A Review of mRNA polyadenylation and a historical perspective of how the field has evolved since the discovery of poly(A) tails in the 1970s.
Keywords: RNA polymerase II, transcriptional termination, cotranscriptional processing, poly(A) signal
Abstract
Polyadenylation [poly(A)] signals (PAS) are a defining feature of eukaryotic protein-coding genes. The central sequence motif AAUAAA was identified in the mid-1970s and subsequently shown to require flanking, auxiliary elements for both 3′-end cleavage and polyadenylation of premessenger RNA (pre-mRNA) as well as to promote downstream transcriptional termination. More recent genomic analysis has established the generality of the PAS for eukaryotic mRNA. Evidence for the mechanism of mRNA 3′-end formation is outlined, as is the way this RNA processing reaction communicates with RNA polymerase II to terminate transcription. The widespread phenomenon of alternative poly(A) site usage and how this interrelates with pre-mRNA splicing is then reviewed. This shows that gene expression can be drastically affected by how the message is ended. A central theme of this review is that while genomic analysis provides generality for the importance of PAS selection, detailed mechanistic understanding still requires the direct analysis of specific genes by genetic and biochemical approaches.
The molecular biology of eukaryotic genes has been transformed in recent years from specific knowledge of how a few eukaryotic genes are expressed to a genome-wide perspective. This has been achieved by ingenious technological advances that afford the accumulation of enormous molecular detail. However, a theme of this review is that much of our current understanding of how gene expression is regulated was laid down in early experiments on specific genes, which is now being confirmed and extended by new genomic analysis. This particularly holds true for polyadenylation [poly(A)] signals (PAS) of eukaryotic protein-coding genes. This review charts our ever-increasing knowledge of the mechanism of formation of the ubiquitous 3′-terminal poly(A) tail, taken as a defining feature of translationally competent messenger RNA (mRNA). I begin by describing early experiments that revealed the presence of poly(A) tails on mRNA and the first clues as to how these nontemplated sequences are added to the right RNA 3′ ends. I also update our knowledge of how the placement of a poly(A) tail at the 3′ end of mRNA influences the expression of so many genes. This is revealed by current genomic data, where sequencing a whole genome now takes less time than it initially took to sequence the 3′ end of a single mRNA. Finally, I describe the host of experiments that place the polyadenylation process at its central point in the gene expression pathway, in particular by defining the extent of mRNA 3′ untranslated regions (UTRs).
Polyadenylation signals and 3′ noncoding RNA (ncRNA) sequences
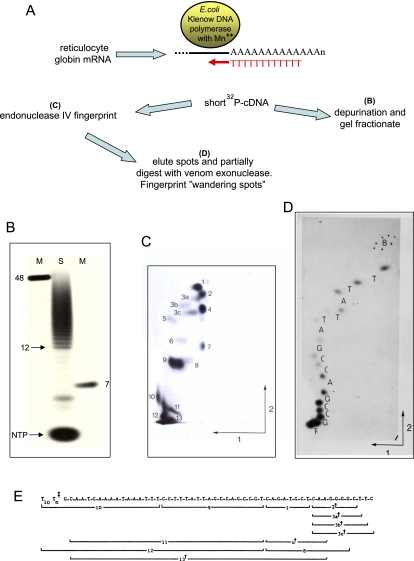
The first clues that mRNA has a unique 3′-terminal tail came from early mammalian cell fractionation experiments that allowed the isolation of translationally active polysome-associated mRNA. RNase digestion (pancreatic RNase cuts at C and U residues, while T1 RNase cuts at G) of this mRNA preparation revealed a resistant fraction presumed to be poly(A) (Lim and Canellakis 1970; Edmonds et al. 1971; Adesnik et al. 1972; Mendecki et al. 1972; Birnboim et al. 1973). Since long poly(A) tracts were not thought to be DNA-templated (Birnboim et al. 1973; Jelinek et al. 1973), a poly(A) polymerase was sought and found that was subsequently shown to be responsible for poly(A) tail formation on mRNA (Winters and Edmonds 1973a,b). The function of mRNA poly(A) could only be guessed at in these initial studies. However, it was certainly very handy as a natural tag to allow isolation of mRNA by oligo(dT) affinity chromatography away from bulk ribosomal RNA that lacks poly(A) segments (Aviv and Leder 1972). These early experiments predated recombinant DNA technology. The only way to isolate individual mRNAs was to select tissue that had pronounced and selective gene expression so that particular mRNA is unusually abundant. Thus, globin mRNA was purified from mammalian red blood cells (Mathews et al. 1971), ovalbumin mRNA was purified from chicken oviduct cells (Rosen et al. 1975), and immunoglobulin mRNA was purified from murine B cells (Brownlee et al. 1973). My own research studies as a graduate student at Cambridge, UK, in the mid-1970s with George Brownlee and Fred Sanger began by using Escherichia coli DNA polymerase to partially reverse-transcribe mRNA (retroviral reverse transcriptase had yet to be purified). This enzyme inefficiently uses an RNA template when Mg2+ is replaced by Mn2+ in the reaction mix. Short stretches of complementary DNA (cDNA) were synthesized by use of oligo(dT) priming on the poly(A) tails of mRNA. In effect, these experiments were among the very first described examples of cDNA synthesis (Proudfoot 1976). I next used a now seemingly primitive, although quite effective, DNA sequencing technique developed in the Sanger laboratory (Brownlee and Sanger 1969; Sanger et al. 1973; Galibert et al. 1974) that involved two-dimensional (2D) chromatographic “fingerprinting.” By this and other nucleic acid analytic techniques (Fig. 1), I was able to piece together six separate mRNA sequences adjacent to the poly(A) tail and showed that each mRNA possessed the common sequence AAUAAA placed close (within 20–30 nucleotides [nt]) to the 3′-terminal poly(A) tail (Fig. 2A). We predicted from this small but then complete set of purified mRNA that AAUAAA was a signal for mRNA polyadenylation as well as a signal to terminate transcription (Proudfoot and Brownlee 1976). As described below, both of these predictions have turned out to be correct. What these early mRNA sequencing experiments also revealed was the fact that the stop codon of mRNA did not define the mRNA 3′ end. Rather, a 3′ ncRNA sequence, subsequently called the 3′ UTR, existed that at its 3′ end possessed the gene's PAS (Proudfoot and Longley 1976).
Figure 1.
(A) Schematic of cDNA synthesis by E. coli DNA polymerase (Klenow subfragment that lacks the 5′–3′exonuclease domain) using oligo(dT)12 as primer (in red), base-paired to mRNA 3′ poly(A) in a reaction mix containing MnCl2 in place of MgCl2 and four deoxyribonucleotides, one α32P-labeled. Short cDNAs produced by this process correspond to the 3′ end of the mRNA 3′ UTR. Purified cDNA was depurinated by formic acid treatment (selectively degrades G and A) releasing oligopyrimidies, especially oligo(dT) copied from the poly(A) sequence of the mRNA. Alternatively cDNA was digested with partially C-specific endonuclease IV, and the endonuclease IV digestion products were then purified and further degraded by partial digestion with venom 5′–3′ exonuclease (Proudfoot 1976). (B) cDNA depurination products were fractionated by denaturing polyacrylamide gel electrophoresis. A series of oligo(dT) products are visible, ranging from dT13–dT50, and correspond to oligo(dT)-primed cDNA generated by oligo(dT), which is base-paired at positions ranging from the 5′ to 3′ ends of the poly(A) tail (known to be about A50 for globin mRNA). Marker (M) and sample (S) lanes are shown, with the positions of oligo(dT)12 and unincorporated α32P dNTP also indicated. (C) 2D fractionation of cDNA endonuclease IV products. The first dimension was by electrophoresis on cellulose acetate strips at pH 3.5. Base composition of oligonucleotide determines mobility. A cellulose acetate strip with fractionated oligonucleotides was blotted onto the bottom of a DEAE cellulose acetate, thin-layer plate using high-salt buffer. Homochromatography was then performed (second dimension) using a buffer (called a homomix) containing partially degraded crude yeast RNA that acts to displace endonuclease IV oligonucleotides from the base of the thin-layer plate. The extent of this displacement depends on molecular size (Brownlee and Sanger 1969). Separated 32P oligonucleotides were then eluted from the thin-layer plate. The first and second dimensions are indicated by arrows. (D) 2D fractionation (as in C) of partial venom exonuclease digested spot 9. A series of products are evident, with adjacent ones varying by 1 nt. The angle of the 2D shift between adjacent spots is characteristic of a specific nucleotide loss. Purine nucleotides (G and A) give a larger shift in the second dimension. T and G give large and small rightward shifts (respectively), while C and A both give small leftward shifts in the first dimension. The sequence TTATT was deduced from observed mobility shifts and corresponds to the PAS AAUAAA. The first T of the PAS cDNA was inferred from depurination data (not shown). (B) Bromophenol blue marker. (E) Separated oligonucleotide sequences were aligned, as some sequences overlap due to partial endonuclease IV digestion. This gave the sequence of the β-globin cDNA adjacent to the poly(A) tail. Endonuclease IV spots not in the β-globin cDNA sequence were inferred to derive from the α-globin cDNA (Proudfoot 1976). († and ‡) Related oligonucleotides from varying 5′ and 3′ ends of cDNA.
Figure 2.
(A) Sequence alignment of the original six mRNA 3′ ends derived from sequencing technology as outlined in Figure 1. The positions of the conserved AAUAAA signals (boxed) and 3′-terminal nucleotides (underlined) originally noted to be conserved are indicated. (Proudfoot and Brownlee 1976). (B) Current general consensus sequences for the mammalian poly(A) signal. Distance variation between different parts of the PAS is indicated. (Red thunderbolt) Cleavage position.
Soon after these early cDNA sequencing experiments, the recombinant DNA era began, and coincidently Fred Sanger developed much more powerful gel-based DNA sequencing techniques (Sanger et al. 1977). Many more mRNAs were cloned and characterized from so-called libraries of cDNA plasmids that were made from various RNA preparations derived from tissues or cell culture sources of different eukaryotes (Maniatis et al. 1976, 1982). It was generally shown to be the case that mRNAs possess 3′ UTRs varying in length from ∼50 nt to several thousand nucleotides with a 3′ proximal AAUAAA PAS or close variant (AU/GUAAA or UAUAAA) (Wickens and Stephenson 1984; Zhao et al. 1999). Recombinant DNA technology also led to site-directed mutagenesis approaches. Thus, mutation analysis of the SV40 late PAS by limited exonuclease degradation from a closely positioned restriction site showed that AAUAAA did indeed form a required part of the mRNA PAS (Fitzgerald and Shenk 1981). Two rare forms of thalassaemia in humans were then characterized with point mutations in the AAUAAA sequence of the mutant gene (AAUAAG in the α2-globin gene and AACAAA in the β-globin gene). In both cases, subcloning of the mutant PAS revealed that it lost its poly(A) formation function (Higgs et al. 1983; Orkin et al. 1985).
Although the AAUAAA sequence was shown to be absolutely required for mRNA 3′-end polyadenylation, other sequence elements were shown to be necessary to fully reconstitute a functional PAS. In particular, the GU-rich sequence (or downstream sequence element, DSE) present just past the mRNA 3′ end in the immediate gene 3′ flanking region was shown to enhance 3′-end formation (Gil and Proudfoot 1984, 1987; McLauchlan et al. 1985). Similarly, the sequence immediately upstream of AAUAAA (upstream sequence element, USE) (Carswell and Alwine 1989; DeZazzo et al. 1991; Valsamakis et al. 1991; Moreira et al. 1995; Brackenridge and Proudfoot 2000; Venkataraman et al. 2005; Danckwardt et al. 2007) can in some cases act as an enhancing element for 3′-end processing efficiency. Finally, the actual nucleotides at the site of 3′-end cleavage can also influence the efficiency of this process (Chen et al. 1995). This proved to be medically relevant for the human prothrombin gene, which normally carries a CG dinucleotide sequence just 5′ to the cleavage position. In ∼1% of Caucasians (myself included), this sequence is mutated to CA, causing a mild thrombophilia phenotype. This is due to a twofold increase in prothrombin gene expression as a result of the acquisition of a more efficient PAS (Gehring et al. 2001; Danckwardt et al. 2008). These defining experiments for mRNA PAS were focused on mammalian mRNA. However, other eukaryotic model systems revealed that the mammalian pattern of USE-AAUAAA-DSE (Fig. 2B) was generally conserved across eukaryotes, even though in budding yeast greater variation in these cis elements appears to be tolerated (Graber et al. 1999). One notable feature of PAS is that, depending on exactly how the AAUAAA and DSE signals are positioned and defined, the actual site of poly(A) addition can vary by several to tens of nucleotides (Zhang et al. 1986; Sheets et al. 1990; Tian et al. 2005). This is especially evident in yeast PAS, which, perhaps due to lower sequence conservation, do not accurately define the position of polyadenylation (Zhao et al. 1999). This is in marked contrast to pre-mRNA splicing, which must always occur at a precise nucleotide position to maintain the correct reading frame of the mRNA for translation.
Genomic analysis of poly(A) sites has strikingly illustrated the generality of the originally laboriously defined eukaryotic PAS. Initially, annotation of genes within complete genome sequences proved hard, as the highly intronic nature of higher eukaryotic genes meant that even defining genes based on their protein-coding capacity was difficult, let alone exactly where the gene transcript starts or stops. The standard approach has been to match transcriptomes to genomes by hybridizing oligo(dT)-primed cDNA from various RNA preparations to genomic microarrays, possible in smaller yeast genomes but still technically challenging for larger mammalian genomes. From these data, many potential mRNA poly(A) sites were assigned (Nam et al. 2002; Brockman et al. 2005). While a good number corresponded to canonical poly(A) sites as defined by the earlier, above-described mutagenesis experiments, many more potential poly(A) sites were apparently devoid of the expected RNA signals. However, several artifacts crept into these bioinformatic analyses. Firstly, oligo(dT) priming on RNA fractions can frequently occur on internal oligo(A) sequences (Nam et al. 2002), especially a problem for genomes such as Caenorhabditis elegans, which possess relatively A-rich nongenic sequence (Jan et al. 2011). Secondly, alternative poly(A) polymerases have been identified in most eukaryotes (Trf4 and Trf5 in Saccharomyces cerevisiae) that cooperate with the nuclear RNA degradation apparatus to promote degradation of unwanted or misprocessed transcripts (Schmid and Jensen 2008). RNA degradation-associated oligo(A) tailing (West et al. 2006; Slomovic et al. 2010) can similarly generate oligo(dT)-primed cDNA that is unrelated to authentic mRNA 3′-terminal poly(A) tails. Finally, reverse transcriptase can, in some cases, “misbehave” by template switching from the original RNA template to the newly synthesized cDNA (Houseley and Tollervey 2010). Consequently, antisense transcription, often a widely attributed feature of transcriptomes, may be less common than many studies have predicted. Such template switching can also result in the apparent fusion of separate RNA templates and may be miscategorized as trans splicing.
Recently, various genomic approaches using massive parallel DNA sequencing technology have been applied that avoid some or all of the above pitfalls. For example, an analysis of mRNA 3′ ends in C. elegans used a clever trick of ligating a biotin-tagged RNA:DNA duplex “splint” onto the 3′ end of poly(A) tails (Jan et al. 2011). The DNA component has a 3′-terminal oligo(dT) overhanging sequence that guides the complementary biotinylated RNA up against the poly(A) 3′ end. Following RNA ligation, partial RNase digestion, biotin selection, and reverse transcription with dTTP and RNase H digestion to remove the mRNA poly(A), authentic mRNA 3′-end fragments lacking their poly(A) tail were then amplified and sequenced. Using this clever “splint” 3′-end sequencing procedure, many new examples of alternate poly(A) sites were defined for C. elegans mRNA. Also, as predicted, a lot of mRNA 3′ ends were shown to be misassigned, presumably derived from priming of oligo(dT) on internal A-rich sequences prevalent in the worm genome. Another approach applied to both yeast and mammalian mRNA is to sequence the nucleotides adjacent to the poly(A) tail by direct sequencing using an approach not dissimilar to my original analysis of the mRNA PAS. Essentially, a bacterial DNA polymerase is used to reverse-transcribe the mRNA poly(A) tail using dTTP. Then, fluorescent, chain terminator-modified (VT) nucleotides (their chemistry is proprietary and therefore hard to fully comprehend) are added one by one, complementary to the mRNA sequence adjacent to the oligo(dT) copy of the poly(A) tail. This process is performed on a matrix so that millions of single mRNA 3′-end sequences can be simultaneously read for up to ∼50 nt from the poly(A) tail of the mRNA population (Ozsolak et al. 2009). By these massive sequencing procedures, the original sequence consensus for the mammalian PAS has been shown to be truly general (Ozsolak et al. 2010). Where deviations from the AAUAAA sequence occur, these can be ascribed to a weaker PAS that may have a particular regulatory purpose. Also VT nucleotide-derived massive sequencing identified new sequence motifs associated with polyadenylated ncRNA, especially a U9 sequence motif just upstream of the polyadenylated 3′ end. It remains to be established by mutagenesis whether this U9 element is actually required for RNA 3′-end formation of particular ncRNA classes.
mRNA 3′-end processing: connections with transcriptional termination
A milestone in our understanding of mRNA 3′-end processing (cleavage and polyadenylation) came from biochemical characterization of the process in mammals, pioneered by the laboratories of J.L. Manley and W. Keller. Fractionation of HeLa cell nuclear extract resulted in the purification of two protein complexes: cleavage and polyadenylation specificity factor (CPSF) and cleavage stimulatory factor (CstF). These proteins together recognize AAUAAA and GU-rich DSEs on synthetic RNA substrates and act to promote RNA cleavage between these cis elements. Furthermore, poly(A) polymerase was shown to be recruited to this 3′ processing complex, resulting in polyadenylation of the 3′ end of the 5′ cleavage product. Hence, mRNA 3′-end processing was successfully reconstituted in a biochemically tractable in vitro system. Other factors were also shown to be required for in vitro 3′-end processing, such as cleavage factors I and II (CFI/II). For detailed accounts and a literature survey on this 3′-end processing mechanism, see many comprehensive reviews (Colgan and Manley 1997; Zhao et al. 1999; Edmonds 2002; Mandel et al. 2008; Millevoi and Vagner 2010). Slightly later, budding yeast biochemical characterization of mRNA 3′-end processing was achieved. This was greatly aided by genetic screens for 3′-end processing that allowed the identification of many 3′-end processing factors, some homologous to mammalian factors and others apparently unique to yeast. Whole-cell extracts isolated from yeast strains lacking particular factors (through growth of temperature-sensitive mutants at restrictive temperature) proved valuable in pinning down particular 3′-end processing functions. Adding back recombinant factors lacking particular protein domains further uncovered the surprising complexity of this process (Zhao et al. 1999; Mandel et al. 2008). Why upward of 50 polypeptides divided between multiple subcomplexes are required to simply cleave the pre-mRNA 3′ end and then couple poly(A) addition remains an enigma to this day. However, the pivotal role of 3′-end polyadenylation in gene expression is well emphasized by the fact that most 3′-end processing factors are encoded by essential genes. Mutation of any such gene is invariably lethal to the organism.
In more recent years, biochemists and structural biologists have got their teeth into the molecular characterization of many of the components of the mRNA 3′-end processing reaction, and details of these studies are reviewed elsewhere (e.g., Mandel et al. 2008). However, a key feature of this reaction is the actual endoribonuclease activity itself, and I outline here the experiments that have led to our current state of understanding of this process. It was originally presumed that the 3′ ends of mRNA are generated by direct transcriptional termination by RNA polymerase II (Pol II) rather than an RNA endonucleolytic cleavage reaction. Such a simple mechanism is well known to occur for prokaryotic polycistronic mRNA and also eukaryotic RNA polymerase III (Pol III) (Richard and Manley 2009). However, the fact that in vitro synthesized RNA, spanning a PAS, could be demonstrably cleaved and polyadenylated in vitro, which was first shown with mammalian nuclear extracts (Moore and Sharp 1985) and subsequently with yeast whole-cell extracts (Butler and Platt 1988), proved that mRNA 3′-end formation and termination were separate, albeit connected, molecular events. Critically, the cleavage and polyadenylation steps could be biochemically separated. For instance, blocking poly(A) polymerase activity by use of ATP inhibitors such as cordycepin (3′ deoxy ATP) or depletion of essential Mg2+ by EDTA treatment allows the visualization of the 3′ cleaved product without poly(A) addition. Similarly, S. cerevisiae temperature-sensitive mutants in PAP1 [encoding poly(A) polymerase] still generate 3′-end-cleaved RNAs (Butler and Platt 1988; Zhao et al. 1999).
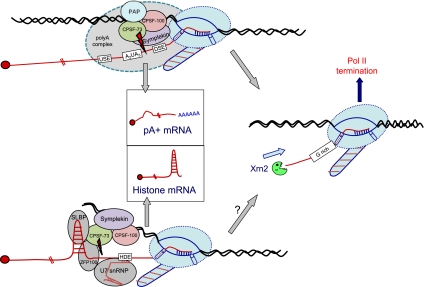
A natural example of cleavage-only mRNA 3′-end formation is found with replication-dependent histone mRNAs (Gick et al. 1986). These mRNAs are formed by the recognition (through direct base-pairing) of a purine-rich sequence downstream from the conserved 3′-terminal hairpin by the small nuclear RNA (snRNA) U7 (Schaufele et al. 1986; Schumperli 1988). This RNA, in association with multiple protein components (forming U7snRNP), is recruited to histone mRNA 3′ ends by interaction with additional factors, including a zinc finger protein and hairpin-binding protein (Marzluff et al. 2008). Additionally, specific components of the cleavage/poly(A) complex are also recruited: CPSF-73, CPSF-100, and a scaffold-like protein, Symplekin (Kolev and Steitz 2005; Sullivan et al. 2009). Importantly, CPSF-73 has a particular metallo β lactamase domain (MLD) with the hallmarks of endoribonuclease activity (Dominski 2007). Both for PAS and histone 3′-end processing, CPSF-73 was shown to cross-link to the exact nucleotide that defines the mRNA 3′ end (Ryan et al. 2004; Dominski et al. 2005), and, at least for histone mRNA, CPSF-73 mediates direct and specific RNA 3′-end cleavage (Kolev et al. 2008). Curiously, CPSF-100 also possesses a clear MLD, although with inactivating amino acid replacements in its active site. From these comparative studies between histone and poly(A)+ mRNA 3′-end formation, it is clear that CPSF-73, aided and abetted by other factors generally specific to the two mRNA classes, mediates the enigmatic 3′ processing reaction to form the authentic mRNA 3′ ends, as shown in Figure 3 (Mandel et al. 2006, 2008).
Figure 3.
Diagram comparing the mechanism of cotranscriptional 3′-end formation of mammalian polyadenylated mRNA versus unpolyadenylated histone mRNA (RNA is in red, and DNA is in black). For poly(A)+ mRNA, only factors conserved with histone pre-mRNA processing are shown. The rest of the poly(A) complex is depicted by a large grey shadow. Pol II (in light blue) is depicted with striped CTD, and the position of the ssDNA bubble is depicted with associated nascent RNA. Histone 3′ processing factors are indicated. The cap at the 5′ end of mRNA is depicted as a red ball. The histone mRNA 3′ hairpin is shown, as is the U7 RNA hairpin and the interaction with the histone downstream element (HDE). Positions of cis RNA 3′-end processing sequence elements are shown in white boxes. mRNA products of poly(A)+ and histone pre-mRNA 3′-end processing are shown (in boxes), as is the subsequent coupled termination of Pol II by Xrn2-mediated torpedo effects [established for poly(A)+ genes, but only inferred for histone genes].
The above-defined RNA processing reaction effectively separates mRNA 3′-end processing from actual Pol II transcriptional termination. Even so, it is long established that 3′-end processing is absolutely required for termination, raising interesting mechanistic questions about how these two processes are connected. Early on when studying the mutated PAS of the α2-globin gene, which is associated with α-thalassaemia, we showed that not only is the 3′ processing mechanism disrupted, but so too is termination (Whitelaw and Proudfoot 1986). In this mutant α2-globin allele, Pol II reads past the normal termination site (∼600 nt into the 3′ flanking region) and actually perturbs the activity of the downstream α1-globin gene by a transcriptional interference process, thus reducing its expression. Soon afterward, the mouse β-globin and SV40 early PAS were also shown to dictate Pol II termination (Logan et al. 1987; Connelly and Manley 1988). These results provided the first evidence that pre-mRNA processing is coupled to transcription. Such coupling has subsequently been shown to be a general feature of all pre-mRNA processing reactions (especially pre-mRNA capping and splicing) (Proudfoot et al. 2002).
The next stage in these experiments was to reveal that the unique heptad repeated C-terminal domain (CTD) of the Pol II large subunit Rpb1 acts to directly recruit cleavage/poly(A) factors to the elongating Pol II complex (McCracken et al. 1997; Zhao et al. 1999). Indeed, in vitro 3′ processing reactions revealed that Pol II CTD significantly enhances this reaction, leading to the view that Pol II CTD itself acts as a component of the 3′ processing machinery (Hirose and Manley 1998). Some cleavage/poly(A) factors, as well as mediating 3′-end processing, also aid subsequent Pol II termination (Birse et al. 1998). A clear example is Pcf11, a component of CFII in mammals and CF1A in yeast. This factor has a CTD-interacting domain (CID) with a preference for CTD Ser 2 phosphorylation, a feature of elongating Pol II (Licatalosi et al. 2002; Meinhart and Cramer 2004). Interestingly, a further 5′–3′ exonucleolytic RNA processing reaction is facilitated that acts to degrade the uncapped residual RNA still attached to elongating polymerase. In yeast, this exonuclease, called Rat1, is part of a complex (comprising Rai1 and Rtt103) that also possesses a CID, presumably explaining how this final RNA processing reaction is also transcriptionally coupled (Kim et al. 2004). In mammals, the Rat1 homolog Xrn2 similarly acts to degrade Pol II-associated RNA (West et al. 2004). In both systems, it is thought that the exonuclease is in kinetic competition with elongating Pol II so that when/if RNA degradation catches up with the elongating Pol II, this will promote termination by inducing conformational changes in the Pol II active site through loss of its associated nascent RNA (Connelly and Manley 1988; Proudfoot 1989; Kuehner et al. 2011). This mechanism is called the torpedo model, and likely acts in consort with Pol II-recruited cleavage/poly(A) factors to promote efficient termination at a distinct 3′ flanking region location, downstream from the gene, poly(A) site (Fig. 3; Richard and Manley 2009; Kuehner et al. 2011).
Two additional classes of termination elements can act to enhance the termination process. One class acts as a Pol II transcription pause site and, by slowing down elongation, allows the exonuclease more time to degrade the nascent RNA and so reach the Pol II complex (Plant et al. 2005; Gromak et al. 2006). These elements may be G-rich in sequence, and recent results show that such G-rich elements are associated with the formation of RNA:DNA hybrids. Indeed, resolution of these hybrids requires the action of dedicated helicases: Sen1 in yeast, and senataxin in mammals (Mischo et al. 2011; Skourti-Stathaki et al. 2011). The other class of terminator elements may occur at more distal 3′ flanking regions and is referred to as a cotranscriptional cleavage (CoTC) sequence (Dye and Proudfoot 2001). CoTC termination may operate when PAS-proximal pause sites are lacking, so that Pol II effectively escapes further into the 3′ flanking region. As the name implies, CoTC RNA sequences are highly unstable, so that as soon as they emanate from the Pol II RNA exit channel, RNA cleavage occurs (West et al. 2008). This will allow Xrn2 to directly degrade the nascent transcript at this rapidly formed uncapped RNA 5′ end. Since this occurs close to the still elongating Pol II, termination quickly ensues. Indeed, in this type of termination, Pol II is released from the chromatin template before cleavage at the PAS occurs. So, in this instance, 3′-end processing occurs post-transcriptionally, but still in association with Pol II. Interestingly, CoTC-mediated termination can greatly increase the yield of mRNA from a gene, as 3′-end processing can occur in the nucleoplasm away from the nuclear RNA degradation apparatus, which appears to be chromatin-associated (West and Proudfoot 2009). As discussed below, defining the actual mRNA 3′ end is a complex interplay between poly(A) site recognition and associated Pol II termination (Fig. 3).
Alternative PAS (APA) define different mRNA 3′ UTRs
The characterization of particular eukaryotic mRNAs has often relied on the long-established Northern blotting technique, which provides a gel fractionation image of a specific gene's mRNA output, giving information relating to both mRNA size and quantity. This technique—unlike its modern replacement of quantitative (real-time) PCR-amplified cDNA (qRT–PCR)—allows visualization of the complete set of mRNA isoforms generated from a particular gene. Smaller genes often yield only one specific mRNA, which is the norm for simpler eukaryotes such as yeast. However, in higher eukaryotes, especially mammals, most of these larger and more complex genes (with multiple exons) generate multiple mRNA isoforms. These are frequently caused by complex alternative splicing patterns. Alternative splicing is now appreciated to regulate both the nature and complexity of mammalian proteomes and, as such, reflects a key aspect of the regulation of gene expression (Black 2003; Johnson et al. 2003; Wang et al. 2008; Chen and Manley 2009). However, a significant part of mRNA size variation derives not from alternative splicing, but rather from alternative PAS selection. Thus, it is calculated that well over half of all mRNAs have variable PAS selection, meaning that they will possess mRNA isoforms differing by the extent of their 3′ UTRs (Edwalds-Gilbert et al. 1997; Tian et al. 2005). Since mRNA 3′-end processing occurs cotranscriptionally and is stimulated by Pol II CTD (Proudfoot 2004), it is clear that once a particular PAS has been selected and mRNA 3′ cleavage occurs with consequent release from chromatin-associated Pol II, then further cleavage of more proximal PAS on the mRNA will not occur. Thus, mRNAs with extended 3′ UTRs are carried through into the cytoplasm, where particular 3′ UTR sequences act to regulate both the stability and translatability of mRNA as described below (for a recent review on APA, see Lutz and Moreira 2011).
The occurrence of alternative PAS selection for mRNA was initially considered to reflect a relatively random process. The failure of one PAS to fully end the mRNA resulted in further downstream cryptic PAS acting to end the rest of the transcripts. Thus, the insertion of multiple, identical PAS at the 3′ end of artificial gene constructs results in all of the PAS working to some degree, yielding mRNA with different-length 3′ UTRs (Denome and Cole 1988). In general, when the PAS is relatively strong (possessing both a canonical AAUAAA and clearly defined USE and/or DSE), then the first PAS dominates, with increasingly reduced usage of downstream PAS. However, with PAS lacking full consensus signals, a more even spread of PAS recognition is evident. However, it is generally the case that the earlier PAS in a series of multiple PAS is used more efficiently, implying a first-come, first-served pattern. Presumably, CTD-bound poly(A) factors will be sequestered onto earlier PAS, excluding the later usage of these factors on subsequent PAS. Another arrangement of PAS tested artificially was to have a weak PAS followed by a strong one. In this situation, the downstream PAS is selectively used. This type of duplicated PAS arrangement allowed the identification of transcription pause sites. When such elements are placed between a weak followed by a strong PAS, higher usage of the upstream weak PAS is promoted. This assay is referred to as a PAS competition assay (Ashfield et al. 1991). With the realization that endonucleolytic cleavage at a poly(A) site allows entry of the exonuclease torpedo (Rat1 or Xrn2) to promote Pol II termination (Kim et al. 2004; West et al. 2004), it is plausible that alternative PAS usage will result in equivalent alternative Pol II termination.
Bioinformatic analysis of PAS usage in higher eukaryotes has revealed the remarkable fact that well over 50% of genes display APA site usage (Tian et al. 2005). In general, where APA is clearly evident, then the downstream PAS appear to have sequence features that more closely match the canonical AAUAAA and DSE sequence elements (Legendre and Gautheret 2003). This sequence specificity is predicted from the above considerations that distal PAS will only be used if they can effectively outcompete proximal PAS. However, the relative usage of tandem PAS is invariably measured by levels of steady-state mRNA possessing particular lengths of 3′ UTRs. Consequently, the relative stability of these different mRNA isoforms will also affect the apparent usage of PAS (Moore 2005).
Several independent studies have revealed the remarkable fact that either levels of cell proliferation or developmental stage can cause a shift in APA from more distal to more proximal PAS selection (Sandberg et al. 2008; Ji et al. 2009; Mayr and Bartel 2009). In particular, rapidly dividing cells, as are often found in cancerous tissues, tend to use proximal PAS, while cells in later developmental stages tend to use more distal PAS. These striking results clearly correlate with the regulation of mRNA stability and translation by microRNAs. Thus, mRNAs possessing longer 3′ UTRs caused by distal PAS selection will have more potential microRNA-binding sites. This view is clearly confirmed by bioinformatics analyses. Even so, recent more comprehensive genomic analysis indicates that the relationship between 3′ UTR length and proliferation stage may be more complex than originally thought (Fu et al. 2011).
The molecular basis of how APA may be differentially regulated remains largely unknown. In general, cleavage/poly(A) factors appear to be constitutively expressed, and little evidence exists for the selective use of factors for one PAS versus another. However, it has been observed that some cleavage/poly(A) factors may be present at lower, limiting levels in some cells, such as CstF-64 in pre-B cells (Takagaki and Manley 1998; Ji et al. 2009). In this situation, stronger PAS will have a significant kinetic advantage (see below). In the case of alternative splicing, regulation of this RNA processing mechanism is often achieved by the enhancement of weak splice sites by so-called splicing enhancers (often in adjacent exons) that are recognized by splicing regulatory factors (usually SR proteins) (Black 2003). It seems plausible that a weaker PAS—particularly proximal PAS—may similarly be enhanced by PAS enhancers, even though evidence for such elements and their associated factors is still lacking.
One mechanism that may regulate APA is through more direct effects of Pol II transcription. Gene promoters may play a role in recruiting factors that subsequently enhance PAS recognition. Thus, CPSF has been shown to associate with the general transcription factor TFIID at gene promoters. Subsequent transfer of CPSF to the CTD may promote 3′-end processing (Dantonel et al. 1997). Communication between the promoter and terminator through gene loop formation (O'Sullivan et al. 2004; Perkins et al. 2008) may also afford efficient transfer of cleavage/poly(A) factors from the 3′ end of the gene back to new Pol II initiation complexes (Glover-Cutter et al. 2008; Mapendano et al. 2010). Also, very recently, specific transcription activators have been shown to enhance 3′-end processing through recruitment of a specific elongation factor complex (PAF1c) that, in turn, enhances PAS recognition (Nagaike et al. 2011). Whether these promoter effects on 3′-end processing factor recruitment play a role in APA remains to be established.
Gene promoters can also determine transcription elongation rates by setting up more or less processive Pol II elongation complexes (Cramer et al. 1999). Alternatively, as mentioned above, in the context of Pol II termination, it seems plausible that specific pause sites positioned within genes may be more or less active in a different cellular context. These could cause localized changes in elongation rate that might favor the use of upstream splice sites or PAS (Roberts et al. 1998; Gromak et al. 2006). In the case of alternative splicing, it is now well known that Pol II processivity set up by specific promoters, by modification of Pol II activity (such as UV-induced hyperphosphorylation of CTD), or by use of an artificial mutant (so-called slow Pol II) can influence alternative splicing patterns (de la Mata et al. 2003; Munoz et al. 2009). Similarly, loss of elongation factors in yeast has been shown to correlate with increased usage of upstream, cryptic PAS present within genes (Cui and Denis 2003). Very recently, similar mechanisms have been shown to exist for Drosophila APA. In the case of the gene Polo, with its two well-defined PAS, a weak proximal and stronger distal PAS are regulated such that the distal PAS is required for higher levels of Polo gene expression. Interestingly, flies expressing a slow Pol II mutant show a clear shift to usage of the proximal Polo PAS as well as several other tested examples of fly APA (Pinto et al. 2011).
Much remains to be learned about APA in eukaryotes. However, it is clear that this is a key regulatory process in eukaryotic gene expression. The ability of APA to generate mRNA with different 3′ UTRs that contain different regulatory cis elements represents a potentially major form of gene regulation. Such regulatory elements may act as targets for microRNAs that regulate mRNA stability or translation (Bartel 2009). Alternatively, they may act as mRNA stability or instability elements recognized by RNA-binding factors such as HuR or TPP. Finally, 3′ UTRs have been shown to contain complex RNA signals (with particular RNA secondary structures) for factors that mediate specific cytoplasmic localization during early development (St Johnston 2005; Lutz and Moreira 2011).
Interplay between poly(A) site selection and splicing
Pre-mRNA processing acts in a highly coordinated manner during transcription. Consequently, mRNA 3′-end processing is closely coordinated with splicing. In particular, the relatively low sequence complexity and redundancy of the PAS argues that inappropriate, premature poly(A) site selection must be prevented. Otherwise, incorrectly shortened mRNAs would form that could translate into truncated proteins with dominant-negative effects on cellular function.
One such example of these interconnections relates to the phenomenon of terminal exon definition. It has been shown that recognition of the 3′ splice site (3′SS) of a gene's last intron strongly enhances the efficiency of the downstream PAS (Niwa and Berget 1991; Niwa et al. 1992; Dye and Proudfoot 1999). In particular, the 3′SS-associated factor U2AF was shown to enhance PAS function by direct molecular contacts with poly(A) polymerase (Vagner et al. 2000). Similarly, protein components of U2snRNP that associate with the 3′SS and nearby lariat branch point help enhance downstream 3′-end processing through interactions with CPSF (Kyburz et al. 2006). This enhancement of terminal exon definition has several other consequences, as PAS recognition also enhances terminal intron splicing, and, furthermore, 3′SS recognition is required for Pol II termination, as this in turn depends on PAS recognition. In eukaryotic genes that lack introns, PAS recognition may require additional as-yet-undetermined selection mechanisms. Interestingly, for mammalian intronless genes, different PAS elements may operate, containing extra DSE (Dalziel et al. 2007; Nunes et al. 2010).
Another situation in which splicing and 3′-end processing must interact is in the few characterized cases in which APA results in the formation of mRNAs with different terminal coding sequences. For the immunoglobulin antibody heavy chain gene, alternative membrane-bound or secreted protein isoforms differ by the presence or absence of a specific hydrophobic C terminus required for membrane retention. An intron-located PAS, if used, must outcompete with the upstream 5′SS, which results in the shorter secreted antibody form found in mature B cells. Alternatively, in pre-B cells, splicing wins so that the intronic PAS is suppressed, and instead a downstream PAS is used with additional terminal exons included in the heavy chain mRNA. This process is regulated by levels of CStF-64, which are more limited in pre-B cells than in mature B cells (Takagaki et al. 1996; Takagaki and Manley 1998). Also, Pol II elongation factors may act to modulate this PAS switch (Martincic et al. 2009). A similar type of APA regulation exists for the calcitonin gene. Here, again, the selective recognition of an intronic PAS results in the formation of a truncated mRNA encoding calcitonin gene-related peptide (CGRP) rather than full-sized calcitonin (Amara et al. 1982; Zhao et al. 1999). Interestingly, genome-wide analysis of this type of APA suggests that the regulated formation of mRNAs encoding different C-terminal protein sequences may be quite widespread (Tian et al. 2005; Wang et al. 2008).
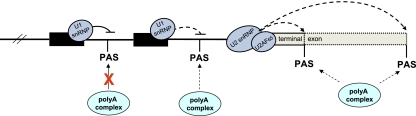
Finally, it has become apparent that PAS commonly present within the body of genes, especially in the much longer introns of higher eukaryotes, must be tightly repressed. Interestingly, this is achieved by the dominant role of the 5′SS and its recognition by U1snRNP, which act to block adjacent PAS recognition. Again, this phenomenon was first described in gene-specific studies (Levitt et al. 1989; Zhao et al. 1999) but has now achieved genome-wide status (Kaida et al. 2010). Early examples of such regulation were found in mammalian viruses, which often need to maximize their gene expression output by selective use of PAS. In bovine papilloma virus, viral gene expression programs require that a particular PAS is only used in late stages of viral infection (Furth et al. 1994). Interestingly, this late PAS was shown to be blocked from use in earlier stages of infection by the presence of a closely positioned, upstream 5′SS. Recognition of this 5′SS by U1snRNP blocks poly(A) polymerase activity at the late PAS by direct interaction with the 70K protein component of U1snRNP (Gunderson et al. 1998). This ability of closely placed 5′SS to repress PAS function has been cleverly manipulated experimentally to allow the specific repression of any gene. Thus, U1snRNA with its 5′ end (that normally binds 5′SS) modified to base-pair with sequence upstream of a particular PAS is expressed in mammalian cells resulting in inhibition of the target gene by blocking PAS function (Fortes et al. 2003). HIV-1 provirus also uses PAS regulation to maximize its gene expression. Like other retroviruses, its duplicated 5′ and 3′ long terminal repeats (LTR) possess identical PAS. While 3′ LTR PAS usage is essential for viral gene expression, use of the 5′ LTR PAS would preclude viral gene expression. Interestingly, cleavage at the 5′ LTR PAS was shown to be repressed by a major viral 5′SS positioned close by, ∼200 base pairs (bp) into the proviral gene sequence. Point mutation of this 5′SS activates the 5′ LTR PAS, causing the provirus to exclusively produce very short 5′ LTR-specific transcripts (Ashe et al. 1995, 1997).
These viral examples of PAS regulation by 5′SS repression have now been generalized to the whole genome by a recent study that tested the effect of blocking U1snRNA by antisense RNA analogs (morpholino) that were electroporated into mammalian cells (Kaida et al. 2010). In addition to blocking the splicing of many gene introns and causing the accumulation of unspliced mRNAs, truncated mRNAs were widely observed caused by the activation of intronic PAS. In other words, these studies imply a second major role of U1snRNA beyond its critical function in 5′SS recognition as a prelude to splicing. This is in the blockage of intronic PAS as first described for viruses. Indeed, a further variation of the mechanism of intronic PAS regulation is found in C. elegans, where cleavage of intronic PAS triggers the process of trans splicing by the small RNA leader sequence SL2 (Blumenthal 2005; Haenni et al. 2009). Overall, it is abundantly clear that PAS recognition is a widely regulated process that dictates multiple regulatory features of eukaryotic genes (Fig. 4).
Figure 4.
Diagram summarizing the selective use of PAS along a Pol II transcribed gene. PAS in introns are recognized by the poly(A) complex, but this is modulated or blocked by U1snRNP binding to adjacent 5′SS. Recognition of PAS at the end of genes is enhanced by splicing factors bound to the terminal intron 3′SS. Different 3′-terminal PAS are competitively and mutually exclusively used, resulting in APA.
Conclusions
This review aims to put current advances in our understanding of how the 3′ ends of mRNAs are selected into historical perspective. This field has accumulated massive detail since the genome-wide era began at the beginning of the second millennium. However, it is striking that many of the mechanistic principles behind PAS selection were already in place long before this time. Indeed, these earlier studies provided actual experimental evidence for the role of specific sequences by measuring the effect of their mutation on biological function. It is abundantly clear that bioinformatic analysis of genomic data has provided invaluable generality to our understanding of PAS function in gene expression. However, current genome-wide analyses often only provide bioinformatic correlations and lack direct functional experimentation. Genomic analysis will only achieve its full potential when bioinformatics can be matched by hypothesis-driven experimental approaches. In spite of the above concerns, it is the case that where and how to end the eukaryotic message is a central regulatory point in the elaborate process of gene expression.
Acknowledgments
I thank my laboratory colleagues past and present for their science and friendship over many years. I also am indebted to the Wellcome Trust for their long-term Programme grant support of my laboratory. I thank Natasha Gromak for help with the figures and Andre Furger and Mick Dye for advice on the manuscript text. Finally, I acknowledge and thank the many laboratories from around the world who work in this research field and have helped it develop into its present fascinating level of complexity, yet with some understanding.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.17268411.
References
- Adesnik M, Salditt M, Thomas W, Darnell JE 1972. Evidence that all messenger RNA molecules (except histone messenger RNA) contain poly (A) sequences and that the poly(A) has a nuclear function. J Mol Biol 71: 21–30 [DOI] [PubMed] [Google Scholar]
- Amara SG, Jonas V, Rosenfeld MG, Ong ES, Evans RM 1982. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature 298: 240–244 [DOI] [PubMed] [Google Scholar]
- Ashe MP, Griffin P, James W, Proudfoot NJ 1995. Poly(A) site selection in the HIV-1 provirus: inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev 9: 3008–3025 [DOI] [PubMed] [Google Scholar]
- Ashe MP, Pearson LH, Proudfoot NJ 1997. The HIV-1 5′ LTR poly(A) site is inactivated by U1 snRNP interaction with the downstream major splice donor site. EMBO J 16: 5752–5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield R, Enriquez-Harris P, Proudfoot NJ 1991. Transcriptional termination between the closely linked human complement genes C2 and factor B: common termination factor for C2 and c-myc? EMBO J 10: 4197–4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv H, Leder P 1972. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci 69: 1408–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim HC, Mitchel RE, Straus NA 1973. Analysis of long pyrimidine polynucleotides in HeLa cell nuclear DNA: absence of polydeoxythymidylate. Proc Natl Acad Sci 70: 2189–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse CE, Minvielle-Sebastia L, Lee BA, Keller W, Proudfoot NJ 1998. Coupling termination of transcription to messenger RNA maturation in yeast. Science 280: 298–301 [DOI] [PubMed] [Google Scholar]
- Black DL 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336 [DOI] [PubMed] [Google Scholar]
- Blumenthal T 2005. Trans-splicing and operons. In WormBook (ed. The C. elegans Research Community). WormBook, doi: 10.1895/wormbook.1.5.1. http://www.wormbook.org [Google Scholar]
- Brackenridge S, Proudfoot NJ 2000. Recruitment of a basal polyadenylation factor by the upstream sequence element of the human lamin B2 polyadenylation signal. Mol Cell Biol 20: 2660–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman JM, Singh P, Liu D, Quinlan S, Salisbury J, Graber JH 2005. PACdb: polyA cleavage site and 3′-UTR database. Bioinformatics 21: 3691–3693 [DOI] [PubMed] [Google Scholar]
- Brownlee GG, Sanger F 1969. Chromatography of 32P-labelled oligonucleotides on thin layers of DEAE-cellulose. Eur J Biochem 11: 395–399 [DOI] [PubMed] [Google Scholar]
- Brownlee GG, Cartwright EM, Cowan NJ, Jarvis JM, Milstein C 1973. Purification and sequence of messenger RNA for immunoglobulin light chains. Nat New Biol 244: 236–240 [DOI] [PubMed] [Google Scholar]
- Butler JS, Platt T 1988. RNA processing generates the mature 3′ end of yeast CYC1 messenger RNA in vitro. Science 242: 1270–1274 [DOI] [PubMed] [Google Scholar]
- Carswell S, Alwine JC 1989. Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol Cell Biol 9: 4248–4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Manley JL 2009. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 10: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, MacDonald CC, Wilusz J 1995. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res 23: 2614–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan DF, Manley JL 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev 11: 2755–2766 [DOI] [PubMed] [Google Scholar]
- Connelly S, Manley JL 1988. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev 2: 440–452 [DOI] [PubMed] [Google Scholar]
- Cramer P, Caceres JF, Cazalla D, Kadener S, Muro AF, Baralle FE, Kornblihtt AR 1999. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol Cell 4: 251–258 [DOI] [PubMed] [Google Scholar]
- Cui Y, Denis CL 2003. In vivo evidence that defects in the transcriptional elongation factors RPB2, TFIIS, and SPT5 enhance upstream poly(A) site utilization. Mol Cell Biol 23: 7887–7901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalziel M, Nunes NM, Furger A 2007. Two G-rich regulatory elements located adjacent to and 440 nucleotides downstream of the core poly(A) site of the intronless melanocortin receptor 1 gene are critical for efficient 3′ end processing. Mol Cell Biol 27: 1568–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S, Kaufmann I, Gentzel M, Foerstner KU, Gantzert AS, Gehring NH, Neu-Yilik G, Bork P, Keller W, Wilm M, et al. 2007. Splicing factors stimulate polyadenylation via USEs at non-canonical 3′ end formation signals. EMBO J 26: 2658–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S, Hentze MW, Kulozik AE 2008. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J 27: 482–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantonel JC, Murthy KG, Manley JL, Tora L 1997. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature 389: 399–402 [DOI] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 12: 525–532 [DOI] [PubMed] [Google Scholar]
- Denome RM, Cole CN 1988. Patterns of polyadenylation site selection in gene constructs containing multiple polyadenylation signals. Mol Cell Biol 8: 4829–4839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZazzo JD, Kilpatrick JE, Imperiale MJ 1991. Involvement of long terminal repeat U3 sequences overlapping the transcription control region in human immunodeficiency virus type 1 mRNA 3′ end formation. Mol Cell Biol 11: 1624–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z 2007. Nucleases of the metallo-β-lactamase family and their role in DNA and RNA metabolism. Crit Rev Biochem Mol Biol 42: 67–93 [DOI] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Marzluff WF 2005. The polyadenylation factor CPSF-73 is involved in histone pre-mRNA processing. Cell 123: 37–48 [DOI] [PubMed] [Google Scholar]
- Dye MJ, Proudfoot NJ 1999. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol Cell 3: 371–378 [DOI] [PubMed] [Google Scholar]
- Dye MJ, Proudfoot NJ 2001. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell 105: 669–681 [DOI] [PubMed] [Google Scholar]
- Edmonds M 2002. A history of poly A sequences: from formation to factors to function. Prog Nucleic Acid Res Mol Biol 71: 285–389 [DOI] [PubMed] [Google Scholar]
- Edmonds M, Vaughan MH Jr, Nakazato H 1971. Polyadenylic acid sequences in the heterogeneous nuclear RNA and rapidly-labeled polyribosomal RNA of HeLa cells: possible evidence for a precursor relationship. Proc Natl Acad Sci 68: 1336–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwalds-Gilbert G, Veraldi KL, Milcarek C 1997. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res 25: 2547–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M, Shenk T 1981. The sequence 5′-AAUAAA-3′ forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell 24: 251–260 [DOI] [PubMed] [Google Scholar]
- Fortes P, Cuevas Y, Guan F, Liu P, Pentlicky S, Jung SP, Martinez-Chantar ML, Prieto J, Rowe D, Gunderson SI 2003. Inhibiting expression of specific genes in mammalian cells with 5′ end-mutated U1 small nuclear RNAs targeted to terminal exons of pre-mRNA. Proc Natl Acad Sci 100: 8264–8269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sun Y, Li Y, Li J, Rao X, Chen C, Xu A 2011. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res 21: 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth PA, Choe WT, Rex JH, Byrne JC, Baker CC 1994. Sequences homologous to 5′ splice sites are required for the inhibitory activity of papillomavirus late 3′ untranslated regions. Mol Cell Biol 14: 5278–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galibert F, Sedat J, Ziff E 1974. Direct determination of DNA nucleotide sequences: structure of a fragment of bacteriophage phiX172 DNA. J Mol Biol 87: 377–407 [DOI] [PubMed] [Google Scholar]
- Gehring NH, Frede U, Neu-Yilik G, Hundsdoerfer P, Vetter B, Hentze MW, Kulozik AE 2001. Increased efficiency of mRNA 3′ end formation: a new genetic mechanism contributing to hereditary thrombophilia. Nat Genet 28: 389–392 [DOI] [PubMed] [Google Scholar]
- Gick O, Kramer A, Keller W, Birnstiel ML 1986. Generation of histone mRNA 3′ ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J 5: 1319–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A, Proudfoot NJ 1984. A sequence downstream of AAUAAA is required for rabbit β-globin mRNA 3′-end formation. Nature 312: 473–474 [DOI] [PubMed] [Google Scholar]
- Gil A, Proudfoot NJ 1987. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit β-globin mRNA 3′ end formation. Cell 49: 399–406 [DOI] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL 2008. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol 15: 71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber JH, Cantor CR, Mohr SC, Smith TF 1999. Genomic detection of new yeast pre-mRNA 3′-end-processing signals. Nucleic Acids Res 27: 888–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromak N, West S, Proudfoot NJ 2006. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol Cell Biol 26: 3986–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson SI, Polycarpou-Schwarz M, Mattaj IW 1998. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell 1: 255–264 [DOI] [PubMed] [Google Scholar]
- Haenni S, Sharpe HE, Gravato Nobre M, Zechner K, Browne C, Hodgkin J, Furger A 2009. Regulation of transcription termination in the nematode Caenorhabditis elegans. Nucleic Acids Res 37: 6723–6736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs DR, Goodbourn SE, Lamb J, Clegg JB, Weatherall DJ, Proudfoot NJ 1983. α-Thalassaemia caused by a polyadenylation signal mutation. Nature 306: 398–400 [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395: 93–96 [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D 2010. Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro. PLoS ONE 5: e12271 doi: 10.1371/journal.pone.0012271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, Bartel DP 2011. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature 469: 97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinek W, Adesnik M, Salditt M, Sheiness D, Wall R, Molloy G, Philipson L, Darnell JE 1973. Further evidence on the nuclear origin and transfer to the cytoplasm of polyadenylic acid sequences in mammalian cell RNA. J Mol Biol 75: 515–532 [DOI] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B 2009. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci 106: 7028–7033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302: 2141–2144 [DOI] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G 2010. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S 2004. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432: 517–522 [DOI] [PubMed] [Google Scholar]
- Kolev NG, Steitz JA 2005. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev 19: 2583–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Yario TA, Benson E, Steitz JA 2008. Conserved motifs in both CPSF73 and CPSF100 are required to assemble the active endonuclease for histone mRNA 3′-end maturation. EMBO Rep 9: 1013–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehner JN, Pearson EL, Moore C 2011. Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol 12: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyburz A, Friedlein A, Langen H, Keller W 2006. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol Cell 23: 195–205 [DOI] [PubMed] [Google Scholar]
- Legendre M, Gautheret D 2003. Sequence determinants in human polyadenylation site selection. BMC Genomics 4: 7 doi: 10.1186/1471-2164-4-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt N, Briggs D, Gil A, Proudfoot NJ 1989. Definition of an efficient synthetic poly(A) site. Genes Dev 3: 1019–1025 [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell 9: 1101–1111 [DOI] [PubMed] [Google Scholar]
- Lim L, Canellakis ES 1970. Adenine-rich polymer associated with rabbit reticulocyte messenger RNA. Nature 227: 710–712 [DOI] [PubMed] [Google Scholar]
- Logan J, Falck-Pedersen E, Darnell JE Jr, Shenk T 1987. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse β maj-globin gene. Proc Natl Acad Sci 84: 8306–8310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CS, Moreira A 2011. Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. Wiley Interdiscip Rev RNA 2: 23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L 2006. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444: 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L 2008. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci 65: 1099–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Kee SG, Efstratiadis A, Kafatos FC 1976. Amplification and characterization of a β-globin gene synthesized in vitro. Cell 8: 163–182 [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritch EF, Sambrook J 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH 2010. Crosstalk between mRNA 3′ end processing and transcription initiation. Mol Cell 40: 410–422 [DOI] [PubMed] [Google Scholar]
- Martincic K, Alkan SA, Cheatle A, Borghesi L, Milcarek C 2009. Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nat Immunol 10: 1102–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 9: 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews MB, Osborn M, Lingrel JB 1971. Translation of globin messenger RNA in a heterologous cell-free system. Nature 233: 206–209 [PubMed] [Google Scholar]
- Mayr C, Bartel DP 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361 [DOI] [PubMed] [Google Scholar]
- McLauchlan J, Gaffney D, Whitton JL, Clements JB 1985. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3′ termini. Nucleic Acids Res 13: 1347–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhart A, Cramer P 2004. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature 430: 223–226 [DOI] [PubMed] [Google Scholar]
- Mendecki J, Lee SY, Brawerman G 1972. Characteristics of the polyadenylic acid segment associated with messenger ribonucleic acid in mouse sarcoma 180 ascites cells. Biochemistry 11: 792–798 [DOI] [PubMed] [Google Scholar]
- Millevoi S, Vagner S 2010. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res 38: 2757–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ 2011. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell 41: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ 2005. From birth to death: the complex lives of eukaryotic mRNAs. Science 309: 1514–1518 [DOI] [PubMed] [Google Scholar]
- Moore CL, Sharp PA 1985. Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell 41: 845–855 [DOI] [PubMed] [Google Scholar]
- Moreira A, Wollerton M, Monks J, Proudfoot NJ 1995. Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogenetically conserved. EMBO J 14: 3809–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz MJ, Perez Santangelo MS, Paronetto MP, de la Mata M, Pelisch F, Boireau S, Glover-Cutter K, Ben-Dov C, Blaustein M, Lozano JJ, et al. 2009. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell 137: 708–720 [DOI] [PubMed] [Google Scholar]
- Nagaike T, Logan C, Hotta I, Rozenblatt-Rosen O, Meyerson M, Manley JL 2011. Transcriptional activators enhance polyadenylation of mRNA precursors. Mol Cell 41: 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam DK, Lee S, Zhou G, Cao X, Wang C, Clark T, Chen J, Rowley JD, Wang SM 2002. Oligo(dT) primer generates a high frequency of truncated cDNAs through internal poly(A) priming during reverse transcription. Proc Natl Acad Sci 99: 6152–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M, Berget SM 1991. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes Dev 5: 2086–2095 [DOI] [PubMed] [Google Scholar]
- Niwa M, MacDonald CC, Berget SM 1992. Are vertebrate exons scanned during splice-site selection? Nature 360: 277–280 [DOI] [PubMed] [Google Scholar]
- Nunes NM, Li W, Tian B, Furger A 2010. A functional human poly(A) site requires only a potent DSE and an A-rich upstream sequence. EMBO J 29: 1523–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Cheng TC, Antonarakis SE, Kazazian HH Jr 1985. Thalassemia due to a mutation in the cleavage-polyadenylation signal of the human β-globin gene. EMBO J 4: 453–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ 2004. Gene loops juxtapose promoters and terminators in yeast. Nat Genet 36: 1014–1018 [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Platt AR, Jones DR, Reifenberger JG, Sass LE, McInerney P, Thompson JF, Bowers J, Jarosz M, Milos PM 2009. Direct RNA sequencing. Nature 461: 814–818 [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM 2010. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143: 1018–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins KJ, Lusic M, Mitar I, Giacca M, Proudfoot NJ 2008. Transcription-dependent gene looping of the HIV-1 provirus is dictated by recognition of pre-mRNA processing signals. Mol Cell 29: 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto PA, Henriques T, Freitas MO, Martins T, Domingues RG, Wyrzykowska PS, Coelho PA, Carmo AM, Sunkel CE, Proudfoot NJ, et al. 2011. RNA polymerase II kinetics in polo polyadenylation signal selection. EMBO J 30: 2431–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant KE, Dye MJ, Lafaille C, Proudfoot NJ 2005. Strong polyadenylation and weak pausing combine to cause efficient termination of transcription in the human Gγ-globin gene. Mol Cell Biol 25: 3276–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ 1976. Sequence analysis of the 3′ non-coding regions of rabbit α- and β-globin messenger RNAs. J Mol Biol 107: 491–525 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ 1989. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem Sci 14: 105–110 [DOI] [PubMed] [Google Scholar]
- Proudfoot N 2004. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr Opin Cell Biol 16: 272–278 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Brownlee GG 1976. 3′ Non-coding region sequences in eukaryotic messenger RNA. Nature 263: 211–214 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Longley JI 1976. The 3′ terminal sequences of human α and β globin messenger RNAs: comparison with rabbit globin messenger RNA. Cell 9: 733–746 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ 2002. Integrating mRNA processing with transcription. Cell 108: 501–512 [DOI] [PubMed] [Google Scholar]
- Richard P, Manley JL 2009. Transcription termination by nuclear RNA polymerases. Genes Dev 23: 1247–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts GC, Gooding C, Mak HY, Proudfoot NJ, Smith CW 1998. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res 26: 5568–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen JM, Woo SL, Holder JW, Means AR, O'Malley BW 1975. Preparation and preliminary characterization of purified ovalbumin messenger RNA from the hen oviduct. Biochemistry 14: 69–78 [DOI] [PubMed] [Google Scholar]
- Ryan K, Calvo O, Manley JL 2004. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA 10: 565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Donelson JE, Coulson AR, Kossel H, Fischer D 1973. Use of DNA polymerase I primed by a synthetic oligonucleotide to determine a nucleotide sequence in phage fl DNA. Proc Natl Acad Sci 70: 1209–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR 1977. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci 74: 5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaufele F, Gilmartin GM, Bannwarth W, Birnstiel ML 1986. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3′ end of H3 messenger RNA. Nature 323: 777–781 [DOI] [PubMed] [Google Scholar]
- Schmid M, Jensen TH 2008. The exosome: a multipurpose RNA-decay machine. Trends Biochem Sci 33: 501–510 [DOI] [PubMed] [Google Scholar]
- Schumperli D 1988. Multilevel regulation of replication-dependent histone genes. Trends Genet 4: 187–191 [DOI] [PubMed] [Google Scholar]
- Sheets MD, Ogg SC, Wickens MP 1990. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res 18: 5799–5805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N 2011. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell 42: 794–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic S, Fremder E, Staals RH, Pruijn GJ, Schuster G 2010. Addition of poly(A) and poly(A)-rich tails during RNA degradation in the cytoplasm of human cells. Proc Natl Acad Sci 107: 7407–7412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D 2005. Moving messages: the intracellular localization of mRNAs. Nat Rev Mol Cell Biol 6: 363–375 [DOI] [PubMed] [Google Scholar]
- Sullivan KD, Steiniger M, Marzluff WF 2009. A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol Cell 34: 322–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL 1998. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell 2: 761–771 [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL 1996. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87: 941–952 [DOI] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 33: 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S, Vagner C, Mattaj IW 2000. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev 14: 403–413 [PMC free article] [PubMed] [Google Scholar]
- Valsamakis A, Zeichner S, Carswell S, Alwine JC 1991. The human immunodeficiency virus type 1 polyadenylylation signal: a 3′ long terminal repeat element upstream of the AAUAAA necessary for efficient polyadenylylation. Proc Natl Acad Sci 88: 2108–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman K, Brown KM, Gilmartin GM 2005. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev 19: 1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Proudfoot NJ 2009. Transcriptional termination enhances protein expression in human cells. Mol Cell 33: 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Gromak N, Proudfoot NJ 2004. Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 432: 522–525 [DOI] [PubMed] [Google Scholar]
- West S, Gromak N, Norbury CJ, Proudfoot NJ 2006. Adenylation and exosome-mediated degradation of cotranscriptionally cleaved pre-messenger RNA in human cells. Mol Cell 21: 437–443 [DOI] [PubMed] [Google Scholar]
- West S, Proudfoot NJ, Dye MJ 2008. Molecular dissection of mammalian RNA polymerase II transcriptional termination. Mol Cell 29: 600–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw E, Proudfoot N 1986. α-Thalassaemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3′ end processing in the human α2 globin gene. EMBO J 5: 2915–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M, Stephenson P 1984. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3′ end formation. Science 226: 1045–1051 [DOI] [PubMed] [Google Scholar]
- Winters MA, Edmonds M 1973a. A poly(A) polymerase from calf thymus. Characterization of the reaction product and the primer requirement. J Biol Chem 248: 4763–4768 [PubMed] [Google Scholar]
- Winters MA, Edmonds M 1973b. A poly(A) polymerase from calf thymus. Purification and properities of the enzyme. J Biol Chem 248: 4756–4762 [PubMed] [Google Scholar]
- Zhang F, Denome RM, Cole CN 1986. Fine-structure analysis of the processing and polyadenylation region of the herpes simplex virus type 1 thymidine kinase gene by using linker scanning, internal deletion, and insertion mutations. Mol Cell Biol 6: 4611–4623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]