A Perspective on the study by D'Andrea and colleagues, discussing newly identified components of a SUMO-like interaction network that orchestrates the Fanconi anemia and replication-coupled repair pathways.
Keywords: Fanconi anemia, deubiquitinating enzymes, PCNA, SUMO-like domains
Abstract
In this issue of Genes & Development, Yang and colleagues (pp. 1847–1858) identify new components of a small ubiquitin-like modifier (SUMO)-like interaction network that orchestrates and fine-tunes the Fanconi anemia (FA) pathway and replication-coupled repair. This new pathway emphasizes the intricate interplay of ubiquitin (Ub) and SUMO networks in the DNA damage response.
Ubiquitin (Ub) and the related small ubiquitin-like modifier (SUMO) (hereafter commonly referred to as ubiquitin-like proteins [UBLs]) are part of sophisticated and complex post-translational modification systems (Deribe et al. 2010). Ub and SUMO share substantial levels of sequence homology and become attached to proteins by similar conjugation machineries (Kerscher et al. 2006; Schulman and Harper 2009; Ye and Rape 2009; Gareau and Lima 2010). Both modifications are recognized, or read, by specialized domains that integrate conjugated UBLs into dynamic and complex interaction networks underlying many biological processes, like DNA repair pathways (Fig. 1A,B; Bergink and Jentsch 2009; Dikic et al. 2009). Ub and SUMO are actively deconjugated from their substrates in a highly controlled spatiotemporal manner, thus assuring the plasticity of UBL-mediated protein–protein interactions (Reyes-Turcu et al. 2009; Grabbe et al. 2011). Importantly, some proteins bear integral UBL domains as part of larger structures, but do not form conjugates with cellular proteins (Grabbe and Dikic 2009). Together, the dynamic nature of Ub- and SUMO-based complex networks form the basis for UBL-mediated cellular signaling events.
Figure 1.
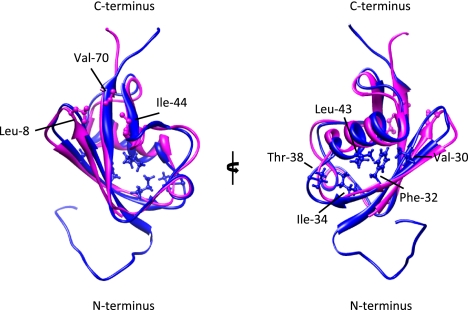
UBD and SIM interaction surfaces on Ub and SUMO are not conserved. Structural alignment of a molecular ribbon representation of Ub (magenta; Protein Data Bank [PDB]: 1aar) and SUMO-3 (cyan; PDB: 2rpq). On Ub, the canonical Leu 8, Ile 44, and Val 70 residues that contact UBDs are indicated (see Dikic et al. 2009). Val 30, Phe 32, Ile 34, Thr 38, and Leu 43 on SUMO3 have been shown to contact a canonical SIM in MCAF1 (the MBD1 [methyl-CpG-binding domain protein 1]-containing chromatin-associated factor 1) (Sekiyama et al. 2008). A similar surface in SUMO1 or SUMO2/3 has also been shown to be involved in binding to the hydrophobic SIM region of PIAS family members (Hecker et al. 2006). Remarkably, positively charged residues in SUMO paralogs, including Lys 33 of SUMO2, which is conserved in the SLD2 of UAF1, contribute to SIM binding. Note that opposite surfaces on SUMO and Ub serve as interaction platforms for the respective binding modules. The image was generated using University of California at San Francisco Chimera (release date May 24, 2011; http://www.cgl.ucsf.edu/chimera).
Ub and SUMO encode multivalent signaling molecules
Ub and SUMO are transcribed as precursor molecules that require processing prior to their conjugation. Subsequently, both modifiers are typically attached to lysine residues of target proteins and can be conjugated either as monomers or as polymeric chains that are generally linked through internal lysine residues (Ikeda and Dikic 2008). Conjugation of Ub and SUMO typically relies on the coordinated activity of the catalytic E1–E2–E3 triad, but compared with Ub, the SUMO conjugation machinery is less complex. SUMO becomes activated by the dimeric UBA2 (Ub-associated domain 2)/AOS1 complex and is subsequently transferred to Ubc9, the only known SUMO E2 (Kerscher et al. 2006; Gareau and Lima 2010). Although Ubc9 is able to transfer SUMO directly to substrates, it typically interacts with SUMO E3 ligases that mediate an optimal positioning of the SUMO-loaded E2 and the substrate to allow for efficient substrate SUMOylation (Gareau and Lima 2010). Well-characterized E3 SUMO ligases are Ran-binding protein 2 (RanBP2) and the family of SP-RING-containing proteins, including the yeast Siz proteins and the human family of protein inhibitor of STAT (PIAS) proteins. In contrast to SUMOylation, the activation and conjugation of Ub involves at least two E1 enzymes, dozens of E2 enzymes, and hundreds of different Ub E3 ligases that specifically interact to select and modify numerous substrates (Schulman and Harper 2009; Ye and Rape 2009). Finally, both Ub and SUMO are actively hydrolyzed and removed from their substrates by specialized cysteine proteases: the deubiquitinating enzymes (DUBs) and the SUMO-specific proteases (SENP1–3), respectively (Reyes-Turcu et al. 2009; Gareau and Lima 2010).
Only one Ub isoform has been found, and modification of substrates with a single ubiquitin (monoubiquitination) has been associated with alterations in protein activity and localization (for example, through endocytosis, meiosis, and transcriptional regulation) (Ikeda and Dikic 2008). PolyUb (polyubiquitination) chains, linked through one or several of Ub's seven lysines, are associated with 26S proteasomal degradation, immune signaling, and DNA repair (Ikeda and Dikic 2008; Grabbe and Dikic 2009). Ub can also be linked into chains via C-terminal diglycine–N-terminal Met-1 linkages (linear or head-to-tail ubiquitination) by the LUBAC E3 ligase complex, and this fulfills essential roles in nuclear factor-κB (NF-κB) signaling pathways (Iwai and Tokunaga 2009). While only a single, highly conserved Ub is found in lower and higher eukaryotes, four SUMO forms (SUMO 1–4) have been identified in humans. Among these, only SUMO1–3 are conjugated to substrates. SUMO2 and SUMO3 share 97% sequence identity and show ∼50% amino acid identity with SUMO1. SUMO2/3 are able to form SUMO chains mainly via lysine residue Lys 11, while modification by SUMO1 preferentially occurs as a monomodification (Geoffroy and Hay 2009; Gareau and Lima 2010). SUMO1 may also be conjugated to the end of an extending SUMO2/3 chain, thereby terminating chain elongation (Gareau and Lima 2010). Like ubiquitination, modification by SUMO generally acts to modulate the dynamics of protein–protein interactions, thereby controlling critical cellular pathways. Distinctive functions of mono- or polymodification with SUMO are just beginning to be uncovered (Ulrich 2008). Interestingly, however, polymeric chains of SUMO2/3 can trigger substrate ubiquitination by recruiting SUMO-targeted ubiquitin ligases (STUbL) such as Slx5/8 proteins in Saccharomyces cerevisiae or RING finger protein 4 (RNF4) in mammals, thus exemplifying the intimate cross-talk of UBL modification systems (Perry et al. 2008; Geoffroy and Hay 2009).
Ub-binding domains (UBDs) and SUMO-binding domains
An important aspect of Ub and SUMO signaling is that they are recognized and coupled to specific cellular responses by interactions with effector proteins containing specialized UBDs and SUMO-interacting motifs (SIMs), respectively (Hecker et al. 2006; Grabbe et al. 2011). Numerous studies have addressed the structures, functions, and Ub specificities of a wide variety of UBDs, of which >20 distinct families have been identified so far (Dikic et al. 2009). These UBDs employ specialized structural and functional characteristics that determine their ability to recognize and bind to different types of modifications. For example, the Pru (Plexstrin receptor for Ub) of the proteasomal receptor Rpn13 or UBA of several shuttling receptors binds preferentially to Lys 48 Ub chains (Raasi et al. 2005; Husnjak et al. 2008), whereas the NZF (Npl4-like zinc finger) domain of TAK1-binding protein 2 (TAB2) preferentially binds to Lys 63-linked polyUb chains with much higher affinity than Lys 48 chains (Kulathu et al. 2009; Sato et al. 2009), and ubiquitin binding by the ABIN and NEMO (UBAN) domain of the IKK adaptor NEMO specifically binds to linear ubiquitin chains (Rahighi et al. 2009). Importantly, UBDs not only recognize ubiquitin conjugates but can also function as readers for proteins, which contain ubiquitin-like domains (Grabbe et al. 2011). A paradigm stems from the Rad23 protein, which has an N-terminal ubiquitin-like domain that binds to the UBD of the 26S proteasomal ubiquitin receptor RPN10/S5 (Dantuma et al. 2009). Rad23 is involved in nucleotide excision repair (NER) via the XPC (xeroderma pigmentosum group C) protein complex. XPC has been shown to interact with RAD23A and RAD23B, which contain an N-terminal ubiquitin-like domain and C-terminal UBA domains (Bergink and Jentsch 2009).
Similar to what has been observed for ubiquitin, signaling by SUMO often relies on the recognition of the post-translational mark by a specialized interaction module, termed the SIM (Hecker et al. 2006). In many cases SIM-mediated interactions direct the assembly of protein complexes, like the SIM-dependent targeting of CENP-E and Daxx to the centromere or PML nuclear bodies, respectively (Lin et al. 2006; Zhang et al. 2008). SIMs have been identified in a wide range of proteins and are typically characterized by a short consensus sequence of hydrophobic residues [(V/I/L)-X-(V/I/L)-(V/I/L) or (V/I/L)-(V/I/L)-X-(V/I/L)], which folds into a small β sheet that becomes inserted between an α helix and a β sheet of the interacting SUMO molecule (Hecker et al. 2006). Sequence variations within the hydrophobic core of some characterized SIMs may allow selective recognition of distinct SUMO paralogs (Gareau and Lima 2010). Notably, in some cases, acidic residues are flanking the hydrophobic core, and NMR studies suggest that the acidic stretch mediates electrostatic interactions with a surface area formed by basic residues of SUMO (Fig. 1; Hecker et al. 2006). Importantly, negative charges that stabilize SUMO/SIM interactions can also be provided by phosphorylation of serine or threonine residues in close proximity to the hydrophobic core. Thus, it has been demonstrated that in a subset of SIM-containing proteins, including the PIAS family proteins or PML, phosphorylation by the serine/threonine kinase 2 (CK2) within a phosphoSIM module enhances the binding to SUMO paralogs (Stehmeier and Muller 2009). This mechanism could act as a molecular switch that modulates the dynamics of SUMO/SIM interactions. A novel aspect in the understanding of SUMO/SIM interactions came from the observation that SUMO2 chains can be recognized by proteins containing several repetitive SIMs, such as the STUbL RNF4 ligase, which contains four N-terminal SIMs (Perry et al. 2008; Geoffroy and Hay 2009). In addition to the identified SIMs, a protein family with internal SUMO-like domains (SLDs) has been identified. Members of the RENi protein family, which is defined by the yeast Rad60 and Esc2 proteins and mammalian NIP45, harbor two tandem-positioned SLDs within their C-terminal regions (Novatchkova et al. 2005). Recent biochemical and structural work indicates that the SLD2 of Rad60 and NIP45 interacts noncovalently with Ubc9 in a similar manner to SUMO1 (Prudden et al. 2009; Sekiyama et al. 2010). It is noteworthy, however, that the SLD2 domains of RENi family members are not recognized by canonical SUMO1- or SUMO2/3-binding SIMs or by the tandem SIMs of RNF4 (Sekiyama et al. 2010). Accordingly, the SLD2 of RENi proteins lacks a cluster of positively charged residues that is part of the SIM-binding pocket in SUMO paralogs (Prudden et al. 2009; Sekiyama et al. 2010).
SUMO and Ub networks in DNA repair pathways
Accumulating evidence indicates that SUMO/Ub-mediated protein–protein interactions are particularly important in the DNA damage response network, which allows cells to cope with different types of genotoxic stress (Bergink and Jentsch 2009; Ulrich and Walden 2010). Upon DNA damage, a cell initiates extensive decision-making processes that balance between repair of the DNA lesion or more rigorous consequences, such as cell death. To correct the DNA lesions, highly complex repair systems have evolved that all rely on the timely and spatially controlled assembly and disassembly of protein complexes. This process is commonly referred to as DNA damage response and is initiated by proteins that recognize the actual DNA damage, followed by recruitment of proteins that mediate repair pathways (Bergink and Jentsch 2009).
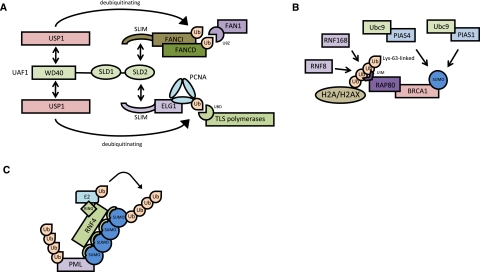
The importance of ubiquitin and SUMO in DNA repair is best illustrated on the proliferating cell nuclear antigen (PCNA), which is loaded as a trimeric ring on DNA strands and functions as a processivity factor for DNA polymerases and as a moving platform for factors that are involved in replication-linked functions (Bergink and Jentsch 2009). In S phase, PCNA is alternatively modified by monoubiquitination, Lys 63-linked polyubiquitination, and SUMOylation at the evolutionarily conserved lysine Lys 164 (Moldovan et al. 2007). PCNA ubiquitination is induced by DNA damage and is essential for DNA repair by the error-free and error-prone branches of the RAD6 pathway. Monoubiquitination, which is catalyzed by the E2 enzyme Rad6 together with the E3 ligase Rad18, facilitates translesion DNA synthesis (TLS) by recruitment of TLS polymerases that contain specific ubiquitin-binding motifs (Fig. 2A; Bienko et al. 2005). Lys 63-linked polyubiquitination, which involves the dimeric E2 Mms2/Ubc13 and the ligase Rad5, directs PCNA to the error-free repair pathway (Hoege et al. 2002). SUMOylation of PCNA inhibits recombination by mediating the recruitment of the anti-recombinogenic factor Srs2 via a canonical SIM in Srs2 (Pfander et al. 2005). The work on PCNA thus exemplifies the concept of an ubiquitin/SUMO switch that allows the dynamic exchange of binding partners.
Figure 2.
Interplay of Ub and SUMO networks in DNA repair pathways. Three different scenarios underscoring the relevance of selective Ub–UBD and SUMO–SIM interactions in DNA repair pathways. (A) The SLD–SLIM-mediated deubiquitination of PCNA-Ub and FANCD2-Ub coordinates HR and TLS. (B) A ubiquitin-SUMO cross-talk mediates the assembly of DNA damage foci. (C) Polymeric chains of SUMO2/3 can trigger substrate ubiquitination by recruiting STUbLs such as RNF4 that contain repetitive SIMs. At the moment, it remains unclear whether RNF4 creates SUMO/Ub mixed chains or modifies substrates with SUMO and Ub on two separate lysine residues. See the text for more detailed descriptions.
Double-strand breaks (DSBs) are repaired by either the homologous recombination (HR) or nonhomologous end-joining (NHEJ) pathways. A characteristic feature of DSB repair in mammals is the rapid accumulation of repair factors, including the recombinase RAD51 and the ubiquitin–ligase BRCA1 in subnuclear foci (Venkitaraman 2004; Yan and Jetten 2008). Recent data demonstrate that the assembly of these structures on the damaged chromatin is triggered by a pathway that integrates SUMOylation and Lys 63-linked ubiquitination-dependent processes. Lys 63-linked chains, which are formed by the RING-type ligases RNF8 and RNF168, serve as an anchor for the ubiquitin-interacting motif (UIM) domain-containing adaptor protein RAP80 that finally brings BRCA1 into repair foci (Fig. 2B; Yan and Jetten 2008; Doil et al. 2009; Stewart et al. 2009). Intriguingly, after exposure of cells to a variety of genotoxic agents, such as hydroxyurea or cisplatin, SUMO as well as PIAS family members are also recruited to DNA damage foci. Moreover, PIAS1 and PIAS4 promote BRCA1 accumulation at these sites and stimulate SUMOylation of BRCA1, which in turn appears to influence the ubiquitin–ligase activity of BRCA1 (Fig. 2B; (Galanty et al. 2009; Morris et al. 2009). The importance of PIAS family members for DNA DSB repair is also supported by an increased sensitivity to DNA-damaging agents and a failure to carry out efficient HR and NHEJ in cells that have compromised expression of PIAS1 or PIAS4 (Galanty et al. 2009; Morris et al. 2009). Although the critical substrates of SUMO at DNA damage foci remain to be defined and the molecular details need to be elucidated, these data substantiate the idea of extensive SUMO/Ub cross-talk in DNA repair pathways.
DSBs are also required for efficient progression of meiosis, V(D)J recombination during early B- and T-cell development, and switching of the mating type in yeast (Bergink and Jentsch 2009). DSB responses are initiated by recruitment of the heterotrimeric MRN complex composed of the DSB repair protein MRE11, DNA repair protein RAD50, and nibrin (Williams et al. 2010). This complex mediates the formation of single-stranded stretches of DNA at the site of lesion that quickly become covered by the trimeric RPA complex. This step is followed by the action of RAD52 and BRCA2, which replace the RPA complex with the RAD51 recombinase that mediates the actual DNA recombination (Liu and Heyer 2010). It has been shown that RAD52 becomes SUMOylated, but only when DSBs are present (Sacher et al. 2006). As a consequence, SUMOylated RAD52 becomes stabilized into foci, possibly interfering with proteasomal degradation.
Complexity rose again with the identification of Slx5 and Slx8 in yeast, two proteins initially linked to genome stability in S. cerevisiae (Perry et al. 2008). Both proteins contain a RING domain commonly found in E3 ligases that mediate heterodimerization. Both Slx5 and Slx8 contain SIM domains that interact with SUMO, and the RING finger of Slx8 mediates ubiquitination of SUMOylated substrates (Perry et al. 2008). The Slx5/Slx8 dimer has recently been implicated in DNA repair at the nuclear pore complex. The mammalian homolog of Slx5/Slx8 is RNF4, which, besides a RING domain, contains four SIM domains (Geoffroy and Hay 2009). RNF4 has been shown to bind strongly to SUMO chains and mediates the subsequent ubiquitination of SUMO chains. The biological significance of RNF4 as a STUbL became clear with the observation that the tripartite motif-containing protein PML and the leukemogenic fusion protein PML-RARα, which causes acute promyelocytic leukemia, are degraded in a SUMO-dependent RNF4-mediated process (Geoffroy and Hay 2009). This pathway can be triggered by administration of the anti-leukemogenic drug arsenic, which induces a rapid polySUMOylation of PML and PML-RARα, after which the SUMOs become recognized by RNF4 (Fig. 2C; Geoffroy and Hay 2009).
Another example of the involvement of SUMO networks relates to the base excision repair (BER) pathway that is activated upon damage to DNA bases, such as alkylation, deamidation, or oxidation. These lesions are recognized by glycosylases that remove the lesion, leading to an abasic site (apurinic or apyrimidinic [AP]). As a consequence, the phosphodiester bond 5′ of the AP site is cleaved by AP endonucleases (APE), followed by repair of the gap. One of these glycosylases is thymine–DNA glycosylase (TDG), which hydrolyzes the N-glycosidic bonds of thymine or uracil in the case when these are mismatched with guanine. TDG has been shown to be modified with SUMO, which results in the dissociation of the enzyme from the affected site (Hardeland et al. 2002). Structural data revealed that the conjugated SUMO interacts with a SUMO-binding motif within the molecule itself, leading to a structural rearrangement of the enzyme that terminates DNA binding (Baba et al. 2005).
Ub-like domains, SLDs, and signaling networks in DNA repair
In this issue of Genes & Development, D'Andrea and coworkers (Yang et al. 2011) further emphasize the interconnectivity and significance of the Ub/SUMO conjugation machineries and recognition of these modifications by UBDs and SIMs in DNA repair pathways. In their study, Yang et al. (2011) focused on two key pathways responsible for sensing DNA damage and initiating repair: the Fanconi anemia (FA) pathway and replication-coupled repair. A key protein in the FA pathways is FANCD2 (FA, complementation group D2 protein), which belongs to the family of FA proteins (Moldovan and D'Andrea 2009). At present, 15 FA proteins have been identified, and mutations in these proteins are the cause of an autosomal recessive and X-linked genetic disorder in which patients suffer from congenital abnormalities, bone marrow failure, and pronounced cancer susceptibility (Moldovan and D'Andrea 2009). One feature of FA cells is their extreme sensitivity to DNA cross-linking agents like mitomycin C and cisplatin. Upon DNA damage, the majority of FA proteins cooperate to form an active E3 ligase that, together with the Ub E2 enzyme UBE2T, catalyzes monoubiquitination of FANCD2 and FANCI. As a direct consequence, monoubiquitinated FANCD2 and FANCI are translocated to specific chromatin foci, where they recruit and colocalize with other DNA repair proteins (Moldovan and D'Andrea 2009).
FANCD2-Ub and PCNA-Ub thus play critical roles in mediating DNA damage responses, and it has been demonstrated that cellular levels of both FANCD2-Ub and PCNA-Ub directly control the extent and duration of HR and TLS repair (Bergink and Jentsch 2009). It is therefore crucial to tightly regulate the dynamics of monoubiquitination on FANCD2 and PCNA and to control the DUB-mediated removal of Ub from these proteins at certain time points. USP1 has been identified as the DUB for FANCD2-Ub and PCNA-Ub, since loss of USP1 leads to accumulation of FANCD2-Ub and PCNA-Ub, even in cases without DNA damage (Nijman et al. 2005). Moreover, cells lacking USP1 are more sensitive to DNA cross-linking agents, as observed in cells derived from FA patients, underscoring the relevance of USP1 and deubiquitination in repair pathways. USP1 interacts with UAF1 (USP1-associated factor 1), an 80-kDa protein that stabilizes and activates USP1 (Cohn et al. 2007).
Previously, it was unclear how the UAF1/USP1 DUB complex was recruited to FANCD2-Ub and PCNA-Ub. Yang et al. (2011) now demonstrate the involvement of SLD/SIM-mediated interactions. The UAF1 protein contains eight WD40 repeats at its N terminus, and Yang et al. (2011) were able to identify two SLDs within the C terminus of UAF1. These SLDs are ∼100 residues in size and share between 50% and 60% sequence similarity with the SUMO2 protein. This is in contrast with SLDs of RENi family members, which are more closely related to SUMO1 (Novatchkova et al. 2005). In addition, the SLDs contain the prototypical C-terminal diglycine present in Ub and SUMO, which has been observed in multiple Ub-like domains and SLDs. The D'Andrea laboratory (Yang et al. 2011) also discovered two SIM-like domains (SLIM) in FANCI and hELG1 that mediate recruitment of UAF1 to FANCD2 or PCNA, respectively (Fig. 2A). Importantly, the interaction between isolated SLD2 and FANCI is dependent on the integrity of its SLIM. Similar to canonical SIMs, SLIMs are characterized by hydrophobic residues and a cluster of acidic residues, which may also be flanked by phosphorylated amino acids. The negatively charged residues in SLIMs likely form electrostatic interactions with conserved lysine residues of SLD2 in a similar way to SUMO–SIM interactions. Importantly, however, SLD2 does not bind to canonical SIMs of PIAS family members, and, vice versa, SUMO paralogs do not interact with the SLIM of FANCI, indicating the specificity and selectivity of these binding modules. Future structural studies will be aimed at elucidating the critical determinants and molecular details underlying this specificity.
As mentioned above, SLDs have been identified in proteins of the RENi family (Novatchkova et al. 2005). For example, the RAD60 DNA repair protein contains two SLDs at its C terminus and is involved in DNA DSB repair, recovery from replication arrest, and cell viability. It has been shown that RAD60 SLD2 is able to mimic SUMO by a noncovalent interaction with the SUMO E2 Ubc9 within the Ubc9 SUMO interaction interface. Disruption of this interaction leads to genomic instability and effects associated with disturbances in SUMO conjugation. Additional SLDs are found in NIP45, another member of the RENi family of proteins. NIP45 also contains two SLDs, and it has been demonstrated that SLD2 is able to compete with a noncovalent SUMO-binding site on the SUMO E2, thereby preventing SUMO chain elongation in vitro. This is consistent with the observation that the SLD2 domain of S. cerevisiae RAD60 inhibits longer SUMO chain formation and that disturbance of the RAD60 SLD2–Ubc9 interaction leads to DNA damage hypersensitivity. Furthermore, the crystal structure of NIP45 SLD2 in complex with Ubc9 revealed conserved residues that are also present in SUMO1, which are well conserved in other RENi family members (Novatchkova et al. 2005; Prudden et al. 2009). One intriguing question that remains is why SLDs are paired and separated by a short linker sequence. Until now, only the second SLD has been shown to interact with SUMO, although the proximal SLD has been reported to interact with the SUMO E1 (Prudden et al. 2009). How the specificity of the SLDs toward SUMO or SLDs could be explained structurally remains to be answered, just like the role of the linker sequence between both SLDs.
Parallels between ubiquitin-like domains and SLDs
In addition to Ub and SUMO, the family of UBL family members has expanded to approximately a dozen members, like F-associated transcript 10 (FAT10) and ISG15, which share sequence homology with Ub. Human leukocyte antigen FAT10 is a small UBL of which no physiological substrate has yet been identified (Hochstrasser 2009). Fusion of FAT10 to artificial substrates, like green fluorescent protein (GFP), leads to degradation by the 26S proteasome, linking FAT10 to proteasomal degradation when covalently conjugated to a substrate (Groettrup et al. 2008). ISG15, on the other hand, is up-regulated upon exposure of cells to type I interferon and is involved in cellular antiviral responses within the innate immune system (Hochstrasser 2009). Both FAT10 and ISG15—like the SLDs identified in UAF1 and the RENi family—contain two ubiquitin-like domains in a tandem array. It might therefore be possible that this unique double pattern could serve as new recognition and binding motif for a new type of ubiquitin-(like)-binding domain.
Conclusions
By identifying a central role for SLD–SLIM mediated protein–protein interactions in DNA repair pathways, this study by Yang et al. (2011) has expanded our insights into the multifaceted roles of the Ub and SUMO interaction networks. The interplay between highly dynamic and often transient UBL modifications at one end and recognition of ubiquitin-(like)-binding domains at the other end allows the creation of flexible and highly complex interconnected signaling networks that mediate essential cellular response programs. These networks are fine-tuned by selective post-translational modifications, such as observed for the phosphoSIM, and possibly by relaxed UBL-binding selectivity in vivo (for example, as discussed by Ikeda et al. 2010). With the discovery of new UBLs and new ubiquitin-(like)-binding domains, these networks will grow in size and complexity, requiring the need for new technologies to study them.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.17593511.
References
- Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, Shirakawa M 2005. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature 435: 979–982 [DOI] [PubMed] [Google Scholar]
- Bergink S, Jentsch S 2009. Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458: 461–467 [DOI] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. 2005. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310: 1821–1824 [DOI] [PubMed] [Google Scholar]
- Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, D'Andrea AD 2007. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell 28: 786–797 [DOI] [PubMed] [Google Scholar]
- Dantuma NP, Heinen C, Hoogstraten D 2009. The ubiquitin receptor Rad23: at the crossroads of nucleotide excision repair and proteasomal degradation. DNA Repair (Amst) 8: 449–460 [DOI] [PubMed] [Google Scholar]
- Deribe YL, Pawson T, Dikic I 2010. Post-translational modifications in signal integration. Nat Struct Mol Biol 17: 666–672 [DOI] [PubMed] [Google Scholar]
- Dikic I, Wakatsuki S, Walters KJ 2009. Ubiquitin-binding domains—from structures to functions. Nat Rev Mol Cell Biol 10: 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136: 435–446 [DOI] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP 2009. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462: 935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareau JR, Lima CD 2010. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 11: 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy MC, Hay RT 2009. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat Rev Mol Cell Biol 10: 564–568 [DOI] [PubMed] [Google Scholar]
- Grabbe C, Dikic I 2009. Functional roles of ubiquitin-like domain (ULD) and ubiquitin-binding domain (UBD) containing proteins. Chem Rev 109: 1481–1494 [DOI] [PubMed] [Google Scholar]
- Grabbe C, Husnjak K, Dikic I 2011. The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol 12: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groettrup M, Pelzer C, Schmidtke G, Hofmann K 2008. Activating the ubiquitin family: UBA6 challenges the field. Trends Biochem Sci 33: 230–237 [DOI] [PubMed] [Google Scholar]
- Hardeland U, Steinacher R, Jiricny J, Schar P 2002. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J 21: 1456–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I 2006. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem 281: 16117–16127 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M 2009. Origin and function of ubiquitin-like proteins. Nature 458: 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I 2008. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453: 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F, Dikic I 2008. Atypical ubiquitin chains: new molecular signals. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep 9: 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F, Crosetto N, Dikic I 2010. What determines the specificity and outcomes of ubiquitin signaling? Cell 143: 677–681 [DOI] [PubMed] [Google Scholar]
- Iwai K, Tokunaga F 2009. Linear polyubiquitination: a new regulator of NF-κB activation. EMBO Rep 10: 706–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22: 159–180 [DOI] [PubMed] [Google Scholar]
- Kulathu Y, Akutsu M, Bremm A, Hofmann K, Komander D 2009. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat Struct Mol Biol 16: 1328–1330 [DOI] [PubMed] [Google Scholar]
- Lin DY, Huang YS, Jeng JC, Kuo HY, Chang CC, Chao TT, Ho CC, Chen YC, Lin TP, Fang HI, et al. 2006. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol Cell 24: 341–354 [DOI] [PubMed] [Google Scholar]
- Liu J, Heyer WD 2010. Who's who in human recombination: BRCA2 and RAD52. Proc Natl Acad Sci 108: 441–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, D'Andrea AD 2009. How the fanconi anemia pathway guards the genome. Annu Rev Genet 43: 223–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S 2007. PCNA, the maestro of the replication fork. Cell 129: 665–679 [DOI] [PubMed] [Google Scholar]
- Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, et al. 2009. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462: 886–890 [DOI] [PubMed] [Google Scholar]
- Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, Bernards R 2005. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell 17: 331–339 [DOI] [PubMed] [Google Scholar]
- Novatchkova M, Bachmair A, Eisenhaber B, Eisenhaber F 2005. Proteins with two SUMO-like domains in chromatin-associated complexes: the RENi (Rad60–Esc2–NIP45) family. BMC Bioinformatics 6: 22 doi: 10.1186/1471-2105-6-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JJ, Tainer JA, Boddy MN 2008. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci 33: 201–208 [DOI] [PubMed] [Google Scholar]
- Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S 2005. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436: 428–433 [DOI] [PubMed] [Google Scholar]
- Prudden J, Perry JJ, Arvai AS, Tainer JA, Boddy MN 2009. Molecular mimicry of SUMO promotes DNA repair. Nat Struct Mol Biol 16: 509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raasi S, Varadan R, Fushman D, Pickart CM 2005. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol 12: 708–714 [DOI] [PubMed] [Google Scholar]
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, et al. 2009. Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell 136: 1098–1109 [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD 2009. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem 78: 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M, Pfander B, Hoege C, Jentsch S 2006. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat Cell Biol 8: 1284–1290 [DOI] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Mimura H, Yamashita M, Yamagata A, Fukai S 2009. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J 28: 2461–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman BA, Harper JW 2009. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol 10: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiyama N, Ikegami T, Yamane T, Ikeguchi M, Uchimura Y, Baba D, Ariyoshi M, Tochio H, Saitoh H, Shirakawa M 2008. Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3. J Biol Chem 283: 35966–35975 [DOI] [PubMed] [Google Scholar]
- Sekiyama N, Arita K, Ikeda Y, Hashiguchi K, Ariyoshi M, Tochio H, Saitoh H, Shirakawa M 2010. Structural basis for regulation of poly-SUMO chain by a SUMO-like domain of Nip45. Proteins 78: 1491–1502 [DOI] [PubMed] [Google Scholar]
- Stehmeier P, Muller S 2009. Phospho-regulated SUMO interaction modules connect the SUMO system to CK2 signaling. Mol Cell 33: 400–409 [DOI] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136: 420–434 [DOI] [PubMed] [Google Scholar]
- Ulrich HD 2008. The fast-growing business of SUMO chains. Mol Cell 32: 301–305 [DOI] [PubMed] [Google Scholar]
- Ulrich HD, Walden H 2010. Ubiquitin signalling in DNA replication and repair. Nat Rev Mol Cell Biol 11: 479–489 [DOI] [PubMed] [Google Scholar]
- Venkitaraman AR 2004. Tracing the network connecting BRCA and Fanconi anaemia proteins. Nat Rev Cancer 4: 266–276 [DOI] [PubMed] [Google Scholar]
- Williams GJ, Lees-Miller SP, Tainer JA 2010. Mre11–Rad50–Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair (Amst) 9: 1299–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Jetten AM 2008. RAP80 and RNF8, key players in the recruitment of repair proteins to DNA damage sites. Cancer Lett 271: 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Moldovan GL, Vincigeurra P, Murai J, Takeda S, D'Andrea AD 2011. Regulation of the Fanconi anemia pathway by a SUMO-like delivery network. Genes Dev (this issue) doi: 10.1101/gad.17020911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Rape M 2009. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 10: 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XD, Goeres J, Zhang H, Yen TJ, Porter AC, Matunis MJ 2008. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol Cell 29: 729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]