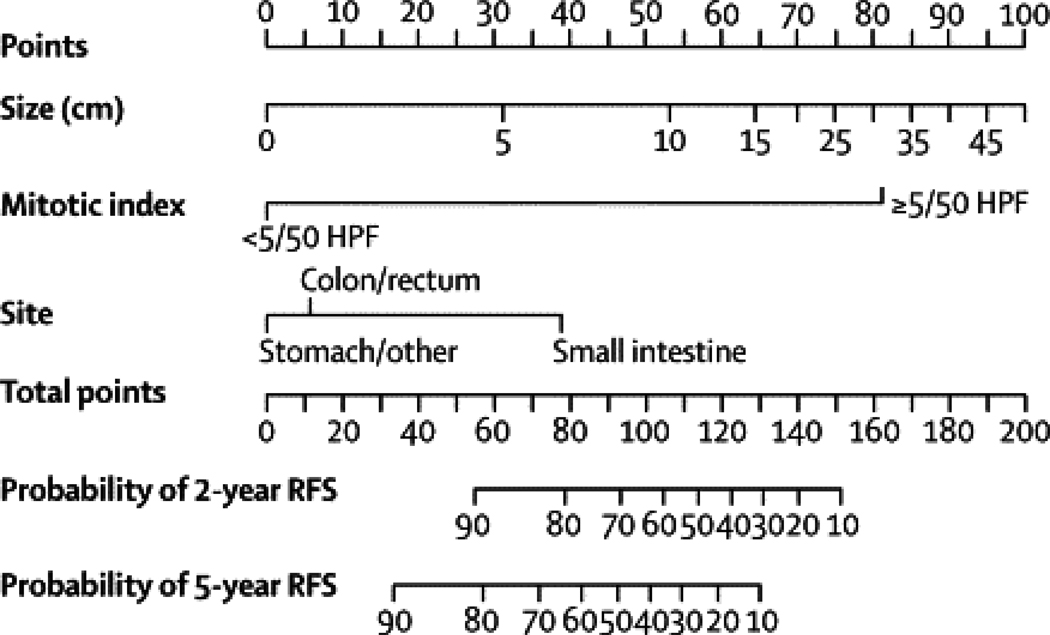
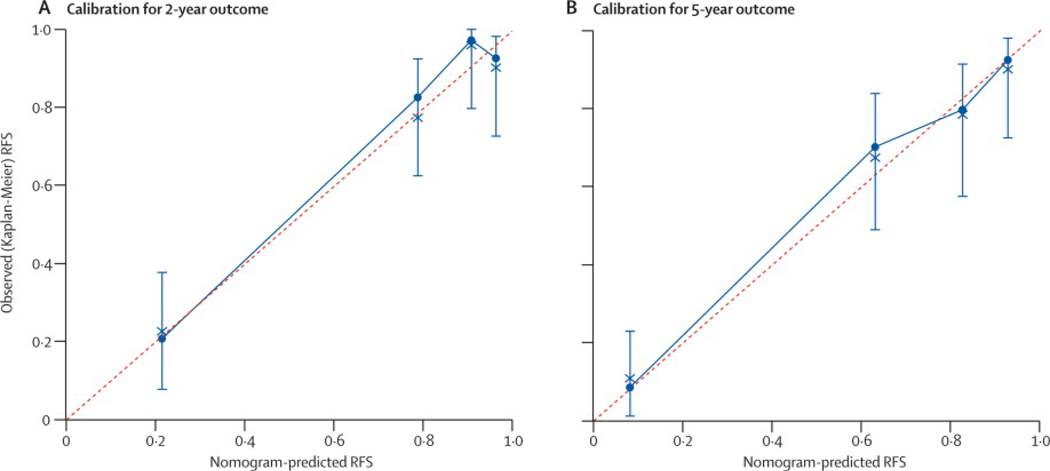
A nomogram was constructed using the 127 patients from MSKCC. The median follow-up of patients free of recurrence in this series was 4.7 years, with 42 patients experiencing recurrence. Recurrence-free survival (RFS) is shown in Figure 1. Nomogram construction was based on previous analysis of this series showing that tumor mitotic rate (with a breakpoint of <5 or ≥5 mitoses per 50 high power fields (HPF)), size (assessed as a continuous variable), and tumor site independently predict RFS.27 The nomogram assigned points based on tumor size in a continuous but nonlinear fashion. Points for tumor site were assigned based on whether the tumor arose in the stomach, small intestine, colon/rectum, or an extra-intestinal location, and for mitotic index based on whether the primary tumor had <5 or ≥5 mitoses per 50 HPF (Figure 2). The total number of points then determined the 2- and 5-year RFS probabilities. The concordance probability of the nomogram was 0.78 (standard error ±0.02) (Table 3). In other words, 78% of the time the nomogram correctly predicted the ordering of the outcome between 2 randomly selected patients. Remodeling the nomogram to include the presence or type of KIT or PDGFRA mutation did not improve its discriminatory ability (data not shown). The nomogram-predicted RFS was well calibrated with the Kaplan-Meier observed RFS (Figure 3A and B).
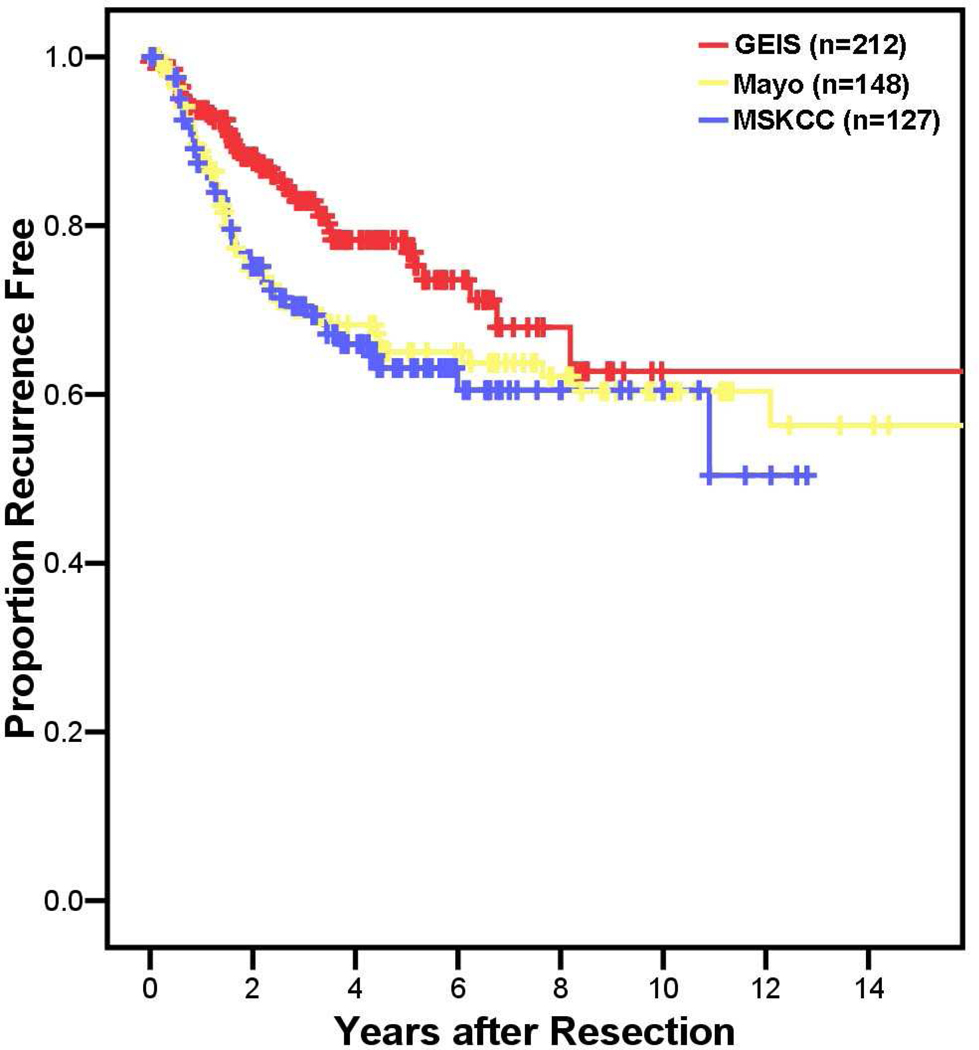
Figure 1. Recurrence-free survival for the 3 patient populations.
Kaplan Meier estimates of recurrence-free survival of localized, primary gastrointestinal stromal tumors after complete surgical resection based on patient series from 2 North American institutions and a Spanish sarcoma registry are shown.
Figure 2. Nomogram to predict the probabilities of 2- and 5-year recurrence-free survival.
Points are assigned for size, mitotic index, and site of origin by drawing a line upward from the corresponding values to the “Points” line. The sum of these 3 points plotted on the “Total Points” line corresponds to predictions of 2- and 5-year RFS.
Table 3.
Concordance probabilities (±standard error) of the nomogram compared with the NIH Workshop staging systems.
| Nomogram | NIH-Fletcher | NIH-Miettinen | AFIP-Miettinen | ||||
|---|---|---|---|---|---|---|---|
| Concordance | Concordance | p-value* | Concordance | p-value* | Concordance | p-value* | |
| MSKCC | 0.78 (±0.02) | 0.72 (±0.03) | 0.03 | 0.56 (±0.04) | <0.01 | 0.76 (±0.004) | 0.33 |
| GEIS | 0.76 (±0.03) | 0.70 (±0.04) | 0.04 | 0.66 (±0.04) | 0.01 | 0.73 (±0.004) | 0.28 |
| Mayo | 0.80 (±0.02) | 0.74 (±0.02) | 0.04 | 0.78 (±0.02) | 0.05 | 0.76 (±0.003) | 0.09 |
p-value vs. nomogram
Figure 3. Performance of the nomogram.
Calibration of nomogram-predicted recurrence-free survival (RFS) with observed RFS is shown at (A) 2 years and (B) 5 years for the Memorial Sloan-Kettering Cancer Center series.
The nomogram was validated using 2 external datasets. Two hundred twelve patients from the GEIS registry and 148 patients from the Mayo Clinic were identified who had sufficient information to utilize the nomogram. The median follow-up of patients free of recurrence is 3.1 years for the GEIS series and 4.8 years for the Mayo series. Forty patients developed tumor recurrence in the GEIS series and 46 patients did so in the Mayo Clinic series. RFS for the 2 validation cohorts is shown in Figure 1. The nomogram was used to assign points to each patient, and a concordance probability of 0.76 (standard error ±0.03) was calculated for the GEIS series and a concordance probability of 0.80 (standard error ±0.02) was calculated for the Mayo Clinic series (Table 3). Calibration of the 2-year and 5-year nomogram predictions for the GEIS and Mayo Clinic series is shown in Table 4. Nomogram predictions of RFS at 2 and 5 years appeared to be well calibrated with actual RFS for both external validation cohorts.
Table 4.
Calibration of the nomogram on the validation cohorts.
| Series | Time Point |
Group | Number of Patients |
Predicted RFS |
Kaplan- Meier Estimated RFS |
|---|---|---|---|---|---|
| GEIS | 2 yrs | 1 | 44 | 50% | 67% |
| 2 | 62 | 85% | 91% | ||
| 3 | 51 | 93% | 93% | ||
| 4 | 55 | 97% | 96% | ||
| 5 yrs | 1 | 64 | 40% | 55% | |
| 2 | 37 | 74% | 79% | ||
| 3 | 56 | 86% | 89% | ||
| 4 | 55 | 93% | 91% | ||
| Mayo | 2 yrs | 1 | 30 | 40% | 29% |
| 2 | 48 | 84% | 75% | ||
| 3 | 35 | 93% | 94% | ||
| 4 | 35 | 96% | 100% | ||
| 5 yrs | 1 | 51 | 39% | 26% | |
| 2 | 25 | 75% | 72% | ||
| 3 | 37 | 86% | 89% | ||
| 4 | 35 | 93% | 100% |
RFS - recurrence-free survival
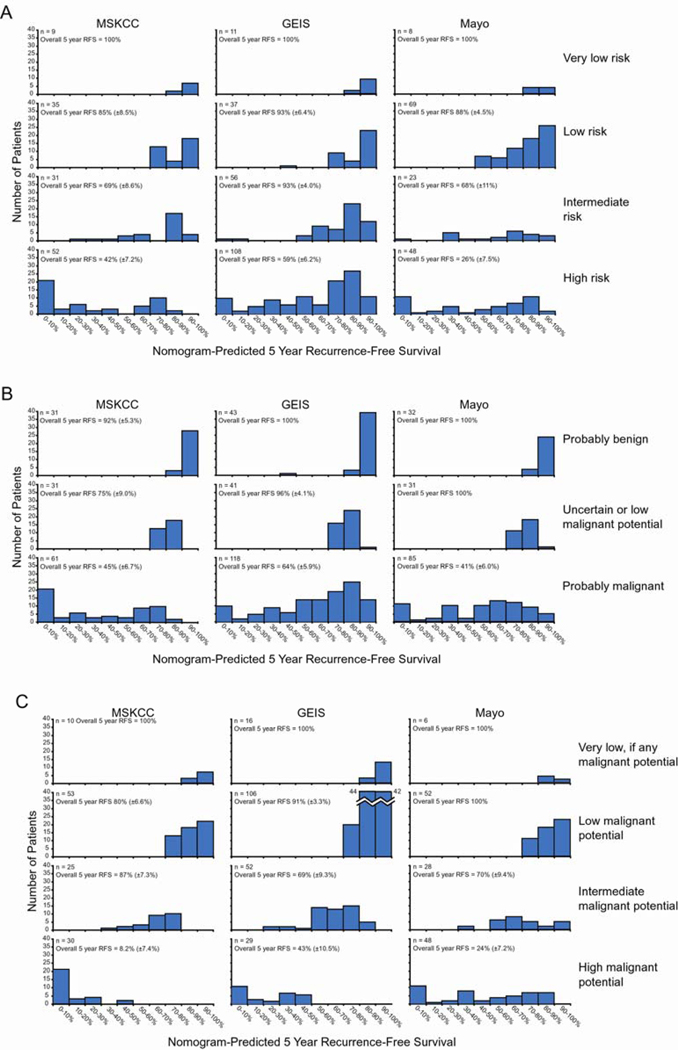
The predictive ability of the nomogram was compared to 3 commonly used staging systems that are used to predict the risk of recurrence following the resection of primary GIST.14,15,18 First, the nomogram was compared to the NIH Workshop Staging Systems proposed in 2001. The concordance probabilities of both these risk stratification schemes were significantly worse than that of the nomogram when tested on the MSKCC patients as well as each validation cohort (Table 3). The ability of these 2 staging systems to predict the risk of recurrence for individual patients as compared to the nomogram is illustrated in Figure 4A and 4B. Notably, the intermediate risk and high risk groupings of the NIH-Fletcher staging system as well as the “probably malignant” grouping of the NIH-Miettinen each encompass a large number of patients that have very heterogeneous outcomes as predicted by the nomogram. The very low risk and low risk groupings of the NIH-Fletcher system as well as the “probably benign” and “uncertain or low malignant potential” groupings of the NIH-Miettinen system do not identify groups of patients with nomogram-predicted outcomes that are distinct from each other.
Figure 4. Nomogram scores compared to commonly used risk groupings.
Nomogram predictions of recurrence-free survival (RFS) for individual patients in all 3 datasets are shown by 10 percentile intervals for each (A) NIH-Fletcher, (B) NIH-Miettinen, and (C) AFIP-Miettinen stage.
Next, the nomogram was compared to the AFIP-Miettinen staging system, a modification of the NIH-Miettinen system that was proposed in 2006. While the nomogram achieved slightly higher concordance probabilities for the MSKCC dataset and the 2 validation cohorts, this did not reach statistical significance (Table 3). As the concordance ability of the nomogram was not significantly superior to that of the AFIP-Miettinen staging system, to further compare these 2 risk stratification methods, the calibration of the AFIP-Miettinen system was tested. Predictions for 2-year and 5-year RFS were assigned for each risk group using the Kaplan-Meier RFS of that group in the MSKCC dataset. These predictions were then compared to observed Kaplan-Meier RFS using the 2 validation cohorts. For comparison, nomogram-predicted RFS was also calculated for each of stage of the AFIP-Miettinen system (Table 5). The predictions of the AFIP-Miettinen staging did not appear as well calibrated as those of the nomogram particularly for the “high malignant potential” risk group. This is likely due to the fact that there is a heterogeneity of outcomes as predicted by the nomogram for the patients in this group (Figure 4C). As the mix of outcomes in the “high malignant potential” stage of the AFIP-Miettinen system varied between the 3 datasets, so did the observed RFS; this group of patients had a 5-year RFS of 8% in the MSKCC series, 43% in the GEIS series, and 24% in the Mayo series.
Table 5.
Calibration of the AFIP-Miettinen staging system compared to that of the nomogram.
| Series | Time Point |
Group | Number of Patients |
AFIP- Miettinen Predicted RFS |
Nomogram Predicted RFS |
Kaplan- Meier Estimated RFS |
|---|---|---|---|---|---|---|
| GEIS | 2 yrs | 1 | 29 | 16% | 44% | 62% |
| 2 | 52 | 100% | 79% | 93% | ||
| 3 | 106 | 92% | 93% | 96% | ||
| 4 | 16 | 100% | 97% | 100% | ||
| 5 yrs | 1 | 29 | 8% | 24% | 43% | |
| 2 | 52 | 87% | 63% | 69% | ||
| 3 | 106 | 80% | 86% | 91% | ||
| 4 | 16 | 100% | 94% | 100% | ||
| Mayo | 2 yrs | 1 | 55 | 16% | 60% | 46% |
| 2 | 28 | 100% | 83% | 84% | ||
| 3 | 54 | 92% | 93% | 100% | ||
| 4 | 9 | 100% | 94% | 100% | ||
| 5 yrs | 1 | 55 | 8% | 45% | 24% | |
| 2 | 28 | 87% | 68% | 70% | ||
| 3 | 54 | 80% | 87% | 100% | ||
| 4 | 9 | 100% | 89% | 100% |
Group 1 = “High malignant potential,” Group 2 = “Intermediate malignant potential,” Group 3 = “Low malignant potential,” Group 4 = “Very low, if any malignant potential”
DISCUSSION
This study describes the development and validation of a prognostic nomogram to predict recurrence-free survival (RFS) following the resection of localized, primary GIST. A nomogram that assigns predictions for 2-year and 5-year RFS based on tumor size, site of origin, and mitotic index was created based on a series of 127 patients from a single institution. The nomogram was shown to have better predictive accuracy as determined by concordance probabilities than 2 commonly used staging systems developed at the United States National Institutes of Health (NIH) GIST Workshop in 2001. The nomogram had a concordance probability that was higher but not statistically different than that of a third staging system (a 2006 modification of one the NIH staging systems). Nomogram predictions appeared better calibrated to actual RFS that those of this third staging system. The nomogram may be useful to select patients for adjuvant imatinib therapy.
The ability to predict the likelihood of postoperative recurrence for any primary cancer that is treated by surgical resection is important for several reasons. Foremost, patients can be counseled appropriately regarding their likely outcome. If effective adjuvant therapy exists, patients can be selected properly for postoperative treatment. Furthermore, physicians can determine the type (e.g., physical examination, blood tests, or radiologic tests) and frequency of postoperative surveillance for tumor recurrence. The aim of the present study was to establish a prognostic tool to predict RFS for individual patients after complete resection of localized, primary GIST in the absence of adjuvant treatment.
Tumor recurrence is a common event for patients with GIST as RFS ranged from 63 ±4.8% to 78 ±3.5% at 5 years in the 3 datasets in this study (Figure 1). Mitotic index and size are the most well validated prognostic variables for determining the likelihood of recurrence after complete surgical resection of GIST. We found on multivariate analysis of the MSKCC patients that mitotic index ≥5 was the dominant predictor of RFS (hazard ratio 14.6, p<0.001).27 In contrast, tumor size ≥10 cm had a hazard ratio of only 2.5. Primary tumor site has also been shown to influence outcome in several large retrospective studies.
The 2 NIH Workshop staging systems were developed empirically based on tumor size and mitotic activity with or without primary tumor site (Table 1).14,15 Neither had been subjected to statistical validation prior to publication. Subsequently a modification of the NIH-Miettinen staging system has been proposed based on observations in a large number of GIST patients (Table 1), but similarly not subjected to statistical validation prior to publication.18 The NIH-Fletcher Staging System has now become the most well studied staging system for GIST.14 The high risk group of that staging system has been reliably associated with an increased risk of recurrence in several reports.20,21,28,29,36,37 It has been noted, however, that the very low risk and low risk groups do not discriminate risk of recurrence.20,21,29 Furthermore, as the high risk group in most studies has a 5-year RFS of approximately 45–50% and often accounts for approximately 50% of the total number of patients,16,20,21,28,29,37 some authors have noted the need for a grouping of very high risk patients.20,21 Our data corroborate these findings (Figure 4A). The very low risk and low risk groups both identify patients with a good prognosis; the majority of patients have a nomogram predicted 5-year RFS of 90–100%. The intermediate and high-risk groups each appear to identify a group of patients with very heterogeneous outcomes, including a large number of patients with nomogram predicted 5-year RFS of 90–100%. Other proposed staging systems, including the one originally proposed by Miettinen15 and its subsequent modifications,18,19 have not been as rigorously evaluated. Likewise, no staging system has been assessed for its ability to assign a quantitative risk of recurrence for individual patients.
In general, prognostic nomograms are better able to predict the likelihood of events for individual patients than staging systems that stratify patients into a few broad groups. Nomograms are based on statistical models that utilize a combination of prognostic variables to determine the likelihood of a certain event. For instance, nomograms for outcome following the resection of gastric24 and pancreatic25 adenocarcinoma are more accurate in predicting disease-specific survival (DSS) than the corresponding American Joint Committee on Cancer (AJCC) staging systems and these findings have been validated on external data sets.38,39
In this study, we created a nomogram based on patients treated at MSKCC to predict the risk of recurrence after complete resection of localized, primary GIST. The nomogram is shown to predict an accurate relative risk of recurrence as validated by 2 independent series of patients. The concordance probability of the nomogram was very acceptable (0.76 and 0.80 on the validation cohorts). For comparison, the MSKCC pancreas adenocarcinoma and gastric adenocarcinoma DSS nomograms have concordance probabilities of 0.80 and 0.64, respectively, on the original datasets,24,25 and 0.77 and 0.62, respectively, on the validation cohorts.38,39
The nomogram was better able to predict the relative risk of recurrence than the 2 commonly used NIH Workshop GIST stratification schemes. The predictive accuracy of the nomogram as determined by the concordance probability was at least as good as that of the AFIP-Miettinen staging system. The present nomogram can assign numeric predictions for the risk of recurrence at 2 years and 5 years. These predictions appear to be accurate in the 3 cohorts presented. In contrast, while predictions for recurrence could be assigned to the AFIP-Miettinen system stages, these predictions, particularly those of the “high malignant potential” group, were not as well calibrated as those of the nomogram. The difference in the observed RFS of the “high malignant potential” risk group between the 3 datasets may be related to the variable mix of heterogeneous patient outcomes that is seen when nomogram predictions are plotted for this risk group (Figure 4C). Based on the datasets in this study, the AFIP-Miettinen staging system defines 2 stages with very good outcomes, one stage with a good outcome, and one stage with a poor outcome. The 5-year RFS by stage was 100% for the “very low, if any malignant potential” group, 80–100% for the “low malignant potential” group, 69–87% for the “intermediate malignant potential” group, and 8–43% for the “high malignant potential” group. Furthermore, the The AFIP-Miettinen staging system is limited in that it can only assign patients to these broad groups. In contrast the nomogram can calculate risk of recurrence for any individual patient. If used for stratification, the nomogram offers flexibility in defining risk groups. For instance, if it is decided to administer adjuvant treatment to those patients with < 75% 5-year RFS or < 50% 5-year RFS, these groups can be defined.
It is possible that the prognostic value of the nomogram could be improved with the incorporation of additional variables. There have been conflicting results about whether KIT and PDGFRA mutation status affects outcome in resected localized, primary GIST.27,28,36,40–52 The discrepancies may be due in part to differences in how mutation status is analyzed (e.g., presence or absence of KIT mutation, exon of mutation, or type of mutation). In the multivariate analysis performed on the cohort used to construct the nomogram, mutation status was not an independent predictor of RFS regardless of how it was analyzed.27 Also, we failed to observe an improvement in the accuracy of the nomogram when mutation status was included. The effect of mutational status on the performance of the nomogram in the validation cohorts was not assessed. Ki-67 staining by immunohistochemistry17,53–57, p16 staining,58 and tumor cellularity28,56 have also been reported to predict independently recurrence in large series of GISTs. One group has proposed a risk stratification system based on Ki-67 staining and tumor size.16 We did not assess the prognostic value of these variables. While the addition of other variables may improve the prognostic ability of the nomogram, the appeal of the current nomogram is that unlike mutational status, Ki-67 staining, and p16 staining, the variables of tumor size, location, and mitotic index are routinely reported by many pathologists and therefore the nomogram should be broadly applicable. Tumor rupture has been described as an adverse prognostic variable.59 Rupture was an infrequent event in the series used to create the nomogram, and thus the association between rupture or spillage and recurrence did not reach statistical significance.
It also should be noted that of the 3 variables used to construct the nomogram, tumor size and mitotic rate might not be measured uniformly across institutions. Tumor size could potentially be influenced by when the specimen is measured in relation to fixation. Mitotic rate is especially subject to variability as it requires the subjective determination of an individual observer about whether an individual cell is undergoing mitosis. Mitotic rate in this study was determined by expert soft tissue pathologists. These variables, nevertheless, appear to be the most important predictors of recurrence in several studies. Despite the potential variability in size and mitotic rate, the nomogram predictions were well calibrated in the 3 datasets in this study.
The standard of care for localized, primary GIST after surgical resection has changed recently based on the results of the ACOSOG Z9001 trial, which showed an increased 1-year RFS in patients assigned to 1 year of imatinib versus placebo.10 The trial was powered on RFS of the entire study population, which was patients with ≥3 cm tumors. Nevertheless, ad hoc analysis of tumor size (which was the only stratification factor) demonstrated significant differences in RFS between the imatinib and placebo arms in each size category (i.e., 3–6 cm, 6–10 cm, and ≥10 cm). Retrospective analysis of mitotic index determined by central pathologic review and tumor location are underway. Once there is additional follow-up and more events, it will be desirable to validate further the nomogram using the patients assigned to the placebo arm in the ACOSOG Z9001 trial. It is likely that patients at low risk of tumor recurrence do not need adjuvant imatinib. On the other hand, patients at high risk of relapse may require longer periods than 1 year of postoperative therapy.
In summary, we report a nomogram to predict RFS for patients with completely resected localized, primary GIST who have not received adjuvant treatment. This tool can provide an accurate prediction of the risk of recurrence for individual patients as validated in 2 external datasets. The nomogram may prove useful in patient care, interpretation of clinical trial results, and selection of patients for adjuvant therapy.
ACKNOWLEDGMENTS
We would like to acknowledge the contributions of Drs. Imran Hassan and Y. Nancy You to the Mayo Clinic patient series.
This work was supported by Public Health Service Grant CA102613 (RPD) and P01 CA47179 (SS) from the National Cancer Institute and a Clinical Investigator Award from The Society of Surgical Oncology (RPD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Presented in part at the American Society of Clinical Oncology 2007 Gastrointestinal Cancers Symposium in Orlando, Florida
AUTHORS’ DISCLOSRES OF POTENTIAL CONFLICTS OF INTEREST.
Drs. Gönen, Martín Broto, García-del-Muro, Maki and DeMatteo report receiving honoraria and consulting fees from Novartis Pharmaceuticals.
AUTHOR CONTRIBUTIONS
Conception and design: Jason S. Gold, Mithat Gönen, Ronald P. DeMatteo
Financial support: Antonio Gutiérrez, Javier Martín Broto, Xavier García-del-Muro, Murray F. Brennan, John H. Donohue, Ronald P. DeMatteo
Administrative support: Antonio Gutiérrez, Javier Martín Broto, Xavier García-del-Muro, Murray F. Brennan, John H. Donohue, Ronald P. DeMatteo
Provision of study materials or patients: Antonio Gutiérrez, Javier Martín Broto, Xavier García-del-Muro, Thomas C. Smyrk, Robert G. Maki, Samuel Singer, Murray F. Brennan, Cristina R. Antonescu, John H. Donohue, Ronald P. DeMatteo
Collection and assembly of data: Jason S. Gold, Mithat Gönen, Antonio Gutiérrez, Javier Martín Broto, Xavier García-del-Muro, Thomas C. Smyrk, Cristina R. Antonescu, John H. Donohue, Ronald P. DeMatteo
Data analysis and interpretation: Jason S. Gold, Mithat Gönen, Antonio Gutiérrez, Javier Martín Broto, Xavier García-del-Muro, Thomas C. Smyrk, Robert G. Maki, Samuel Singer, Murray F. Brennan, Cristina R. Antonescu, John H. Donohue, Ronald P. DeMatteo
Manuscript writing: Jason S. Gold, Mithat Gönen, Antonio Gutiérrez, Javier Martín Broto, Xavier García-del-Muro, Thomas C. Smyrk, Robert G. Maki, Samuel Singer, Murray F. Brennan, Cristina R. Antonescu, John H. Donohue, Ronald P. DeMatteo
Final approval of manuscript: Jason S. Gold, Mithat Gönen, Antonio Gutiérrez, Javier Martín Broto, Xavier García-del-Muro, Thomas C. Smyrk, Robert G. Maki, Samuel Singer, Murray F. Brennan, Cristina R. Antonescu, John H. Donohue, Ronald P. DeMatteo
REFERENCES
- 1.Miettinen M, Monihan JM, Sarlomo-Rikala M, et al. Gastrointestinal stromal tumors/smooth muscle tumors (GISTs) primary in the omentum and mesentery: clinicopathologic and immunohistochemical study of 26 cases. Am J Surg Pathol. 1999;23:1109–1118. doi: 10.1097/00000478-199909000-00015. [DOI] [PubMed] [Google Scholar]
- 2.Reith JD, Goldblum JR, Lyles RH, et al. Extragastrointestinal (soft tissue) stromal tumors: an analysis of 48 cases with emphasis on histologic predictors of outcome. Mod Pathol. 2000;13:577–585. doi: 10.1038/modpathol.3880099. [DOI] [PubMed] [Google Scholar]
- 3.Hirota S, Isozaki K, Moriyama Y, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 4.Heinrich MC, Corless CL, Duensing A, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- 5.Heinrich MC, Owzar K, Corless CL, et al. Correlation of kinase genotype and clinical outcome in the North American Intergroup Phase III Trial of imatinib mesylate for treatment of advanced gastrointestinal stromal tumor: CALGB 150105 Study by Cancer and Leukemia Group B and Southwest Oncology Group. J Clin Oncol. 2008;26:5360–5367. doi: 10.1200/JCO.2008.17.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanke CD, Demetri GD, von Mehren M, et al. Long-term results from a randomized phase II trial of standard- versus higher-dose imatinib mesylate for patients with unresectable or metastatic gastrointestinal stromal tumors expressing KIT. J Clin Oncol. 2008;26:620–625. doi: 10.1200/JCO.2007.13.4403. [DOI] [PubMed] [Google Scholar]
- 7.Verweij J, Casali PG, Zalcberg J, et al. Progression-free survival in gastrointestinal stromal tumours with high-dose imatinib: randomised trial. Lancet. 2004;364:1127–1134. doi: 10.1016/S0140-6736(04)17098-0. [DOI] [PubMed] [Google Scholar]
- 8.Demetri GD, van Oosterom AT, Garrett CR, et al. Efficacy and safety of sunitinib in patients with advanced gastrointestinal stromal tumour after failure of imatinib: a randomised controlled trial. Lancet. 2006;368:1329–1338. doi: 10.1016/S0140-6736(06)69446-4. [DOI] [PubMed] [Google Scholar]
- 9.DeMatteo RP, Lewis JJ, Leung D, et al. Two hundred gastrointestinal stromal tumors: recurrence patterns and prognostic factors for survival. Ann Surg. 2000;231:251–258. doi: 10.1097/00000658-200001000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dematteo RP, Ballman KV, Antonescu CR, et al. Adjuvant imatinib mesylate after resection of localised, primary gastrointestinal stromal tumour: a randomised, double-blind, placebo-controlled trial. Lancet. 2009 doi: 10.1016/S0140-6736(09)60500-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. http://www.fda.gov/cder/Offices/OODP/whatsnew/imatinib_mesylate.htm.
- 12. http://www.emea.europa.eu/pdfs/human/opinion/Glivec_65949508en.pdf.
- 13.Franquemont DW. Differentiation and risk assessment of gastrointestinal stromal tumors. Am J Clin Pathol. 1995;103:41–47. doi: 10.1093/ajcp/103.1.41. [DOI] [PubMed] [Google Scholar]
- 14.Fletcher CD, Berman JJ, Corless C, et al. Diagnosis of gastrointestinal stromal tumors: A consensus approach. Hum Pathol. 2002;33:459–465. doi: 10.1053/hupa.2002.123545. [DOI] [PubMed] [Google Scholar]
- 15.Miettinen M, El-Rifai W, L HLS, et al. Evaluation of malignancy and prognosis of gastrointestinal stromal tumors: a review. Hum Pathol. 2002;33:478–483. doi: 10.1053/hupa.2002.124123. [DOI] [PubMed] [Google Scholar]
- 16.Nilsson B, Bümming P, Meis-Kindblom JM, et al. Gastrointestinal stromal tumors: the incidence, prevalence, clinical course, and prognostication in the preimatinib mesylate era--a population-based study in western Sweden. Cancer. 2005;103:821–829. doi: 10.1002/cncr.20862. [DOI] [PubMed] [Google Scholar]
- 17.Bucher P, Egger JF, Gervaz P, et al. An audit of surgical management of gastrointestinal stromal tumours (GIST) Eur J Surg Oncol. 2006;32:310–314. doi: 10.1016/j.ejso.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 18.Miettinen M, Lasota J. Gastrointestinal stromal tumors: review on morphology, molecular pathology, prognosis, and differential diagnosis. Arch Pathol Lab Med. 2006;130:1466–1478. doi: 10.5858/2006-130-1466-GSTROM. [DOI] [PubMed] [Google Scholar]
- 19.Miettinen M, Lasota J. Gastrointestinal stromal tumors: pathology and prognosis at different sites. Semin Diagn Pathol. 2006;23:70–83. doi: 10.1053/j.semdp.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Huang HY, Li CF, Huang WW, et al. A modification of NIH consensus criteria to better distinguish the highly lethal subset of primary localized gastrointestinal stromal tumors: a subdivision of the original high-risk group on the basis of outcome. Surgery. 2007;141:748–756. doi: 10.1016/j.surg.2007.01.024. [DOI] [PubMed] [Google Scholar]
- 21.Goh BK, Chow PK, Yap WM, et al. Which is the optimal risk stratification system for surgically treated localized primary GIST? Comparison of three contemporary prognostic criteria in 171 tumors and a proposal for a modified Armed Forces Institute of Pathology risk criteria. Ann Surg Oncol. 2008;15:2153–2163. doi: 10.1245/s10434-008-9969-z. [DOI] [PubMed] [Google Scholar]
- 22.Kattan MW, Wheeler TM, Scardino PT. Postoperative nomogram for disease recurrence after radical prostatectomy for prostate cancer. J Clin Oncol. 1999;17:1499–1507. doi: 10.1200/JCO.1999.17.5.1499. [DOI] [PubMed] [Google Scholar]
- 23.Kattan MW, Leung DH, Brennan MF. Postoperative nomogram for 12-year sarcoma-specific death. J Clin Oncol. 2002;20:791–796. doi: 10.1200/JCO.2002.20.3.791. [DOI] [PubMed] [Google Scholar]
- 24.Kattan MW, Karpeh MS, Mazumdar M, et al. Postoperative nomogram for disease-specific survival after an R0 resection for gastric carcinoma. J Clin Oncol. 2003;21:3647–3650. doi: 10.1200/JCO.2003.01.240. [DOI] [PubMed] [Google Scholar]
- 25.Brennan MF, Kattan MW, Klimstra D, et al. Prognostic nomogram for patients undergoing resection for adenocarcinoma of the pancreas. Ann Surg. 2004;240:293–298. doi: 10.1097/01.sla.0000133125.85489.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kattan MW, Giri D, Panageas KS, et al. A tool for predicting breast carcinoma mortality in women who do not receive adjuvant therapy. Cancer. 2004;101:2509–2515. doi: 10.1002/cncr.20635. [DOI] [PubMed] [Google Scholar]
- 27.Dematteo RP, Gold JS, Saran L, et al. Tumor mitotic rate, size, and location independently predict recurrence after resection of primary gastrointestinal stromal tumor (GIST) Cancer. 2008;112:608–615. doi: 10.1002/cncr.23199. [DOI] [PubMed] [Google Scholar]
- 28.Martín J, Poveda A, Llombart-Bosch A, et al. Deletions affecting codons 557–558 of the c-KIT gene indicate a poor prognosis in patients with completely resected gastrointestinal stromal tumors: a study by the Spanish Group for Sarcoma Research (GEIS) J Clin Oncol. 2005;23:6190–6198. doi: 10.1200/JCO.2005.19.554. [DOI] [PubMed] [Google Scholar]
- 29.Hassan I, You YN, Shyyan R, et al. Surgically managed gastrointestinal stromal tumors: a comparative and prognostic analysis. Ann Surg Oncol. 2008;15:52–59. doi: 10.1245/s10434-007-9633-z. [DOI] [PubMed] [Google Scholar]
- 30.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–462. [Google Scholar]
- 31.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 32.Kattan MW, Eastham JA, Stapleton AM, et al. A preoperative nomogram for disease recurrence following radical prostatectomy for prostate cancer. J Natl Cancer Inst. 1998;90:766–771. doi: 10.1093/jnci/90.10.766. [DOI] [PubMed] [Google Scholar]
- 33.Gönen M, Heller G. Concordance probability and discriminatory power in proportional hazards regression. Biometrika. 2005;92:965–970. [Google Scholar]
- 34.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 35.Efron B, Tibshirani T. An Introduction to the Bootstrap. London: Chapman & Hall; 1993. [Google Scholar]
- 36.Rutkowski P, Nowecki ZI, Michej W, et al. Risk criteria and prognostic factors for predicting recurrences after resection of primary gastrointestinal stromal tumor. Ann Surg Oncol. 2007;14:2018–2027. doi: 10.1245/s10434-007-9377-9. [DOI] [PubMed] [Google Scholar]
- 37.Mucciarini C, Rossi G, Bertolini F, et al. Incidence and clinicopathologic features of gastrointestinal stromal tumors. A population-based study. BMC Cancer. 2007;7:230. doi: 10.1186/1471-2407-7-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peeters KC, Kattan MW, Hartgrink HH, et al. Validation of a nomogram for predicting disease-specific survival after an R0 resection for gastric carcinoma. Cancer. 2005;103:702–707. doi: 10.1002/cncr.20783. [DOI] [PubMed] [Google Scholar]
- 39.Ferrone CR, Kattan MW, Tomlinson JS, et al. Validation of a postresection pancreatic adenocarcinoma nomogram for disease-specific survival. J Clin Oncol. 2005;23:7529–7535. doi: 10.1200/JCO.2005.01.8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ernst SI, Hubbs AE, Przygodzki RM, et al. KIT mutation portends poor prognosis in gastrointestinal stromal/smooth muscle tumors. Lab Invest. 1998;78:1633–1636. [PubMed] [Google Scholar]
- 41.Taniguchi M, Nishida T, Hirota S, et al. Effect of c-kit mutation on prognosis of gastrointestinal stromal tumors. Cancer Res. 1999;59:4297–4300. [PubMed] [Google Scholar]
- 42.Singer S, Rubin BP, Lux ML, et al. Prognostic value of KIT mutation type, mitotic activity, and histologic subtype in gastrointestinal stromal tumors. J Clin Oncol. 2002;20:3898–3905. doi: 10.1200/JCO.2002.03.095. [DOI] [PubMed] [Google Scholar]
- 43.Kim TW, Lee H, Kang YK, et al. Prognostic significance of c-kit mutation in localized gastrointestinal stromal tumors. Clin Cancer Res. 2004;10:3076–3081. doi: 10.1158/1078-0432.ccr-03-0581. [DOI] [PubMed] [Google Scholar]
- 44.Koay MH, Goh YW, Iacopetta B, et al. Gastrointestinal stromal tumours (GISTs): a clinicopathological and molecular study of 66 cases. Pathology. 2005;37:22–31. doi: 10.1080/00313020400023628. [DOI] [PubMed] [Google Scholar]
- 45.Liu XH, Bai CG, Xie Q, et al. Prognostic value of KIT mutation in gastrointestinal stromal tumors. World J Gastroenterol. 2005;11:3948–3952. doi: 10.3748/wjg.v11.i25.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iesalnieks I, Rummele P, Dietmaier W, et al. Factors associated with disease progression in patients with gastrointestinal stromal tumors in the pre-imatinib era. Am J Clin Pathol. 2005;124:740–748. doi: 10.1309/AKK3-VFF6-10CW-M566. [DOI] [PubMed] [Google Scholar]
- 47.Cho S, Kitadai Y, Yoshida S, et al. Deletion of the KIT gene is associated with liver metastasis and poor prognosis in patients with gastrointestinal stromal tumor in the stomach. Int J Oncol. 2006;28:1361–1367. [PubMed] [Google Scholar]
- 48.Andersson J, Bümming P, Meis-Kindblom JM, et al. Gastrointestinal stromal tumors with KIT exon 11 deletions are associated with poor prognosis. Gastroenterology. 2006;130:1573–1581. doi: 10.1053/j.gastro.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 49.Tzen CY, Wang MN, Mau BL. Spectrum and prognostication of KIT and PDGFRA mutation in gastrointestinal stromal tumors. Eur J Surg Oncol. 2008;34:563–568. doi: 10.1016/j.ejso.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 50.Kontogianni-Katsarou K, Dimitriadis E, Lariou C, et al. KIT exon 11 codon 557/558 deletion/insertion mutations define a subset of gastrointestinal stromal tumors with malignant potential. World J Gastroenterol. 2008;14:1891–1897. doi: 10.3748/wjg.14.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keun Park C, Lee EJ, Kim M, et al. Prognostic stratification of high-risk gastrointestinal stromal tumors in the era of targeted therapy. Ann Surg. 2008;247:1011–1018. doi: 10.1097/SLA.0b013e3181724f9d. [DOI] [PubMed] [Google Scholar]
- 52.Yamaguchi U, Nakayama R, Honda K, et al. Distinct gene expression-defined classes of gastrointestinal stromal tumor. J Clin Oncol. 2008;26:4100–4108. doi: 10.1200/JCO.2007.14.2331. [DOI] [PubMed] [Google Scholar]
- 53.Wong NA, Young R, Malcomson RD, et al. Prognostic indicators for gastrointestinal stromal tumours: a clinicopathological and immunohistochemical study of 108 resected cases of the stomach. Histopathology. 2003;43:118–126. doi: 10.1046/j.1365-2559.2003.01665.x. [DOI] [PubMed] [Google Scholar]
- 54.Nakamura N, Yamamoto H, Yao T, et al. Prognostic significance of expressions of cell-cycle regulatory proteins in gastrointestinal stromal tumor and the relevance of the risk grade. Hum Pathol. 2005;36:828–837. doi: 10.1016/j.humpath.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 55.Bümming P, Ahlman H, Andersson J, et al. Population-based study of the diagnosis and treatment of gastrointestinal stromal tumours. Br J Surg. 2006;93:836–843. doi: 10.1002/bjs.5350. [DOI] [PubMed] [Google Scholar]
- 56.Wu TJ, Lee LY, Yeh CN, et al. Surgical treatment and prognostic analysis for gastrointestinal stromal tumors (GISTs) of the small intestine: before the era of imatinib mesylate. BMC Gastroenterol. 2006;6:29. doi: 10.1186/1471-230X-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang HY, Huang WW, Lin CN, et al. Immunohistochemical expression of p16INK4A, Ki-67, and Mcm2 proteins in gastrointestinal stromal tumors: prognostic implications and correlations with risk stratification of NIH consensus criteria. Ann Surg Oncol. 2006;13:1633–1644. doi: 10.1245/s10434-006-9188-4. [DOI] [PubMed] [Google Scholar]
- 58.Steigen SE, Bjerkehagen B, Haugland HK, et al. Diagnostic and prognostic markers for gastrointestinal stromal tumors in Norway. Mod Pathol. 2008;21:46–53. doi: 10.1038/modpathol.3800976. [DOI] [PubMed] [Google Scholar]
- 59.Ng EH, Pollock RE, Munsell MF, et al. Prognostic factors influencing survival in gastrointestinal leiomyosarcomas. Implications for surgical management and staging. Ann Surg. 1992;215:68–77. doi: 10.1097/00000658-199201000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]