Abstract
Circadian genes have the potential to influence a variety of cancer-related biological pathways, including immune regulation, which may influence susceptibility to non-Hodgkin’s lymphoma (NHL). However, few studies have examined the role of circadian genes in lymphomagenesis. The current study examined Cryptochrome 2 (CRY2), a core circadian gene and transcriptional repressor, as a potential circadian biomarker for NHL. We first performed genetic association analyses of tagging SNPs in CRY2 and NHL risk using DNA samples from a population-based case-control study (N= 455 cases and 527 controls). Three SNPs were found to be significantly associated with risk of NHL when combining all subtypes (dbSNP IDs, odds ratios (ORs), and 95% confidence intervals: rs11038689, OR=2.34 (1.28-4.27), P=0.006; rs7123390, OR=2.40 (1.39-4.13), P=0.002; and rs1401417, OR=2.97 (1.57-5.63), P=0.001). Each of these associations remained significant when restricting the analysis to B-Cell cases and when further restricting to follicular lymphomas. An analysis of CRY2 diplotypes confirmed these significant findings. To further determine the functional impact of CRY2, we silenced the gene in vitro and performed a whole genome expression microarray. A pathway-based analysis showed that genes significantly altered by CRY2 knockdown formed networks associated with immune response and hematological system development. In addition, these genes were predicted to have significant impacts on several disease processes, including cancer (B-H P-value=3.75E-9) and hematological disease (B-H P=8.01E-8). In conclusion, both genetic association and functional analyses suggest that the circadian gene CRY2 may play an important role in NHL development.
Keywords: CRY2, NHL, Circadian Genetics
Introduction
The human circadian rhythm is a fundamental aspect of human physiology, and a wide range of biological processes are influenced by the circadian clock, including body temperature, energy metabolism, hormone secretion, and sleep-wake cycles (1). Several observational studies indicate that individuals who do not maintain a normal sleep/wake cycle may be at increased risk for several cancer types, and after considering evidence from epidemiologic and experimental studies, the International Agency for Research on Cancer (IARC) recently concluded that shift work that involves circadian disruption is “probably carcinogenic to humans” (2). Although much of the current epidemiologic evidence has focused on breast cancer, it has recently been hypothesized that circadian disruption may also affect risk for NHL, possibly through its influence on immune regulation (3).
Limited indirect evidence suggests that genetic components of the circadian system may have a role in processes relevant for NHL tumorigenesis. For example, reduced expression of the circadian gene PER2 has been detected in lymphoma cell lines and in samples drawn from patients with acute myeloid leukemia (AML) (4), and a recent genetic association study demonstrated that a non-synonymous polymorphism in the core circadian gene NPAS2 is associated with decreased risk of NHL, especially B-cell lymphoma (5). In addition, several studies have established an important role for circadian rhythm in the maintenance of proper immune function. First, it has been shown that several key components of the immune system are under circadian regulation, with circadian rhythmicity present in nearly all aspects of immune response (6-11). Specifically, circadian rhythms have been observed in natural killer (NK) cells, which are an essential component of the innate immune system against infections and cancerous growth (12). Secondly, disruption of circadian rhythms can cause aberrant immune cell trafficking and abnormal cell proliferation cycles (13). Moreover, disruption of the circadian rhythms in NK cells and phagocytic activity has been observed in malignant melanoma cells, leading to a discoordination between the two immune system components that is not observed in healthy humans (14).
Overall, these preliminary studies suggest that circadian disruption has the potential to significantly impact a number of mechanisms that may influence NHL susceptibility, most notably through its role in influencing immune response. However, while immune dysfunction remains the only well-established risk factor for NHL (15, 16), immunodeficiency is seen only in a subset of NHL patients. As such, if an association between circadian disruption and lymphomagenesis can be firmly established, there remains the additional question of whether the relationship is maintained outside of pathways related to immune system function. Further study into these associations is therefore warranted and are apropos to investigations into the potential for circadian gene variants to serve as a novel panel of NHL risk biomarkers.
The current study investigates the role of the core circadian gene CRY2 in NHL tumorigenesis. CRY2 is essential to the maintenance of circadian rhythm through its role in the negative arm of the circadian feedback loop, and may have a broader regulatory role as a transcriptional repressor (17, 18). CRY2 has also been shown to be involved in cell cycle regulation, including roles in DNA damage checkpoint control (19) and regulation of genes important for cell cycle progression (20). Here, we report findings from an epidemiological analysis of the association between genetic variants in CRY2 and risk of NHL. In addition, we performed a whole genome expression microarray to determine the effect of CRY2 silencing on the expression of cancer-related genes, and to determine whether CRY2 influences biological pathways which may be relevant for lymphomagenesis.
Patients, materials, and methods
Case-control study of NHL
The study population has previously been described (21). Briefly, all participants were female residents of Connecticut, and cases were incident, histologically-confirmed NHL (ICD-O, M-9590-9642, 9690-9701, 9740-9750) identified through Yale Cancer Center’s Rapid Case Ascertainment (RCA) between 1996 and 2000. Population-based controls younger than age 65 were recruited by random digit dialing (RDD), and controls older than 65 were identified through Health Care Financing Administration files. Five year age strata were constructed, and controls were frequency matched to cases by intermittently adjusting the number of controls selected from each stratum. Participation rates were: 72% for cases, 69% for RDD controls, and 47% for controls identified by health care financing records.
Data collection
The study was approved by Institutional Review Boards at Yale University, the Connecticut Department of Public Health, and the National Cancer Institute. Participation was voluntary, and written informed consent was obtained. Those who agreed were interviewed by trained study nurses either at the subject’s home or at a convenient location, and following the administration of a questionnaire, subjects provided a 10 ml peripheral blood sample. Genomic DNA was isolated from peripheral blood lymphocytes for each study subject.
SNP selection and genotyping
SNPs were identified using the Tagger algorithm (22), which is implemented in the Haploview interface (23) of HapMap’s genome browser, Release 22 (http://www.hapmap.org/cgi-perl/gbrowse/hapmap22_B36/ accessed on January 15, 2008). Five SNPs (rs10838524, rs11038689, rs11605924, rs2292912, and rs7123390) were identified as representative of all variations found within the exonic and intronic regions of the CRY2 gene using the CEU population returning SNPs with MAF ≥ 0.2 and r2 ≥ 0.8. In addition, one intronic SNP (rs1401417) which had been identified as significantly associated with prostate cancer risk in a previous study (24) was also included in the genotyping pool. Genotyping for all SNPs was performed at Yale University’s W.M. Keck Foundation Biotechnology Research Laboratory using the Sequenom MassARRAY multiplex genotyping platform (Sequenom, Inc., San Diego, CA) according to the manufacturer’s protocol. Duplicate samples from 100 study subjects and 40 replicate samples from each of two blood donors were interspersed throughout each batch for all genotyping assays. Genotyping failed for one SNP (rs10838524). The concordance rates for QC samples were over 95% for the remaining assays. Genotyping call rates were: 97.0% for rs11038689, 97.5% for rs11605924, 97.7% for rs2292912, 96.6% for rs7123390, and 97.8% for rs1401417. All genotyping scores, including quality control data, were re-checked by different laboratory personnel and the accuracy of each assay was confirmed.
Cell culture and treatments
Human breast adenocarcinoma cells (MCF-7) were used to determine the impact of CRY2 knockdown on pathways related to lymphomagenesis. MCF-7 cells were chosen rather than cells derived from lymphoma tissue, as lymphoma cells would likely begin with aberrant immune signaling, thus causing difficulty in interpreting the effects of CRY2 knockdown on immune regulation. Cells were obtained from American Type Culture Collection (Manassas, VA), and were maintained in Dulbecco’s modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Invitrogen), 0.01 mg/ml bovine insulin, and 1% penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO). siRNA oligos targeting CRY2 (Sense: 5’-UGCUUCAUUCGUUCAAUGUUAAGCCGG-3’ Antisense: 5’-GGCUUAACAUUGAACGAAUGAAGCA-3’) and a scrambled sequence negative control siRNA (Sense: 5’-CUUCCUCUCUUUCUCUCCCUUGUGA-3’, Antisense: 5’-UCACAAGGGAGAGAAAGAGGGAAGGA-3’) were designed and manufactured by Integrated DNA Technologies (IDT, Coralville, IA). Each oligo was diluted in OPTI-MEM serum-free medium (Invitrogen), complexed with Lipofectamine RNAiMax transfection reagent (Invitrogen), and reverse transfected with approximately 100,000 cells in 12 well plates at a final concentration of 10nM in growth medium without penicillin/streptomycin. Cells were harvested 48 hours after transfection for subsequent analysis.
RNA isolation and quantitation
CRY2 silencing efficiency was determined by qPCR of RNA samples isolated using the RNA Mini Kit (Qiagen, Valencia, CA), with on-column DNA digestion, according to the manufacturer’s instructions for mammalian cells. First-strand cDNA was synthesized from purified RNA using the AffinityScript cDNA kit (Stratagene, La Jolla, CA) with oligo-dT primers. Quantitative real-time PCR conditions were prepared using a SYBR Green PCR master mix (Applied BioSystems, Foster City, CA) with gene-specific primers, and a standard thermal cycling procedure on an ABI 7500 instrument (Applied BioSystems). The primers used for CRY2 amplification were: (L: ACCGGGGACTCTGTCTACTG, R: GCCTGCACTGCTCATGCT). CRY2 knockdown was assessed using the 2-ΔΔCt method with RNA content normalized to the housekeeping gene HPRT1.
Gene expression microarray and pathway analysis
Gene expression differences in CRY2 knockdown and normal cells were interrogated by whole genome microarray (Agilent, Inc 41k chip, performed by MoGene, LC, St Louis, MO). RNA was isolated in duplicate (biological replicates) from cells treated with CRY2-targeting or scrambled negative siRNA. Transcripts were identified as “differentially expressed” if they fit the criteria of Benjamini-Hochberg adjusted P-value < 0.01 in both biological replicates and mean fold change > |2|. Transcripts with differential expression were investigated for network and functional inter-relatedness using the Ingenuity Pathway Analysis software tool (Ingenuity Systems, www.ingenuity.com). This software scans the set of input genes to identify networks using information in the Ingenuity Pathways Knowledge Base, an extensive, manually curated database of functional interactions extracted from peer-reviewed publications (25). A Fisher’s exact test, based on the hypergeomtric distribution, is then performed to determine the likelihood of obtaining at least the same number of molecules by chance (i.e. from a random input set), as actually overlap between the input gene set and the genes present in each identified network. All microarray data has been uploaded to the Gene Expression Omnibus (GEO) database. Raw and processed data related to this experiment can be accessed via their website (http://www.ncbi.nlm.nih.gov/projects/geo/) by referencing accession #GSE14617.
Statistical analysis
All statistical analyses were performed using the SAS statistical software (SAS Institute, Cary, NC), unless otherwise noted. For the case-control analyses, allelic distributions for all SNPs were tested by goodness-of-fit chi-square for compliance with Hardy-Weinberg equilibrium (HWE). A chi-square test was also used to determine whether any of the variants were associated with case-control status, using either the full table (homozygous wild-type, heterozygous, and homozygous variant), a dominant model, or a recessive model. Odds ratios and 95% confidence intervals were determined for each SNP-disease association by unconditional multivariate logistic regression, including the following covariates: age, race, and family history of cancer in a first-degree relative. Haplotype estimates were calculated by the PHASE program, which reconstructs haplotypes from population genotyping data (26). Diplotypes were constructed based on the pair of haplotypes estimated for each individual, and odds ratios and 95% confidence intervals for each diplotype were determined by unconditional multivariate logistic regression, using the same covariates as the main effects model, but with all other diplotypes as the referent category.
Due to the multiple comparisons inherent in the microarray analysis, adjustments were made to control for false discoveries. A multiple comparisons correction was applied to each observation using the Benjamini-Hochberg method, as previously described (27), to obtain an adjusted p-value (B-H P-value). Alpha was set at 0.01, and in order to be considered statistically significant.
Results
Association between CRY2 variants and NHL risk
Compared to controls, NHL cases reported a higher proportion of NHL and other cancers among first-degree relatives. There were no significant differences in age and race between cases and controls (Table 1). Genotypic frequencies were determined at each locus, and no allelic distributions significantly departed from the values expected under HWE among the controls (p<0.01). Three of the five SNPs: rs11038689, rs7123390, and rs1401417 were found to be significantly associated with case-control status in the overall three-by-two table of allele distribution vs. case-control status (Supp. Table 1), with p-values of 0.005, 0.003, and 0.004, respectively for the two degrees of freedom chi-square test. Each of the three SNPs significantly associated with disease status in the full table was also significant when a recessive model was assumed, but not when assuming a dominant model. As such, odds ratios and confidence intervals were determined by unconditional multivariate logistic regression under the assumption of a recessive model. Variant alleles in the same three SNPs (rs11038689, rs7123390, and rs1401417) which were associated with case-control status in the unadjusted chi-square analysis were also significantly associated with increased risk of NHL in the adjusted logistic regression model, with ORs (95%CIs) of 2.34 (1.28-4.27, p=0.006), 2.40 (1.39-4.13, p=0.002), and 2.97 (1.57-5.63, p=0.001), respectively (Table 2).
Table 1.
Distribution of selected characteristics by case-control status
| Variable | Cases (N=455) | Controls (N=527) | P-Value |
|---|---|---|---|
| N (%) | N (%) | ||
| Mean age (years) | 61.88 | 62.34 | 0.607 |
| Race | |||
| Caucasian | 438 (96.26) | 496 (94.12) | |
| African-American | 13 (2.86) | 14 (2.66) | |
| Other | 4 (0.88) | 17 (3.22) | 0.108 |
| Family history of cancer in first degree relatives | |||
| No | 96 (21.10) | 130 (24.67) | |
| NHL | 9 (1.98) | 2 (0.38) | |
| Other cancer | 350 (76.92) | 395 (74.95) | 0.030 |
| Case pathology | |||
| All B cell | 365 (80.22) | ||
| Diffuse large B-cell | 135 (36.99) | ||
| Follicular | 105 (28.77) | ||
| SLL/CLL | 54 (14.79) | ||
| Marginal Zone | 30 (8.22) | ||
| Other | 41 (11.23) | ||
| All T cell | 33 (7.25) | ||
| NOS | 58 (12.75) | ||
Table 2.
Association of CRY2 variants with NHL risk
| Genotype | All | B-Cell | T-Cell | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Cases N | Controls N | OR† (95% CI) | Cases N | Controls N | OR† (95% CI) | Cases N | Controls N | OR† (95% CI) | |
| rs11038689 | |||||||||
| A/A or A/G | 408 | 495 | - | 326 | 495 | - | 29 | 495 | - |
| G/G | 33 | 17 | 2.34 (1.28-4.27) | 26 | 17 | 2.35 (1.25-4.41) | 3 | 17 | 2.69 (0.74-9.84) |
| P-value | 0.006 | 0.008 | 0.135 | ||||||
| rs11605924 | |||||||||
| A/A or A/C | 327 | 365 | - | 263 | 365 | - | 25 | 365 | - |
| C/C | 114 | 151 | 0.82 (0.62-1.10) | 89 | 151 | 0.79 (0.58-1.08) | 8 | 151 | 0.78 (0.34-1.77) |
| P-value | 0.182 | 0.142 | 0.550 | ||||||
| rs2292912 | |||||||||
| C/C or C/G | 418 | 489 | - | 334 | 489 | - | 32 | 489 | - |
| G/G | 23 | 29 | 1.00 (0.55-1.82) | 18 | 29 | 1.03 (0.54-1.96) | 1 | 29 | 0.48 (0.06-4.07) |
| P-value | 0.990 | 0.938 | 0.502 | ||||||
| rs7123390 | |||||||||
| G/G or G/A | 396 | 491 | - | 315 | 491 | - | 29 | 491 | - |
| A/A | 41 | 21 | 2.40 (1.39-4.13) | 34 | 21 | 2.54 (1.44-4.46) | 3 | 21 | 2.11 (0.59-7.54) |
| P-value | 0.002 | 0.001 | 0.253 | ||||||
| rs1401417 | |||||||||
| G/G or G/C | 409 | 503 | - | 331 | 503 | - | 28 | 503 | - |
| C/C | 34 | 14 | 2.97 (1.57-5.63) | 26 | 14 | 2.87 (1.47-5.58) | 4 | 14 | 4.64 (1.41-15.34) |
| P-value | 0.001 | 0.002 | 0.012 | ||||||
Adjusted for age (continuous), race, and family history of cancer in 1st or 2nd degree relatives.
Since NHL is comprised of various subtypes with the potential for distinct etiologies, we also performed a stratified analysis including B-cell and T-cell lymphomas only. These results were qualitatively and quantitatively similar to those obtained in the overall analysis with ORs (95% CIs) for homozygous variants of rs11038689, rs7123390, and rs1401417 among B-cell lymphomas only of: 2.35 (1.25-4.41, p=0.008), 2.54 (1.44-4.46, p=0.001), and 2.87 (1.47-5.58, p=0.002), respectively. Although similar point estimates for the odds ratios were observed among T-cell lymphomas only, only rs1401417 reached statistical significance (OR=4.64 (1.41-15.34, p=0.012)) due to the small sample size. Further stratification was performed to analyze four common subtypes of B-cell lymphoma (Table 3). Each of the SNPs significantly associated with lymphoma in the full population (rs11038689, rs7123390, and rs1401417), was also significantly associated with follicular lymphoma (FL), with ORs (95% CIs) of: 3.17 (1.39-7.19, p=0.006), 3.67 (1.79-7.56, p<0.001), and 3.06 (1.24-7.54, p=0.015), respectively. Two SNPs, rs7123390 and rs1401417 were significantly associated with B-cell chronic lymphocytic leukemia/prolymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL), with ORs (95% CIs) of: 3.38 (1.35-8.45, p=0.009) and 4.23 (1.53-11.67, p=0.005), respectively. Only rs1401417 was significantly associated with diffuse large B-cell lymphoma (DLBCL; OR=2.67 (1.13-6.33, p=0.026)). Very few patients were classified as having marginal zone B-cell lymphoma (MZBL), and no CRY2 SNPs were significantly associated with this NHL subtype.
Table 3.
Association of CRY2 variants with NHL risk by B-Cell subtype
| Genotype | DLBCL | FL | CLL/SLL | MZBL | |||||
|---|---|---|---|---|---|---|---|---|---|
| Controls N | Cases N | OR† (95% CI) | Cases N | OR† (95% CI) | Cases N | OR† (95% CI) | Cases N | OR† (95% CI) | |
| rs11038689 | |||||||||
| A/A or A/G | 495 | 126 | - | 90 | - | 48 | - | 27 | - |
| G/G | 17 | 6 | 1.41 (0.54-3.65) | 10 | 3.17 (1.39-7.19) | 5 | 2.75 (0.96-7.86) | 2 | 2.30 (0.50-10.61) |
| P-value | 0.484 | 0.006 | 0.060 | 0.285 | |||||
| rs11605924 | |||||||||
| A/A or A/C | 365 | 93 | - | 79 | - | 31 | - | 19 | - |
| C/C | 151 | 39 | 0.98 (0.64-1.50) | 21 | 0.61 (0.36-1.04) | 12 | 0.72 (0.36-1.40) | 10 | 1.21 (0.54-2.70) |
| P-value | 0.931 | 0.068 | 0.329 | 0.643 | |||||
| rs2292912 | |||||||||
| C/C or C/G | 489 | 127 | - | 97 | - | 52 | - | 27 | - |
| G/G | 29 | 4 | 0.53 (0.17-1.65) | 4 | 0.95 (0.31-2.93) | 1 | 0.27 (0.03-2.26) | 2 | 1.60 (0.34-7.60) |
| P-value | 0.274 | 0.932 | 0.227 | ||||||
| rs7123390 | |||||||||
| G/G or G/A | 491 | 124 | - | 85 | - | 45 | - | 27 | - |
| A/A | 21 | 7 | 1.31 (0.54-3.16) | 14 | 3.67 (1.79-7.56) | 7 | 3.38 (1.35-8.45) | 2 | 1.86 (0.41-8.43) |
| P-value | 0.547 | <0.001 | 0.009 | 0.423 | |||||
| rs1401417 | |||||||||
| G/G or G/C | 503 | 122 | - | 95 | - | 47 | - | 29 | - |
| C/C | 14 | 9 | 2.67 (1.13-6.33) | 8 | 3.06 (1.24-7.54) | 6 | 4.23 (1.53-11.67) | 1 | 1.30 (0.16-10.33) |
| P-value | 0.026 | 0.015 | 0.005 | 0.806 | |||||
Adjusted for age (continuous), race, and family history of cancer in 1st or 2nd degree relatives.
DLBCL: diffuse large B-cell lymphoma; FL: follicular lymphoma; MZBL: marginal zone B-cell lymphoma
CLL/SLL: B-cell chronic lymphocytic leukemia/prolymphocytic leukemia/small lymphocytic lymphoma
Association between CRY2 diplotypes and NHL risk
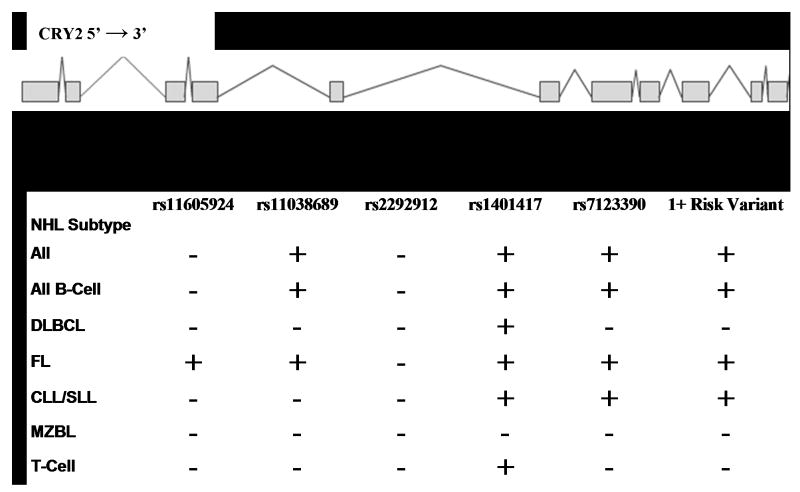
To further explore the relationship among SNPs, haplotypes were estimated for these five SNPs and thirteen different haplotypes were identified among all subjects (Supp. Table 2). Nine haplotypes carried at least one risk allele (i.e. a variant associated with increased risk in the main effects analysis). Since the individual risk estimates indicated a recessive disease model, we used the haplotype information to construct diplotypes for each individual. Figure 1 depicts the location of each marker within the CRY2 gene, and shows the NHL subtypes which were significantly associated with each homozygous variant diplotype. Full results are provided in the supplementary material (Supp. Table 3). ORs (95% CIs) for homozygous variant diplotypes of rs11065924, rs11038689, rs2292912, rs1401417, and rs7123390 using the full population were: 0.81 (0.61-1.08), 2.32 (1.27-4.24), 0.96 (0.53-1.75), 2.81 (1.51-5.23), and 2.36 (1.39-4.02), respectively. The odds ratio associated with having the homozygous variant diplotype at one or more of the risk loci (rs11038689, rs1401417, or rs7123390) was 2.72 (1.61-4.59).
Figure 1. Diplotype analysis and location of CRY2 variants.
Haplotypes were estimated for each individual using the PHASE program, and diplotypes were constructed by combining the two haplotype estimates for each individual. Since the main effects associations indicated a recessive disease model, risk estimates were calculated for all combinations of haplotypes resulting in a homozygous variant diplotype at each locus, with all other diplotypes as the reference category. Estimates were obtained for each NHL subtype represented in the population, and homozygous diplotypes which were significantly associated with each subtype (p<0.05) are depicted with a plus symbol, while nonsignifcant associations are shown as a minus. “1+ risk variant” refers to any haplotype combination which resulted in a homozygous variant diplotype for at least one of the following makers: rs11038689, rs1401417, or rs7123390.
Microarray reveals several cancer- and immune-related genes which are influenced by CRY2
In order to determine which genes and biological pathways might be influenced by CRY2, we silenced the gene in vitro, and performed a whole genome array comparing gene expression in cells with reduced CRY2 to cells with normal CRY2 levels. 734 differentially expressed transcripts were identified (B-H P-value<0.01 in both biological replicates and mean fold change>|2|), and these genes were further explored through pathway and functional analyses using the IPA software package. 23 networks were identified as significantly associated with the differentially expressed genes at P less than 1.0E-10. Among these, 7 were associated with Cancer, 6 networks were important for Immune Response, and 4 were important for Hematological System Development or Hematological Disease (Supp. Table 4).
The top network operative in Hematological System Development and Function (p=1.0E-36, Figure 2) also contained several cancer-related transcripts, including molecules important for DNA repair, cell migration and metastasis, apoptosis, cell proliferation, and angiogenesis. The details of each transcript in the network, including fold changes, adjusted P-values, and a short description of function with relevant citations, can be found in the supplementary material (Supp. Table 5). Briefly, 35 genes were present in the network, 33 of which could be uniquely associated with a single transcript in the microarray. Of these, 27 were significantly upregulated following CRY2 silencing, 4 were not significantly altered, and only 2 were significantly downregulated; which is consistent with CRY2’s role as a transcriptional repressor. The probability of obtaining at least this many differentially expressed molecules in one network by chance alone is reflected by the P-value for the network (P=1.0E-36), indicating that it is very unlikely that the network was obtained randomly, and therefore CRY2 is likely to be important for immune response coordination and processes related to hematological system development. At the center of this network is interleukin-6 (IL-6), an immunoregulatory cytokine that is important for cell growth. This transcript was strongly upregulated following CRY2 silencing, with a mean fold increase of 8.17 (B-H P=0). In addition, a member of the vascular endothelial growth factor family (VEGFC), which is also important for cell growth, as well as angiogenesis, was upregulated more than 6-fold (B-H P=6.99E-4). Interestingly, a number of transcripts with pro-apoptotic, anti-proliferative, or other tumor suppressor capabilities were also upregulated in this network. These include Interferon Beta 1 (IFNB1) which was upregulated more than 3-fold (B-H P=2.96E-9), thioredoxin interacting protein (TXNIP), greater than 11-fold upregulated (B-H P=2.47E-4), retinoic acid receptor responder 3 (RARRES3), greater then 2-fold upregulated (B-H P=1.86E-4), and two members of the chemokine (C-X-C motif) ligand family (CXCL9 and CXCL11), greater than 10- and 4-fold upregulated, respectively (B-H P=1.01E-5 and=3.04E-6).
Figure 2. Network of interactions among genes differentially expressed following CRY2 knockdown.
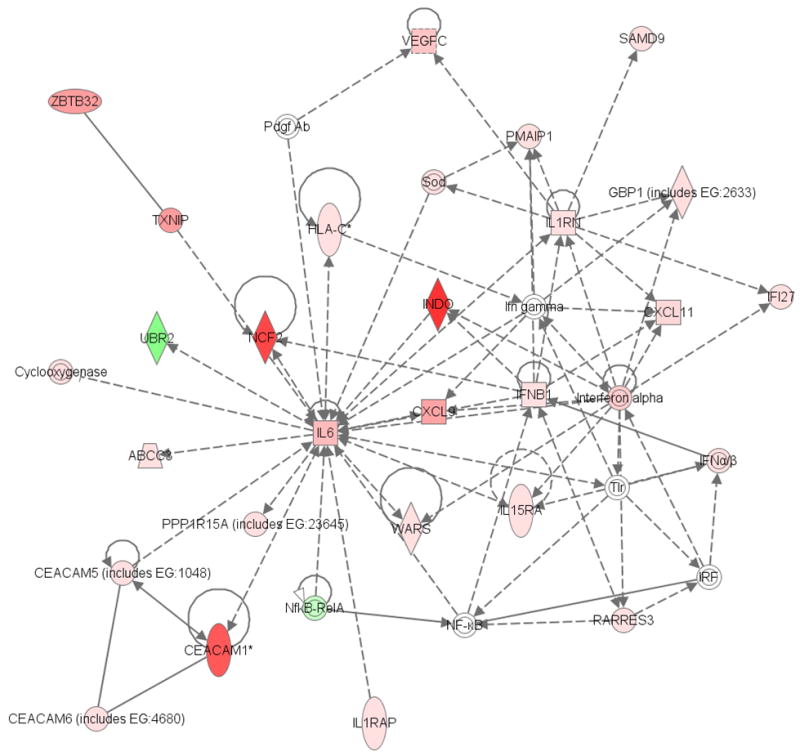
This network was identified as operative in Immune Response, Cell-to-Cell Signaling and Interaction, and Hematological System Development and Function, as determined by Ingenuity Pathway Analysis software. Each shaded transcript was significantly altered following CRY2 silencing, with a mean fold change>|2| and B-H P-value <0.01 in both biologic replicates of the whole genome microarray. Genes depicted in red are upregulated, while those in green are downregulated. Higher intensity color corresponds to greater magnitude of the fold change. Of the 29 molecules in the network which were significantly altered 27 were upregulated, while only 2 were downregulated; which is consistent with CRY2’s role as a transcriptional repressor.
Apart from the transcripts in this network, several additional genes which may be relevant for lymphomagenesis were identified as significantly altered following CRY2 silencing. These include three members of the chemokine (C-C motif) ligand family (CCL3, CCL4, and CCL5), which are important for immune regulation and inflammatory processes; several additional members of the interleukin family of cytokines (including IL-11, IL-15, IL-18, IL-28a, IL-28b, and IL-29,); two insulin-like growth factor binding proteins (IGFBP3 and IGFBP6); and genes in the major histocompatibility complex, class I (HLA-A, HLA-B, HLA-C and HLA-E). Of note, CRY2 itself was significantly downregulated in both microarray replicates (B-H P<0.05), with exactly the same fold change of -3.17 observed in both samples.
The program also computes a P-value based on the likelihood of obtaining the observed number of differentially expressed molecules in a given process by chance alone. The top three disease processes associated with the differentially expressed gene set were “Cancer” (B-H P=3.75E-9), “Inflammatory Disease” (B-H P=5.19E-8), and “Hematological Disease” (B-H P=8.01E-8) (Figure 3). In addition, 144 of the differentially expressed transcripts were related to “Tumorigenesis” (B-H P=4.57E-9), 64 molecules were associated with “Immune Response” (B-H P=7.03E-10), and 49 were involved in “Proliferation of Lymphocytes” (B-H P=6.76E-9). The large number of cancer- and immune-related molecules influenced by CRY2 knockdown provides further evidence suggesting a role for CRY2 in lymphoproliferative processes.
Figure 3. Diseases most strongly associated with the significantly altered genes.
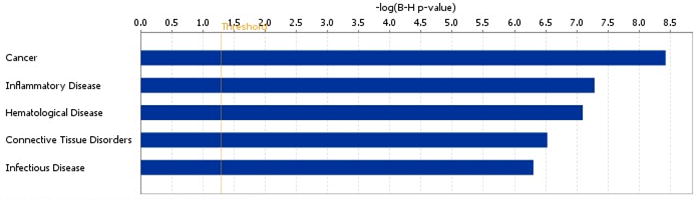
The Ingenuity software scans its Knowledge Base of manually curated relationships for instances in the literature which can link a specific molecule to a functional disease process. The software also assigns a P-value based on the likelihood of obtaining the observed number of disease-related molecules in a given dataset by chance alone. Due to the large number of disease processes represented in the database, B-H P-values are presented to correct for multiple comparisons. The threshold line indicates a B-H P-value of 0.05.
Discussion
The observed connection between circadian disruption (e.g. shift work) and cancer risk in epidemiologic studies has led to the circadian gene hypothesis, which suggests that genetic variants in genes responsible for maintaining circadian rhythm may affect an individual’s susceptibility to human cancers. This hypothesis is supported by results from recent genetic association studies of breast cancer (28, 29), prostate cancer (24), and NHL (5). The findings from the current study of CRY2 provide more evidence demonstrating a role for the circadian genes in NHL susceptibility.
To the best of our knowledge, genetic variants in CRY2 have not been previously examined in NHL, and only one of the SNPs genotyped in the case-control portion of this analysis had been studied previously; in a population-based case-control study conducted in China, which showed a significant association between the variant allele of rs1401417 and increased risk of prostate cancer (24). This finding is consistent with the significant association we observed in the NHL population, and further investigations into the nature of these relationships are warranted in order to determine whether CRY2 has a global impact on cancer susceptibility.
The observed associations between CRY2 and NHL risk provide additional molecular epidemiologic evidence supporting the proposed role of circadian genes as tumor suppressors (30). Circadian genes have been shown to affect expression of 2–10% of mammalian genes (31) including many cancer-related genes (30). Emerging data from animal models have further demonstrated a substantial impact of circadian genes on tumor-related biological pathways such as cell proliferation, cell cycle control, DNA damage response, and apoptosis (30). Mice with mutations in the circadian gene PER2 have deficiencies in DNA damage response and are more prone to tumorigenesis (32). Altered expression of circadian genes also occurs in human tumors; as studies have shown that the period (PER) genes fail to maintain daily rhythmic expression patterns in cancer cells (33). Although CRY2 has also been shown to be involved in cell cycle regulation (34), explicit mechanisms for its role in cancer susceptibility, especially NHL tumorigenesis, are currently unknown.
The microarray analysis, which implicated CRY2 as having the potential to influence gene expression in a number of pathways, including those with relevance for cancer and immune function, provides the first mechanistic evidence suggesting that CRY2 may be important for NHL susceptibility. The findings relative to IL-6 are of particular interest. It has been demonstrated that IL-6 can inhibit DNA synthesis by preventing cell cycle progression into S phase (35). It has also been fairly well established that IL-6 plays an important role in B-cell proliferation (36), and antibodies against IL-6 or its receptor have been explored as treatments for B-lymphoproliferative disorder (37) and inflammatory autoimmune diseases (38). In addition, a previous study has demonstrated that mice expressing an IL-6 transgene exhibit lymphoproliferation and plasmacytosis, and nearly a third of these mice developed follicular and diffuse large cell B-cell lymphomas (39). Moreover, serum levels of IL-6 and VEGF have been shown to be of value in predicting clinical outcome in some patients with NHL (40, 41). The observation that silencing of CRY2 results in more than 8-fold induction of IL-6, and 6-fold induction of VEGFC, is therefore highly relevant in understanding the etiology of hematologic malignancies, and may have important implications for predicting NHL prognosis. Furthermore, cell signaling by IL-6 is partially mediated by its effects on the JAK/STAT3 pathway (42). While JAK was unaffected following CRY2 silencing, STAT3 was upregulated more than twofold (B-H P=7.06E-5). Since IL-6 has been shown to confer increased survival and chemotherapeutic resistance on lymphoma cells, which is at least partially mediated by STAT3; STAT3 has been proposed as a potential therapeutic target for patients with NHL (43). As such, although the pleiotropic biological effects of CRY2 may make it a poor candidate to be directly targeted by therapeutic agents, its effects on both IL-6 and STAT3 may lead future investigations to consider circadian timing when administering cytokine-targeting chemotherapies.
Apart from IL-6, several other interleukins were also significantly altered in CRY2 knockdown cells. IL-18, which was upregulated by more than 10 fold, may play an important, but paradoxical role in cancer risk (44). It has been implicated in cancer protection, through its role in activating immune cells to eliminate sporadic cancers, but may also promote tumor progression by encouraging angiogenesis, tumor growth and local invasion. Interestingly all three members of a newly described class of interferon lambdas (IL-28a, IL-28b, and IL-29) were significantly upregulated following CRY2 silencing. These immunoregulatory cytokines have antiviral and antitumor activity, and may also have potential as therapeutic agents in the treatment of cancer (45).
Three members of the chemokine (C-C motif) ligand family (CCL3, CCL4, and CCL5) were also upregulated in CRY2 knockdown cells. These genes are clustered together on the long arm of chromosome 17, and are important for immune regulation and inflammation (46). CCL3, which was upregulated more than 15 fold in CRY2 knockdown cells, has been shown to be elevated in patients with multiple myeloma (MM) and other hematologic cancers compared to healthy controls; and increased serum levels of CCL3 were associated with advanced MM stage (47) and poorer prognosis in MM (48). Of additional interest was the identification of four genes in the major histocompatibility complex, class I (HLA-A, HLA-B, HLA-C, and HLA-E), which were all significantly upregulated following CRY2 silencing. Apart from being central to immune regulation, these genes have been associated with several cancers, including Hodgkin’s disease (49) and non-Hodgkin’s lymphoma (50). Taken together, these results suggest that reductions in CRY2 have the potential to significantly impact processes relevant for lymphomagenesis, and while they represent a first step in understanding the mechanism by which SNPs in the CRY2 gene might influence NHL susceptibility.
It is important to recognize some of the study’s limitations. First, it is unclear how closely the effect of CRY2 silencing in vitro might mimic the conditions which would arise in vivo. At the level of the organism, circadian rhythmicity is made even more complex by environmental cues, such as light exposure, and hormonal pathways, which may influence circadian gene expression. In addition, since one aim of the study was to examine the influence of CRY2 silencing on immune response pathways, we chose not to use cells derived from lymphoma tissue, which may begin with some level of aberrant immune-related gene expression. As such, it will be necessary for future studies to confirm that the findings we observed are applicable to lymphocytes, and that the changes in gene expression following CRY2 silencing that we observed at the cellular level, are representative of those that would occur at the organismal level.
In summary, the findings from our case-control analysis suggest a novel association between the circadian gene CRY2 and risk of NHL; supporting the hypothesis that genetic variations in circadian genes may confer inherited susceptibility to NHL. The subsequent loss-of-function analysis and whole genome expression microarray suggest that the observed association could potentially be attributed to the impact of CRY2 on several genes important for cancer in general, as well as a number of genes with known relevance for hematological malignancies. Our findings provide a novel panel of promising biomarkers for NHL risk and prognosis, which warrant further investigation. In addition, since previous studies have suggested that circadian-related environmental exposures, such as light at night or rotating shift work, may influence cancer susceptibility, future investigations into potential interactions between circadian gene variants and environmental exposures may be of interest in developing cancer prevention strategies.
Supplementary Material
Acknowledgments
We thank Irina Tikhonova at at Yale University’s W.M. Keck Foundation Biotechnology Research Laboratory for Sequenom genotyping analysis. This work was supported by the National Institutes of Health (grants CA122676, CA110937, and CA108369).
Footnotes
Conflict-of-interest disclosure The authors declare no competing financial interests.
References
- 1.Kondratov RV, Gorbacheva VY, Antoch MP. The role of mammalian circadian proteins in normal physiology and genotoxic stress responses. Curr Top Dev Biol. 2007;78:173–216. doi: 10.1016/S0070-2153(06)78005-X. [DOI] [PubMed] [Google Scholar]
- 2.IARC monographs on the evaluation of carcinogenic risks to humans. Lyon: International Agency for Research on Cancer; 2007. IARC Shift-work, painting and fire-fighting. [Google Scholar]
- 3.Zhu Y, Zheng T. Clock-cancer connection in non-Hodgkin’s lymphoma. Med Hypotheses. 2008;70:788–792. doi: 10.1016/j.mehy.2007.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gery S, Gombart AF, Yi WS, Koeffler C, Hofmann WK, Koeffler HP. Transcription profiling of C/EBP targets identifies Per2 as a gene implicated in myeloid leukemia. Blood. 2005;106:2827–2836. doi: 10.1182/blood-2005-01-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu Y, Leaderer D, Guss C, Brown HN, Zhang Y, Boyle P, et al. Ala394Thr polymorphism in the clock gene NPAS2: a circadian modifier for the risk of non-Hodgkin’s lymphoma. Int J Cancer. 2007;120:432–435. doi: 10.1002/ijc.22321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arjona A, Sarkar DK. Circadian oscillations of clock genes, cytolytic factors, and cytokines in rat NK cells. J Immunol. 2005;174:7618–7624. doi: 10.4049/jimmunol.174.12.7618. [DOI] [PubMed] [Google Scholar]
- 7.Arjona A, Sarkar DK. Evidence supporting a circadian control of natural killer cell function. Brain Behav Immun. 2006;20:469–476. doi: 10.1016/j.bbi.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Chacon F, Cano P, Lopez-Varela S, Jimenez V, Marcos A, Esquifino AI. Chronobiological features of the immune system. Effect of calorie restriction. Eur J Clin Nutr. 2002;56(Suppl 3):S69–72. doi: 10.1038/sj.ejcn.1601491. [DOI] [PubMed] [Google Scholar]
- 9.Esquifino AI, Cano P, Jimenez-Ortega V, Fernandez-Mateos P, Cardinali DP. Neuroendocrine-immune correlates of circadian physiology: studies in experimental models of arthritis, ethanol feeding, aging, social isolation, and calorie restriction. Endocrine. 2007;32:1–19. doi: 10.1007/s12020-007-9009-y. [DOI] [PubMed] [Google Scholar]
- 10.Esquifino AI, Selgas L, Arce A, Maggiore VD, Cardinali DP. Twenty-four-hour rhythms in immune responses in rat submaxillary lymph nodes and spleen: effect of cyclosporine. Brain Behav Immun. 1996;10:92–102. doi: 10.1006/brbi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 11.Seres J, Herichova I, Roman O, Bornstein S, Jurcovicova J. Evidence for daily rhythms of the expression of proopiomelanocortin, interleukin-1-beta and interleukin-6 in adenopituitaries of male long-evans rats: effect of adjuvant arthritis. Neuroimmunomodulation. 2004;11:316–322. doi: 10.1159/000079412. [DOI] [PubMed] [Google Scholar]
- 12.Arjona A, Sarkar DK. Evidence supporting a circadian control of natural killer cell function. Brain Behav Immun. 2005 doi: 10.1016/j.bbi.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Mormont MC, Levi F. Circadian-system alterations during cancer processes: a review. Int J Cancer. 1997;70:241–247. doi: 10.1002/(sici)1097-0215(19970117)70:2<241::aid-ijc16>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 14.Gamaleia NF, Skivka LM, Fedorchuk AG, Shishko ED. Circadian rhythms of cytotoxic activity in peripheral blood mononuclear cells of patients with malignant melanoma. Exp Oncol. 2006;28:54–60. [PubMed] [Google Scholar]
- 15.Filipovich AH, Mathur A, Kamat D, Shapiro RS. Primary immunodeficiencies: genetic risk factors for lymphoma. Cancer Res. 1992;52:5465s–5467s. [PubMed] [Google Scholar]
- 16.Kersey JH, Shapiro RS, Filipovich AH. Relationship of immunodeficiency to lymphoid malignancy. Pediatr Infect Dis J. 1988;7:S10–12. [PubMed] [Google Scholar]
- 17.Griffin EA, Jr, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- 18.Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci U S A. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol. 2005;25:3109–3116. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Holford TR, Leaderer B, Boyle P, Zahm SH, Owens PH, et al. Blood transfusion and risk of non-Hodgkin’s lymphoma in Connecticut women. Am J Epidemiol. 2004;160:325–330. doi: 10.1093/aje/kwh233. [DOI] [PubMed] [Google Scholar]
- 22.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nature Genetics. 2005;37:1217–1223. doi: 10.1038/ng1669. see comment. [DOI] [PubMed] [Google Scholar]
- 23.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Chu LW, Zhu Y, Yu K, Zheng T, Yu H, Zhang Y, et al. Variants in circadian genes and prostate cancer risk: a population-based study in China. Prostate Cancer Prostatic Dis. 2007 doi: 10.1038/sj.pcan.4501024. [DOI] [PubMed] [Google Scholar]
- 25.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 26.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 28.Zhu Y, Brown HN, Zhang Y, Stevens RG, Zheng T. Period3 structural variation: a circadian biomarker associated with breast cancer in young women. Cancer Epidemiol Biomarkers Prev. 2005;14:268–270. [PubMed] [Google Scholar]
- 29.Zhu Y, Stevens RG, Leaderer D, Hoffman A, Holford T, Zhang Y, et al. Non-synonymous polymorphisms in the circadian gene NPAS2 and breast cancer risk. Breast Cancer Res Treat. 2007 doi: 10.1007/s10549-007-9565-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–361. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 31.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 32.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 33.You S, Wood PA, Xiong Y, Kobayashi M, Du-Quiton J, Hrushesky WJ. Daily coordination of cancer growth and circadian clock gene expression. Breast Cancer Res Treat. 2005;91:47–60. doi: 10.1007/s10549-004-6603-z. [DOI] [PubMed] [Google Scholar]
- 34.Gauger MA, Sancar A. Cryptochrome, circadian cycle, cell cycle checkpoints, and cancer. Cancer Res. 2005;65:6828–6834. doi: 10.1158/0008-5472.CAN-05-1119. [DOI] [PubMed] [Google Scholar]
- 35.Shen WH, Zhou JH, Broussard SR, Freund GG, Dantzer R, Kelley KW. Proinflammatory cytokines block growth of breast cancer cells by impairing signals from a growth factor receptor. Cancer Res. 2002;62:4746–4756. [PubMed] [Google Scholar]
- 36.Kishimoto T. The biology of interleukin-6. Blood. 1989;74:1–10. [PubMed] [Google Scholar]
- 37.Haddad E, Paczesny S, Leblond V, Seigneurin JM, Stern M, Achkar A, et al. Treatment of B-lymphoproliferative disorder with a monoclonal anti-interleukin-6 antibody in 12 patients: a multicenter phase 1-2 clinical trial. Blood. 2001;97:1590–1597. doi: 10.1182/blood.v97.6.1590. [DOI] [PubMed] [Google Scholar]
- 38.Ding C, Jones G. Anti-interleukin-6 receptor antibody treatment in inflammatory autoimmune diseases. Rev Recent Clin Trials. 2006;1:193–200. doi: 10.2174/157488706778250168. [DOI] [PubMed] [Google Scholar]
- 39.Kovalchuk AL, Kim JS, Park SS, Coleman AE, Ward JM, Morse HC, 3rd, et al. IL-6 transgenic mouse model for extraosseous plasmacytoma. Proc Natl Acad Sci U S A. 2002;99:1509–1514. doi: 10.1073/pnas.022643999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurzrock R. Cytokine deregulation in hematological malignancies: clinical and biological implications. Clin Cancer Res. 1997;3:2581–2584. [PubMed] [Google Scholar]
- 41.Pedersen LM, Klausen TW, Davidsen UH, Johnsen HE. Early changes in serum IL-6 and VEGF levels predict clinical outcome following first-line therapy in aggressive non-Hodgkin’s lymphoma. Ann Hematol. 2005;84:510–516. doi: 10.1007/s00277-005-1020-x. [DOI] [PubMed] [Google Scholar]
- 42.Brasier AR. Expanding role of cyclin dependent kinases in cytokine inducible gene expression. Cell Cycle. 2008;7:2661–2666. doi: 10.4161/cc.7.17.6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alas S, Bonavida B. Inhibition of constitutive STAT3 activity sensitizes resistant non-Hodgkin’s lymphoma and multiple myeloma to chemotherapeutic drug-mediated apoptosis. Clin Cancer Res. 2003;9:316–326. [PubMed] [Google Scholar]
- 44.Dinarello CA. The paradox of pro-inflammatory cytokines in cancer. Cancer Metastasis Rev. 2006;25:307–313. doi: 10.1007/s10555-006-9000-8. [DOI] [PubMed] [Google Scholar]
- 45.Lasfar A, Lewis-Antes A, Smirnov SV, Anantha S, Abushahba W, Tian B, et al. Characterization of the mouse IFN-lambda ligand-receptor system: IFN-lambdas exhibit antitumor activity against B16 melanoma. Cancer Res. 2006;66:4468–4477. doi: 10.1158/0008-5472.CAN-05-3653. [DOI] [PubMed] [Google Scholar]
- 46.Wolpe SD, Davatelis G, Sherry B, Beutler B, Hesse DG, Nguyen HT, et al. Macrophages secrete a novel heparin-binding protein with inflammatory and neutrophil chemokinetic properties. J Exp Med. 1988;167:570–581. doi: 10.1084/jem.167.2.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi SJ, Cruz JC, Craig F, Chung H, Devlin RD, Roodman GD, et al. Macrophage inflammatory protein 1-alpha is a potential osteoclast stimulatory factor in multiple myeloma. Blood. 2000;96:671–675. [PubMed] [Google Scholar]
- 48.Terpos E, Politou M, Szydlo R, Goldman JM, Apperley JF, Rahemtulla A. Serum levels of macrophage inflammatory protein-1 alpha (MIP-1alpha) correlate with the extent of bone disease and survival in patients with multiple myeloma. Br J Haematol. 2003;123:106–109. doi: 10.1046/j.1365-2141.2003.04561.x. [DOI] [PubMed] [Google Scholar]
- 49.Diepstra A, Niens M, te Meerman GJ, Poppema S, van den Berg A. Genetic susceptibility to Hodgkin’s lymphoma associated with the human leukocyte antigen region. Eur J Haematol Suppl. 2005:34–41. doi: 10.1111/j.1600-0609.2005.00452.x. [DOI] [PubMed] [Google Scholar]
- 50.Choi HB, Roh SY, Choi EJ, Yoon HY, Kim SY, Hong YS, et al. Association of HLA alleles with non-Hodgkin’s lymphoma in Korean population. Int J Hematol. 2008;87:203–209. doi: 10.1007/s12185-008-0040-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.