This study unravels a novel mechanism underlying mutually antagonistic ventralizing Wnt and dorsalizing Shh signaling during embryonic brain patterning. Induced by Shh, the Vax2 transcription factor activates an internal promoter within Tcf7l2, thereby expressing a truncated dnTcf7l2 unable to bind β-catenin. This dominant-negative dnTcf7l2 acts as a Wnt antagonist, resulting in the restriction of Wnt signaling to the dorsal regions of the developing nervous system.
Keywords: Vax genes, Sonic hedgehog, canonical Wnt signaling, brain development
Abstract
Axial patterning of the embryonic brain requires a precise balance between canonical Wnt signaling, which dorsalizes the nervous system, and Sonic hedgehog (Shh), which ventralizes it. The ventral anterior homeobox (Vax) transcription factors are induced by Shh and ventralize the forebrain through a mechanism that is poorly understood. We therefore sought to delineate direct Vax target genes. Among these, we identify an extraordinarily conserved intronic region within the gene encoding Tcf7l2, a key mediator of canonical Wnt signaling. This region functions as a Vax2-activated internal promoter that drives the expression of dnTcf7l2, a truncated Tcf7l2 isoform that cannot bind β-catenin and that therefore acts as a potent dominant-negative Wnt antagonist. Vax2 concomitantly activates the expression of additional Wnt antagonists that cooperate with dnTcf7l2. Specific elimination of dnTcf7l2 in Xenopus results in headless embryos, a phenotype consistent with a fundamental role for this regulator in forebrain development.
The powerful morphogens of the Wnt family function through multiple stages of embryonic development and beyond. At the onset of vertebrate embryogenesis, they and their inhibitors contribute to specification of the primary anterior–posterior (AP) body axis (Petersen and Reddien 2009). Later, they regulate axial patterning and differentiation in a wide variety of tissues, including the limbs (Capdevila and Izpisúa Belmonte 2001). Utimately, they function as homeostatic regulators in many cells in adult tissues, where in some settings they promote the development of cancer (Hovanes et al. 2001).
Canonical Wnt signaling plays a prominent role in patterning the dorsal–ventral (DV) axis of the nervous system (Ulloa and Marti 2010). Several members of the Wnt family, including Wnt1, Wnt3, and Wnt3a, are localized at or near the roof plate—the dorsal pole of the developing nervous system (Parr et al. 1993)—where, in concert with bone morphogenetic proteins (BMPs), they dorsalize the embryonic spinal cord and brain (Jessell 2000; Sousa and Fishell 2010). In the context of DV specification, canonical Wnt signaling is frequently opposed by Sonic hedgehog (Shh). This protein is produced first by the notochord and prechordal mesoderm, and then by the floor plate—the ventral pole of the neural tube (Echelard et al. 1993). Shh spreads dorsally from these foci and acts as a ventralizing morphogen. Although several lines of experiment have demonstrated that Wnt and Shh signaling are mutually antagonistic during DV patterning (Ulloa and Marti 2010), the molecular mechanisms that orchestrate this antagonism are incompletely understood. It is nonetheless appreciated that central to these mechanisms are morphogen-induced transcription factors that inhibit opponent morphogen pathways (Ulloa and Marti 2010). This form of regulation requires that such transcription factors be expressed in a DV gradient, and many transcription factors involved in DV patterning of the embryonic spinal cord and brain indeed display steeply graded DV expression (Jessell 2000; Sousa and Fishell 2010).
The homeodomain transcription factors ventral anterior homeobox 1 (Vax1) and Vax2, which are expressed specifically in the ventral forebrain, are exemplars of such proteins. They are induced by Shh that is secreted from the prechordal mesoderm and ventral midline of the forebrain (Take-uchi et al. 2003; Kim and Lemke 2006), where they display a steep high-ventral to low-dorsal gradient of expression. In the absence of Shh, Vax1 and Vax2 are not expressed. Genetic studies in mice, chicks, and zebrafish demonstrate that they control DV patterning of the ventral forebrain and its derivatives, including the corpus callosum, optic chiasm, optic nerve, and retina, and that they drive the differentiation of these ventral structures by inhibiting the differentiation of dorsal structures (Schulte et al. 1999; Take-uchi et al. 2003; Mui et al. 2005). Ectopic expression of Vax2 in the dorsal retina, for example, leads to its ventralization, as shown by the loss of dorsal markers (e.g., Tbx5) and the acquisition of ventral markers (e.g., EphB2) (Schulte et al. 1999). At the same time, when canonical Wnt signaling is repressed in the developing eye, expression of the dorsal markers is lost and expression of Vax2 expands dorsally (Veien et al. 2008; Zhou et al. 2008; Fujimura et al. 2009). Perturbation of Wnt signaling in the eye has also been shown to result in aberrant axonal projections to the brain that resemble those seen in Vax2−/− mice (Barbieri et al. 1999; Mui et al. 2002; Veien et al. 2008; Wiley et al. 2008; Zhou et al. 2008; Paridaen et al. 2009).
These observations led us to hypothesize that the Vax proteins might function in DV specification of the forebrain through their ability to induce antagonists of canonical Wnt signaling. We therefore sought to identify Vax target genes that might function as Wnt signaling regulators. We found that Vax2 indeed controls an entire suite of general Wnt antagonists. More interestingly, we found that Vax2 binds at several sites within an ∼1-kb region of DNA that is among the most highly conserved noncoding sequences across vertebrate evolution in the entire genome. This region lies within the fifth intron of the gene encoding Tcf7l2, one of four Tcf/Lef nuclear mediators of canonical Wnt signaling. Vax2 activates an internal promoter contained within this conserved region, from which it drives transcription of a set of Tcf7l2 mRNAs that lack the first five Tcf7l2 exons. The first of these exons encodes the β-catenin interaction domain of Tcf7l2, and so the resulting truncated protein, which we show to be abundant in the developing forebrain, cannot bind β-catenin. Since β-catenin binding is absolutely required for nuclear transduction of canonical Wnt signaling, the truncated protein—designated dnTcf7l2—functions as a global dominant-negative Wnt antagonist.
We identified an equivalent dnTcf7l2 isoform in Xenopus laevis. Like mouse dnTcf7l2, this XdnTcf7l2 isoform is expressed in the embryonic forebrain, after gastrulation. We found that injection of specific loss-of-function morpholinos—which eliminate only XdnTcf7l2, while leaving expression of full-length Tcf7l2 intact—results in embryos with little or no anterior head, a phenotype consistent with dnTcf7l2 acting as a naturally occurring, dominant-negative Wnt antagonist in vivo. Our results illuminate a new mechanism for the inhibition of Wnt signaling, in which a single Shh-induced transcription factor activates expression of a battery of Wnt antagonists and, at the same time, a potent dominant-negative Tcf effector with which these antagonists cooperate.
Results
Vax2 binds to an intronic region of the Tcf7l2 gene that displays extraordinary sequence conservation through vertebrate evolution
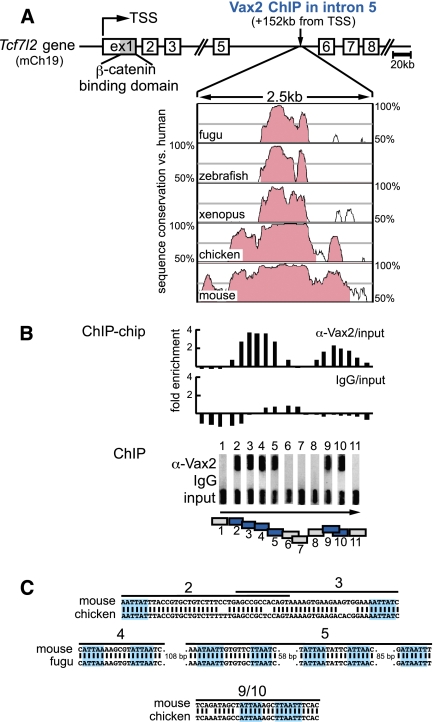
We used chromatin immunoprecipitation (ChIP) to identify direct Vax target genes in the embryonic day 13.5 (E13.5) mouse eye. Although we used an antibody specific to Vax2 (Mui et al. 2005), we anticipate that most of our results will also extend to Vax1, since Vax1 and Vax2 have identical homeodomains, partially overlapping expression profiles early in forebrain development, and additive loss-of-function phenotypes. Among the Vax2-bound regions identified in a ChIP–chip analysis was a DNA segment located 152 kb downstream from the conventional transcription start site (TSS) of the Tcf7l2 gene—in the fifth intron of this gene (Fig. 1A).
Figure 1.
Vax2 binds to an extremely conserved region in Tcf7l2 intron 5. (A) The first three exons and exons 5–8 of the mouse Tcf7l2 gene (Weise et al. 2010) are shown. Exon 1 encodes the β-catenin-binding domain. The Vax2-bound intron 5 region 152 kb downstream from the Tcf7l2 TSS is extraordinarily conserved through evolution, as shown by the Vista Browser plots. Regions in which a 100-bp sequence window exhibits >70% conservation are shaded (Mayor et al. 2000). (B, top bar graph) ChIP–chip analysis of the intron 5 region demonstrates enrichment by the Vax2 antibody relative to input sample. (Lower bar graph) No enrichment is observed with rabbit IgG. (Middle ChIP panel) ChIP confirms binding of Vax2 to regions covered by primer pairs 2–5 and 9–10. Agarose gel panels containing ChIP-PCR products amplified with primer pairs 1–11 are turned 90° from running direction (indicated by the arrow) to allow for alignment. (Bottom box diagram) Vax2-bound ChIP-PCR-positive regions are shown in blue. The sequence conservation plots of A, the ChIP–chip graphs, the ChIP-PCR panels 1–11, and the bottom box diagram of B are all in exact alignment. (C) Potential homeodomain-binding sites in intron 5 segments bound by Vax2 are highlighted. Numbers correspond to primer pairs/box diagram in B.
The nucleotide sequence of this region is extraordinarily highly conserved across vertebrate evolution. Over 710 base pairs (bp), it displays 99.2% identity between humans and chickens, and 85.8% identity between humans and the puffer fish Takifugu rubripes (Supplemental Fig. S1). This degree of conservation over such an extended length of DNA is exceptional in the extreme. Indeed, in the entirety of the human genome, we identified only 23 noncoding regions ≥500 bp in length that exhibit sequence conservation of 85% or higher when compared with fugu. When ranked by length, the Tcf7l2 intron 5 region is the fourth longest of these regions (Supplemental Fig. S1). Such hyperconservation argues strongly for a fundamental role for the Tcf7l2 intron 5 DNA.
To validate our ChIP–chip data, we designed 11 PCR primer pairs that span 2.1 kb of the intron 5 region, and used them in an independent ChIP assay. This assay confirmed binding of Vax2 to two segments of DNA, one of which encompasses primer pairs 2–5, and a second that encompasses pairs 9 and 10 (Fig. 1B). No Vax2 binding was detected using primer pairs 1, 6, 7, 8, or 11. No PCR products were obtained for any primer pair when DNA was immunoprecipitated with rabbit IgG (Fig. 1B) or when template DNA was immunoprecipitated from Vax2−/− tissue (data not shown).
The Vax proteins are Emx-related transcription factors that bind to a degenerate homeodomain-binding motif containing an ATTA core sequence (Mui et al. 2005). We searched for ATTA sequences within the Vax2-bound intron 5 region and found 11, of which seven display 100% conservation between mice and fugu (Fig. 1C). All of these sequences lie in the regions bound by Vax2 (Fig. 1B,C).
Truncated mRNAs initiate in the fifth intron of the Tcf7l2 gene
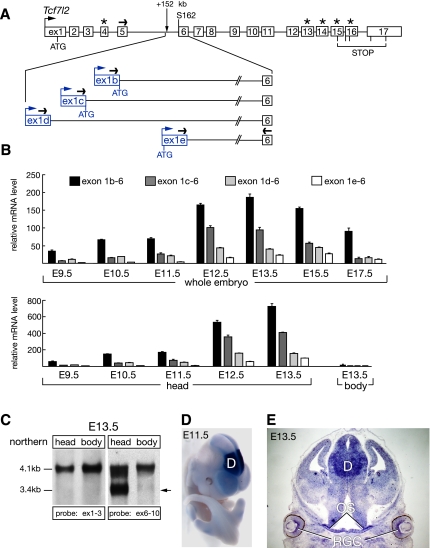
We examined the Vax2-bound region in silico and identified several mouse ESTs whose 5′-most exons lie in Tcf7l2 intron 5, with their second exons corresponding to Tcf7l2 exon 6. The first exons of two ESTs (GenBank: CB244473.1 and CA750374.1) match the first exon of a RIKEN E16 head cDNA clone (RefSeq: NM_001142924.1) whose downstream exons are identical to those of full-length Tcf7l2 mRNAs (see Fig. 2A). These observations suggest that the intron 5 region functions as an enhancer/promoter for alternative Tcf7l2 mRNAs that lack the first five exons of the gene (Fig. 2A). To investigate this possibility, we performed 5′ RACE from exon 8 (see Supplemental Table S2) to amplify the 5′ ends of all transcripts sharing this exon, using mRNA isolated from an E13.5 mouse head. In addition to amplifying the conventional Tcf7l2 5′ end (exons 1–8), we obtained four shorter PCR products. Sequencing of these products revealed that all contained exons 6–8, but that their 5′ ends matched the 5′-most exons of the ESTs located in Tcf7l2 intron 5. We designate these alternative first exons 1b, 1c, 1d, and 1e (Figs. 2A; Supplemental S2A). Exons 1b and 1c contain an ATG start codon that is in-frame with Tcf7l2 Ser 162 (S162) in exon 6 and is located at their very 3′ end, while exon 1e contains two ATGs in-frame with S162 (Supplemental Fig. S2A). Each of these exons initiates within intron 5; that is, none are part of the full-length Tcf7l2 mRNA that is initiated 152 kb upstream.
Figure 2.
Truncated Tcf7l2 mRNAs originate in Tcf7l2 intron 5. (A) 5′ RACE reveals the existence of four alternative Tcf7l2 first exons in Tcf7l2 intron 5. Exons 1–17 of the mouse Tcf7l2 gene are shown in white (Weise et al. 2010). Alternative exons 4, 13, 14, 15, and 16 are marked with asterisks. Alternative first exons of the truncated Tcf7l2 mRNAs are shown in blue. Black arrows depict primers used in the qRT–PCR experiments, and blue arrows depict alternative TSSs for exons 1b–e. (B) qRT–PCR analysis of the expression of the alternative Tcf7l2 mRNAs 1b, 1c, 1d, and 1e during embryogenesis, relative to Hprt. Error bars are mean ± SD (n = 3). (C) Northern analysis of Tcf7l2 mRNAs at E13.5. (Left two lanes) A probe against exons 1–3 detects only the full-length Tcf7l2 mRNAs (∼4.1 kb) in both the E13.5 head and body. (Right two lanes) A probe against exons 6–10 detects the full-length mRNAs in both the head and body, and also the truncated Tcf7l2 mRNAs (∼3.4 kb, arrow), but only in the E13.5 head. (D) Whole-mount in situ hybridization with a probe against exon 1b reveals a strong diencephalic expression (D) of the alternative Tcf7l2 mRNA 1b at E11.5. (E) In situ hybridization on a coronal section through an E13.5 head reveals the expression of the Tcf7l2 mRNA isoform 1b in ganglion cells of the developing retina (RGC), the optic stalk (OS), and diencephalon (D).
We assessed expression of these alternative mRNAs during mouse embryogenesis, and also in adult tissues, by quantitative RT–PCR (qRT–PCR). We found that their embryonic expression is first detected around E9 and peaks at E13.5, with the 1b and 1c mRNA isoforms most strongly expressed (Fig. 2B). These mRNAs are virtually absent from the embryonic body, but are abundant in the head (Fig. 2B). Neither mRNA is appreciably expressed in the whole embryo at E7.5, nor in adult kidney, liver, lung, spleen, lymph node, smooth muscle, testis, stomach, prostate, salivary gland, heart, pancreas, small intestine, thymus, or spinal cord (data not shown). In contrast, the truncated Tcf7l2 mRNAs are readily detected in the adult brain, with strongest expression in the thalamus and hypothalamus (Supplemental Fig. S3).
We also performed a northern analysis with poly(A)+ RNA from an E13.5 mouse head and body. Using an exon 1–3 probe, we detected the full-length (∼4.1 kb) Tcf7l2 mRNA in both the head and body (Fig. 2C). An exon 6–10 probe again detected this full-length form, but also truncated mRNAs of ∼3.4 kb, which were present only in the E13.5 head (Fig. 2C). In situ hybridization with an exon 1b probe revealed that mRNAs containing this exon are strongly and specifically expressed in the diencephalon of the brain at E11.5 (Fig. 2D) and E13.5, with notable expression in the Vax-positive retina and optic stalk (Fig. 2E). As noted above, the mRNAs that contain exons 1b–e are not part of the full-length Tcf7l2 transcript, and thus are not generated by alternative splicing.
The truncated Tcf7l2 mRNAs encode a dominant-negative Tcf7l2 repressor
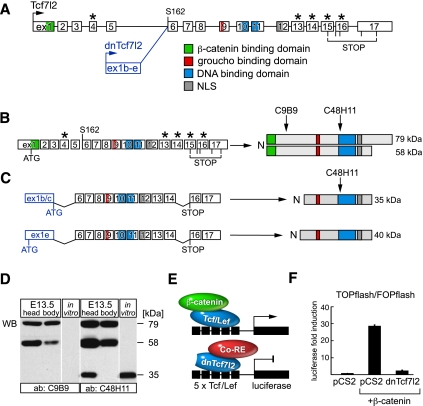
A complex alternative usage of exons 13–16 results in production of several different Tcf7l2 proteins, which differ at their C terminus but fall into two groups based on their molecular weight: long isoforms of ∼79 kDa, and short isoforms of ∼58 kDa (Fig. 3A,B; Struewing et al. 2010; Weise et al. 2010). We found that the truncated Tcf7l2 mRNAs containing alternative first exons 1b–e undergo a similarly complex splicing of conventional exons 13–16 and identified the following structure as the most abundant at E13.5: alternative first exon + exons 6–14 + exons 16–17. The 1b, 1c, and 1e mRNAs thus lack the first five exons and alternative exon 15. Their translation is terminated by a stop codon in exon 16, which leads to a protein of ∼35 kDa for the 1b and 1c isoforms and 40 kDa for the much less abundant 1e isoform (Fig. 3C). In vitro translation of the 1b, 1c, and 1e mRNAs results in proteins of the expected sizes, although mRNAs containing exon 1b may be less efficiently translated in vivo due to the presence of upstream in-frame ATGs (see Supplemental Fig. S2). No protein is produced in vitro from exon 1d-containing mRNAs or from mRNAs with mutated exon 1b/c/e start codons (Supplemental Fig. S2B). To detect the 35-kDa and 40-kDa proteins in vivo, we performed a Western analysis with two Tcf7l2 antibodies: C9B9 and C48H11. With C9B9, which recognizes an N-terminal epitope around Glu81 (Fig. 3B), we detected both the 79-kDa and 58-kDa isoforms in both the E13.5 head and body (Fig. 3D). With C48H11, which recognizes an epitope near Leu331 (Fig. 3B,C), we again detected these full-length isoforms, and also an ∼35-kDa protein present in the E13.5 head but not the E13.5 body (Fig. 3D; Supplemental S2C). As expected from the qRT–PCR expression profiling (Fig. 2B), the 40-kDa 1e isoform was present at very low levels (Supplemental Fig. S2C). Truncated Tcf7l2 proteins are therefore abundantly expressed in the E13.5 brain.
Figure 3.
The truncated Tcf7l2 mRNAs encode dominant-negative Tcf7l2. (A) Tcf7l2 and dnTcf7l2 exon structure, with important protein domains highlighted. Asterisks mark alternatively spliced exons. (B) A complex alternative usage of exons 13–16 gives rise to two major groups of Tcf7l2 isoforms (long proteins of ∼79 kDa and short proteins of ∼58 kDa), all of which are recognized by both the C9B9 and the C48H11 antibodies. (C) The alternative dnTcf7l2 mRNAs 1b and 1c encode a protein of ∼35 kDa, while the rare 1e isoform encodes a protein of ∼40 kDa. All of these truncated proteins are recognized only by the C48H11 antibody. (D) C9B9 antibody recognizes the 79-kDa and 58-kDa proteins in both the E13.5 head and body (first two lanes), but no protein translated in vitro from the truncated Tcf7l2 mRNA (third lane). The C48H11 antibody detects the 79-kDa and 58-kDa proteins in both the E13.5 head and body (fourth and fifth lanes) and also the 35-kDa protein, but only in the E13.5 head (fourth lane). (Sixth lane) This same 35-kDa protein is detected by C48H11 in protein translated in vitro from the pCS2-exon 1b + exon 6–14 + exon 16–17 expression vector. (E) Diagram of the TOPflash Wnt-responsive reporter construct, consisting of five Tcf/Lef-binding sites fused to a minimal promoter and the luciferase gene. (Top) Expression of this reporter is activated by β-catenin binding to full-length Tcf/Lef proteins. (Bottom) The truncated 35-kDa Tcf7l2 protein acts as a dominant-negative antagonist (dnTcf7l2) that cannot bind β-catenin; it instead interacts with corepressors. (F) Activation of the TOPflash luciferase reporter by transfected β-catenin is abolished by dnTcf7l2. β-Catenin expression vector was transfected into HEK293 cells in the absence (second bar) or presence (third bar) of the same amount (50 ng) of the dnTcf7l2 expression construct used for the in vitro transcription/translation reactions in D. Fold induction is the ratio between the normalized luciferase activity of the TOPflash and FOPflash constructs. Error bars are mean ± SD (n = 3).
Tcf7l2 is an HMG-box transcription factor that functions as an essential mediator of canonical Wnt/β-catenin signaling (Korinek et al. 1998b; Hurlstone and Clevers 2002). (In much of the literature, Tcf7l2 is referred to as “Tcf4.” However, this name has now been assigned to a helix–turn–helix transcription factor that is unrelated to Tcf7l2 and Wnt signaling.) Binding of a Wnt ligand to its receptors—Frizzled and Lrp5/6—leads to the accumulation of β-catenin in the nucleus (Logan and Nusse 2004). This protein interacts with the N-terminal β-catenin-binding domain of Tcf7l2, which displaces transcriptional corepressors and promotes β-catenin/Tcf7l2 interaction with coactivators that drive transcription of Wnt target genes. Alternatively, if β-catenin does not bind to Tcf7l2, Tcf7l2 instead interacts with the corepressors (proteins such as groucho [Grg] and histone deacetylases [Hdacs]), which repress transcription (Hurlstone and Clevers 2002; MacDonald et al. 2009).
The truncated Tcf7l2 protein encoded by the 1b, 1c, and 1e mRNAs lacks the β-catenin-binding domain and cannot be activated. This protein nonetheless retains the ability to (1) translocate to the nucleus via its nuclear localization signal (NLS) (Fig. 3A; Supplemental S2C), (2) bind DNA through its HMG domain, and (3) interact with corepressors through its groucho domain (Fig. 3C). It should therefore act as a natural dominant-negative antagonist of Tcf7l2, which we designate dnTcf7l2 (Fig. 3E). To investigate this possibility, we performed a Wnt reporter assay in human embryonic kidney (HEK) 293 cells using the TOPflash reporter construct, in which the luciferase gene is driven by a minimal promoter and five Tcf/Lef-binding sites (Korinek et al. 1997). As a negative control, we used the FOPflash construct that contains five mutated Tcf/Lef-binding sites. We found that the dnTcf7l2 protein very potently represses β-catenin activation of the TOPflash reporter (Fig. 3F).
Vax proteins exhibit activator activity
Since dnTcf7l2 is encoded by mRNAs that start in the Vax2-bound region (Fig. 1B), we hypothesized that Vax2 activates dnTcf7l2 transcription through this region. Although Vax1 and Vax2 have been shown to repress the Pax6 retinal enhancer (Mui et al. 2005), and the expression of some genes (e.g., ephrin-B1) is acquired in Vax2−/− mutant retina, the expression of other genes (e.g., EphB2) is lost in these mutants (Mui et al. 2002). To clarify the nature of Vax2 regulatory activity more generally, we assayed Vax2 for the presence of repression and/or activation domains. We fused the Vax2 domains shown in Supplemental Figure S4A with the DNA-binding domain of the transcription factor Gal4 (Gal4BD), and cotransfected these fusion constructs with a Gal4-responsive luciferase reporter into HEK293 cells. We used both a 5xUAS-tkP-luc reporter with high basal activity and a 5xUAS-minP-luc reporter with low activity (Supplemental Fig. S4B). None of the fusion constructs exhibited repressor activity in the Gal4-luciferase assays. In contrast, the Vax2 C-terminal region strongly activated the luciferase gene from both constructs (Supplemental Fig. S4C). We delimited the Vax2 activation domain to the most C-terminal region of the protein (Supplemental Fig. S4C,D), whose removal completely abolished activation function (Supplemental Fig. S4E). This region contains a motif (SAFEPY) that is 100% conserved from fugu to humans, and also between Vax2 and Vax1 (Supplemental Fig. S4D). We found that the Vax1 C terminus also exhibits activation activity, which is lost when its conserved SAFEPY motif is deleted (Supplemental Fig. S4E).
Vax2-dependent expression of dnTcf7l2 and transcriptional corepressors in the eye
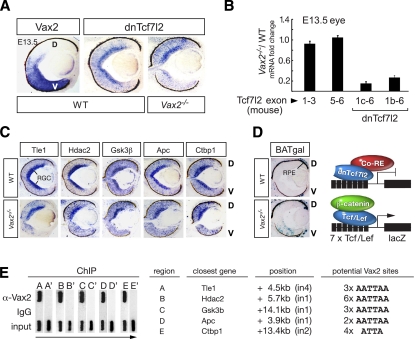
At E13.5, Vax2 is expressed in the ventral neural retina (Mui et al. 2002), with high expression in retinal ganglion cells (RGCs) (Fig. 4A). Using in situ hybridization, we found that the 1b dnTcf7l2 mRNA is expressed throughout the retinal DV axis, with highest expression in the RGC layer, and that this expression is lost specifically in the ventral neural retina of Vax2−/− mutants (Fig. 4A). Consistent with these in situ results, a comparative qRT–PCR analysis on mRNA from E13.5 embryonic eyes of wild-type and Vax2−/− mutants revealed that the 1b and 1c dnTcf7l2 mRNAs were both strongly down-regulated in the Vax2−/− mutants (Fig. 4B). At the same time, this analysis detected no difference in expression of the full-length Tcf7l2 mRNAs (Fig. 4B).
Figure 4.
Vax2-dependent expression of dnTcf7l2 and transcriptional corepressors. (A) In situ hybridization reveals that the Vax2 (left) and dnTcf7l2 1b (middle) mRNAs are coexpressed in the wild-type ventral retina at E13.5. (Right) dnTcf7l2 expression is lost in this region in the Vax2−/− mutant retina. (D) Dorsal; (V) ventral. (B) Truncated Tcf7l2 mRNAs 1b and 1c are down-regulated in Vax2−/− eyes, as determined by qRT–PCR. Expression of the full-length Tcf7l2 mRNA is unchanged, as determined using primers spanning exons 1–3 and exons 5–6. Fold change is the ratio between the relative mRNA expression normalized to Gapdh in the Vax2−/− mutant versus wild-type eyes. (C) Tle1, Hdac2, Gsk3β, Apc, and Ctbp1 mRNAs are expressed in the wild-type (WT) retina at E13.5, with strongest expression in the RGC layer, as shown by in situ hybridization. Their expression is lost in the Vax2−/− ventral retina. (D) Dorsal; (V) ventral. (D, left, top) Wnt signaling is not active in the E13.5 wild-type retina, except in the dorsal RPE, as shown by X-Gal staining of retinal sections from a BATgal reporter mouse. (Left, bottom) The BATgal reporter is derepressed in the ventral neural retina of a Vax2−/−/BATgal mouse. A schematic of the BATgal reporter transgene containing seven Tcf/Lef consensus binding sites fused to the siamois promoter and the lacZ gene (Maretto et al. 2003) in repressed (right, top) and activated (right, bottom) states. (E) Vax2 binds to the potential regulatory regions of the Wnt antagonists (A–E), as shown by ChIP analysis. Gel panels containing ChIP-PCR products are turned 90° from running direction, as indicated by the arrow. Candidate regulatory regions A–E are located at the indicated positions and contain the indicated clusters of potential Vax2 (homeodomain)-binding sites. A′–E′ indicate negative control regions surrounding each gene.
We asked whether Vax2 might also regulate Wnt antagonists that interact with Tcf/Lef proteins or β-catenin. We found that Tle1, Hdac2, Ctbp1, Apc, and Gsk3β are all expressed in the E13.5 eye, with the strongest expression in the RGC layer, where dnTcf7l2 and Vax2 are also most strongly expressed, and that expression of all of these corepressors is also lost specifically in the ventral neural retina of Vax2−/− mice (Fig. 4C). Expression of the general corepressor Kdm1a is unchanged in the Vax2−/− retina (data not shown). To test whether the loss of Vax2 affects Wnt signaling, we used the BATgal Wnt reporter mouse line, which expresses β-galactosidase driven by seven Tcf/Lef-binding sites (Maretto et al. 2003). We crossed this line with Vax2−/− mice and analyzed Vax2−/−/BATgal retinae at E13.5. We observed activated expression of β-gal, visualized by X-gal staining, specifically in the ventral neural retina of the Vax2−/− mice (Fig. 4D), indicating that canonical Wnt signaling is derepressed in the absence of Vax2. Wild-type mice carrying the reporter exhibited no β-gal expression in the E13.5 neural retina (Fig. 4D), which is consistent with the fact that Wnt signaling is only active in the most dorsal retinal region, the RPE, at this time (Fujimura et al. 2009). Thus, loss of Shh-induced Vax2 leads to the loss of dnTcf7l2 (Fig. 4A,B) and additional Wnt antagonists (Fig. 4C) and to the concomitant derepression of canonical Wnt signaling (Fig. 4D).
We examined the results from our ChIP–chip experiment for Vax2-bound DNA in the regulatory regions (−8.2/+3 kb) of the genes encoding Tle1, Hdac2, Ctbp1, Apc, and Gsk3β, but did not identify any. We therefore scanned their introns for clustered homeodomain (A)ATTA(A)-binding sites lying in or near cis-regulatory modules (Sharov et al. 2006). We identified candidate regions for Tle1 (A), Hdac2 (B), Gsk3β (C), and Apc (D) using the AATTAA sequence, and for Ctbp1 (E) using the ATTA sequence (Fig. 4E). We designed primers flanking these regions (designated A–E) and also negative control regions (designated A′–E′) and tested these in an independent ChIP-PCR experiment for Vax2 binding. Regions A–E yielded a strong signal with the Vax2 antibody, while we did not obtain PCR products for negative control regions A′–E′ or for regions A–E immunoprecipitated with rabbit IgG (Fig. 4E). PCR products were not amplified from Vax2−/− tissue (data not shown). Although we cannot rule out the possibility that Vax2 regulates the Wnt antagonists through other sites, the intronic regions we identified as Vax2-bound are good candidates for such regulatory elements.
Vax2 activates dnTcf7l2 expression and Wnt repressor activity in HEK293 cells
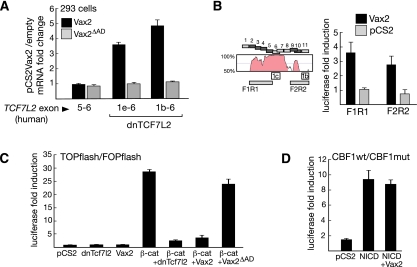
We identified a set of human dnTCF7l2 mRNAs whose first exons also lie in intron 5 and whose second exons are identical to exon 6 of the full-length TCF7L2 mRNA. The human dnTCF7l2 mRNAs 1b and 1e are represented in public databases by human thalamic mRNAs with GenBank accession numbers AK296305.1 and AK316111.1, respectively, and correspond to the mouse dnTcf7l2 mRNAs 1b and 1e, respectively. Like the mouse dnTcf7l2 mRNAs, human dnTCF7L2 exons 1b and 1e contain one and two ATGs, respectively, which are in-frame with Ser 162 in exon 6. Using primers specific for these dnTCF7L2 mRNAs, we tested the ability of Vax2 to induce their expression in HEK293 cells. We found that wild-type Vax2 stimulates expression of endogenous dnTCF7L2 mRNAs 1b and 1e (Fig. 5A, 1e-6, 1b-6, black bars), while the expression of full-length TCF7L2 mRNA is unaffected by Vax2 (Fig. 5A, 5-6). A mutant Vax2 that lacks the C-terminal activation domain (amino acids 265–292; Vax2ΔAD) is unable to induce the human dnTCF7L2 mRNAs (Fig. 5A, gray bars).
Figure 5.
Vax2 activates endogenous dnTCF7L2 and represses Wnt signaling. (A) Expression of full-length TCF7L2 mRNA (exons 5–6) is unaffected by overexpressing mouse Vax2 in HEK293 cells, as determined by qRT–PCR. In contrast, the truncated TCF7L2 mRNAs 1b and 1e are up-regulated by full-length Vax2, but not by Vax2 lacking the activation SAFEPY motif (Vax2ΔAD). Fold change is the ratio between expression normalized to GAPDH in cells transfected with pCS2-Vax2 versus cells transfected with empty pCS2 vector. Error bars are mean ± SD (n = 3). (B) Vax2 up-regulates the enhancer activity of the F1R1 and F2R2 Vax2-bound regions in a luciferase reporter assay. The Vista Browser plot shows sequence conservation of the dnTcf7l2 regulatory regions between humans and chickens. The alternative Tcf7l2 first exons 1b and 1c lie immediately downstream from the Vax2-bound regions (blue squares). Fold induction is the ratio between the normalized luciferase activity of the insert containing reporter construct activated with Vax2 and that activated with empty expression vector. Error bars are mean ± SD (n = 3). (C) Both dnTcf7l2 and Vax2 are strong repressors of β-catenin activation of Wnt targets. While full-length Vax2 is able to repress the β-catenin-activated Wnt reporter with a potency comparable with dnTcf7l2, Vax2 lacking the SAFEPY activation motif (Vax2ΔAD) is not. Fold induction is the ratio between the normalized luciferase activity of the TOPflash and FOPflash reporter constructs. Error bars are mean ± SD (n = 3). (D) Vax2 is unable to repress an activated Notch-responsive CBF1-pGL2 reporter containing eight CBF1-binding sites. The reporter was activated by overexpressing the Notch intracellular domain (NICD). Fold induction is the ratio between the normalized luciferase activity of the wild-type and mutated reporter. Error bars are mean ± SD (n = 3).
To demonstrate that Vax2 regulates dnTcf7l2 through the conserved intron 5 region, we assayed segments of this region in a luciferase reporter assay in HEK293 cells. We found that segments immediately upstream of mouse exon 1b (F1R1) and 1c (F2R2), which are bound by Vax2 (Fig. 5B, dark blue), are activated by Vax2 in this assay (Fig. 5B). Taken together, all of the above results support the hypothesis that Vax2 functions as a direct activator of dnTcf7l2 transcription.
We therefore tested whether Vax2 is itself capable of repressing a β-catenin-activated Wnt reporter (Fig. 5C). We did not see any activation when we cotransfected the TOPflash reporter with the empty expression vector, Vax2, or dnTcf7l2 in HEK293 cells. In contrast, we saw robust activation with β-catenin (Fig. 5C). This β-catenin-mediated activation was strongly repressed by both dnTcf7l2 (Figs. 3F, 5C) and Vax2 (Fig. 5C), and Vax2-mediated repression was lost if the cotransfection was performed with Vax2 lacking its C-terminal activation domain (Fig. 5C). This Vax2-repressive effect is specific for Tcf/Lef targets, since induction of a Notch-responsive luciferase reporter by the Notch intracellular domain (NICD) was unaffected by Vax2 (Fig. 5D).
Elimination of dnTcf7l2 in Xenopus results in headless embryos
Taken together, the above results demonstrate that dnTcf7l2 is a novel Shh/Vax-triggered antagonist of canonical Wnt signaling in the developing eye. To probe this activity more generally, we sought to perform loss-of-function experiments in a developing embryo in which the activities of canonical Wnt signaling have been previously studied. Since Wnt function has been extensively analyzed in X. laevis embryos, and since loss-of-function experiments are readily performed in these embryos using antisense morpholinos, we chose to assess the effect of specific ablation of dnTcf7l2 on early Xenopus development.
Given the extreme conservation of the intron 5 promoter of dnTcf7l2, we hypothesized that this antagonist should indeed be present in Xenopus. We used 5′ RACE reverse primers in exon 6 of the Xenopus Tcf7l2 (XTcf7l2) mRNA to identify all possible 5′ ends of this transcript in stage 24 embryos, and cloned two distinct 5′ RACE products, one of which corresponds to the full-length XTcf7l2 mRNA. The sequence of the other product (Supplemental Fig. S5), when aligned against the Xenopus tropicalis genome, is positioned in a highly conserved region in XTcf7l2 intron 5, and its position corresponds precisely to that of mouse Tcf7l2 exon 1e. Despite its positional similarity to mouse exon 1e, we designate this single XTcf7l2 alternative first exon 1b. We identified two ATGs in this exon 1b that are in-frame with Ser 180 in XTcf7l2 exon 6 (and that are not followed by a stop codon) (Supplemental Fig. S5). We also found that this alternative XTcf7l2 mRNA 1b undergoes complex alternative splicing of conventional exons 13–16 during development, similar to that seen for mouse dnTcf7l2 mRNAs.
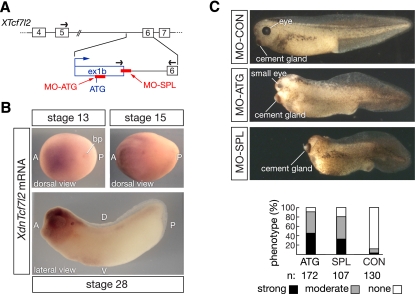
We used primers spanning XTcf7l2 exons 5 and 6 to detect full-length XTcf7l2 mRNA by qRT–PCR, and primers spanning exon 1b and 6 to detect the truncated XdnTcf7l2 transcript (Fig. 6A). We found that XdnTcf7l2 mRNA is first expressed in whole embryos at stage 13, after the completion of gastrulation (Supplemental Fig. S6). There is no expression in the Spemann organizer or any other structure prior to stage 13. XdnTcf7l2 mRNA expression plateaus by stage 20, during the course of neural tube formation, and remains elevated through stage 28 (Supplemental Fig. S6). This truncated transcript is expressed almost exclusively in the embryonic head (Supplemental Fig. S6). The timing and localization of XdnTcf7l2 expression are thus very similar to mouse dnTcf7l2 (Figs. 2B–E). Whole-mount in situ hybridizations confirmed these qRT–PCR data, and demonstrated that XdnTcf7l2 mRNA is first detected in the anterior region of the embryo at stage 13 and remains highly restricted to anterior neural tissue thereafter (Fig. 6B). This expression pattern is again concordant with the forebrain expression of mouse dnTcf7l2 mRNA (Fig. 2D,E).
Figure 6.
DnTcf7l2 is essential for Xenopus forebrain development. (A) A 5′ RACE experiment in X. laevis reveals the existence of an alternative XTcf7l2 first exon in intron 5, which contains an ATG start codon in-frame with Ser 180 in XTcf7l2 exon 6. The positions of the translation-blocking (MO-ATG) and splicing-blocking (MO-SPL) morpholinos are indicated. Arrows represent the primers used for qRT–PCR. (B) In situ hybridization analysis of dnTcf7l2 expression at stages 13, 15, and 28 of X. laevis development. (A) Anterior; (P) posterior; (D) dorsal; (V) ventral; (bp) blastopore. (C) Knockdown of XdnTcf7l2 with MO-ATG or MO-SPL leads to embryos with severely truncated anterior head regions at stage 40. Phenotype ranges from moderate (gray bars), showing small head and small eyes, to strong (black bars), where no head and no eyes are present.
We determined the sequence of the XdnTcf7l2 exon 1b–intron 5 boundary and used this to design a morpholino antisense oligonucleotide, designated MO-SPL, which blocks splicing of exon 1b to exon 6 (Fig. 6A; Supplemental Fig. S5). We also used the sequence of exon 1b to design an additional, nonoverlapping morpholino, designated MO-ATG, which blocks translation initiated from the downstream ATG in exon 1b; this ATG has a stronger Kozak consensus than its upstream counterpart (Fig. 6A; Supplemental Fig. S5). These two morpholinos were designed to inhibit the expression of only XdnTcf7l2 without affecting expression of full-length XTcf7l2. X. laevis embryos injected with a control morpholino (MO-CON) did not display developmental abnormalities (Fig. 6C). In marked contrast, injection of either the MO-ATG or MO-SPL morpholinos lead to drastic developmental defects in the anterior head region (Fig. 6C), the site at which XdnTcf7l2 is normally expressed (Fig. 6B). These morphants displayed a headless phenotype whose severity ranged from moderate, with a small head and very small eyes, to strong, with no head and no eyes (Fig. 6C). The expression of full-length XTcf7l2 mRNA was unaffected by injection of the splicing MO-SPL morpholino, while XdnTcf7l2 mRNA was completely eliminated by this morpholino (Supplemental Fig. S7). Neither mRNA was affected by the control morpholino (Supplemental Fig. S7).
Although both XdnTcf7l2 morpholinos consistently yield forebrain-ablated Xenopus embryos (Fig. 6C), they do not perturb initial AP patterning of the embryo. XdnTcf7l2 is not expressed prior to stage 13, when these early patterning events occur, and the expression of chordin, a marker of the Spemann organizer, is unperturbed at stage 10.5 in embryos treated with either the MO-ATG or MO-SPL XdnTcf7l2 morpholinos (Supplemental Fig. S8). Similarly, at stage 12.5, before the initial appearance of XdnTcf7l2, expression of Otx2, which is first expressed in the organizer and then later in the overlying anterior neuroectoderm, is also unaffected by either morpholino (Supplemental Fig. S8). In marked contrast, at stage 20, when XdnTcf7l2 mRNA is maximal (Supplemental Fig. S6), both the broad anterior expression of Otx2 and the more restricted expression of En2, which marks the midbrain–hindbrain boundary, are severely reduced in the XdnTcf7l2 morphants (Supplemental Fig. S8). The forebrain deletion in our Xenopus morphants is similar to the forebrain deletions seen in loss-of-function mutants for Wnt signaling inhibitors in zebrafish and mice (see below), and strongly suggests that XdnTcf7l2 functions as a naturally occurring dominant-negative Wnt antagonist in vivo. Consistent with this hypothesis, overexpression of XdnTcf7l2 results in developmental anomalies associated with the aberrant early inhibition of canonical Wnt signaling (Supplemental Fig. S9).
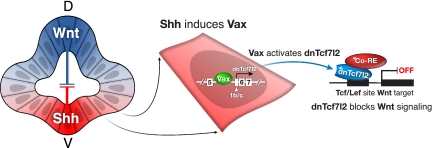
As discussed below, these results, together with those detailed above, are consistent with a powerful new mechanism for the regulation of Wnt signaling in the developing eye and forebrain. In this model (Fig. 7), a morphogen (e.g., Shh) induces a transcription factor (e.g., Vax) that drives the expression of a truncated dnTcf7l2 protein that then acts as a global antagonist of canonical Wnt signaling.
Figure 7.
A transcriptional mechanism through which Sonic hedgehog may antagonize Wnt/β-catenin signaling in the developing eye. In the developing ventral diencephalon and eye, Vax proteins are induced by Shh (left), and these transcription factors in turn activate expression of the truncated, dominant-negative isoform of Tcf7l2 (middle). (Right) DnTcf7l2 then shuts down genes that are targets of canonical Wnt signaling. The extreme conservation of the dnTcf7l2 intron 5 enhancer/promoter suggests that additional transcription factors and signaling pathways may impinge on and regulate its activity elsewhere in the developing forebrain.
Discussion
A transcriptional mechanism for the global antagonism of Wnt/β-catenin signaling
Morphogen-induced transcription factors that inhibit key components of an antagonistic pathway are prime candidates for mediating reciprocal morphogen repression (Jessell 2000; Ulloa and Marti 2010). The Vax proteins are two such factors: They are induced by Shh released from the prechordal mesoderm and ventral midline (Take-uchi et al. 2003; Kim and Lemke 2006), are restricted to ventral forebrain domains, and are steeply graded in their expression. Genetic analyses in mice, chicks, and zebrafish have demonstrated that Vax1 and Vax2 drive the development of ventral forebrain structures by inhibiting dorsal development (Schulte et al. 1999; Take-uchi et al. 2003; Mui et al. 2005). For example, Vax2−/− mice exhibit dorsalized RGC projections to the midbrain and flattened expression gradients of retinal DV markers (Mui et al. 2002), Vax1−/− mice exhibit a dorsalization of the optic stalk and the loss of anterior midline commissures (Bertuzzi et al. 1999; Hallonet et al. 1999), and Vax1−/−Vax2−/− double mutants exhibit a complete transformation of the optic nerve (the most ventral optic structure) into neural retina (the next most dorsal structure) (Mui et al. 2005). Expression of dorsal neural genes (e.g., Pax6 and ephrinB1) is acquired in these mutants, while that of ventral genes (e.g., Pax2 and EphB2) is lost (Barbieri et al. 1999; Hallonet et al. 1999; Mui et al. 2002 , 2005). Conversely, when Vax2 is ectopically expressed in the dorsal retina, it ventralizes this region (Schulte et al. 1999).
Dorsal markers are also lost, and Vax2 is acquired, when canonical Wnt signaling is repressed in the developing eye (Veien et al. 2008; Zhou et al. 2008). When Wnt/β-catenin signaling is repressed in the RPE, the most dorsal structure of the eye, Vax2 is activated (Fujimura et al. 2009). The reciprocally antagonistic nature of Vax2 expression and canonical Wnt signaling is further illustrated by the finding that the latter stimulates retinal proliferation (Logan and Nusse 2004), while Vax2 suppresses it (Mui et al. 2005; Liu et al. 2008). All of these observations led us to propose that the Vax proteins ventralize the anterior forebrain by antagonizing canonical Wnt signaling.
We show that in the embryonic eye, this is achieved straightforwardly—through the ability of Vax2 to drive the expression of the novel global Wnt antagonist dnTcf7l2. Vax2 does so by binding to a remarkably conserved internal promoter within the fifth intron of the Tcf7l2 gene and thereby activating transcription of truncated Tcf7l2 mRNAs. The expression of the dnTcf7l2 isoform encoded by these mRNAs is lost in the ventral retina of Vax2−/− mice (Fig. 4A).
Coordinate induction of Wnt antagonists by Vax2
DnTcf7l2 is a potent antagonist of Wnt signaling, even in the face of a fully activated canonical Wnt pathway. The presence of a stoichiometrically significant nuclear level of this inhibitor (Fig. 3D; Supplemental Fig. S2C), which cannot interact with β-catenin but can bind to the regulatory elements of Wnt target genes, ensures that these target genes remain repressed (Fig. 3E). Cells expressing dnTcf7l2 are therefore “blind” to Wnt signaling. Conversely, losing the activity of a single critical component of the Wnt repression machinery—e.g., Apc or Gsk3β—leads to derepression of Wnt signaling even in the absence of a Wnt ligand (Wiley et al. 2008; Paridaen et al. 2009). Our results demonstrate that Vax2 orchestrates these two flanks of Wnt repression in the ventral neural retina, in that it both activates the expression of dnTcf7l2 and also drives the expression of a set of Wnt antagonists with which this truncated protein cooperates. Loss of Vax2 leads to the loss of the groucho-related protein Tle1, the histone deacetylase Hdac2, the repressor Ctbp1, the tumor suppressor Apc, and the kinase Gsk3β (Fig. 4C). The absence of dnTcf7l2 and these corepressors in the ventral retina of the Vax2−/− mice indeed results in the derepression of Wnt signaling (Fig. 4D). Retinal disruption of the repressor complex through loss-of-function mutation of Apc or Gsk3β has previously been shown to result in Wnt signaling derepression and aberrantly patterned axonal projections from the eye to the brain (Wiley et al. 2008; Paridaen et al. 2009) that are similar to the Wnt derepression (Fig. 4D) and axon projection phenotypes seen in Vax2−/− mutants (Barbieri et al. 1999; Mui et al. 2002). While the expression of Wnt antagonists is Vax2-dependent in the ventral neural retina at E13.5, their expression in the dorsal neural retina, which is Vax2-negative, clearly is not (Fig. 4A,C). Dorsal expression of these antagonists must therefore be under the control of an additional transcription factor or factors. As discussed below, this must also be the case for dnTcf7l2 expression in other non-Vax-expressing regions of the developing brain.
Dominant-negative Tcf/Lef variants as a general mechanism of Wnt repression
Although the four Tcf/Lef proteins—Lef1, Tcf7 (Tcf1), Tcf7l1 (Tcf3), and Tcf7l2 (Tcf4)—are all related DNA-binding proteins that mediate canonical Wnt signaling, they nonetheless exhibit distinct transcriptional properties (Hurlstone and Clevers 2002). The complexity of Tcf/Lef-mediated regulation is further augmented by the existence of multiple alternative isoforms with differences at both their N and C termini, which are generated by alternative splicing and promoter usage (Arce et al. 2006). These include N-terminal truncations of Lef1 and Tcf7 that have been detected in the intestine (Van de Wetering et al. 1996; Hovanes et al. 2001). Like the dnTcf7l2 protein we describe here, these dominant-negative Lef1 and Tcf7 isoforms lack the N-terminal β-catenin interaction domain, and therefore cannot be activated even in the presence of a Wnt ligand. As opposed to the dnTcf7l2 mRNAs, the truncated dnLef1 and dnTcf7 mRNAs are initiated in more upstream regions (closer to the conventional TSS), and the signals regulating their expression are not currently known. Nonetheless, the inability to produce the dominant-negative variants of Lef1 and Tcf7, which balance the activity of the corresponding full-length Tcf/Lef proteins, is strongly associated with derepression of Wnt signaling and the development of colon cancer (de Lau and Clevers 2001; Hovanes et al. 2001; Hoppler and Kavanagh 2007).
DnTcf7l2 and forebrain development
Although Tcf7l2 is highly expressed in the embryonic forebrain (Cho and Dressler 1998; Korinek et al. 1998b), complete mouse Tcf7l2 knockouts (Korinek et al. 1998a) display only a subtle brain phenotype, the most prominent feature of which is an enlargement of the anterior pituitary (Brinkmeier et al. 2007). This pituitary enlargement is interesting in light of the recent demonstration that Vax1−/− mice have a duplicated anterior pituitary (Bharti et al. 2011). While the dramatic difference in phenotypic severity between the complete Tcf7l2 mouse knockouts and our Xenopus morphants may reflect differences in Tcf redundancy between mammals and amphibians, we suggest an alternative possibility. Since the Tcf7l2−/− mice were prepared by disrupting the Tcf7l2 HMG domain (Korinek et al. 1998a), and since dnTcf7l2 uses this same DNA-binding domain (Fig. 3A–C), both full-length Tcf7l2 and dnTcf7l2 are eliminated in these mouse mutants. This is not the case for the Xenopus morphants, in which expression of full-length Tcf7l2 is preserved (Supplemental Fig. S7). Given that Tcf7l2 and dnTcf7l2 are coexpressed in the embryonic brain (Figs. 2C–E; Supplemental Fig. S6), dnTcf7l2 may be necessary to counterbalance the activity of Tcf7l2 and prevent excessive Wnt signaling. We suggest that the consequences of removing both Tcf7l2 isoforms may therefore be less severe than the specific deletion of only the dnTcf7l2 antagonist or only full-length Tcf7l2. Recently published Tcf7l2 (“Tcf4”) exon 1 mouse knockouts, which may be full-length-specific, have heads; however, their brain phenotypes have not been assessed (Angus-Hill et al. 2011). These mice were analyzed with respect to their intestinal phenotype, which indeed differs from that seen in the Tcf7l2 knockouts that lack both Tcf7l2 isoforms (Korinek et al. 1998a).
Consistent with this idea is the finding that (1) ablation of only full-length XTcf7l2 (XTcf4) in Xenopus, using morpholinos that block translation from the ATG of the full-length isoform, leads to defects in isthmus and midbrain development that are not observed in the complete Tcf7l2 mouse knockouts (Kunz et al. 2004), and (2) our elimination of only dnTcf7l2 in Xenopus leads to morphant embryos that lack a forebrain (Fig. 6C). It is important to note that these “headless” dnTcf7l2 Xenopus morphants are similar to (1) the “headless” zebrafish embryos that result from loss-of-function mutation of the fish Tcf7l1 (Tcf3) Wnt repressor (Kim et al. 2000) and (2) the “headless” mouse embryos that result from loss-of-function mutation of the dickkopf1 Wnt inhibitor (Mukhopadhyay et al. 2001). All three Wnt antagonist mutants display similarly sharp truncations of the embryonic nervous system anterior to the midbrain, and all display mutant phenotypes that reflect the loss of Wnt antagonists subsequent to initial AP patterning of the embryo. Just like the dnTcf7l2 Xenopus morphants (Supplemental Fig. S8), the zebrafish Tcf7l1 mutants have an intact organizer, as marked by chordin and dickopf expression (Kim et al. 2000), and the mouse dickkopf1 mutants display normal Otx2 expression in anterior neural tissue immediately after gastrulation (Mukhopadhyay et al. 2001).
The dnTcf7l2 regulatory region as a critical regulatory node
Vax1−/−Vax2−/− mice exhibit a complete transformation of the optic nerve into fully differentiated retina, which represents an overt dorsalization of this structure (Mui et al. 2005). They also exhibit multiple forebrain and craniofacial defects, but their brains are nonetheless relatively intact (Bertuzzi et al. 1999; Mui et al. 2005). In contrast, global elimination of dnTcf7l2 in Xenopus leads to a missing anterior forebrain (Fig. 6C). This difference may be explained by the fact that the Vax proteins drive the expression of dnTcf7l2 only in the regions of their expression, and that there are many other locations in the developing diencephalon where dnTcf7l2 is expressed (Fig. 2D,E) but Vax1 and Vax2 are not. Although our evidence for the model of Figure 7 is established only in the developing ventral diencephalon and eye, it is likely that transcription factors other than the Vax proteins, some of them morphogen-induced, drive dnTcf7l2 expression at these other locations. These transcription factors may not play an overt role in DV specification. The nearly unprecedented evolutionary conservation of the dnTcf7l2 regulatory region, together with its extended length, suggests that it is an especially important regulatory node, upon which many additional transcription factors—both activators and repressors—are likely to impinge.
Conclusions
Our results link the activity of a basic developmental morphogen (Shh) to the antagonism of one of its key regulatory opponents (the canonical Wnt pathway). This linkage is achieved through Shh induction of a transcription factor (Vax2) that drives the expression of a potent Wnt antagonist (dnTcf7l2). The regulatory region through which dnTcf7l2 expression is activated and controlled is among the most stringently conserved DNA sequences across vertebrate evolution. These results are likely to be significant not only to our understanding of morphogen regulatory networks in development, but also to the operation of these same networks in adult tissues and organs and in cancer.
Materials and methods
Mice
The Vax2−/− mice have been described previously (Mui et al. 2002). C57BL/6J and BATgal mice were purchased from Jackson Laboratory.
ChIP assay
ChIP and ChIP–chip assays were performed as described previously (Lee et al. 2006; Mui et al. 2005), using pooled E13.5 C57Bl6 and Vax2−/− eyes as the starting material and α-Vax2 antibody (Mui et al. 2005) or rabbit IgG (Vector Laboratories). The ChIP–chip hybridization was performed using NimbleGen (2.1M Mouse Deluxe Promoter array, Roche), and data were analyzed with CisGenome software (Ji et al. 2008). ChIP primers are listed in Supplemental Table S1.
RNA isolation, 5′ RACE, and qRT–PCR
Total RNA was isolated using Trizol, reverse transcription was performed using SuperScript III, and 5′ RACE was performed using the GeneRacer kit (all Invitrogen). qRT–PCR reactions were performed in triplicate using SYBR Green PCR master mix (Applied Biosystems) and normalized against expression of Hprt and GAPDH. The 5′ RACE and mouse qPCR primers are listed in Supplemental Tables S2 and S3, respectively. Human qPCR primers are listed in Supplemental Table S4.
In situ hybridization and X-Gal staining
Probes were prepared using DIG RNA labeling mix (Roche) and RT–PCR products amplified from C57Bl6 E13.5 head RNA with primers listed in Supplemental Table S6. In situ hybridizations and X-Gal staining were performed as described previously (Fujimura et al. 2009).
Reporter assays
One-hundred-twenty nanograms of firefly luciferase reporter constructs and 170 ng of expression constructs were cotransfected into HEK293 cells (48-well plate) using Fugene6 (Roche) together with 10 ng of the renilla luciferase pGL4.74 plasmid (Promega) as a transfection efficiency control. Firefly and renilla luciferase activity was measured 48 h after transfection, using the Dual-Glo Luciferase assay system (Promega). Values are mean ± standard deviation for three experiments.
DNA constructs
The F1R1 and F2R2 regions were amplified from C57Bl6 genomic DNA and cloned into pGL4.23 (Promega). Coding regions of Vax2, Vax1, dnTcf7l2, and β-catenin were amplified by RT–PCR from C57Bl6 E13.5 head RNA and cloned into pCS2(+)MT (6xmyc tag) or pCS2(+). Truncated forms of Vax2 and Vax1 were cloned into pCMVBD (Stratagene) and pCS2(+)MT. Constructs were verified by sequencing. The primers are listed in Supplemental Tables S7 and S8.
Northern
Poly(A)+ RNA was isolated from total RNA using Oligotex mRNA kit (Qiagen). Probes were prepared using Biotin RNA labeling mix (Roche) and RT–PCR products amplified from C57Bl6 E13.5 head RNA with primers listed in Supplemental Table S5. Northerns were carried out with 1 μg of poly(A)+ RNA per lane with NorthernMax kit (Ambion) and Chemiluminescent Detection Module (ThermoScientific).
Western
Whole-cell lysates were prepared using Cell Lysis buffer (Cell Signaling) supplemented with protease inhibitors (Roche). DnTcf7l2 proteins were prepared in vitro from pCS2(+)-dnTcf7l2 using TNT SP6 Quick Coupled Transcription/Translation system (Promega). The following antibodies were used: α-TCF4 (C9B9) and α-TCF4 (C48H11) (Cell Signaling).
X. laevis total RNA, genomic DNA, 5′ RACE, and qRT–PCR
Total RNA was isolated from pooled embryos as described previously (Stubbs et al. 2008). Genomic DNA was isolated using the QIAamp tissue kit (Qiagen). 5′ RACE; qRT–PCR was performed as described for mice. Each reaction was normalized against the expression of XOdc. Xenopus primers used for 5′ RACE, qPCR, and analysis of the exon 1b–intron 5 boundary are listed in Supplemental Table S9.
X. laevis morpholino microinjections and in situ hybridization
Xenopus embryos were obtained and treated as described previously (Stubbs et al. 2008). Phenotype was scored based on the DA index (Kao and Elinson 1988). Embryos were injected in all four quadrants of the animal pole at the two-cell stage with the following morpholinos (GeneTools): 60–90 ng of MO-ATG, 30–45 ng of MO-SPL, and 75–100 ng of MO-CON. Morpholinos used in this study are listed in Supplemental Table S9. Whole-mount in situ hybridization with chordin, Otx2, and En2 probes were carried out as described previously (Stubbs et al. 2008).
X. laevis mRNA microinjections
Xenopus embryos were obtained and treated as described previously (Stubbs et al. 2008). Embryos were injected at the four-cell stage with capped, synthetic mRNAs at a concentration of 1–5 ng per embryo. Injections were targeted to the two dorsal blastomeres or to one ventral blastomere. At the desired stage, embryos were fixed in 3.7% formaldehyde and then stained with X-gal to mark the injected region of the embryo.
Acknowledgments
We thank Chris Kintner for conceptual and experimental guidance, for pCS2(+)MT-NICD, wild-type, and mutant 8xCBF1-pGL2, and for the use of frog facilities; Dannielle Engle for the TOPflash and FOPflash plasmids; and Joseph Hash for superb technical support. This work was supported by a grant from the NIH (EY017478 to G.L.) and by the Salk Institute.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.17227011.
References
- Angus-Hill ML, Elbert KM, Hidalgo J, Capecchi MR 2011. T-cell factor 4 functions as a tumor suppressor whose disruption modulates colon cell proliferation and tumorigenesis. Proc Natl Acad Sci 108: 4914–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce L, Yokoyama NN, Waterman ML 2006. Diversity of LEF/TCF action in development and disease. Oncogene 25: 7492–7504 [DOI] [PubMed] [Google Scholar]
- Barbieri AM, Lupo G, Bulfone A, Andreazzoli M, Mariani M, Fougerousse F, Consalez GG, Borsani G, Beckmann JS, Barsacchi G, et al. 1999. A homeobox gene, vax2, controls the patterning of the eye dorsoventral axis. Proc Natl Acad Sci 96: 10729–10734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertuzzi S, Hindges R, Mui SH, O'Leary DD, Lemke G 1999. The homeodomain protein vax1 is required for axon guidance and major tract formation in the developing forebrain. Genes Dev 13: 3092–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti K, Gasper M, Bertuzzi S, Arnheiter H 2011. Lack of the ventral anterior homeodomain transcription factor VAX1 leads to induction of a second pituitary. Development 138: 873–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmeier ML, Potok MA, Davis SW, Camper SA 2007. TCF4 deficiency expands ventral diencephalon signaling and increases induction of pituitary progenitors. Dev Biol 311: 396–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila J, Izpisúa Belmonte JC 2001. Patterning mechanisms controlling vertebrate limb development. Annu Rev Cell Dev Biol 17: 87–132 [DOI] [PubMed] [Google Scholar]
- Cho EA, Dressler GR 1998. TCF-4 binds β-catenin and is expressed in distinct regions of the embryonic brain and limbs. Mech Dev 77: 9–18 [DOI] [PubMed] [Google Scholar]
- de Lau W, Clevers H 2001. LEF1 turns over a new leaf. Nat Genet 28: 3–4 [DOI] [PubMed] [Google Scholar]
- Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, McMahon AP 1993. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 75: 1417–1430 [DOI] [PubMed] [Google Scholar]
- Fujimura N, Taketo MM, Mori M, Korinek V, Kozmik Z 2009. Spatial and temporal regulation of Wnt/β-catenin signaling is essential for development of the retinal pigment epithelium. Dev Biol 334: 31–45 [DOI] [PubMed] [Google Scholar]
- Hallonet M, Hollemann T, Pieler T, Gruss P 1999. Vax1, a novel homeobox-containing gene, directs development of the basal forebrain and visual system. Genes Dev 13: 3106–3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppler S, Kavanagh CL 2007. Wnt signalling: variety at the core. J Cell Sci 120: 385–393 [DOI] [PubMed] [Google Scholar]
- Hovanes K, Li TW, Munguia JE, Truong T, Milovanovic T, Lawrence Marsh J, Holcombe RF, Waterman ML 2001. β-Catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat Genet 28: 53–57 [DOI] [PubMed] [Google Scholar]
- Hurlstone A, Clevers H 2002. T-cell factors: turn-ons and turn-offs. EMBO J 21: 2303–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessell TM 2000. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet 1: 20–29 [DOI] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH 2008. An integrated software system for analyzing ChIP–chip and ChIP-seq data. Nat Biotechnol 26: 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao KR, Elinson RP 1988. The entire mesodermal mantle behaves as Spemann's organizer in dorsoanterior enhanced Xenopus laevis embryos. Dev Biol 127: 64–77 [DOI] [PubMed] [Google Scholar]
- Kim JW, Lemke G 2006. Hedgehog-regulated localization of Vax2 controls eye development. Genes Dev 20: 2833–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Oda T, Itoh M, Jiang D, Artinger KB, Chandrasekharappa SC, Driever W, Chitnis AB 2000. Repressor activity of Headless/Tcf3 is essential for vertebrate head formation. Nature 407: 913–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H 1997. Constitutive transcriptional activation by a β-catenin–Tcf complex in APC−/− colon carcinoma. Science 275: 1784–1787 [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H 1998a. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet 19: 379–383 [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Willert K, Molenaar M, Roose J, Wagenaar G, Markman M, Lamers W, Destree O, Clevers H 1998b. Two members of the Tcf family implicated in Wnt/β-catenin signaling during embryogenesis in the mouse. Mol Cell Biol 18: 1248–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz M, Herrmann M, Wedlich D, Gradl D 2004. Autoregulation of canonical Wnt signaling controls midbrain development. Dev Biol 273: 390–401 [DOI] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA 2006. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc 1: 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Liu Y, Lupo G, Lan L, Barsacchi G, He R 2008. A role for Xvax2 in controlling proliferation of Xenopus ventral eye and brain progenitors. Dev Dyn 237: 3387–3393 [DOI] [PubMed] [Google Scholar]
- Logan CY, Nusse R 2004. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20: 781–810 [DOI] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X 2009. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev Cell 17: 9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S 2003. Mapping Wnt/β-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci 100: 3299–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I 2000. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046–1047 [DOI] [PubMed] [Google Scholar]
- Mui SH, Hindges R, O'Leary DD, Lemke G, Bertuzzi S 2002. The homeodomain protein Vax2 patterns the dorsoventral and nasotemporal axes of the eye. Development 129: 797–804 [DOI] [PubMed] [Google Scholar]
- Mui SH, Kim JW, Lemke G, Bertuzzi S 2005. Vax genes ventralize the embryonic eye. Genes Dev 19: 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay M, Shtrom S, Rodriguez-Esteban C, Chen L, Tsukui T, Gomer L, Dorward DW, Glinka A, Grinberg A, Huang SP, et al. 2001. Dickkopf1 is required for embryonic head induction and limb morphogenesis in the mouse. Dev Cell 1: 423–434 [DOI] [PubMed] [Google Scholar]
- Paridaen JT, Danesin C, Elas AT, van de Water S, Houart C, Zivkovic D 2009. Apc1-mediated antagonism of Wnt/β-catenin signaling is required for retino-tectal pathfinding in the zebrafish. Zebrafish 6: 41–47 [DOI] [PubMed] [Google Scholar]
- Parr BA, Shea MJ, Vassileva G, McMahon AP 1993. Mouse Wnt genes exhibit discrete domains of expression in the early embryonic CNS and limb buds. Development 119: 247–261 [DOI] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW 2009. Wnt signaling and the polarity of the primary body axis. Cell 139: 1056–1068 [DOI] [PubMed] [Google Scholar]
- Schulte D, Furukawa T, Peters MA, Kozak CA, Cepko CL 1999. Misexpression of the Emx-related homeobox genes cVax and mVax2 ventralizes the retina and perturbs the retinotectal map. Neuron 24: 541–553 [DOI] [PubMed] [Google Scholar]
- Sharov AA, Dudekula DB, Ko MS 2006. CisView: a browser and database of cis-regulatory modules predicted in the mouse genome. DNA Res 13: 123–134 [DOI] [PubMed] [Google Scholar]
- Sousa VH, Fishell G 2010. Sonic hedgehog functions through dynamic changes in temporal competence in the developing forebrain. Curr Opin Genet Dev 20: 391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struewing I, Boyechko T, Barnett C, Beildeck M, Byers SW, Mao CD 2010. The balance of TCF7L2 variants with differential activities in Wnt-signaling is regulated by lithium in a GSK3β-independent manner. Biochem Biophys Res Commun 399: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs JL, Oishi I, Izpisua Belmonte JC, Kintner C 2008. The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat Genet 40: 1454–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Take-uchi M, Clarke JD, Wilson SW 2003. Hedgehog signalling maintains the optic stalk-retinal interface through the regulation of Vax gene activity. Development 130: 955–968 [DOI] [PubMed] [Google Scholar]
- Ulloa F, Marti E 2010. Wnt won the war: antagonistic role of Wnt over Shh controls dorso–ventral patterning of the vertebrate neural tube. Dev Dyn 239: 69–76 [DOI] [PubMed] [Google Scholar]
- Van de Wetering M, Castrop J, Korinek V, Clevers H 1996. Extensive alternative splicing and dual promoter usage generate Tcf-1 protein isoforms with differential transcription control properties. Mol Cell Biol 16: 745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veien ES, Rosenthal JS, Kruse-Bend RC, Chien CB, Dorsky RI 2008. Canonical Wnt signaling is required for the maintenance of dorsal retinal identity. Development 135: 4101–4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise A, Bruser K, Elfert S, Wallmen B, Wittel Y, Wohrle S, Hecht A 2010. Alternative splicing of Tcf7l2 transcripts generates protein variants with differential promoter-binding and transcriptional activation properties at Wnt/β-catenin targets. Nucleic Acids Res 38: 1964–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley A, Edalat K, Chiang P, Mora M, Mirro K, Lee M, Muhr H, Elul T 2008. GSK-3β and α-catenin binding regions of β-catenin exert opposing effects on the terminal ventral optic axonal projection. Dev Dyn 237: 1434–1441 [DOI] [PubMed] [Google Scholar]
- Zhou CJ, Molotkov A, Song L, Li Y, Pleasure DE, Pleasure SJ, Wang YZ 2008. Ocular coloboma and dorsoventral neuroretinal patterning defects in Lrp6 mutant eyes. Dev Dyn 237: 3681–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]